Abstract
Caudal regression syndrome (sacral agenesis), which impairs development of the caudal region of the body, occurs with a frequency of about 2 live births per 100 000 newborns although this incidence rises to 1 in 350 infants born to mothers with gestational diabetes. The lower back and limbs can be affected as well as the genitourinary and gastrointestinal tracts. The axial skeleton is formed during embryogenesis through the process of somitogenesis in which the paraxial mesoderm periodically segments into bilateral tissue blocks, called somites. Somites are the precursors of vertebrae and associated muscle, tendons and dorsal dermis. Vertebral anomalies in caudal regression syndrome may arise through perturbation of somitogenesis or, alternatively, could result from defective bone formation and patterning. We discovered that MBTPS1/SKI-1/S1P, which proteolytically activates a class of transmembrane transcription factors, plays a critical role in somitogenesis and the pathogenesis of lumbar/sacral vertebral anomalies. Conditional deletion of Mbtps1 yields a viable mouse with misshapen, fused and reduced number of lumbar and sacral vertebrae, under-developed hind limb bones and a kinky, shortened tail. We show that Mbtps1 is required to (i) maintain the Fgf8 ‘wavefront’ in the presomitic mesoderm that underpins axial elongation, (ii) sustain the Lfng oscillatory ‘clock’ activity that governs the periodicity of somite formation and (iii) preserve the composition and character of the somitic extracellular matrix containing fibronectin, fibrillin2 and laminin. Based on this spinal phenotype and known functions of MBTPS1, we reason that loss-of-function mutations in Mbtps1 may cause the etiology of caudal regression syndrome.
Introduction
The spine provides structural support for the body, flexibility for movement and protection for the spinal cord and nerves. The vertebral column consists of bone segments separated by intervertebral disk joints supported by ligaments and axial muscles. Collectively this constitutes the musculoskeletal component of the spine. The precise integration of bone, cartilage, nerves, muscles and tendons is essential for proper spine function and birth defects or degenerative conditions that perturb these tissues and their developmental integration result in congenital vertebral malformations. Caudal regression syndrome (sacral agenesis) is one such birth defect, and occurs with a frequency of about 2 in 100 000 live births, although the frequency rises to 1 in 350 in mothers with gestational diabetes (1,2). While presentation can be variable, multiple vertebrae of the lower spine are absent or misshapen in this syndrome that can also exhibit under-developed lower limb bones in a frog-leg position.
The vertebral column is formed through a process known as somitogenesis (3). During embryogenesis, the paraxial mesoderm periodically segments into bilateral blocks of tissue called somites, which are the precursors of the vertebrae and their associated muscle, tendons, ligaments and dorsal dermis. The periodicity of somite formation is governed by a molecular oscillator termed the segmentation clock which is evident in the form of cyclic patterns of FGF6, WNT and NOTCH activity in the presomitic mesoderm (PSM). The actual process of somite boundary formation and thus axial segmentation involves a mesenchymal to epithelial transformation (MET) of the PSM and this coincides with intersection of oscillatory gene activity with the determination front. The determination front is thought to be specified primarily by a gradient of FGF8 and WNT signaling in the caudal PSM that diminishes rostrally. However, in contrast to this established model, recently it was proposed that somites may have the capacity for self-organization independent of any clock and wavefront mechanism (4).
Vertebral anomalies may result from perturbation of somitogenesis or alternatively via defective bone formation and patterning. Mutations causing autosomal-recessive spondylocostal dysostosis which affects the spinal vertebrae and ribs have been found in LFNG (5), HES7 (6), DLL3 (7) and MESP2 (8), which play a critical role in regulating different steps of somitogenesis. In contrast, the cause of caudal regression syndrome, which predominantly affects the lower spine and hind limbs, is unclear. However, exposure to retinoic acid (RA) (9), telomere dysfunction (10), Brachyury (T) loss-of-function (11), double transgenic over-expression of Islet-1 under a Hoxc8 promoter (12) and inactivation of PCSK5 (13) produce these symptoms in mice. Since known mutations only account for a minor fraction of congenital vertebral anomaly cases, there is a need to identify additional causative loci to clarify their etiology and pathogenesis.
Here we describe a novel role for MBTPS1/SKI-1/S1P (membrane bound transcription factor protease, subtilisin kexin isozyme-1, or site 1 protease) in somite segmentation and in the pathogenesis of vertebral anomalies. MBTPS1 is an auto-catalytically activatable member of the proprotein convertase family of serine proteases (14). MBTPS1 activates a small family of transmembrane spanning b-ZIP transcription factors (15–17) and it is essential for mineralization of osteoblastic cultures (18,19). Mbtps1 null mice die prior to implantation due to a failure to form an epiblast layer (20,21), hence the exact developmental requirements for SKI-1/S1P in vivo remain unknown. To better understand the role of SKI-1 in osteogenic differentiation and skeletal development, we conditionally deleted Mbtps1 during embryogenesis using Cre recombinase driven by the rat 3.6 kb Col1a1 promoter (3.6 Col1).
We discovered that this conditional Mbtps1 loss-of-function mouse model exhibits phenotypic changes localized to the lumbar/sacral vertebral region (decreased vertebral number, vertebral fusion and kinky tail) which mimic those in caudal regression syndrome. Consistent with this phenotype, we show that Mbtps1 plays critical roles in regulating somitogenesis.
Results
Mbtps1 is expressed ubiquitously in the post-implantation and mid-gestation mouse embryo
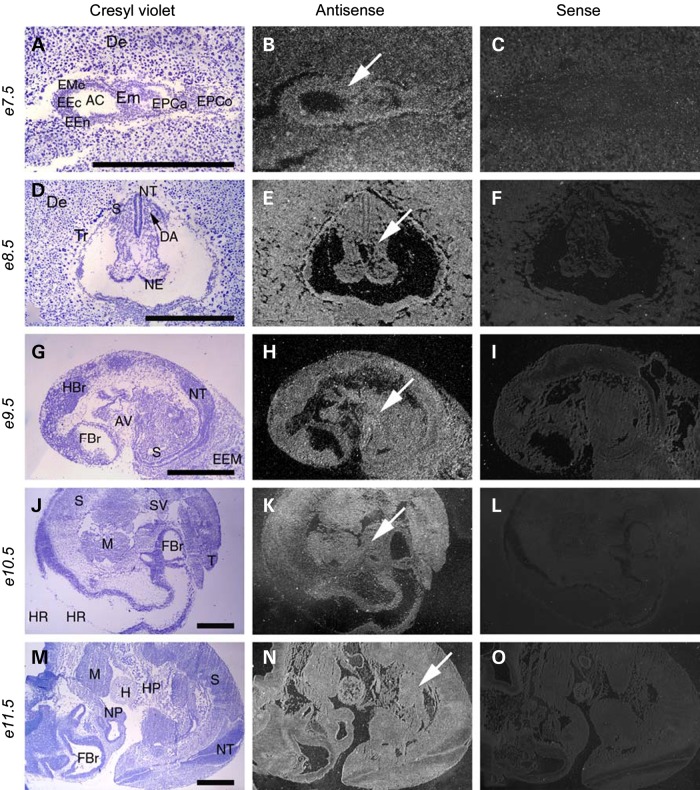
As a first step in determining the developmental function of MBTPS1, we characterized its spatiotemporal activity in embryonic day (E) 7.5–11.5 embryos via section in situ hybridization. At E7.5, Mbtps1 mRNA was detected throughout the embryo, the ectoplacental cone and in the decidua (Fig. 1A–C). In E8.5 embryos, Mbtps1 was highly expressed in the neural tube and in the newly formed somites (Fig. 1D–F). These domains of expression continue from E9.5 until E11.5 (Fig. 1G–O). Although the level of activity varies between different tissues, Mbtps1 is expressed ubiquitously throughout post-implantation and mid-gestation mouse embryos.
Figure 1.
Mbtps1 is expressed ubiquitously in the post-implantation and mid-gestation mouse embryo. Left column shows embryo sections stained with Cresyl Violet; middle column shows the same sections with comparative antisense hybridization labeling seen as bright under darkfield illumination (arrows) and right column shows control sense hybridization. (A–C) Comparison of in situ hybridization with sense and antisense probes reveals a broad expression of Mbtps1 in the uterine decidua and in the embryo at E7.5 (arrow). E7.5 early embryo sagittal section seen within uterine cavity near site of implantation. Presence of Mbtps1 mRNA labeling is evident within embryo, ectoplacental cone and decidua. (D–F) E8.5 embryo coronal section. (G–I) E9.5 embryo sagittal section. (J–L) E10.5 embryo sagittal section. (M–O) E11.5 embryo sagittal section. AC, amniotic cavity; AV, atrioventricular canal; DA, dorsal aorta, primordium; De, decidua; EEc, embryonic ectoderm; EEM, extraembryonic membranes; EEn, embryonic endoderm; Em, embryo; EMe, embryonic mesenchyma; EPCa, ectoplacental cavity; EPCo, ectoplacental cone; FBr, forebrain; H, heart; HBr, hindbrain; HP, hepatic primordium; HR, hinbrain roof; M, mandible; NE, neuroepithelium; NP, nasal pit; NT, neural tube; S, somite; SV, sinus venosum; T, tail; and Tr, trophoblasts. Magnifications: (A–C) ×36; (D–F) ×24; (G–I) ×15 and (J–O) ×10. Scale bar: 1 mm.
