Abstract
The risk of Hirschsprung disease (HSCR) is ∼15/100 000 live births per newborn but has been reported to show significant inter-individual variation from the effects of seven common susceptibility alleles at the RET, SEMA3 and NRG1 loci. We show, by analyses of these variants in 997 samples from 376 HSCR families of European ancestry, that significant genetic risk can only be detected at RET (rs2435357 and rs2506030) and at SEMA3 (rs11766001), but not at NRG1. RET rs2435357 also showed significant frequency differences by gender, segment length of aganglionosis and familiality. Further, in combination, disease risk varied >30-fold between individuals with none and up to 6 susceptibility alleles. Thus, these polymorphisms can be used to stratify the newborn population into distinct phenotypic classes with defined risks to understand HSCR etiology.
Introduction
Hirschsprung disease (HSCR: MIM 142623) is a congenital disorder of the enteric nervous system (ENS) characterized by complete absence of neuronal ganglia in the myenteric and sub-mucosal plexuses from contiguous segments of the intestinal tract (1). This developmental defect arises from a failure of the cranio-caudal migration, proliferation, differentiation or colonization of precursor enteric neural crest cells (ENCCs) in the gastrointestinal tract. A hallmark of HSCR is the marked variation in the resulting length of the contiguous aganglionic segment. Short-segment HSCR (S-HSCR), where aganglionosis is limited up to the upper sigmoid colon, is observed in 80% of HSCR cases; long-segment HSCR (L-HSCR), where aganglionosis extends up to splenic flexure and beyond, is observed in 15–20% of HSCR cases; while total colonic aganglionosis (TCA), where aganglionosis affects the entire large intestine, is observed in ∼5% of HSCR cases (1). With an incidence of ∼15 in 100 000 live births, HSCR is the most common manifestation of a functional intestinal obstruction in neonates (2,3). The phenotype is isolated or non-syndromic in ∼80% of patients and, in the rest, HSCR manifests along with known chromosomal anomalies, such as trisomy 21, or recognized syndromes, such as Mowat–Wilson syndrome, or presents with additional congenital anomalies (1,4).
HSCR is a multifactorial genetic disorder with high heritability (>80%), significant gender bias (4:1 affected males: females), high sibling recurrence risk (4%) and complex inheritance patterns (2). Numerous molecular genetic studies have identified rare, coding, high penetrance variants in fourteen genes (RET [MIM 164761], GDNF [MIM 600837], NRTN [MIM 602018], SOX10 [MIM 602229], EDNRB [MIM 131244], EDN3 [MIM 131242], ECE1 [MIM 600423], ZFHX1B [MIM 605802], TCF4 [MIM 602272], PHOX2B [MIM 603851], KBP [MIM 609367], L1CAM [MIM 308840], SEMA3C [MIM 602645], SEMA3D [MIM 609907]) that are the primary causal factors in syndromic or familial cases (4–7, Jiang et al. under review). Among these, RET, encoding a receptor tyrosine kinase, harbors the largest number (>80%) of HSCR-associated loss-of-function variants (4,5). These genes have uncovered critical ENS developmental pathways, defects which lead to HSCR, but these pathogenic alleles cumulatively occur in <20% of cases.
In ∼80% of HSCR cases, a common, low penetrance, noncoding RET variant (rs2435357) underlies disease risk in cases with both European and Asian ancestry (8,9). rs2435357 maps to intron 1 of RET and the disease-associated allele disrupts binding of SOX10, a key ENS transcription factor, to a gut-specific RET enhancer (9). Analogously, NRG1 is a second susceptibility locus identified through a genome-wide association study (GWAS) in Asian subjects with two common, independent risk variants, rs7835688 and rs16879552, both of which map to intron 1 of NRG1 (10–12). This locus encodes Neuregulin 1, a signaling glycoprotein that binds to ERBB receptor tyrosine kinases and plays a major role in the development and maintenance of the ENS (13). In a more recent GWAS of European (the USA, Italy, France, Spain and the Netherlands) ancestry HSCR cases, we identified an independent association at rs2506030, ∼125 kb upstream of RET, and a novel common noncoding variant rs12707682 mapping within the class 3 Semaphorin (SEMA3) gene cluster (SEMA3 A/C/D/E) (Jiang et al. under review). Class 3 Semaphorins encode secreted, transmembrane or GPI-linked proteins that play a significant role in development and homeostasis across tissues types, and have been broadly studied for their roles in axonal guidance, neuronal plasticity/connectivity and cardiac development; they represent a new gene for HSCR (14,15). The causal molecular identities of these associations, excepting RET rs2435357, are unknown and the identified variants are likely surrogate markers and not causal variants.
