Abstract
A powerful, cost-effective new method for studying single-nucleotide polymorphisms (SNPs) is described. This method is based on the use of hairpin-shaped primers (HP), which give a sensitive and specific PCR amplification of each specific allele, without the use of costly fluorophore-labeled probes and any post-PCR manipulation. The amplification is monitored in real-time using SYBR Green I dye and takes only 2 h to yield results. The HP assay has a simple design and utilizes a conventional real-time PCR apparatus. The −44 C→G transversion in the DEFB1 gene (which encodes human β-defensin 1) has been previously associated with Candida carriage in oral epithelia. In this study, we analyzed the association between early-onset periodontal disease (EOP) and the −44 SNP. We used an HP assay to study the distribution of the −44 SNP in 264 human DNAs obtained from two cohorts of EOP patients and healthy controls from different ethnic backgrounds. The results indicate that the −44 SNP has a similar distribution between EOP and healthy patients, suggesting that it is not associated with the disease.
The completion of the Human Genome Project has led to researchers becoming increasingly interested in studying single-nucleotide polymorphisms (SNPs). SNPs are the most abundant form of genetic variation between individuals; some have been observed to cause several diseases (e.g., sickle cell anemia [28]) and have been proven to be useful tools for identifying genes involved in common polygenic and multifactorial diseases (8). SNPs have also proven to be useful tools for understanding the pathogenesis of several diseases (e.g., (4).
Most of the methods available for SNP detection, such as techniques based on mass spectrometry (20), high-pressure liquid chromatography (23), microarray analysis (6), Taqman probes (16), or molecular beacons (25), are generally costly, time consuming, and/or difficult to automate, hindering their implementation in most laboratories.
The recently developed hairpin-shaped primer (HP) assay is a feasible alternative method for SNP detection, which is easy to implement in a wide variety of laboratories (10). This method offers an improved specificity over conventional amplification refractory mutation system (ARMS) assays (22) because it uses primers that carry a 5′-end nucleotide tail complementary to the 3′ end of the SNP-detecting primer, creating a hairpin structure. This prevents nonspecific priming. The 3′ end of the HP hybridizes to the SNP residue. Two HPs are designed, one complementary to each allele, together with a common linear primer. Two PCRs are performed in parallel, each with one of the two HPs, using the same DNA template, and the amplification is monitored in real time, by fluorescence increase, using the free fluorophore SYBR green I. An HP that is fully complementary to the target DNA will yield a more efficient amplification than a mismatched reaction, leading to an earlier threshold cycle (Ct). The greater the difference between the Cts of the mismatched and matched primer-template reactions (ΔCt) for each HP, the more robust the assay is. The technique has been developed and proven to be effective in prokaryotic contexts (10). This study presents the first report of the application of a HP assay to SNP genotyping in a human disease.
Early-onset periodontal disease (EOP) is a complex multifactorial disorder triggered by colonization of the oral epithelia by bacteria and yeasts, which leads to a gingival inflammation, loss of alveolar bone, and tooth loss (27). The importance of the protective role played by β-defensins against pathogenic microorganisms in oral epithelia has been discussed by several authors (5, 15, 18). Moreover, a recent observation by Putsep et al. on patients suffering from morbus Kostmann (24) highlights the role of antimicrobial peptides in periodontal disease. Epidemiological studies clearly demonstrate that genetic factors play a significant role in EOP (9, 21), underscoring the importance of applying genetics approaches to study EOP. The C→G transversion at position −44 of the DEFB1 gene (which encodes human β-defensin 1) has been associated with Candida sp. carriage in the mouth (12) and with the risk of human immunodeficiency virus type 1 infection in the Caucasian population (1).
In order to establish if the −44 SNP is associated with EOP, we studied its distribution in EOP patients and healthy controls from two different ethnic backgrounds, using the HP assay.
MATERIALS AND METHODS
DNA samples.
Anonymous, historically acquired DNA from two cohorts coming from different ethnic backgrounds was analyzed. DNA samples from 76 Caucasian EOP patients and 44 African-American EOP patients were included in this study. As controls, DNA samples from 80 ethnically and geographically matched healthy subjects were included in the study.
PCRs.