Col1-Cre mediated Mbtps1 knockout mice survive until at least P10 but exhibit severe vertebral defects
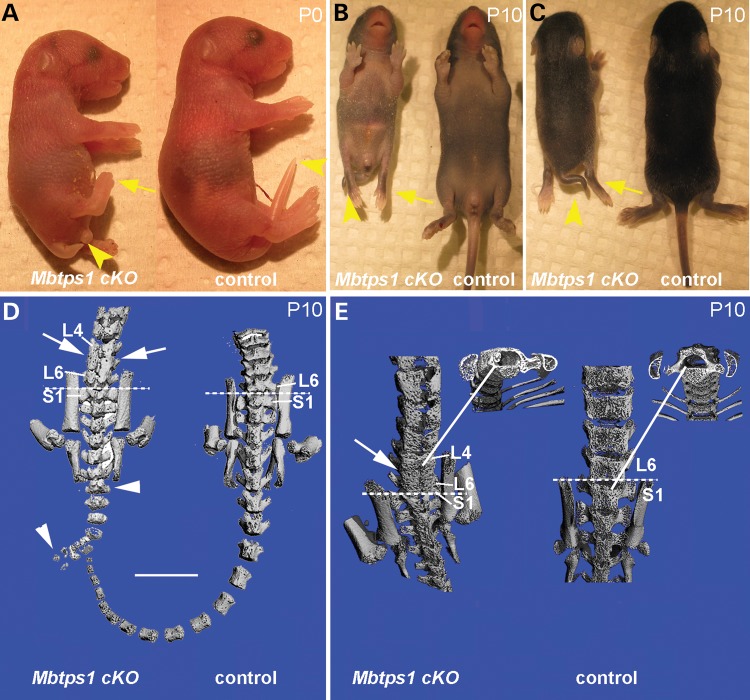
Mbtps1 was previously conditionally deleted in cartilage by using the Col2-Cre recombinase (22). These conditional mice have a shortened body axis, severe chondrodysplasia and die during or shortly after birth. Since we wanted to investigate the role of Mbtps1 both embryonically and postnatally during osteogenesis, we decided to conditionally excise it using the Col1-Cre. In order to do that we crossed heterozygous male Mbtps1 floxed mice carrying one copy of the 3.6 Col1-Cre transgene (Mbtps1flx/+: 3.6 Col1-Cre+/−) with female mice homozygous for the Mbtps1 floxed allele (Mbtps1flx/flx). Offspring genoytpes were consistent with expected Mendelian ratios. P0 heterozygous (Mbtps1flx/+: 3.6 Col1-Cre+/−) pups were visually indistinguishable from wild-type littermates. In contrast, P0 homozygous conditional knockout mice (Mbtps1flx/flx: 3.6 Col1-Cre+/−), which we call Mbtps1cKO, were all postnatal viable, but were smaller than their control siblings with a short kinky tail and smaller hind limbs (Fig. 2A). By P10 the difference in size was particularly evident [an average body weight of 6.5 gm for wild-type males versus 4.7 gm for male Mbtps1cKO (P = 0.001) littermates] along with severe hypertrophy and paralysis of the hind limbs (Fig. 2B and C). Signifying the severity of the phenotype, about 60% of the Mbtps1cKO pups were cannibalized by their mothers shortly after birth. MicroCT scans of spines from P10 Mbtps1cKO mice revealed extensive developmental anomalies in vertebral bones compared with normal littermate controls. Figure 2D and E depict a representative sampling of two Mbtps1cKO mice which display abnormal lumbar and sacral vertebrae. Abnormal bone patterning was consistently observed on the posterior faces of lumbar (L) 4 to 6 vertebrae (Fig. 2D and E). This ectopic mineralized bone obscured the boundaries of individual vertebrae, demonstrating that these abnormal vertebrae are effectively fused. However, cross-sectional views at this location did not show an obstruction of the spinal column (Fig. 2E). These spinal defects resemble congenital vertebral anomalies such as caudal dysgenesis and spondylocostal dysostosis, which are, respectively, characterized by severe malformations of the vertebral column including a shortened trunk due to loss of vertebral segments and fusion of vertebrae during development (23).
Figure 2.
Mbtps1cKO mice exhibit a severe caudal truncation and changes in vertebrae patterning. (A) At P0, Mbtps1cKO mice exhibit caudal axial truncation (arrowheads) and hypotrophy of the hind limbs (arrows). (B and C) At P10, Mbtps1cKO mice are smaller than normal littermates and the caudal axial truncation, as well as the hypotrophy of the hind limbs, are even more evident. B, ventral and C, dorsal view. (D and E) Micro-CT images of control and Mbtps1cKO mice at P10. (Each micro-CT image represents a separate mouse.). (D) Posterior comparison of Mbtps1cKO spine with that for control littermate. Arrowheads denote abnormal tail vertebrae and arrows demark abnormal bone patterning and fusion of lower lumbar vertebrae. Scale bar: 5 mm. (E) Sacral-lumbar comparison. Arrow denotes region with abnormal bone patterning and fusion of L4-L6 vertebrae in Mbtps1cKO. Solid lines identify regions from which respective cross-sections were taken. Dashed lines denote the sacral-lumbar border. S, sacral. L, lumbar.
Expression of Cre recombinase under the control of the 3.6 Col1 promoter
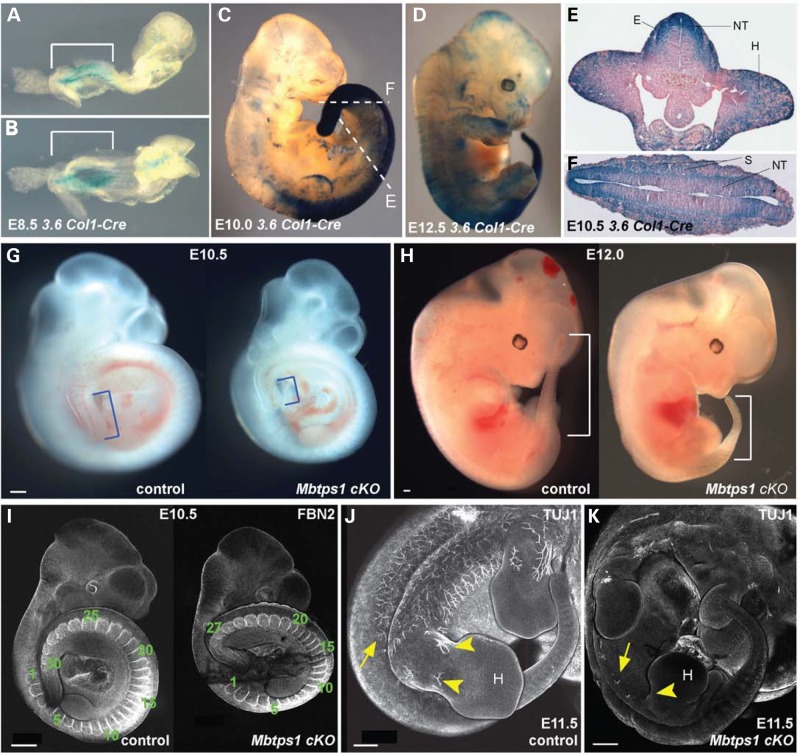
Vertebral anomalies observed in conditional Mbtps1cKO mutant newborn pups were surprising given the reported expression of 3.6 Col1-Cre in adult calvaria, long bone, tendon and skin (24). This suggested firstly that there may be an earlier, as yet unappreciated, 3.6 Col1-Cre activity during embryogenesis and secondly that MBTPS1 may play a critical role in somitogenesis, the process in which the paraxial mesoderm is segmented into somites. Therefore, we crossed 3.6 Col1-Cre transgenic mice to ROSA26 reporter mice (25). The latter expresses β-galactosidase in the presence of Cre recombinase after excision of a floxed STOP cassette that inhibits the expression of the reporter gene. We observed expression of 3.6 Col1-Cre as determined by β-galactosidase activity in whole-mount embryos beginning around E8.5 (Fig. 3A and B). Interestingly, β-galactosidase expression was highly enriched in the caudal region of the embryo, particularly in the PSM (Fig. 3A and D). Transverse sectioning of labeled embryos revealed widespread recombination among tissues that had not been previously appreciated (Fig. 3E and F) (24). In particular, there are high levels of expression in the ectoderm, the neuroepithelium and hind limb buds. Therefore, these findings indicate that 3.6 Col1-Cre-mediated excision of the floxed Mbtps1 allele initially takes place around E8.5 in the caudal region of the embryo. Beginning in the PSM at an axial level posterior to the future forelimb, the labeled cells contribute to a variety of tissues including the paraxial mesoderm that will give rise to the embryonic somites and consequently to the adult spine.
Figure 3.
The Cre recombinase expression in 3.6 Col1-Cre mice nicely correlates with the caudal phenotype of the Mbtps1cKO mice. (A–D) Whole-mount LacZ staining of 3.6 Col1-Cre;ROSA26 mice at E8.5 (A,B), E10.0 (C) and E12.5 (D) reveals expression of the Cre recombinase prominently in the posterior tail region (brackets). (E and F) LacZ stained transverse sections of E10.5 3.6 Col- Cre: ROSA26 embryos showing strong expression of the Cre recombinase in the somites (S), the neural tube (NT), the hind limb buds (H) and the ectoderm (E). Sections were taken at the axial levels denoted in 3C. (G and H) A caudal axial truncation is evident in homozygous Mbtps1cKO embryos (right) by E10.5 compared to their wild type (left) littermates (G, brackets). The caudal axial truncation becomes more prevalent by E12.0 (H, brackets). (I) At E10.5, Fibrillin2 (FBN2) antibody staining reveals a reduced number of somites in Mbtps1cKO embryos (right) compared to their littermate control (left) (27 instead of 30 in these embryos). (J and K) Whole mount antibody staining of neuron-specific class III β tubulin (TUJ1) at E11.5 in Mbtps1cKO embryo (K) and control littermate (J) reveals a general failure of neuronal projections in the Mbtps1cKO embryos (arrows). Interestingly, no neuronal projections are seen in the hind limb bud (H) and spines of Mbtps1cKO embryos (arrowheads). Scale bar: 500 μm.
Skeletal preparations support vertebral abnormalities
Consistent with the early expression of 3.6 Col1-Cre in the PSM at E8.5, we observed a caudal axial truncation in Mbtps1cKO embryos as early as E10.5 compared with control littermates (Fig. 3G and H). Immunostaining for fibrillin2 (FBN2), which is a structural component of myofibrils that demarcates individual somites, demonstrated that the axial truncation is consistent with a significantly (P<0.012) reduced number of somites (26.4 ± 2.1 STD) in E10.5 Mbtps1cKOembryos compared with each control genotypic group [30.1 ± 1.5 STD (Mbtps1flx+/−: 3.6ColCre+); 30.5 ± 0.9 (Mbtps1flxflx); 28.9 ± 1.1 (Mbtps1flx+/−)] (Fig. 3I). Caudal axial truncation was more prevalent by E12.0 and progressively worsened throughout embryogenesis (Fig. 3H). Consistent with Mbtps1 expression and 3.6 Col1-Cre activity in the neuroepithelium, we also observed a striking neuronal phenotype of E11.5 Mbtps1cKO embryos. Immunostaining with neuron-specific antibodies against type III β-tubulin revealed a lack of neuronal projections from the spinal cord into the limb in contrast to controls (Fig. 3J and K). This embryonic neuronal phenotype likely presages the hind limb paralysis exhibited by postnatal Mbtps1cKO mice.