Except for RET rs2435357, the other markers either have not been studied in independent samples or replicated in European-ancestry subjects. Nevertheless, given their high frequency and large effects in S-HSCR, the commonest subclass, they are expected to be larger contributors to disease risk than all known rare coding variants: indeed, RET rs2435357 explains 10-fold greater disease liability than do all RET coding variants (8). Finally, this variant is positively associated with male gender, S-HSCR, and simplex cases (familiality) (9), revealing some novel genotype–phenotype correlations. However, several questions remain unanswered. First, are the genetic associations at RET, NRG1 and SEMA3 universal to HSCR or dependent on the phenotypes of patients studied in European-ancestry subjects? Second, can genotype–phenotype correlations be demonstrated for all identified variants? Third, what are the combined effects of all significant associations? The main rationale for examining their total (combined) effect is that unlike rare variants, HSCR patients can harbor multiple common disease variants so that we can test whether disease risk increases additively or synergistically with susceptibility allele dosage. For a multifactorial disorder, two extreme genetic risk scenarios can be postulated: one in which affected individuals vary from low to high risk propensity, and, a second in which clinical affection results only when an individual crosses some allele dosage threshold (truncate selection) (16). To resolve these questions we require that all known disease-associated polymorphisms be examined in the same group of patients.
We conducted this study, in a large number of European-ancestry HSCR probands, and their parents, all ascertained using the same criteria within the USA, to assess the individual and combined quantitative roles of seven reported common variants at the RET, NRG1 and SEMA3 loci, using both case–control and family-based association studies. We further wanted to quantify their genotype–phenotype correlations with respect to gender, segment length of aganglionosis and familiality. We used both population- and family-based association analyses to demonstrate that RET and SEMA3 but not NRG1 variants display persistent associations and are not the result of population stratification. We also demonstrate that HSCR risk increases as a logistic function of the total number of risk alleles and differs by >30-fold across the spectrum, showing significant gender differences. Our study suggests that there is great heterogeneity in HSCR risk in the human newborn population and that known susceptibility polymorphisms can distinguish and stratify these individuals for research into additional genetic or environmental differences between these groups.
Results
Defining HSCR-marker associations at RET, SEMA3 and NRG1
We studied seven variants at RET, SEMA3 and NRG1 to test whether they were significantly associated in 355 European-ancestry HSCR subjects and 379 controls plus the untransmitted pseudo-controls from 254 HSCR trios. We used population-based case–control analysis and we compared allele and genotype frequencies (Table 1 and Supplementary Material, Table S4). At both RET SNPs, the risk allele frequencies (T at rs2435357 and G at rs2506030) were significantly higher in cases when compared with controls (58% versus 26% at rs2435357, P = 4.3 × 10−44; 56% versus 41% at rs2506030, P = 4.7 × 10−10). At the genotype level, a significant excess of risk allele homozygotes at both RET SNPs was observed in cases versus controls (42% versus 7% at rs2435357, P = 1.5 × 10−41; 30% versus 18% at rs2506030, P = 5.3 × 10−9). The polymorphisms rs2435357 and rs2506030 at RET have different allele frequencies in controls (0.26 versus 0.41; Table 1) and show a small level of linkage disequilibrium (LD) (r2 = 0.08) in controls but a small and somewhat higher correlation (r2 = 0.16; cases versus controls P = 0.05) in our cases (Supplementary Material, Table S3b). These results suggest that the two RET markers define different, independent causal factors that are weakly correlated in controls by virtue of their physical proximity; the higher correlation in cases likely represents their interaction in specifying HSCR risk. The current samples estimate the genetic effects of these SNPs on HSCR risk as significant with odds ratio (OR) of 3.9 (P = 4.3 × 10−44) and 1.8 (P = 4.7 × 10−10) for rs2435357 and rs2506030, respectively (Table 1), comparable with those observed previously for rs2435357 [OR = 4 (8), OR = 5.3 (9), OR = 4.3 (Jiang et al. under review)] and for rs2506030 [OR = 1.5 (Jiang et al. under review)]. Thus, we now confirm the evidence of multiple genetic associations within RET (Table 1).
Table 1.