Primers were designed using the Primer Express 2.0 software (ABI, Foster City, Calif.), with reference to the published human genome sequence (GenBank accession number U50930). A tail was added to the 5′ end of the SNP-specific primers in order to produce a stem with a melting temperature (Tm) of 67 to 70°C and a variation of free energy of between −0.5 and −2.0, using the mfold software (http://www.bioinfo.rpi.edu/applications/mfold/old/dna/).
Sequences of the forward C- and G-allele-specific HPs are GGCTGGACCTCCAAtGGAGCCAGCC and CGCTGGACCTCCAAtGGAGCCAGCG, respectively, where the underlined residues correspond to the 5′-end tails, the bold upper-case corresponds to the 3′ SNP-specific residue, and the lowercase corresponds to a secondary mutation inserted to improve the discriminatory power of the assay. When used with the common reverse primer (CAGGATTTCAGGAACTGGGGAG), the PCR yields a 45-bp amplicon. For each DNA sample, two real-time PCRs of 10 μl were run in parallel, one with the C-allele-specific HP and the other with the G-allele-specific HP. Each reaction contained 1× PCR buffer II (ABI); 200 μM concentration each of dATP, dCTP, dGTP, and dTTP; 0.3 U of AmpliTaq Gold (ABI); 2 mM MgCl2; 5 pmol (each) primer; 1× SYBR Green I; 1.75 ng of 6-carboxy X-rhodamine; succinimidyl ester (Molecular Probes, Eugene, Ore.); and 10 ng of template DNA. All PCRs were run in a Stratagene MX4000 real-time PCR instrument (Stratagene, La Jolla, Calif.). Thermal cycling conditions were as follows: stage 1, 95°C for 10 min, 70°C for 30 s; stage 2, 72°C for 30 s, 95°C for 20 s, 69°C for 30 s (lowering 1°C each cycle) for 10 cycles; stage 3, 72°C for 30 s, 95°C for 20 s, 60°C for 30 s for 40 cycles. Data were collected in the last step of stage 3 in order to calculate the Ct of each amplification curve.
As controls, we designed two 45-bp single-stranded oligonucleotide artificial templates, which match the target sequence of each allele: tcagcctccaaaggagccagcC/Gtctccccagttcctgaaatcctg (the residues shown in bold uppercase correspond to the SNP residues). For homozygous controls, each oligonucleotide was used separately as the template for a PCR (using 106 molecules/reaction). A 1:1 mix of both oligonucleotides was used as a C/G heterozygous control, combining 5 × 105 molecules of each per reaction.
DNA sequencing reactions.
PCR amplification of the specific region containing −44 C→G was performed under the conditions previously described by Jurevic et al. (13). DNA sequences were obtained with the Big Dye Terminator sequencing kit (ABI), following the manufacturer's instructions.
Statistical analyses.
The mean Cts and the 95% confidence intervals (CI) were calculated using Microsoft Excel (Microsoft Corporation, Redmond, Wash.).
Gene frequencies were calculated from the observed number of genotypes. The significance of differences in allelic and genotypes frequencies was calculated by a χ2 test by using 2 × 2 and 3 × 2 contingency tables.
RESULTS
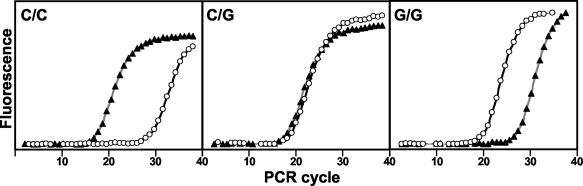
For the HP assay, the three possible genotypes were identified as follows: (i) Allele C homozygous samples gave an earlier Ct with the C HP; (ii) allele G homozygous samples gave an earlier Ct with the G HP; (iii) heterozygous samples gave similar Cts in both reactions. Figure 1 demonstrates that the HP assay allows a clear discrimination of the three different genotypes, using human DNA samples.
FIG. 1.
HP assay for detection of the −44 DEFB1 SNP. Real-time PCR results using chromosomal DNA extracted from blood from two homozygous (C/C and G/G) patients and a heterozygous (C/G) patient. Amplifications using the C-allele HP are represented by closed triangles (▴), and the G-allele HP is represented by open circles (○).