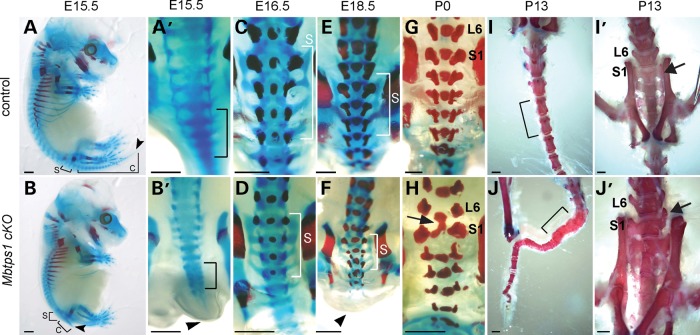
To investigate the developmental cause of the axial truncation phenotype and its correlation with postnatal malformations, we characterized bone and cartilage differentiation via Alizarin red and Alcian blue staining, respectively, in E15.5 to P13 individuals. At E15.5, we observed defects in the developing spine of Mbtps1cKO embryos in the form of hypoplastic cartilage pre-vertebrae particularly in the sacral and coccygeal regions (Fig. 4A and B). A day later at E16.5, we observed a general delay in ossification in association with vertebral irregularities in Mbtps1cKO embryos compared with wild-type littermates (Fig. 4C and D). By P0, more severe defects were consistently observed in the form of fused vertebrae in the sacral and the coccygeal regions together with infrequently altered anterior–posterior vertebral identity, particularly at the lumbar-sacral boundary (Fig. 4G and H, arrows). Thus, the irregular formation or ‘stacking’ of the developing cartilaginous vertebrae in the coccyx observed at E15.5 presages the fused vertebrate subsequently observed in that same region in P13 Mbtps1cKO mice (Fig. 4B’ and J, brackets).
Figure 4.
Skeletal preparations show that Mbtps1 deficiency disrupts the development of the lumbar/sacral spine. Alcian blue and Alizarin red staining reflect cartilage and mineralized bone, respectively. (A and B) Control (A) and Mbtps1cKO (B) skeletons at E15.5 reveal a severe caudal axial truncation (arrowheads). Brackets mark the sacral (S) and coccygeal (C) vertebrae. (A’ and B’) Dorsal views of higher magnification of the caudal region of control and Mbtps1cKO embryos at E15.5. Note the fused vertebrae precursors that resemble “stacking” (B’, bracket) and the short tail (B’ arrowhead) in the Mbtps1cKOembryo. (C–F) Dorsal views of E16.5 (C and D) and E18.5 (E and F) control and Mbtps1cKO skeletons reveal delayed mineralization of sacral vertebrae (compare the alizarin red staining). Brackets denote the sacral (S) vertebrae. (G and H) Fusion of L6 and S1 vertebrae in Mbtps1cKO embryos is observed at P0 (arrow). (I and J) Tails of Mbtps1cKO mice routinely exhibited multiple fused vertebrae (compare bracketed regions). (I’ and J’) Some S1 vertebrae in Mbtps1cKO spines displayed asymmetric abnormal differentiation (see arrow) resembling that for L6. Scale bar: 2 mm.
In summary, conditional knockout of Mbtps1 using the 3.6 Col1-Cre recombinase results in axial truncation, vertebral fusions and occasional unilateral fate changes of sacral and lumbar vertebrae. These defects demonstrate that MBTPS1 is required during embryogenesis for proper vertebral segmentation and development.
MBTPS1 is required for somitogenesis during development
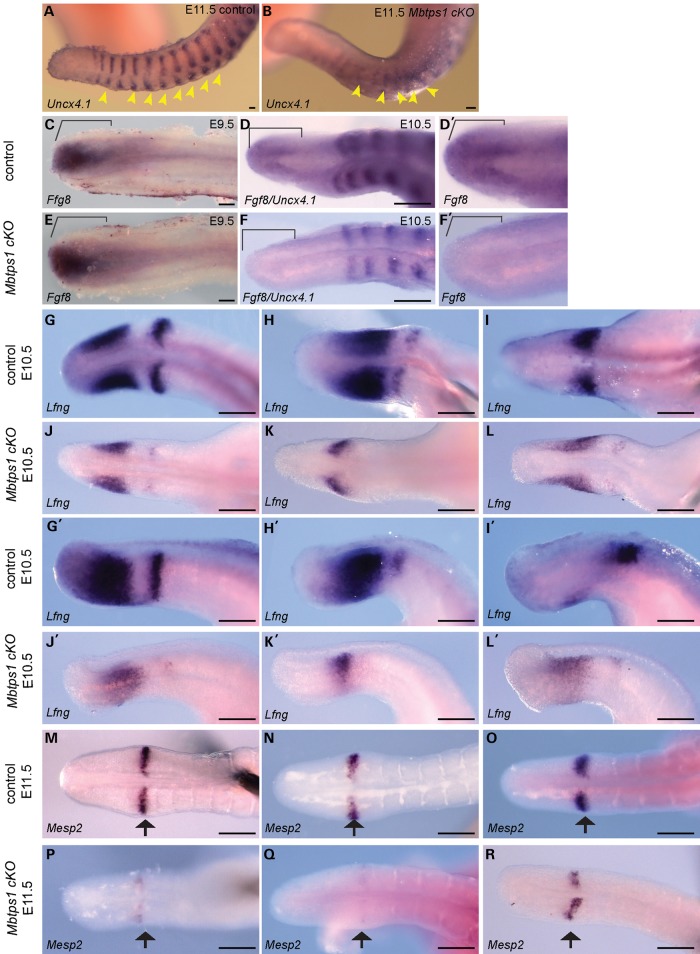
Based on the expression of Mbtps1, and the activity of 3.6 Col1-Cre, we hypothesized that the axial truncation phenotype observed as early as E10.5 in Mbtps1cKO embryos was consistent with a perturbation of somitogenesis. To test this idea we characterized the expression of Uncx4.1 which demarcates the posterior boundary of newly formed somites and provides a readout of the regularity and bilateral symmetry of somite formation as well as proper polarity or anterior–posterior patterning (26,27). In E11.5 control embryos, in situ hybridization revealed strong segmental expression of Uncx4.1 in the caudal region of each newly formed and maturing somite (Fig. 5A). In contrast, in Mbtps1cKO embryos, the segmental pattern was perturbed (Fig. 5B). Thus Mbtps1 loss-of-function results in irregular and indistinguishable somite boundaries together with abnormally spaced and fused somites. This pattern of disorganized somite segmentation is consistent with the vertebral fusions and axial truncation observed in Mbtps1cKO embryos and postnatal mice.
Figure 5.
Expression of key markers of somitogenesis including Fgf8 and Lunatic Fringe (Lfng) are dramatically decreased in the presomitic mesoderm of Mbtps1cKO embryos. Whole mount in situ hybridization was performed at E9.5 (C and E), E10.5 (D, F and G–L) or E11.5 (A, B and M–R) in control and Mbtps1cKO embryos. (A and B) Expression of Uncx4.1 reveals a general disorganization of the somites including loss of somite polarity in Mbtps1cKO embryos, 3 days after the onset of the 3.6 Col1-Cre recombinase. (C–F) Expression of Fgf8, normally restricted to the posterior-most region of the PSM at E9.5 and E10.5 (brackets) (C and D), is absent in Mbtps1cKO embryos at E10.5 (F), but still present a day earlier in Mbtps1cKO embryos at E9.5 (E). (G–L) Lfng expression is greatly reduced and its cyclic expression disturbed in E10.5 Mbtps1cKO embryos as demonstrated by dorsal (G–L) and lateral (G’–L’) views of the non-segmented PSM. (M–R) As illustrated by a representative sampling of three embryos, Mesp2 expression displays a range of expression in the determination front of Mbtps1cKO embryos which is consistently less than that for controls. Scale bar: 100 µm.
Several models have been proposed to describe the periodicity of somitogenesis and integral to each of these models is an oscillator that drives segmentation (28–32). A negative feedback loop mechanism involving the NOTCH, WNT and FGF signaling pathways constitutes the core of this oscillatory clock. One of the genes whose oscillatory expression is required for proper progression of the clock is Lunatic fringe (Lfng) (33–37). LFNG is a glycosyltransferase which is both a transcriptional target and a negative regulator of the NOTCH signaling pathway. Lfng is normally expressed in distinct phases that progress dynamically from the tail bud through the PSM to the boundary of the newly forming somite (Fig. 5G–I). In contrast, representative E10.5 Mbtps1cKO embryos exhibited reduced levels and diminished domains of Lfng activity in the PSM (Fig. 5J–L). Additionally, Lfng was only infrequently expressed at the boundary of any newly formed somite. Collectively, our results illustrate a perturbation of cyclic expression of Lfng in Mbtps1cKO embryos which could subsequently impact somite segmentation. Consistent with these observations, it was shown that the segmentation clock and consequently posterior skeletal development are sensitive to LFNG dosage during embryogenesis (38). Furthermore, these results are indicative of a disruption of the NOTCH signaling pathway. NOTCH signaling is a key component of the molecular oscillator, the perturbation of which disrupts the cyclic activity of genes such as Lfng and Hes7, resulting in perturbed somite segmentation (39). Thus an alteration of Lfng expression in Mbtps1cKO embryos demonstrates that Mbtps1 loss of function disrupts the segmentation clock.
To further validate this idea, we examined the activity of Mesp2, a basic helix–loop–helix transcription factor that suppresses NOTCH activity through induction of Lfng and which is required for the initiation of somite formation (40–42). In control embryos at E11.5, Mesp2 is expressed in bilateral single bands that correspond with the anterior half or border of the next presumptive somite to form (Fig. 5M–O). By comparison, in Mbtps1cKO siblings, expression of Mesp2 is substantially reduced in two out of three representative samples and slightly reduced in the third (Fig. 5P–R). Overall, all embryos showed a disruption in Mesp2 activity which is consistent with abnormal somite segmentation and the subsequent vertebral fusions and anomalies observed in Mbtps1cKO mice.