Case–control and transmission disequilibrium (TDT) association tests of RET, SEMA3 and NRG1 polymorphisms in HSCR
| Gene | SNP ID and risk/non-risk allele | Case–control |
TDT |
|||||
|---|---|---|---|---|---|---|---|---|
| Risk allele (case–control frequency) | Odds ratio (95% CI) | P | Risk allele transmitted/un-transmitted (T/U) | Odds ratio(95% CI) | P | Transmission rate (τ ± sd) | ||
| RET | rs2435357: T/C | 0.58/0.26 | 3.9 (3.2–4.7) | 4.3 × 10−44 | 219/50 | 4.4 (3.2–6.0) | 6.8 × 10−25 | 0.82 ±0.02 |
| RET | rs2506030: G/A | 0.56/0.41 | 1.8 (1.5–2.2) | 4.7 × 10−10 | 164/93 | 1.8 (1.4–2.3) | 9.5 × 10−6 | 0.63 ±0.03 |
| SEMA3 | rs11766001: C/A | 0.22/0.15 | 1.6 (1.3–2.0) | 1.0 × 10−4 | 96/55 | 1.7 (1.3–2.4) | 8.5 × 10−4 | 0.64 ±0.04 |
| SEMA3 | rs12707682: C/T | 0.30/0.24 | 1.3 (1.1–1.6) | 0.01 | 114/92 | 1.2 (0.9–1.6) | 0.13 | 0.57 ±0.03 |
| SEMA3 | rs1583147: T/C | 0.28/0.23 | 1.3 (1.1–1.6) | 0.01 | 115/86 | 1.3 (1.0–1.8) | 0.04 | 0.57 ±0.03 |
| NRG1 | rs16879552: C/T | 0.97/0.96 | 1.2 (0.7–2.1) | 0.43 | 13/15 | 0.9 (0.4–1.8) | 0.71 | 0.50 ±0.09 |
| NRG1 | rs7835688: C/G | 0.49/0.47 | 1.1 (0.9–1.3) | 0.44 | 134/124 | 1.1 (0.8–1.4) | 0.53 | 0.53 ±0.03 |
For case–control association, the risk allele frequency in cases and controls, odds ratio with 95% confidence interval (CI) and the significance value of association (P) are provided. For TDT, the counts of risk allele transmitted and un-transmitted from heterozygous parents, odds ratio with 95% CI, the significance value of association (P) and the estimated transmission rate (τ) with its standard deviation (SD) are provided. The transmission rate (τ) was estimated from all trios and duos using a maximum likelihood method (8).
The values in bold are statistically significant findings.
At SEMA3, allelic associations were strongest at rs11766001 where the risk allele C was at a significantly higher frequency in cases (22%) than in controls (15%) (P = 1.0 × 10−4). The risk allele frequencies at the two other SEMA3 SNPs, C at rs12707682 and T at rs1583147, were also higher in cases than in controls (30% versus 24% at rs12707682, P = 0.01; 28% versus 23% at rs1583147, P = 0.01) but showed borderline significance after adjusting for multiple tests. Similarly, at the genotype level, significant association of HSCR with rs11766001 was observed (P = 6.4 × 10−4), with borderline associations at rs12707682 and rs1583147 (P = 0.03 for both) after multiple testing corrections. Finally, the estimated genetic effects of the SEMA3 SNPs on HSCR were with odds ratios of 1.6 (P = 1.0 × 10−4), 1.3 (P = 0.01) and 1.3 (P = 0.01) for rs11766001, rs12707682 and rs1583147, respectively, comparable with those observed in a smaller study [OR = 1.5 for both rs11766001 and rs12707682; Jiang et al. under review]. The current larger dataset therefore confirms an independent genetic susceptibility at SEMA3 rs11766001; we conclude that the effects at rs12707682 and rs1583147 are owing to LD (Table 1).
In contrast, we found no evidence of genetic association between HSCR and either NRG1 SNPs, rs16879552 and rs7835688, at either the allele or the genotype levels, in our European-ancestry subjects (Table 1 and Supplementary Material, Table S4). Association of HSCR with NRG1 was originally reported in Chinese HSCR patients (10) and has indeed been replicated in other Asian-ancestry cases (11,12). However, association of HSCR with these common NRG1 SNPs was not replicated in a recent study of Spanish HSCR patients either (17). And using European-ancestry HSCR probands collected from the USA, Italy, the Netherlands, France and Spain, we also failed to detect association with rs4541858, a variant in near perfect LD with the NRG1 rs7835688 variant (Jiang et al. under review). We therefore conclude, based on the larger sample presented here, that NRG1 is not a susceptibility polymorphism for European-ancestry HSCR.
Population-based association studies are notorious for being confounded with cryptic population structure. Consequently, we also performed family-based association analyses using the transmission disequilibrium test (TDT) in 254 both parents-child trios and 72 one parent-child duos to guard against this possibility (Table 1). First, at both RET SNPs, the known risk alleles (T at rs2435357 and G at rs2506030) are significantly over-transmitted to probands with transmission rates (τ) of 0.82 and 0.63 for rs2435357 (P = 6.8 × 10−25) and rs2506030 (P = 9.5 × 10−6), respectively (Table 1). Among the three SEMA3 SNPs evaluated, TDT analyses showed significant association only at rs11766001, where the risk allele C was transmitted more often than expected by chance (τ = 0.64, P = 8.5 × 10−4) (Table 1). Once again, although the same trend was observed for the risk alleles C at rs12707682 and T at rs1583147, they were not statistically significant (τ = 0.57, P = 0.13 for rs12707682 and τ = 0.57, P = 0.04 for rs1583147). Finally, TDT analysis did not show any evidence of association of NRG1 SNPs, rs16879552 and rs7835688, with HSCR (Table 1). The availability of family data provided us the opportunity to test for parent-of-origin effects by comparing transmission frequency of the risk variant from fathers versus mothers at all seven SNPs, but we failed to detect any statistically significant differences (Supplementary Material, Table S5). We conclude that the three associations detected at rs2435357, rs2506030 and rs11766001 represent independent pervasive disease associations for European-ancestry HSCR. Importantly, the magnitude of their associations with disease (OR) are highly concordant between the case–control and TDT analyses.