Samples were analyzed in sets of up to 44 in 96-well plates, together with the three artificial templates and a no-template negative control, in less than 2 h. The HP assay was able to identify C/C, C/G, and G/G genotypes with average ΔCts of 11.6, 0.3, and 7.7, respectively. Results are summarized in Table 1.
TABLE 1.
Average Cts for the two different HPs for the three genotypes on human genomic DNAa
| Genotype | C-Allele HP Ct (CI) | G-Allele HP Ct (CI) | ΔCt |
|---|---|---|---|
| C/C | 17.4 (17.3-17.5) | 29.0 (28.8-29.2) | 11.6 |
| C/G | 17.7 (17.5-17.9) | 18.0 (17.9-18.1) | 0.3 |
| G/G | 26.4 (25.7-27.1) | 18.7 (18.2-19.3) | 7.7 |
CI, 95% confidence intervals; ΔCt, Ct mismatched primer − Ct matched primer.
Genotypes were confirmed by direct PCR sequencing of the region containing the C→G transversion at position −44 of the DEFB1 gene. Five samples for each genotype were selected for this purpose. Sequencing data agrees with the HP assay in all cases (data not shown).
Genotype and allele frequencies for the four populations studied are shown in Table 2. All the cohorts agree with the Hardy-Weinberg law, further confirming the reliability of the assay. No significant differences were observed in the allele or genotype frequencies at position −44 of the DEFB1 gene among patients suffering from EOP versus healthy controls in the two ethnic populations studied. The C-allele frequency for the Caucasian (23%) and African American (5%) healthy controls agrees with previously reported data by Jurevic et al. (13).
TABLE 2.
Allele and genotype frequencies in the populations studied
| Allele or genotype | Frequency (no. found/no. analyzed) per population
|
|||
|---|---|---|---|---|
| EOP patients
|
Healthy controls
|
|||
| Caucasians (n = 76) | African-Americans (n = 44) | Caucasians (n = 80) | African-Americans (n = 80) | |
| Alleles | ||||
| C | 0.79 (120/152) | 0.92 (81/88) | 0.77 (123/160) | 0.95 (152/160) |
| G | 0.21 (32/152) | 0.08 (7/88) | 0.23 (37/160) | 0.05 (8/160) |
| Genotypes | ||||
| C/C | 0.62 (47/76) | 0.84 (37/44) | 0.60 (48/80) | 0.90 (72/80) |
| C/G | 0.34 (26/76) | 0.16 (7/44) | 0.34 (27/80) | 0.10 (8/80) |
| G/G | 0.04 (3/76) | 0 (0/44) | 0.07 (5/80) | 0 (0/80) |
DISCUSSION
We have developed a useful assay for medium-scale association studies of human populations based on an ARMS principle. The HP assay uses unlabeled hairpin-shaped primers, designed following the conditions and the parameters recommended for the design of molecular beacons (26). Kaboev et al. (14) reported that HPs increase the specificity and the sensitivity of PCRs by strongly diminishing the formation of primer dimers and nonspecific products. The superiority of these primers over conventional linear primers for SNP detection has previously been demonstrated (10). However, this is the first report describing the use of HPs to increase the specificity of an ARMS assay for human DNA samples.
The HP assay was designed using the conditions recommended by Hazbón and Alland (10) and required no further modifications, illustrating the robustness of the technique. A secondary mutation was introduced in the HPs to avoid an undesired secondary structure. Secondary mutations have also been reported to improve the discriminatory power of the assay (3, 10, 22, 29).
The use of the SYBR Green I chemistry for the detection of the amplicons in a real-time PCR instrument, instead of expensive fluorophore-labeled probes, decreases the cost of the assay to an estimated $0.27 per genotype. Recently, new techniques based on the analysis of the melting profile of the amplicon have been developed for single-tube genotyping, using SYBR Green I chemistry in a real-time instrument. However, most of them require post-PCR manipulation (17) (increasing the risk of cross contamination) or a time-consuming data handling process (11). Germer and Higuchi (7) proposed a protocol for SNP frequency studies, based on ARMS, followed by a melting profile of the amplicons, but it requires a demanding optimization process, which our technique does not.