The translation of the temporal periodicity of cyclic gene expression, as driven by the segmentation clock, into the spatial periodicity of somite formation is mediated by two dynamic antagonizing gradients: a caudorostral FGF8/WNT gradient and a rostrocaudal RA gradient (3,43). The interface between these gradients is thought to define a threshold level of FGF8 signaling activity that establishes the position of the ‘determination’ or ‘maturation’ wavefront. Cells in the PSM are competent to respond to the segmentation program during axis elongation only when exposed to a specific threshold of FGF8 activity. Fgf8 is expressed in the posterior region of the PSM and is critical for the spatial positioning of the determination front (44–46). In E9.5 and E10.5 control embryos, we observed strong expression of Fgf8 in the caudal PSM (Fig. 5C, D and D’) which is consistent with previous reports (44). In contrast, Mbtps1cKO embryos fail to exhibit any measurable expression of Fgf8 in the PSM at E10.5 (Fig. 5F and F’). The FGF8 gradient is thought to be required for body axis elongation by controlling cell motility in the PSM (47). The loss of Fgf8 expression as a consequence of Mbtps1 loss-of-function is consistent with the truncated posterior body axis observed in Mbtps1cKO mice. Consistent with this rationale, conditional knockout of the FGF receptor 1 (Fgfr1), the only FGF receptor expressed in the paraxial mesoderm, results in abnormal somite segmentation and vertebrae development (48,49) similar to Mbtps1cKO mice. These similarities in phenotype suggest that an absence of FGF8 signaling in the PSM mechanistically underpins much of the Mbtps1cKO phenotype. Furthermore, a day earlier, at E9.5, Fgf8 expression in the PSM of Mbtps1cKO mice is unchanged compared with control littermates (Fig. 5E). This is consistent with normal somite development observed at this stage and correlates with the latency of effect of Cre recombinase on the spatiotemporal excision of Mbtps1. This further supports the idea of altered Fgf8 expression being a cause of the phenotype. In summary, vertebral defects observed in Mbtps1cKO embryonic and newborn mice are due to perturbation of somitogenesis during development as evidenced by the defects in the segmentation clock, the determination front, and the spatiotemporal lack of Fgf8 expression in the PSM.
Cell death is increased in the PSM in Mbtps1cKO embryos
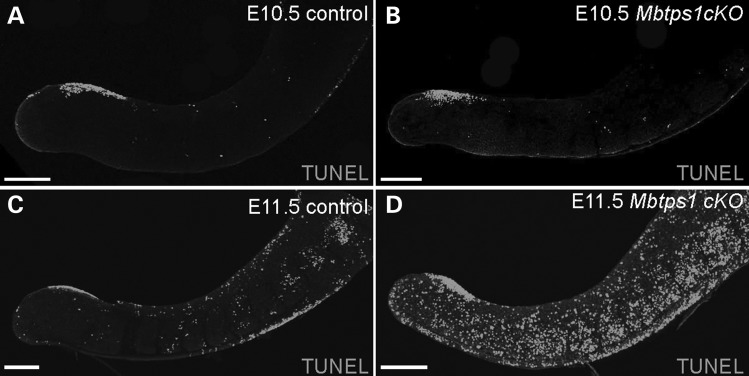
Since FGF8 is known to be required for the proliferative maintenance of the PSM (46), we explored whether there were alterations in cell survival in Mbtps1cKOmice that correlate with the axial truncation phenotype. TUNEL staining of E10.5 mouse embryos revealed a comparable basal level of cell death in the caudal trunk of control and Mbtps1cKO embryos (Fig. 6A and B). However, by E11.5, Mbtps1cKO embryos showed a dramatic increase in cell death throughout the PSM and in the newly formed somites compared with their control littermates (Fig. 6C and D). The loss of Fgf8 expression is consistent with elevated apoptosis in the PSM, which may negatively impact the proliferation of cells during axial elongation and somite development.
Figure 6.
Cell death is evident in the caudal region of Mbtps1cKO embryos. (A–D) Whole mount TUNEL staining reveals increased cell death in the caudal region of the Mbtps1cKO embryos at E11.5 (C and D), but no change at E10.5 (A and B). Scale bar: 200 µm.
Extracellular matrix components are down-regulated and are highly disorganized in the caudal region of Mbtps1cKO embryos
In order for a somite to form, mesenchymal cells in the PSM move rostrally and, upon reaching the determination front, undergo an MET (23). These types of cell movements and MET require a properly organized extracellular matrix (ECM). We have previously shown that inactivation of MBTPS1 protease in osteoblastic cells resulted in down-regulation of ECM genes including Fn1, Fbn2 and Lama (19). Interestingly, Fn1 knockout mice fail to form any somites (50,51) and the importance of the fibronectin matrix in somite formation has also been demonstrated during zebrafish, chick and frog embryogenesis (52–55). Therefore, we examined the ECM in the PSM of Mbtps1cKO mouse embryos for perturbations of fibronectin, consistent with disrupted somitogenesis.
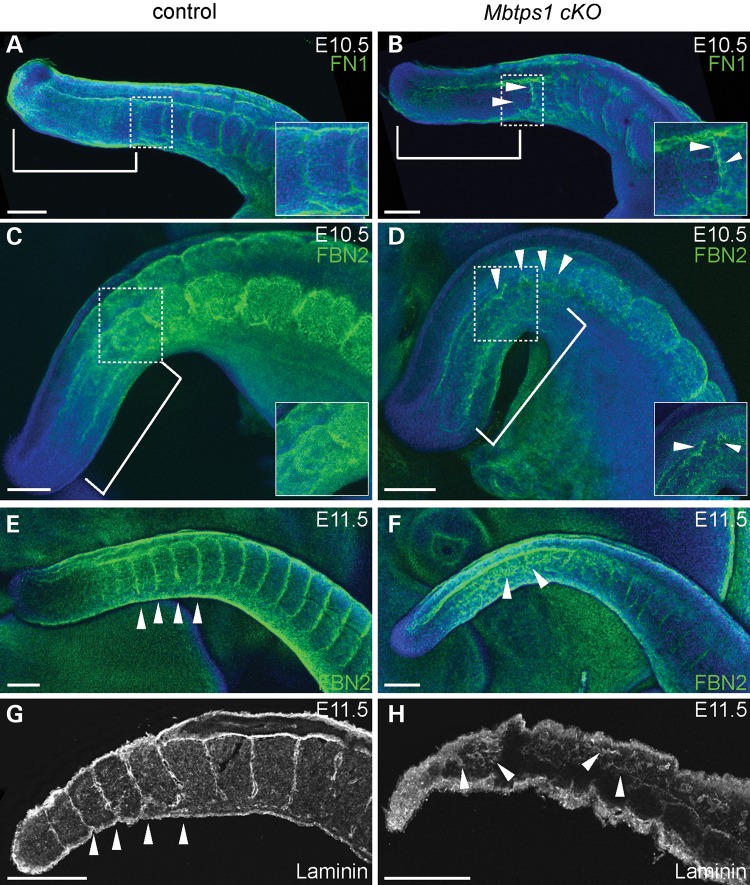
In wild-type embryos, fibronectin localizes to the medial interface of the non-segmented PSM and neural tube, while more rostrally, fibronectin surrounds each formed somite (Fig. 7A). In contrast, Mbtps1cKO embryos exhibit a considerable reduction in fibronectin in the PSM validating our previous microarray and qPCR results obtained after MBTPS1 inactivation in cultures of osteoblast-like cells (Fig. 7B) (19). Additionally, we observed a reduction in the fibronectin content of the matrix surrounding each somite (see arrowheads, Fig. 7B) along with fibronectin-containing fibrils forming abnormally near the determination front. Our results are consistent with a role for MBTPS1 protease in regulation of fibronectin expression and suggest that insufficient ECM proteins in Mbtps1cKO embryos limit assembly of the highly organized fibronectin matrix resulting in the formation of ectopic fibrils in the rostral PSM.
Figure 7.
Mbtps1 deficiency disrupts the organized distributions of Fibronectin, Fibrillin2, and Laminin, within the extracellular matrix of the developing spine. Immunofluorescence staining was carried out in whole mount embryos at E10.5 (A–D) or E11.5 (E and F) and in sagittal sections of E11.5 mice (G and H). (A and B) Fibronectin (FN1) is normally localized in the non-segmented PSM (bracket) and around each formed somite (A). In Mbtps1cKO embryos, FN1 is reduced in the PSM (bracket) and a general disorganization of the ECM is observed (arrowheads). The inserts are higher magnifications of the boxed area. (C and D) Fibrillin2 (FBN2) normally surrounds each formed somite (C), but it is abnormally expressed and distributed in the Mbtps1cKO embryos (arrowheads). A reduction of FBN2 is also observed in the somites of Mbtps1cKOembryos. The inserts are higher magnifications of the boxed area. The insert in D was enhanced to emphasize the disorganization of the fibrils. (E and F), A day later, expression of FBN2 reveals an even more disorganized extracellular matrix in the Mbtps1cKO embryos (arrowheads, F). (G and H) Laminin normally demarcates the somite boundaries (G), but its localization pattern is greatly disturbed in the Mbtps1cKO embryos (H). Arrowheads demark the positions of somite boundaries within the developing tail (G). Arrowheads also point at disturbed patterns of Laminin in the PSM and the presumptive somites (H). Scale bar: 200 µm.
To further validate the association of perturbed ECM with somitogenesis defects, we examined fibrillin2, which is one of the major structural components of extracellular microfibrils that serve as a scaffold for deposition of elastin. In control E10.5 mouse embryos, fibrillin2 demarcates the boundaries of each somite, which is consistent with what has been shown previously in avian embryos (Fig. 7C) (56). Unlike fibronectin, however, fibrillin2 does not accumulate in the non-segmented PSM. In sibling Mbtps1cKO embryos, fibrillin2 containing microfibrils are highly disorganized in the anterior PSM from where the newly forming somites will emerge (see arrowheads, Fig. 7D). In addition, fibrillin2 localizes in the posterior PSM in a similar highly disorganized fashion. The disorganization of the ECM is even more evident at E11.5 (compare Fig. 7E and F). Also, the levels of fibrillin2 in the somites may also be reduced in Mbtps1cKO embryos compared with the control siblings.
Lastly, we examined the distribution of laminins as they are major components of the basal lamina, one of the ECM layers. The distribution of laminin in sections of E11.5 wild-type mouse embryos resembled that of fibrillin2 and fibronectin, in that it demarcates the boundaries of each somite (Fig. 7G). In Mbtps1cKO littermate embryos, however, this restricted localization is absent, replaced by a disorganized distribution of extracellular laminins in the non-segmented PSM as well as in the newly formed somites (see arrowheads, Fig. 7H).