Genotype–phenotype associations at HSCR
We have previously reported that the RET enhancer variant rs2435357 shows genotype–phenotype associations with male gender, short- or long-segment aganglionosis and isolated case; in contrast, the mirror image HSCR features, female gender, TCA and multiplex case are associated with RET coding variants (9). Consequently, we wished to test whether all three significant polymorphisms showed such patterns or whether they were restricted to specific markers by comparing frequencies of risk alleles within cases classified by these features (Supplementary Material, Table S6). Since in our previous report the effect of RET rs2435357 was not significantly different between S-HSCR and L-HSCR (9), we grouped these two classes together and compared them with TCA HSCR cases. For gender, the risk allele at RET rs2435357 was more common in males than females (61% versus 48%, P = 0.001). However, no significant difference by gender was observed at RET rs2506030 (P = 0.34) and SEMA3 rs11766001 (P = 0.25). For segment length of aganglionosis, the risk allele at RET rs2435357 was observed to be more common in S-HSCR/L-HSCR when compared with TCA HSCR (63% versus 51%, P = 0.02). Likewise, no significant difference by segment length was observed at RET rs2506030 (P = 0.34) and SEMA3 rs11766001 (P = 0.30). Finally, for familiality, the risk allele at RET rs2435357 was also observed to be more common in simplex when compared with multiplex HSCR (60% versus 52%, P = 0.05). Once again, no significant difference by familiality was observed at RET rs2506030 (P = 0.65) and SEMA3 rs11766001 (P = 0.14). Thus, among the three HSCR-associated polymorphisms, only RET rs2435357 showed significant genotype–phenotype associations (Supplementary Material, Table S6).
Combined effect of RET and SEMA3 variants
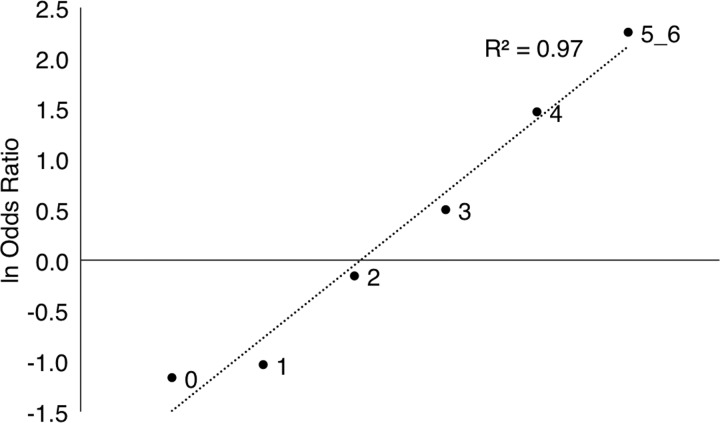
Given that RET and SEMA3 are early ENS development genes with common polymorphisms that impart high risk, we enquired whether combinations of such alleles were synergistic with respect to risk. Thus, we compared the total number of risk alleles at RET rs2435357 and rs2506030 and SEMA3 rs11766001 between HSCR cases and controls (Table 2). HSCR risk, measured in terms of the odds ratio, was directly and convincingly related to risk allele dosage (Table 2) and demonstrated three classes of individuals. First, disease risk was highly significant in individuals with three or more risk alleles (OR = 1.6, P = 0.003; OR = 4.3, P = 1.2 × 10−11; OR = 9.5, P = 8.3 × 10−8 for three, four and five or more risk alleles, respectively). Second, possessing one or no risk allele was significantly protective (OR = 0.4, P = 8.2 × 10−9; OR = 0.3, P = 1.2 × 10−6 for one or no risk allele, respectively). Third, individuals with 2 risk alleles were neither protected nor susceptible (OR = 0.9, P = 0.311). The risk of HSCR owing to these genetic variants increased steadily from 0.3 to 9.5, a 32-fold change, as the number of risk variants increased from 0 to 6. This relationship is clearly described by a logistic function with a highly significantly fit to the observations (R2 = 0.97) (Fig. 1). Interestingly, these risk features also demonstrate variation by gender with similar patterns but with a 65- and 14-fold change for males (range 0.2 to 13.0) and females (0.5 to 6.8), respectively, as the number of risk variants increased from 0 to 6 (Supplementary Material, Table S7). Once again, these relationships are also described by a logistic function with a highly significantly fit to the observations (R2 = 0.90 for females and 0.97 for males) (Supplementary Material, Fig. S1).