Sequencing data confirmed the results obtained with our HP assay. Using a 96-well format, we were able to test the 264 samples in only 6 PCR runs (each of which lasts 2 h). These results confirm the potential of the HP assay for medium- to large-scale association studies of complex human diseases.
We applied this technique to study whether there was an association between the C→G transversion at position −44 in the DEFB1 gene and EOP. The results do not support a direct association of this SNP with EOP. It is possible that other SNPs in the DEFB1 gene could be associated with EOP. However, there are no reported associations of other polymorphisms in the DEFB1 gene with an increased risk of infection, with the exception of a nonsynonymous mutation, associated with chronic obstructive pulmonary disease, that has been reported only in a Japanese population (19). It is also possible that human beta-defensin 1 is not active against periodontal bacteria and does not play a role in this disease. Supporting this hypothesis, it has been demonstrated that Treponema denticola is resistant to human beta-defensin 1 (2). Unfortunately, no data are available regarding the biological activity of this peptide against Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis, two key oral pathogens implicated in periodontal disease.
In conclusion, we have designed and optimized a simple, reliable assay for examining SNP frequencies that provides several major advantages over traditional SNP-detecting techniques. We have successfully applied this technique to examine, for the first time, the associations between the SNP at position −44 in the DEFB1 gene and EOP.
Acknowledgments
This work was supported in part by a grant from the National Institute of Dental and Craniofacial Research, Health Disparities Programme, number R1DE14997A, and in part by NIH grant AI46669. M. Boniotto is supported by a Department of Defense appropriation. J. Eskdale and G. Lennon are supported by the New Jersey Dental School.
REFERENCES
- 1.Braida, L., M. Boniotto, A. Pontillo, P. A. Tovo, A. Amoroso, and S. Crovella. A single-nucleotide polymorphism (SNPs) in human beta-defensin 1 gene is associated with HIV-1 infection in Italian children. AIDS, in press. [DOI] [PubMed]
- 2.Brissette, C. A., and S. A. Lukehart. 2002. Treponema denticola is resistant to human beta-defensins. Infect Immun. 70:3982-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha, R. S., H. Zarbl, P. Keohavong, and W. G. Thilly. 1992. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 2:14-20. [DOI] [PubMed] [Google Scholar]
- 4.Doherty, T. M., L. A. Fitzpatrick, A. Shaheen, T. B. Rajavashisth, and R. C. Detrano. 2004. Genetic determinants of arterial calcification associated with atherosclerosis. Mayo Clin. Proc. 79:197-210. [DOI] [PubMed] [Google Scholar]
- 5.Dunsche, A., Y. Acil, R. Siebert, J. Harder, J. M. Schroder, and S. Jepsen. 2001. Expression profile of human defensins and antimicrobial proteins in oral tissues. J. Oral Pathol. Med. 30:154-158. [DOI] [PubMed] [Google Scholar]
- 6.Flavell, A. J., V. N. Bolshakov, A. Booth, R. Jing, J. Russell, T. H. Ellis, and P. Isaac. 2003. A microarray-based high throughput molecular marker genotyping method: the tagged microarray marker (TAM) approach. Nucleic Acids Res. 31:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Germer, S., and R. Higuchi. 1999. Single-tube genotyping without oligonucleotide probes. Genome Res. 9:72-78. [PMC free article] [PubMed] [Google Scholar]
- 8.Gray, I. C., D. A. Campbell, and N. K. Spurr. 2000. Single nucleotide polymorphisms as tools in human genetics. Hum. Mol. Genet. 9:2403-2408. [DOI] [PubMed] [Google Scholar]
- 9.Hart, T. C. 1994. Genetic considerations of risk in human periodontal disease. Curr. Opin. Periodontol. 1994:3-11. [PubMed] [Google Scholar]