Overall, the ECM in the PSM of Mbtps1cKO embryos is highly disorganized with aberrant localization of key components, e.g. fibronectin, fibrillin2, and laminin. The level of expression of fibronectin protein is also lower than controls which is consistent with results in cultured osteoblastic cells showing that Fn1 transcription is MBTPS1 dependent (19) and furthermore suggests that proper fibronectin deposition is critical for normal somite formation.
Discussion
We previously discovered that MBTPS1 is essential for bone mineralization in vitro (19). However, the developmental requirement for MBTPS1 in vivo remains unknown. Here we showed that Mbtps1 is broadly expressed as early as E7.5 during mouse embryogenesis. Since Mbtps1 null mice die during embryogenesis prior to implantation (20,21), we conditionally deleted Mbtps1 using Cre recombinase under the control of a 3.6 kb fragment of the rat Col1a1 gene promoter (3.6 Col1 Cre). In postnatal mice, this promoter is active predominantly in bone and tail tendon, but is also expressed in non-osseous tissues (24,57). However, we discovered that Cre recombinase expression under the 3.6kb Col1a1 promoter occurs earlier than previously thought (24) and is spatially restricted primarily to the posterior region of the embryo. Importantly this also coincides with domains of enriched Mbtps1 expression in the posterior region of young embryos identified here by in situ hybridization and with the lumbar/sacral spinal defects we observe in Mbtps1cKO mice. After excision of Mbtps1 starting at about E8.5, all homozygous Mbtps1cKO mice survive until birth and consistently exhibit short, kinky tails and paralyzed hind limbs. Skeletal preparations and micro-CT scans revealed fewer than normal lumbar, sacral and tail vertebrae along the anterior/posterior axis with a general truncation of the tail. Histologic analyses of embryonic development indicated the early onset of these defects was concentrated specifically in the PSM and somites.
Defects in somitogenesis lead to embryonic and postnatal lumbar/sacral vertebral and hind limb defects in Mbtps1cKO mice
To date, no definitive cause for caudal regression syndrome has been identified; however, environmental influences such as hyperglycemia, vascular malformation and environmental toxins have been proposed to underlie its pathogenesis (13). Interestingly, Mbtps1cKO mice exhibit skeletal defects that are localized to the lower lumbar, sacral and caudal vertebrae resembling caudal regression syndrome. Mbtps1cKOmice consistently display axial truncation of their spinal columns together with misshapen and fused lower lumbar vertebrae.
Although somitogenesis initiates normally, the onset of 3.6 Col1-Cre-mediated Mbtps1 deletion within the caudal region of Mbtps1cKOembryos around E8.5 results in the loss of Fgf8 in the caudal PSM by E10.5 (Fig. 5D and F). It is well known that a gradient of FGF8 is critical for establishing the determination front and maintaining cell proliferation in the PSM, and consistent with this role, TUNEL staining revealed a dramatic increase in cell death in the PSM of Mbtps1cKOembryos in association with the loss of FGF8. Mechanistically this helps to account for the axial shortening observed in Mbtps1cKO mice.
FGF8 may also represent a common target of environmental factors known to increase the incidence of caudal agenesis, e.g. RA and gestational diabetes. Intriguingly, RA is known to directly repress the expression of Fgf8 through a specific RA receptor binding site element (RARE) in its promoter (58). Furthermore, caudal regression syndrome phenotypes similar to that observed in Mbtps1cKO mice are associated with elevated RA exposure within the same temporal E8-9 window (9,59). Consistent with these observations, deletion of Cyp26a1, which normally degrades RA, leads to increased retinoid signaling, which in turn diminishes FGF8 activity resulting in caudal dysgenesis associated (60) axial truncations (59). Moreover, Cyp26a1 was mis-regulated in KD1-T mice which exhibit apoptosis in the PSM and axial truncation in association with suppression of Brachyury (T) function (61). Hyperglycemia associated with gestational diabetes was also shown to increase the teratogenicity of RA leading to an increased incidence of caudal regression in mouse embryos (60). We propose that the axial truncation defects observed in Mbtps1cKO mice and in RA treated diabetic mice may share a common mechanism due primarily to increased cell death in the PSM in association with diminished FGF8 signaling. Currently, an association between in utero RA exposure and the risk for caudal regression has not been established for humans.
During the formation of a new somite every 2 h in the mouse, Lfng is normally expressed in a dynamic and cyclic fashion throughout the PSM that proceeds caudorostrally through the PSM (35,36). However, in Mbtps1cKO embryos, the expression of Lfng was reduced and its cyclic expression was perturbed. Additionally, expression of Mesp2, a gene that defines the segmental boundary and the rostrocaudal identity of each somite as it forms, was also considerably reduced. Subsequently, the perturbed expression of Uncx.4.1 as well as of fibronectin, fibrillin-2 and laminin proteins within the prospective lumbar and sacral vertebral regions of Mbtps1cKO embryos, provides a clear temporal record of the progression of somitogenesis defects as a result of Mbtps1 loss-of-function.
Relationship of Mbtps1 deficiency to other mutant or transgenic strains with similar spinal phenotypes
Somites bud off sequentially from the rostral end of the PSM while more undifferentiated presomitic cells are continuously supplied from the paraxial mesoderm in the caudal region of the tail bud. The phenotype of Mbtps1cKO mice resembles that of the Pudgy mutant which has a point mutation in Dll3 which disrupts NOTCH signaling, giving rise to a similarly disrupted pattern of Uncx4.1 expression (62). The randomized polarity of somites in Pudgy mutants leads to misshapen vertebral bodies which resemble those in the Mbtps1cKO mice. Embryonic manipulation and gene expression studies have shown that rostrocaudal polarity is established in the anterior PSM before the initiation of segmentation.
The axial skeleton (vertebral bodies and intervertebral discs) develops from a mesenchymal cell mass derived from the ventral halves of the somites while the lateral regions of the sclerotome form the vertebral arches and ribs. Uncx4.1 expression is restricted to the caudal half of each newly formed somite and future sclerotome (63), and is required for maintenance and differentiation of particular elements of the axial skeleton. Mutations in Uncx4.1 result in the absence of pedicles, transverse processes and proximal ribs, along the entire vertebral column (27). In Mbtps1cKO embryos, Uncx4.1 expression was dramatically disrupted and lacked its normal polarity. This implies that MBTPS1 deficiency can affect the formation of the vertebral arch and contributes to the abnormally shaped sacral S1 and lumbar L6 processes observed in Mbtps1cKO embryos.
In addition to formation of the bony vertebrae, somitogenesis also produces the organized development and segmental organization of associated muscles, nerves and blood vessels of the body. Fibronectin, fibrillin2 and laminin all play important developmental roles in the formation of these tissues. Fibronectin is essential for proper development of embryonic mesenchymal tissues and is required for development of the notochord and formation of somites (51,64). Fibrillin2 surrounds the developing notochord, the somites and the neural tube, and has been suggested to regulate spinal patterning via long-range mechanical tension fields via its microfibrils (65). Consistent with this idea, Fbn2 null mice exhibit bone and tendon defects (66,67). Laminin, a major component of the basal lamina, is localized at both the notochord/somite and the intersomitic boundaries (68). In the frog, inhibition of dystroglycan, a cell adhesion receptor for laminin, with morpholino antisense oligonucleotides or via dominant-negative approaches affects the number, size and integrity of the somites (69). In this work we have shown that Mbtps1cKOembryos exhibit perturbed fibronectin, fibrillin2 and laminin patterns in the trunk region. Therefore, we conclude that the phenotypes observed in Mbtps1cKO mice are due in part to defects in the ECM during somitogenesis. Not only is the clock and wavefront mechanism underpinning the spatiotemporal periodicity of somitogenesis affected as a consequence of Mbtps1cKO loss-of-function, but so too is the ECM which is critical for proper signaling and tissue morphogenesis during somitogenesis (70–72).
How can MBTPS1 act as a new upstream mediator of somitogenesis?
A major impediment to unraveling the signaling networks which control the clock mechanism has been the apparent individuality with which key genes are regulated in a cyclical and positional manner. We hypothesize that MBTPS1 protease could function as an upstream transcriptional regulator for a number of genes that govern somitogenesis.
Mechanistically, proteolytically active MBTPS1 is required for activation of SREBP-1, SREBP-2 and CREB/ATF family transcription factors and we have previously shown that transcription of Fn1, Fbn2, six Lama isoforms, and ∼140 more genes in osteoblastic cells requires activation and nuclear import of CREB-H, SREBP-1 and SREBP-2 (19). Importantly, MBTPS1 activated CREB/ATF family members CREB-H, CREB4, CREB3L2 (OASIS), ATF-6 as well as non-membrane derived CREB and ATF-4 recognize the cyclic-AMP response element sequence (73,74). We speculate that once MBTPS1 activated SREBP-1, SREPB-2 and CREB/ATF factors reach the nucleus they regulate the transcription of responsive genes such as Fn1, Fbn2, Lama and others by binding to CRE and/or SRE enhancer sequences within the respective promoters. Consistent with this paradigm, it was recently shown that dynamic CREB activity drives segmentation and posterior polarity specification during mammaliam somitogenesis (75). Furthermore, CREB loss-of-function results in perturbation of oscillatory (Lfng), segmentation (Mesp2) and ECM (laminin) activity which resembles the phenotype observed in Mbtps1cKO mice. Similarly, mutations in the zebrafish CREB homolog result in skeletal defects due to defective chondrocyte development (76,77). While there remains much to be learned about how CREB co-ordinates segmentation, it is known that CREB is associated with regulating Notch signaling and that CREB confers maturation to the PSM at the determination front (75). Given that it has been well established that MBTPS1 is required to activate transmembrane forms of CREB, MBTPS1 appears to lie upstream of CREB in the regulation of somitogenesis. Thus, we propose that MBTPS1 protease functions as a new upstream mediator of somitogenesis by regulating the transcription of Fgf8, Lfng, Fn1, Fbn2 and Lama within the PSM.
While a complete loss of Mbtps1 in humans would likely be embryonically lethal, coding mutations that generate hypomorphic alleles or cis-regulatory mutations that affect the spatiotemporal activity of MBTPS1 may indeed elicit a caudal regression phenotype. Additionally, expression of Mbtps1 may be further regulated by a complex mix of environmental factors such as fatty acids, cholesterol, polyunsaturated fatty acids, hyperglycemia, oxysterols and insulin. However, a causative role for Mbtps1 in human caudal regression syndrome remains speculative at this time.