Table 2.
Odds ratio and population-level risk (penetrance) of HSCR as a function of the number of risk-increasing variants at RET and SEMA3
| Number of risk alleles | Number (%) of cases | Number (%) of controls | Odds ratio (95% CI) | P | Penetrance |
|---|---|---|---|---|---|
| 0 | 23 (6.7) | 115 (18.6) | 0.3 (0.2–0.5) | 1.2 × 10−6 | 5.4 |
| 1 | 47 (13.3) | 190 (30.7) | 0.4 (0.2–0.5) | 8.2 × 10−9 | 6.6 |
| 2 | 84 (24.3) | 169 (27.3) | 0.9 (0.6–1.2) | 0.311 | 13.4 |
| 3 | 84 (24.3) | 101 (16.3) | 1.6 (1.2–2.3) | 0.003 | 22.3 |
| 4 | 73 (21.4) | 36 (5.8) | 4.3 (2.8–6.6) | 1.2 × 10−11 | 54.5 |
| 5 or 6 | 34 (9.9) | 7 (1.1) | 9.5 (4.2–21.8) | 8.3 × 10−8 | 130.5 |
The numbers of cases and controls classified by the number of risk alleles at RET SNPs rs2435357 and rs2506030 and SEMA3 SNP rs11766001, odds ratio with 95% confidence interval (CI), statistical significance of association (P) and population penetrance are provided. The penetrance value shown is the expected number of cases in 100 000 live births assuming a total population incidence of 15 cases per 100 000 live births.
The values in bold are statistically significant findings.
Figure 1.
Logistic function relating HSCR risk with the number of risk alleles at RET (rs2435357, rs2506030) and SEMA3 (rs11766001).
These results can be used to estimate the population penetrance (population probability of being affected given each genotype) for each risk allele count (classes: 0-6) using the observed background frequency of each class in controls and assuming a population HSCR incidence of 15 cases per 100 000 live births. These values, shown in Table 2 as the expected number of cases per 100 000 live births, vary between 5.4 cases and 130.5 cases for zero risk allele count to five or more risk allele counts, respectively. Thus, the lowest risk class (allele count 0) has a population incidence of ∼1/20 000 live births while the highest risk class (allele count 5 or 6) has a population incidence of ∼1/800 live births. The highest risks emanate from those with three or more risk alleles (Table 2).
We finally tested whether the combined genetic effects of RET rs2435357 and rs2506030 and SEMA3 rs11766001 are similar across the known HSCR risk features of gender, segment length of aganglionosis and familiality. HSCR risk was significantly higher in males when compared with females (P = 0.016; Supplementary Material, Table S8); risk was also higher by segment length of aganglionosis and familiality but the effects were not statistically significant (P = 0.36 and P = 0.17, respectively; Supplementary Material, Table S8). As expected from individual SNP analysis (above), the gender effect was primarily driven by RET rs2435357 and was marginal when counting risk alleles only at RET rs2506030 and SEMA3 rs11766001 (P = 0.09).
Discussion
We present here data and genetic analyses of seven polymorphisms at the RET, NRG1 and SEMA3 loci in the largest collection of 997 samples from 376 HSCR families of European ancestry to assess the significance and patterns of susceptibility associations in HSCR. The data here represent nearly twice the number of HSCR cases and families studied before, and do so simultaneously for all known common variants in a common sample. Our study now firmly establishes that two RET variants, rs2435357 and rs2506030, and one SEMA3 variant, rs11766001, are common susceptibility alleles in European-ancestry HSCR subjects.
Our results demonstrate and reiterate that RET rs2435357, with an OR of 3.9 (Table 1), is the single largest known genetic risk factor. Based on TDT, we also find that rs2435357 is significantly over-transmitted (82% transmission rate, Table 1). RET rs2506030, with an OR of 1.8 (Table 1), nearly half of that for rs2435357, is the second largest known genetic risk factor. By TDT, rs2506030 is also significantly over-transmitted (63% transmission rate, Table 1). These two RET SNPs have very low LD in European-ancestry controls thereby indicating the presence of two independent genetic effects at RET with distinct effect sizes (Supplementary Material, Table S3b). The small but somewhat higher correlation between these same markers in HSCR cases (Supplementary Material, Table S3b) suggests an interaction between their genetic effects. We have previously shown that rs2435357, located within RET intron 1, is located within an SOX10-dependent transcription enhancer for the gastrointestinal tract whose activity is lost in the risk allele. The functional basis for the effect at rs2506030, ∼125 kb upstream of RET is unknown but preliminary annotation suggests that it too may be a transcriptional enhancer of RET. However, this hypothesis needs to be functionally proven, particularly to explain the postulated interaction with rs2435357.