- 10.Hazbon, M. H., and D. Alland. 2004. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J. Clin. Microbiol. 42:1236-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hladnik, U., L. Braida, M. Boniotto, D. Pirulli, F. Gerin, A. Amoroso, and S. Crovella. 2002. Single-tube genotyping of MBL-2 polymorphisms using melting temperature analysis. Clin. Exp. Med. 2:105-108. [DOI] [PubMed] [Google Scholar]
- 12.Jurevic, R. J., M. Bai, R. B. Chadwick, T. C. White, and B. A. Dale. 2003. Single-nucleotide polymorphisms (SNPs) in human beta-defensin 1: high-throughput SNP assays and association with Candida carriage in type I diabetics and nondiabetic controls. J. Clin. Microbiol. 41:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jurevic, R. J., P. Chrisman, L. Mancl, R. Livingston, and B. A. Dale. 2002. Single-nucleotide polymorphisms and haplotype analysis in beta-defensin genes in different ethnic populations. Genet. Test. 6:261-269. [DOI] [PubMed] [Google Scholar]
- 14.Kaboev, O. K., L. A. Luchkina, A. N. Tret'iakov, and A. R. Bahrmand. 2000. PCR hot start using primers with the structure of molecular beacons (hairpin-like structure). Nucleic Acids Res. 28:E94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krisanaprakornkit, S., A. Weinberg, C. N. Perez, and B. A. Dale. 1998. Expression of the peptide antibiotic human beta-defensin 1 in cultured gingival epithelial cells and gingival tissue. Infect. Immun. 66:4222-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, L. G., K. J. Livak, B. Mullah, R. J. Graham, R. S. Vinayak, and T. M. Woudenberg. 1999. Seven-color, homogeneous detection of six PCR products. BioTechniques 27:342-349. [DOI] [PubMed] [Google Scholar]
- 17.Lipsky, R. H., C. M. Mazzanti, J. G. Rudolph, K. Xu, G. Vyas, D. Bozak, M. Q. Radel, and D. Goldman. 2001. DNA melting analysis for detection of single nucleotide polymorphisms. Clin. Chem. 47:635-644. [PubMed] [Google Scholar]
- 18.Mathews, M., H. P. Jia, J. M. Guthmiller, G. Losh, S. Graham, G. K. Johnson, B. F. Tack, and P. B. McCray, Jr. 1999. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect. Immun. 67:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsushita, I., K. Hasegawa, K. Nakata, K. Yasuda, K. Tokunaga, and N. Keicho. 2002. Genetic variants of human beta-defensin-1 and chronic obstructive pulmonary disease. Biochem. Biophys. Res. Commun. 291:17-22. [DOI] [PubMed] [Google Scholar]
- 20.Meyer, K., A. Fredriksen, and P. M. Ueland. 2004. High-level multiplex genotyping of polymorphisms involved in folate or homocysteine metabolism by matrix-assisted laser desorption/ionization mass spectrometry. Clin. Chem. 50:391-402. [DOI] [PubMed] [Google Scholar]
- 21.Michalowicz, B. S. 1994. Genetic and heritable risk factors in periodontal disease. J. Periodontol. 65:479-488. [DOI] [PubMed] [Google Scholar]
- 22.Newton, C. R., A. Graham, L. E. Heptinstall, S. J. Powell, C. Summers, N. Kalsheker, J. C. Smith, and A. F. Markham. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17:2503-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ou-Yang, H., L. Hua, Q. H. Mo, and X. M. Xu. 2004. Rapid, accurate genotyping of the common -alpha(4.2) thalassaemia deletion based on the use of denaturing HPLC. J. Clin. Pathol. 57:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Putsep, K., G. Carlsson, H. G. Boman, and M. Andersson. 2002. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet 360:1144-1149. [DOI] [PubMed] [Google Scholar]
- 25.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 26.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 27.Wara-aswapati, N., T. H. Howell, H. L. Needleman, and N. Karimbux. 1999. Periodontitis in the child and adolescent. ASDC J. Dent. Child. 66:167-174,154. [PubMed] [Google Scholar]
- 28.Waterfall, C. M., and B. D. Cobb. 2001. Single tube genotyping of sickle cell anaemia using PCR-based SNP analysis. Nucleic Acids Res. 29:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, D. Y., L. Ugozzoli, B. K. Pal, and R. B. Wallace. 1989. Allele-specific enzymatic amplification of beta-globin genomic DNA for diagnosis of sickle cell anemia. Proc. Natl. Acad. Sci. USA 86:2757-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]