We believe the paralytic phenotype observed in Mbtps1cKO mice is a consequence of the combined transcriptional effects of blocking Mbtps1 expression, which leads to the failure of axons to project into the hind limbs. Furthermore, we envision that the absence of an FGF8 gradient within the PSM inhibits the proliferation of mesenchymal cells and perturbs the determination front. Subsequently, disorganization of the ECM in the anterior PSM in Mbtps1cKO embryos disrupts the process of segmentation. The organization of cables containing fibrillin2, which normally coalesce near the midline before somite formation, is considerably disrupted. Fibrillin2 containing cables have been proposed to participate in mechanical tension fields linked to patterning events such as somitogenesis (65).
Materials and Methods
Skeletal preparations
E15.5–E18.5 embryos and P0–P13 postnatal mice were fixed in 99% ethanol and stained for bone and cartilage as previously described (78). A total of five E15.5–E16.5 Mbtps1cKO embryos, five E18.5–P0 Mbtps1cKO embryos and two P10 Mbtps1cKO mice were analyzed after skeletal preparation.
Immunohistochemistry and apoptosis assay
E10.5–E11.5 mouse embryos were fixed with 4% paraformaldehyde in PBS overnight at 4°C and dehydrated in a graded series of methanol solutions. Dehydrated embryos were then bleached in MeOH:DMSO:30%H2O2 (4:1:1, Dents Bleach) for 2 h at room temperature and rehydrated sequentially in 75% MeOH/PBS, 50% MeOH/PBS and in 25% MeOH/PBS for 10 min each followed by two final washes in PBS for 5 min each. Embryos were then blocked in 3% BSA for 2 h at room temperature before adding the primary antibody [fibrillin2, (Rabbit, Sigma, Cat. #HPA012853, 1:50), fibronectin, (Rabbit, Sigma, Cat. #F3648, 1:100), or Laminin (Chicken, Abcam, Cat. #ab14055, 1:200)]. Embryos were then incubated in primary antibody at 4°C overnight and the next day were washed 5 times for 1 h each in PBS. Rabbit secondary AlexaFluor 488 conjugated antibody (Invitrogen, 1:500) or chicken secondary AlexaFluor conjugated 488 antibody (Invitrogen, 1:500) was then added and incubated overnight at 4°C in the dark. Both primary and secondary antibodies were diluted in 3% BSA. Stained embryos were then washed in PBS 3 × 5 min and 3 × 1 h at room temperature and were either counterstained with DAPI (1 µg/ml) for 20 min at room temperature or immersed in Vectashield with DAPI to label individual nuclei. To analyze cell death, E11.5 embryos were subjected to TUNEL staining using the FITC Cell Death Detection Kit (Roche) according to the manufacturer's protocol after the dehydration and rehydration step.
For the sections, we fixed E10.5–E11.5 mouse embryos with 4% paraformaldehyde in PBS for 2 h at room temperature and then immersed them in 15% sucrose in PBS at 4○C overnight, followed by immersion in 30% sucrose in PBS at 4°C overnight. Embryos were then embedded in OCT and stored at −80°C until used. Cryosections of 10 µm were then generated, washed in PBS for 10 min, blocked in PBS containing 3% BSA, 0.15% glycine and 0.1% Triton-X100 for 30 min. Primary antibodies were then used as mentioned above and washes were performed in PBS 3 × 10 min the next day. Secondary antibodies (as above) were then applied and incubated for 2 h at room temperature. After 3 × 10 min washes, sections were mounted in Vectashield with DAPI. Fluorescence microscopy was performed on an LSM5 PASCAL confocal microscope (Carl Zeiss) using 2.5×, 5× and 10× objective lenses. Confocal optical slices were collected and maximum-intensity projection stacks were made with Zeiss LSM5 software. A total of two Mbtps1cKO embryos were analyzed for each antibody staining, with the exception of the E12.0 TUJ1 staining for which one embryo was analyzed. For the TUNEL staining, a total of four Mbtps1cKO mice were analyzed.
In situ hybridization
E7.5–E11.5 embryos were fixed in 4% paraformaldehyde in PBS overnight at 4°C and dehydrated in a graded series of methanol. In situ hybridization on whole mount embryos was performed as previously described using digoxigenin-labeled probes for Lfng, Mesp2, Uncx4.1 (gift of Dr Olivier Pourquie), Fgf8 (gift of Dr Ivor Mason), and NBT-BCIP detection (79). A total of three Mbtps1cKO embryos were analyzed for Fgf8 and Uncx4.1 at E10.5, a total of four Mbtps1cKO embryos were analyzed for Fgf8 at E9.5, a total of five Mbtps1cKO embryos were analyzed for Lfng and a total of five Mbtps1cKOembryos were analyzed for Mesp2.
For analysis of Mbtps1, embryo stages E7.5 to E11.5 were examined. Tissues were frozen and sectioned with cryostat into 8–10 μm sections and mounted on microscope slides. Tissues were fixed in 4% formaldehyde and hybridized with 35S-labeled-Mbtps1 cRNA antisense and sense probes overnight at 55°C as previously described (80). After hybridization, slides were exposed to X-ray film for 4 days, then dipped in Kodak NTB nuclear track emulsion, and exposed for 14 days in light-tight boxes with desiccant at 4°C. Photographic development was undertaken with Kodak D-19. Slides were lightly counterstained with Cresyl Violet and analyzed under both light- and darkfield optics. Antisense probes that are complimentary to mRNAs generate ISH labeling while control sense RNA probes that are identical to mRNAs always gave background levels of the hybridization signal. Values obtained by subtraction of sense labeling from antisense labeling were considered as specific.
Mating and animal strains
All animals were maintained in the UMKC Laboratory Animal Research Center under an approved protocol, an Association for Assessment and Accreditation of Laboratory Animal Care accredited facility. Mbtps1flx/flx mice were obtained from Dr. Jay Horton, Southwest Medical School and Dr. Linda Sandell, Washington University (21,22); 3.6 Col1-Cre mice were obtained from Dr. Barbara Kream, Univ. of Connecticut Health Center (24). ROSA26 female mice [Jax Strain 003474 B6.129S4-Gt(ROSA)26Sortm1Sor/J] were obtained from the Jackson Laboratory.
Males [Mbtps1+/flx: 3.6 Col1-Cre+/−] were crossed to females [Mbtps1flx/flx] to obtain Mbtps1cKO [Mbtps1flx/flx: 3.6 Col1-Cre+/−] offspring; our strategy deletes exon 2 of the Mbtps1 gene and should be effective in ablating expression of all expressed forms containing the catalytic site (21). Alternatively, male [Mbtps1+/flx: 3.6 Col1-Cre+/−] mice were mated with female homozygous B6.129S4-Gt(ROSA)26Sortm1Sor/J JAX (Jackson Labs, Inc.) mice to produce [3.6 Col1-Cre+/−: ROSA+/−] offspring expressing LacZ as a marker of the 3.6 Col1-Cre recombinase. Mice were maintained on standard mouse chow with free access to food and water and housed in ventilated cages with filter bonnets in rooms with a regulated 12 h light and 12 h dark cycle. Timed pregnancies were monitored and embryos and neonates harvested at days E9 to P10. All offspring (a total of 185 embryos and 68 Mbtps1cKOwere produced for this study) were found to be viable throughout the pregnancy period. Phenotypic characteristics for P10 mice were based on observations of 24 Mbtps1cKO mice along with a comparable number of control littermates. All conditional knock out mice were euthanized before they reached 14 days of age.
Genotyping of tail or yolk sac DNA
Yolk sack or tail segments were digested in 10 mm Tris–HCl pH 8.0 containing 25 mm EDTA, 0.5% SDS, 50 mm NaCl and 0.5 mg/ml proteinase K. A saturated solution of sodium chloride was then added to the digests to precipitate residual contaminating protein. Extracts were then centrifuged for 25 min at room temperature and the DNA containing supernatant was precipitated using 100% ethanol. Resulting pellets were then washed with 70% ethanol and allowed to dry. Dried pellets were hydrated with 0.01 m Tris–HCl buffer (pH 8.0) containing 1 mm EDTA and the DNA concentration was determined by UV spectrophotometry.
DNA samples were genotyped using PCR with gene specific primers for Mbtps1 (21), 3.6 Col1-Cre (81), ROSA26 (82) and Jarid ½ (83), a marker of sex. Primers used were: Mbtps1 floxed locus [P1, 5′-GAGAGCTGCAGATGACAGGGGACACAG-3′ and P2, 5′-GCCCAATCCACCGCTCTG TAGCGGAC-3′; floxed product = 434 bp, wild type product = 380 bp]; and, for 3.6 Cre [P1, 5′-ACTTGGTCGTGGCAGCCCGG-3′; P2, 5′-GGGGGCTGGGTGGACTCCTTT-3′; 700 bp product]. For ROSA26 genotyping assay we used three primers [L221, 5′-CTTGTGATCCGCCTCGGAGTATT-3′; L238, 5′-CGCGCCGCTGTAAAGTGTTACGT-3′; and R316, 5′-GGAGCGGGAGAAATGGATATG-3′] to generate 186 bp (wild-type) and 400 bp (LacZ allele) bands. For sexing, two primers were used (83): [P1, 5′-CTGAAGCTTTTGGCTTTGAG-3′ and P2, 5′-CCACTGCCAAATTCTT TGG-3′; male = 331 and 302 bp, female = 331 bp)]. PCR products were evaluated by electrophoresis on 2.1% agarose gels followed by staining with SYBR-Safe or Fast Blast DNA stain, and imaging with a Fuji LAS4000 CCD scanner.
Embryo collection
Females were checked daily for semen plugs and separated from male mice once a plug was detected. Pregnant mice were euthanized on embryonic day 7.5, 8.5, 9.5, 10.5, 11.5, 12, and P0 to P14 using carbon dioxide gas and embryos were immediately removed for genotyping and analysis.
MicroCT
P10-P14 mice were subjected to whole-body scans and/or dissected limbs and spines were scanned using a calibrated VivaCT40 (Scanco) instrument. Scans followed a standard protocol with a resolution of 10–19 μm (55 kV, 85 μA, 1000 projections per 180°, 500 ms integration time). Image processing included Gaussian filtering and segmentation: Σ = 0.8, support = 1, and a threshold of 140–220 or ∼22% of maximum grey scale. Three-dimensional reconstructions were made and plotted as TIF files.
Statistics
Numbers of somites in E10.5 MbtpscKO and control embryos were counted and subjected to a one-way ANOVA analysis using SigmaPlot (version 12.5). Data passed both a normality and an equal variance test. Pairwise comparisons revealed somite numbers for MbtpscKOembryos were significantly different than all three control genotypes (P < 0.012).