At the SEMA3 locus, all three variants evaluated are significantly associated with HSCR risk by case–control (OR = 1.3–1.6, Table 1) analyses; however, only rs11766001 is significantly associated by TDT (64% transmission rate, Table 1). Thus, only rs11766001 is considered a third and RET-independent HSCR susceptibility factor. Unlike the RET rs2435357 polymorphism, the identity of functional variant(s) and causal gene(s) underlying this association remains unknown; however, functional data clearly implicate the SEMA3C or SEMA3D genes or both (Jiang et al. under review). As mentioned earlier, the SEMA3 locus also harbors additional members of class 3 Semaphorins, namely SEMA3A and SEMA3E. Based on up-regulation of SEMA3A expression observed in the aganglionic smooth muscle layer of the colon in HSCR subjects, increased SEMA3A expression is a likely risk factor in a subset of HSCR subjects (18). Mutation screening in HSCR cases and controls have also identified rare coding variants in SEMA3 A/C/D associated with HSCR (6,7, Jiang et al. under review) indicating that more than one class 3 Semaphorin could be responsible for the underlying HSCR pathology.
Our most significant findings relate to the risk propensity of individuals possessing various numbers of susceptibility alleles. Our observations clearly show that genetic risk increases with the numbers of susceptibility alleles as a logistic function. Individuals with two risk alleles, which comprise 27.3% of the general population (Table 2), have essentially background risk at ∼15/100 000 live births. However, individuals with three or more risk alleles, which comprise 23.3% of the general population, have elevated risk varying from 1.6- to 9.5-fold. The highest risk, those with five or six risk alleles, have an estimated risk of ∼1/800 live births. In contrast, those with one or fewer risk alleles, which comprise ∼50% of the general population, have an estimated risk of ∼1/20 000 live births, typical of many Mendelian disorders. This 32-fold range in risk, OR from 0.3 to 9.5 between those with 0 versus 5 or 6 alleles, is statistically highly significant and, to the best of our knowledge, unprecedented for any heritable genetic disorder. As we have also shown, these variations are more marked when individuals are classified by gender (Supplementary Material, Table S8). Consequently, understanding the molecular basis of this sex-biased risk variation is important since it appears to be a property of multiple risk alleles. One hypothesis to explain these results is that each risk allele, as we have shown for rs2435357 (9), is a partial loss-of-function allele (hypomorph) of either RET or a gene that interacts with RET function in early ENS development, as SEMA3D/C do (Jiang et al. under review). Consequently, increasing numbers of risk alleles correlate with decreasing levels of RET and, consequently, higher HSCR risk. Notwithstanding this hypothesis, the highest risk estimated is ∼1/800 so that most newborns with 5 or 6 risk alleles escape from HSCR. Therefore, how aganglionosis and clinical affection develops remains an enigma. We envision two possibilities: the vast majority of newborns with HSCR get aganglionosis simply by stochastic effects during ENS development or consequent to other hits, be they genetic, such as rare coding mutations, or an environmental insult. We suggest that exome sequencing of a diverse set of HSCR patients may inform these hypotheses.
Our observations argue for the use of these three significant polymorphisms as a basis for risk stratification of HSCR since ∼50% of the newborn population is protected, ∼27% has average risk while the remaining 23% have elevated risk. This type of stratification may lead to an improved search for biological and environmental factors that precipitate clinical disease. More importantly, such risk stratification may help in our understanding of the causes of clinical complications, such as enterocolitis, that are a significant cause of morbidity and mortality in HSCR. Finally, since these polymorphisms are simply inherited we infer that probands with varying numbers of susceptibility alleles will also predict significantly different recurrence risk to their siblings and other relatives.
Materials and Methods
Patient samples
We analyzed HSCR patients and their family members we ascertained within the USA through participating centers, mostly hospitals and clinical units, by review of existing patient medical records or from referrals by practicing physicians, genetic counselors or family members. Diagnosis of HSCR was based on surgical reports, pathological examination of rectal biopsies or other medical records. Patients were categorized by segment length of aganglionosis as defined earlier; the extent of aganglionosis was indeterminable in ∼30% of cases. All individuals, primarily of self-described European-American ancestry, were ascertained under written informed consent approved by the Institutional Review Board of the Johns Hopkins School of Medicine. For genetic analysis, we used genomic DNA isolated from peripheral blood, buccal swabs or salvia samples from participants using standard protocols.
We studied a total of 997 samples from 376 HSCR families. Supplementary Material, Table S1 provides the phenotypic features of these 376 probands by gender, segment length of aganglionosis and familiality. Of these, 515 subjects from 187 families have been previously studied for some of these markers in a primary GWAS and its replication (Jiang et al. under review); we have now added new markers, 482 new samples and genotypes from 189 additional families. A smaller set of these individuals have been previously studied for rs2435357 in conjunction with samples from the HSCR International Consortium (9).
SNP genotyping and quality control
We analyzed seven SNPs reported as significantly associated with HSCR: rs2435357 (8) and rs2506030 (Jiang et al. under review) at RET; rs12707682 and rs11766001 at SEMA3 (Jiang et al. under review); and rs16879552 and rs7835688 at NRG1 (10) (Supplementary Material, Table S2). Additionally, rs1583147 at the SEMA3 locus was genotyped, although not reported by Jiang et al. (under review), since it is in high LD with rs12707682 [r2 = 0.89; 1000 Genomes European-ancestry samples (EUR) and untransmitted pseudo-controls from HSCR trios] (Supplementary Material, Table S3b). DNA samples were genotyped for each SNP using TaqMan Human Pre-Designed genotyping assays following the manufacturer's protocol (Applied Biosystems). The assays IDs are as follows: C__16017524_10 (rs2435357), C__26742714_10 (rs2506030), C__30936238_10 (rs12707682), C__11238335_10 (rs11766001), C__7528379_10 (rs1583147), C__32689001_10 (rs16879552) and C__32689004_10 (rs7835688). The end-point fluorescence measurements were performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems) and analyzed using Sequence Detection System Software v.2.1 (Applied Biosystems). For controls, we used genotypes from the 1000 Genomes (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20110521/) EUR samples (n = 379) and pseudo-controls from 254 HSCR trios generated using PseudoCons (19) (http://www.staff.ncl.ac.uk/richard.howey/pseudocons/index.html/).
Genotyping was performed in a total of 997 samples that included 376 probands. We filtered out 31 samples (3.1% failure) that failed ≥4 SNPs leaving 966 samples: the distribution of missing data and genotyping success rate for each SNP are provided in Supplementary Material, Table S2. The final genotyping success rate was ≥99.0% per SNP leaving a total of 355 probands of which 254 had genotypes on both parents (trios) and 72 had genotypes from only one parent (duos). We used 355 probands for population-based case–control association studies and 254 trios and 72 duos for family-based association studies. Genotypes at each SNP, from all unrelated probands and controls, were tested separately for Hardy–Weinberg equilibrium (HWE): none were significant except rs2435357 in HSCR cases (P = 6.3 × 10−12; Supplementary Material, Table S3a), as expected, owing to the known population-level association with HSCR (8,9).
Statistical genetic analysis
Population-based case–control association analyses were performed, for alleles and genotypes, using standard contingency χ2 tests. Standard methods were used for calculations of odds ratios (OR), their confidence limits and statistical significance of deviation from no effect (OR = 1). Family-based association tests were performed in trios using the transmission disequilibrium test (TDT) (20) for single SNPs as implemented in PLINK (21) (http://pngu.mgh.harvard.edu/~purcell/plink/). A maximum likelihood method was used to estimate the risk allele transmission rate (τ) from the trio and duo genotypes (8). Owing to moderate-to-high LD between the three SEMA3 SNPs (Supplementary Material, Table S3b), they were not considered as independent; thus, statistical significance thresholds were adjusted for multiple testing following the Bonferroni correction for five tests or P < 0.01. Pairwise LD (r2) were compared between cases and controls using Fisher's r-to-z transformation. For analyzing the combined effects of SNPs, we considered only variants significantly associated with HSCR risk and counted the total number of risk alleles in each individual (range: 0–6). Population-level disease penetrance for each risk allele count was estimated using Bayes' theorem with the observed background control frequency and a disease incidence of 15 cases per 100 000 live births (9).
Supplementary Material
Funding
The studies reported here were supported by a grant from the US National Institutes of Health (MERIT Award HD28088 to A.C.).
Supplementary Material
Acknowledgements
We thank the numerous patients, their families and referring physicians that have contributed to these studies, and particularly Erick Kaufmann, Jennifer (Scott) Bubb, Maura Kenton and Julie Albertus for family ascertainment and genetic counseling. We gratefully acknowledge the critical review and suggestions of two anonymous reviewers that greatly improved the presentation.
Conflict of Interest statement. A.C. is on the Scientific Advisory Board of Biogen Idec and this potential competing interest is managed by the policies of the Johns Hopkins University, School of Medicine.