Funding
This work was supported by the Scoliosis Research Society (J.P.G. and P.A.T.); National Institutes of Health [grant R01-AR052775 (J.P.G.)]; the Patton Foundation and Kansas City Life Science Institute (J.P.G. and P.A.T.); the University of Missouri Research Board (J.P.G.); the Stowers Institute for Medical Research (P.A.T);. and an American Association of Anatomists post-doctoral fellowship (A.A.); Canada Chair 216684 (NGS); and CIHR 102741 (NGS).
Acknowledgements
J.P.G. wishes to thank Mr. Mark Dallas for his help with the micro-CT scans and their analysis. J.P.G. and P.A.T. appreciate the members of their respective laboratories for their critical input during the completion of this work.
Conflict of Interest statement. None declared.
References
- 1.Boulas M.M. (2009) Recognition of caudal regression syndrome. Adv. Neonatal. Care 9, 61–69 quiz 70-61. [DOI] [PubMed] [Google Scholar]
- 2.Thottungal A.D., Charles A.K., Dickinson J.E., Bower C. (2010) Caudal dysgenesis and sirenomelia-single centre experience suggests common pathogenic basis. Am. J. Med. Genet. A, 152A, 2578–2587. [DOI] [PubMed] [Google Scholar]
- 3.Pourquie O. Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell, 145, 650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dias A.S., de Almeida I., Belmonte J.M., Glazier J.A., Stern C.D. (2014) Somites without a clock. Science, 343, 791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sparrow D.B., Chapman G., Wouters M.A., Whittock N.V., Ellard S., Fatkin D., Turnpenny P.D., Kusumi K., Sillence D., Dunwoodie S.L. (2006) Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am. J. Hum. Genet., 78, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sparrow D.B., Guillen-Navarro E., Fatkin D., Dunwoodie S.L. (2008) Mutation of hairy-and-enhancer-of-Split-7 in humans causes spondylocostal dysostosis. Hum. Mol. Genet., 17, 3761–3766. [DOI] [PubMed] [Google Scholar]
- 7.Bulman M.P., Kusumi K., Frayling T.M., McKeown C., Garrett C., Lander E.S., Krumlauf R., Hattersley A.T., Ellard S., Turnpenny P.D. (2000) Mutations in the human delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat. Genet, 24, 438–441. [DOI] [PubMed] [Google Scholar]
- 8.Whittock N.V., Sparrow D.B., Wouters M.A., Sillence D., Ellard S., Dunwoodie S.L., Turnpenny P.D. (2004) Mutated MESP2 causes spondylocostal dysostosis in humans. Am. J. Hum. Genet., 74, 1249–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padmanabhan R. (1998) Retinoic acid-induced caudal regression syndrome in the mouse fetus. Reprod. Toxicol., 12, 139–151. [DOI] [PubMed] [Google Scholar]
- 10.Vlangos C.N., O'Connor B.C., Morley M.J., Krause A.S., Osawa G.A., Keegan C.E. (2009) Caudal regression in adrenocortical dysplasia (acd) mice is caused by telomere dysfunction with subsequent p53-dependent apoptosis. Dev. Biol., 334, 418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pennimpede T., Proske J., Konig A., Vidigal J.A., Morkel M., Bramsen J.B., Herrmann B.G., Wittler L. (2012) In vivo knockdown of Brachyury results in skeletal defects and urorectal malformations resembling caudal regression syndrome. Dev. Biol., 372, 55–67. [DOI] [PubMed] [Google Scholar]
- 12.Muller Y.L., Yueh Y.G., Yaworsky P.J., Salbaum J.M., Kappen C. (2003) Caudal dysgenesis in Islet-1 transgenic mice. FASEB J., 17, 1349–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szumska D., Pieles G., Essalmani R., Bilski M., Mesnard D., Kaur K., Franklyn A., El Omari K., Jefferis J., Bentham J., et al. (2008) VACTERL/caudal regression/Currarino syndrome-like malformations in mice with mutation in the proprotein convertase Pcsk5. Genes Dev., 22, 1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seidah N.G., Prat A. (2012) The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov, 11, 367–383. [DOI] [PubMed] [Google Scholar]
- 15.Pasquato A., Pullikotil P., Asselin M.C., Vacatello M., Paolillo L., Ghezzo F., Basso F., Di Bello C., Dettin M., Seidah N.G. (2006) The proprotein convertase SKI-1/S1P. In vitro analysis of Lassa virus glycoprotein-derived substrates and ex vivo validation of irreversible peptide inhibitors. J. Biol. Chem., 281, 23471–23481. [DOI] [PubMed] [Google Scholar]
- 16.Seidah N.G., Mayer G., Zaid A., Rousselet E., Nassoury N., Poirier S., Essalmani R., Prat A. (2008) The activation and physiological functions of the proprotein convertases. Int. J. Biochem. Cell. Biol., 40, 1111–1125. [DOI] [PubMed] [Google Scholar]
- 17.Brown M.S., Goldstein J.L. (1999) A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA, 96, 11041–11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorski J.P., Huffman N.T., Cui C., Henderson E.P., Midura R.J., Seidah N.G. (2009) Potential role of proprotein convertase SKI-1 in the mineralization of primary bone. Cell. Tissues Organ., 189, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorski J.P., Huffman N.T., Chittur S., Midura R.J., Black C., Oxford J., Seidah N.G. (2011) Inhibition of proprotein convertase SKI-1 blocks transcription of key extracellular matrix genes regulating osteoblastic mineralization. J. Biol. Chem., 286, 1836–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell K.J., Pinson K.I., Kelly O.G., Brennan J., Zupicich J., Scherz P., Leighton P.A., Goodrich L.V., Lu X., Avery B.J., et al. (2001) Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat. Genet., 28, 241–249. [DOI] [PubMed] [Google Scholar]
- 21.Yang J., Goldstein J.L., Hammer R.E., Moon Y.A., Brown M.S., Horton J.D. (2001) Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. Proc. Natl Acad. Sci. USA, 98, 13607–13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patra D., Xing X., Davies S., Bryan J., Franz C., Hunziker E.B., Sandell L.J. (2007) Site-1 protease is essential for endochondral bone formation in mice. J. Cell. Biol., 179, 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckalbar W.L., Lasku E., Infante C.R., Elsey R.M., Markov G.J., Allen A.N., Corneveaux J.J., Losos J.B., DeNardo D.F., Huentelman M.J., et al. (2012) Somitogenesis in the anole lizard and alligator reveals evolutionary convergence and divergence in the amniote segmentation clock. Dev. Biol., 363, 308–319. [DOI] [PubMed] [Google Scholar]
- 24.Liu F., Woitge H.W., Braut A., Kronenberg M.S., Lichtler A.C., Mina M., Kream B.E. (2004) Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int. J. Dev. Biol, 48, 645–653. [DOI] [PubMed] [Google Scholar]
- 25.Soriano P. (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet., 21, 70–71. [DOI] [PubMed] [Google Scholar]
- 26.Mansouri A., Yokota Y., Wehr R., Copeland N.G., Jenkins N.A., Gruss P. (1997) Paired-related murine homeobox gene expressed in the developing sclerotome, kidney, and nervous system. Dev. Dyn., 210, 53–65. [DOI] [PubMed] [Google Scholar]
- 27.Leitges M., Neidhardt L., Haenig B., Herrmann B.G., Kispert A. (2000) The paired homeobox gene Uncx4.1 specifies pedicles, transverse processes and proximal ribs of the vertebral column. Development, 127, 2259–2267. [DOI] [PubMed] [Google Scholar]
- 28.Pourquie O. (2012) Development: looking to the future. Development, 139, 1893–1894. [DOI] [PubMed] [Google Scholar]
- 29.Oginuma M., Niwa Y., Chapman D.L., Saga Y. (2008) Mesp2 and Tbx6 cooperatively create periodic patterns coupled with the clock machinery during mouse somitogenesis. Development, 135, 2555–2562. [DOI] [PubMed] [Google Scholar]
- 30.Aulehla A., Wehrle C., Brand-Saberi B., Kemler R., Gossler A., Kanzler B., Herrmann B.G. (2003) Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell, 4, 395–406. [DOI] [PubMed] [Google Scholar]
- 31.Aulehla A., Pourquie O. (2008) Oscillating signaling pathways during embryonic development. Curr. Opin. Cell Biol., 20, 632–637. [DOI] [PubMed] [Google Scholar]
- 32.Diez del Corral R., Olivera-Martinez I., Goriely A., Gale E., Maden M., Storey K. (2003) Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron, 40, 65–79. [DOI] [PubMed] [Google Scholar]
- 33.Zhang N., Gridley T. (1998) Defects in somite formation in lunatic fringe-deficient mice. Nature, 394, 374–377. [DOI] [PubMed] [Google Scholar]
- 34.Evrard Y.A., Lun Y., Aulehla A., Gan L., Johnson R.L. (1998) lunatic fringe is an essential mediator of somite segmentation and patterning. Nature, 394, 377–381. [DOI] [PubMed] [Google Scholar]
- 35.McGrew M.J., Dale J.K., Fraboulet S., Pourquie O. (1998) The lunatic fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr. Biol., 8, 979–982. [DOI] [PubMed] [Google Scholar]
- 36.Forsberg H., Crozet F., Brown N.A. (1998) Waves of mouse Lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. Curr. Biol., 8, 1027–1030. [DOI] [PubMed] [Google Scholar]
- 37.Aulehla A., Johnson R.L. (1999) Dynamic expression of lunatic fringe suggests a link between notch signaling and an autonomous cellular oscillator driving somite segmentation. Dev. Biol., 207, 49–61. [DOI] [PubMed] [Google Scholar]
- 38.Williams D.R., Shifley E.T., Lather J.D., Cole S.E. (2014) Posterior skeletal development and the segmentation clock period are sensitive to Lfng dosage during somitogenesis. Dev. Biol., 388, 159–169. [DOI] [PubMed] [Google Scholar]
- 39.Bessho Y., Sakata R., Komatsu S., Shiota K., Yamada S., Kageyama R. (2001) Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev., 15, 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morimoto M., Takahashi Y., Endo M., Saga Y. (2005) The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature, 435, 354–359. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi Y., Koizumi K., Takagi A., Kitajima S., Inoue T., Koseki H., Saga Y. (2000) Mesp2 initiates somite segmentation through the Notch signalling pathway. Nat. Genet., 25, 390–396. [DOI] [PubMed] [Google Scholar]