References
- 1.Chakravarti A., Lyonnet S. (2001) Hirschsprung disease. In Scriver C.R., Beaudet A.L., Valle D., Sly W.S., Childs B., Kinzler K., Vogelstein B. (eds.), The Metabolic and Molecular Bases of Inherited Disease, 8th edn McGraw-Hill, New York, pp. 6231–6255. [Google Scholar]
- 2.Badner J.A., Sieber W.K., Garver K.L., Chakravarti A. (1990) A genetic study of hirschsprung disease. Am. J. Hum. Genet., 46, 568–580. [PMC free article] [PubMed] [Google Scholar]
- 3.Parisi M.A., Kapur R.P. (2000) Genetics of hirschsprung disease. Curr. Opin. Pediatr., 12, 610–617. [DOI] [PubMed] [Google Scholar]
- 4.Amiel J., Sproat-Emison E., Garcia-Barcelo M., Lantieri F., Burzynski G., Borrego S., Pelet A., Arnold S., Miao X., Griseri P., et al. (2008) Hirschsprung disease, associated syndromes and genetics: a review. J. Med. Genet., 45, 1–14. [DOI] [PubMed] [Google Scholar]
- 5.Alves M.M., Sribudiani Y., Brouwer R.W., Amiel J., Antinolo G., Borrego S., Ceccherini I., Chakravarti A., Fernandez R.M., Garcia-Barcelo M.M., et al. (2013) Contribution of rare and common variants determine complex diseases—hirschsprung disease as a model. Dev. Biol., 382, 320–329. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Q., Turner T., Sosa M.X., Rakha A., Arnold S., Chakravarti A. (2012) Rapid and efficient human mutation detection using a bench-top next-generation DNA sequencer. Hum. Mutat., 33, 281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luzon-Toro B., Fernandez R.M., Torroglosa A., de Agustin J.C., Mendez-Vidal C., Segura D.I., Antinolo G., Borrego S. (2013) Mutational spectrum of semaphorin 3A and semaphorin 3D genes in Spanish hirschsprung patients. PLoS One, 8, e54800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emison E.S., McCallion A.S., Kashuk C.S., Bush R.T., Grice E., Lin S., Portnoy M.E., Cutler D.J., Green E.D., Chakravarti A. (2005) A common sex-dependent mutation in a RET enhancer underlies hirschsprung disease risk. Nature, 434, 857–863. [DOI] [PubMed] [Google Scholar]
- 9.Emison E.S., Garcia-Barcelo M., Grice E.A., Lantieri F., Amiel J., Burzynski G., Fernandez R.M., Hao L., Kashuk C., West K., et al. (2010) Differential contributions of rare and common, coding and noncoding ret mutations to multifactorial hirschsprung disease liability. Am. J. Hum. Genet., 87, 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Barcelo M.M., Tang C.S., Ngan E.S., Lui V.C., Chen Y., So M.T., Leon T.Y., Miao X.P., Shum C.K., Liu F.Q., et al. (2009) Genome-wide association study identifies NRG1 as a susceptibility locus for hirschsprung's disease. Proc. Natl Acad. Sci. USA., 106, 2694–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phusantisampan T., Sangkhathat S., Phongdara A., Chiengkriwate P., Patrapinyokul S., Mahasirimongkol S. (2012) Association of genetic polymorphisms in the RET-protooncogene and NRG1 with hirschsprung disease in Thai patients. J. Hum. Genet., 57, 286–293. [DOI] [PubMed] [Google Scholar]
- 12.Gunadi Kapoor A., Ling A.Y., Rochadi Makhmudi A., Herini E.S., Sosa M.X., Chatterjee S., Chakravarti A. (2014) Effects of RET and NRG1 polymorphisms in indonesian patients with hirschsprung disease. J. Pediatr. Surg., 49, 1614–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Britsch S. (2007) The neuregulin-I/ErbB signaling system in development and disease. Adv. Anat. Embryol. Cell Biol., 190, 1–65. [PubMed] [Google Scholar]
- 14.Kolodkin A.L., Matthes D.J., Goodman C.S. (1993) The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell, 75, 1389–1399. [DOI] [PubMed] [Google Scholar]
- 15.Kruger R.P., Aurandt J., Guan K.L. (2005) Semaphorins command cells to move. Nat. Rev. Mol. Cell Biol., 6, 789–800. [DOI] [PubMed] [Google Scholar]
- 16.Chakravarti A. (1999) Population genetics—making sense out of sequence. Nat. Genet., 21, 56–60. [DOI] [PubMed] [Google Scholar]
- 17.Luzon-Toro B., Torroglosa A., Nunez-Torres R., Enguix-Riego M.V., Fernandez R.M., de Agustin J.C., Antinolo G., Borrego S. (2012) Comprehensive analysis of NRG1 common and rare variants in hirschsprung patients. PLoS One, 7, e36524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L.L., Fan Y., Zhou F.H., Li H., Zhang Y., Miao J.N., Gu H., Huang T.C., Yuan Z.W. (2011) Semaphorin 3A expression in the colon of hirschsprung disease. Birth Defects Res. A Clin. Mol. Teratol., 91, 842–847. [DOI] [PubMed] [Google Scholar]
- 19.Cordell H.J., Barratt B.J., Clayton D.G. (2004) Case/pseudocontrol analysis in genetic association studies: A unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet. Epidemiol., 26, 167–185. [DOI] [PubMed] [Google Scholar]
- 20.Spielman R.S., McGinnis R.E., Ewens W.J. (1993) Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am. J. Hum. Genet., 52, 506–516. [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.