- 42.Saga Y., Hata N., Koseki H., Taketo M.M. (1997) Mesp2: a novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev., 11, 1827–1839. [DOI] [PubMed] [Google Scholar]
- 43.Zhao X., Duester G. (2009) Effect of retinoic acid signaling on Wnt/beta-catenin and FGF signaling during body axis extension. Gene Expr Patterns, 9, 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crossley P.H., Martin G.R. (1995) The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development, 121, 439–451. [DOI] [PubMed] [Google Scholar]
- 45.Dubrulle J., McGrew M.J., Pourquie O. (2001) FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell, 106, 219–232. [DOI] [PubMed] [Google Scholar]
- 46.Naiche L.A., Holder N., Lewandoski M. (2011) FGF4 and FGF8 comprise the wavefront activity that controls somitogenesis. Proc. Natl Acad. Sci. USA, 108, 4018–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benazeraf B., Francois P., Baker R.E., Denans N., Little C.D., Pourquie O. (2010) A random cell motility gradient downstream of FGF controls elongation of an amniote embryo. Nature, 466, 248–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niwa Y., Masamizu Y., Liu T., Nakayama R., Deng C.X., Kageyama R. (2007) The initiation and propagation of Hes7 oscillation are cooperatively regulated by Fgf and notch signaling in the somite segmentation clock. Dev. Cell, 13, 298–304. [DOI] [PubMed] [Google Scholar]
- 49.Wahl M.B., Deng C., Lewandoski M., Pourquie O. (2007) FGF signaling acts upstream of the NOTCH and WNT signaling pathways to control segmentation clock oscillations in mouse somitogenesis. Development, 134, 4033–4041. [DOI] [PubMed] [Google Scholar]
- 50.George E.L., Georges-Labouesse E.N., Patel-King R.S., Rayburn H., Hynes R.O. (1993) Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development, 119, 1079–1091. [DOI] [PubMed] [Google Scholar]
- 51.Georges-Labouesse E.N., George E.L., Rayburn H., Hynes R.O. (1996) Mesodermal development in mouse embryos mutant for fibronectin. Dev. Dyn., 207, 145–156. [DOI] [PubMed] [Google Scholar]
- 52.Koshida S., Kishimoto Y., Ustumi H., Shimizu T., Furutani-Seiki M., Kondoh H., Takada S. (2005) Integrinalpha5-dependent fibronectin accumulation for maintenance of somite boundaries in zebrafish embryos. Dev. Cell., 8, 587–598. [DOI] [PubMed] [Google Scholar]
- 53.Julich D., Geisler R., Holley S.A. (2005) Integrinalpha5 and delta/notch signaling have complementary spatiotemporal requirements during zebrafish somitogenesis. Dev. Cell., 8, 575–586. [DOI] [PubMed] [Google Scholar]
- 54.Kragtorp K.A., Miller J.R. (2007) Integrin alpha5 is required for somite rotation and boundary formation in Xenopus. Dev. Dyn., 236, 2713–2720. [DOI] [PubMed] [Google Scholar]
- 55.Rifes P., Carvalho L., Lopes C., Andrade R.P., Rodrigues G., Palmeirim I., Thorsteinsdottir S. (2007) Redefining the role of ectoderm in somitogenesis: a player in the formation of the fibronectin matrix of presomitic mesoderm. Development, 134, 3155–3165. [DOI] [PubMed] [Google Scholar]
- 56.Rongish B.J., Drake C.J., Argraves W.S., Little C.D. (1998) Identification of the developmental marker, JB3-antigen, as fibrillin-2 and its de novo organization into embryonic microfibrous arrays. Dev. Dyn., 212, 461–471. [DOI] [PubMed] [Google Scholar]
- 57.Kalajzic I., Kalajzic Z., Kaliterna M., Gronowicz G., Clark S.H., Lichtler A.C., Rowe D. (2002) Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J. Bone Miner. Res., 17, 15–25. [DOI] [PubMed] [Google Scholar]
- 58.Kumar S., Duester G. (2014) Retinoic acid controls body axis extension by directly repressing Fgf8 transcription. Development, 141, 2972–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abu-Abed S., Dolle P., Metzger D., Beckett B., Chambon P., Petkovich M. (2001) The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev., 15, 226–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leung M.B., Choy K.W., Copp A.J., Pang C.P., Shum A.S. (2004) Hyperglycaemia potentiates the teratogenicity of retinoic acid in diabetic pregnancy in mice. Diabetologia, 47, 515–522. [DOI] [PubMed] [Google Scholar]
- 61.Vidigal J.A., Morkel M., Wittler L., Brouwer-Lehmitz A., Grote P., Macura K., Herrmann B.G. (2010) An inducible RNA interference system for the functional dissection of mouse embryogenesis. Nucleic Acids Res., 38, e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saga Y., Takeda H. (2001) The making of the somite: molecular events in vertebrate segmentation. Nat. Rev. Genet 2, 835–845. [DOI] [PubMed] [Google Scholar]
- 63.Mansouri A., Voss A.K., Thomas T., Yokota Y., Gruss P. (2000) Uncx4.1 is required for the formation of the pedicles and proximal ribs and acts upstream of Pax9. Development, 127, 2251–2258. [DOI] [PubMed] [Google Scholar]
- 64.Perkinson R.A., Norton P.A. (1997) Expression of the mouse fibronectin gene and fibronectin-lacZ transgenes during somitogenesis. Dev. Dyn., 208, 244–254. [DOI] [PubMed] [Google Scholar]
- 65.Czirok A., Rongish B.J., Little C.D. (2004) Extracellular matrix dynamics during vertebrate axis formation. Dev. Biol. 268, 111–122. [DOI] [PubMed] [Google Scholar]
- 66.Boregowda R., Paul E., White J., Ritty T.M. (2008) Bone and soft connective tissue alterations result from loss of fibrillin-2 expression. Matrix. Biol., 27, 661–666. [DOI] [PubMed] [Google Scholar]
- 67.Arteaga-Solis E., Sui-Arteaga L., Kim M., Schaffler M.B., Jepsen K.J., Pleshko N., Ramirez F. (2011) Material and mechanical properties of bones deficient for fibrillin-1 or fibrillin-2 microfibrils. Matrix. Biol., 30, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ostrovsky D., Sanger J.W., Lash J.W. (1988) Somitogenesis in the mouse embryo. Cell. Differ., 23, 17–25. [DOI] [PubMed] [Google Scholar]
- 69.Hidalgo M., Sirour C., Bello V., Moreau N., Beaudry M., Darribere T. (2009) In vivo analyzes of dystroglycan function during somitogenesis in Xenopus laevis. Dev. Dyn., 238, 1332–1345. [DOI] [PubMed] [Google Scholar]
- 70.Rifes P., Thorsteinsdottir S. (2012) Extracellular matrix assembly and 3D organization during paraxial mesoderm development in the chick embryo. Dev. Biol., 368, 370–381. [DOI] [PubMed] [Google Scholar]
- 71.Martins G.G., Rifes P., Amandio R., Rodrigues G., Palmeirim I., Thorsteinsdottir S. (2009) Dynamic 3D cell rearrangements guided by a fibronectin matrix underlie somitogenesis. PLoS One, 4, e7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Julich D., Mould A.P., Koper E., Holley S.A. (2009) Control of extracellular matrix assembly along tissue boundaries via Integrin and Eph/Ephrin signaling. Development, 136, 2913–2921. [DOI] [PubMed] [Google Scholar]
- 73.Schubert S.W., Abendroth A., Kilian K., Vogler T., Mayr B., Knerr I., Hashemolhosseini S. (2008) bZIP-Type transcription factors CREB and OASIS bind and stimulate the promoter of the mammalian transcription factor GCMa/Gcm1 in trophoblast cells. Nucleic Acids Res., 36, 3834–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mayr B., Montminy M. (2001) Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol., 2, 599–609. [DOI] [PubMed] [Google Scholar]
- 75.Lopez T.P., Fan C.M. (2013) Dynamic CREB family activity drives segmentation and posterior polarity specification in mammalian somitogenesis. Proc. Natl Acad. Sci. USA, 110, E2019–E2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chandhoke T.K., Huang Y.F., Liu F., Gronowicz G.A., Adams D.J., Harrison J.R., Kream B.E. (2008) Osteopenia in transgenic mice with osteoblast-targeted expression of the inducible cAMP early repressor. Bone, 43, 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Melville D.B., Montero-Balaguer M., Levic D.S., Bradley K., Smith J.R., Hatzopoulos A.K., Knapik E.W. (2011) The feelgood mutation in zebrafish dysregulates COPII-dependent secretion of select extracellular matrix proteins in skeletal morphogenesis. Dis. Model. Mech., 4, 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Webb G.N., Byrd R.A. (1994) Simultaneous differential staining of cartilage and bone in rodent fetuses: an alcian blue and alizarin red S procedure without glacial acetic acid. Biotech. Histochem., 69, 181–185. [DOI] [PubMed] [Google Scholar]
- 79.Nagy A., Gertsenstein M., Vintersten K., Behringer R.R. (2003) Manipulating the Mouse Embryo. 3rd edition Cold Spring Harbor Laboratory, Cold Spring Harbor. [Google Scholar]
- 80.Seidah N.G., Mowla S.J., Hamelin J., Mamarbachi A.M., Benjannet S., Toure B.B., Basak A., Munzer J.S., Marcinkiewicz J., Zhong M., et al. (1999) Mammalian subtilisin/kexin isozyme SKI-1: a widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc. Natl. Acad. Sci. USA, 96, 1321–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen X.L., Grey J.Y., Thomas S., Qiu F.H., Medford R.M., Wasserman M.A., Kunsch C. (2004) Sphingosine kinase-1 mediates TNF-alpha-induced MCP-1 gene expression in endothelial cells: upregulation by oscillatory flow. Am. J. Physiol. Heart Circ. Physiol., 287, H1452–H1458. [DOI] [PubMed] [Google Scholar]
- 82.Kohn M.J., Leung S.W., Criniti V., Agromayor M., Yamasaki L. (2004) Dp1 is largely dispensable for embryonic development. Mol. Cell Biol., 24, 7197–7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clapcote S.J., Roder J.C. (2005) Simplex PCR assay for sex determination in mice. Biotechniques, 38, 702, 704, 706. [DOI] [PubMed] [Google Scholar]