Abstract
Purpose of review
In contrast to many other human diseases, the use of genome-wide association studies (GWAS) to identify genes for heart failure (HF) has had limited success. We will discuss the underlying challenges as well as potential new approaches to understanding the genetics of common forms of HF.
Recent findings
Recent research using intermediate phenotypes, more detailed and quantitative stratification of HF symptoms, founder populations and novel animal models has begun to allow researchers to make headway toward explaining the genetics underlying HF using GWAS techniques.
Summary
By expanding analyses of HF to improved clinical traits, additional HF classifications and innovative model systems, the intractability of human HF GWAS should be ameliorated significantly.
Keywords: animal model, genome-wide association studies, heart failure, intermediate phenotype
INTRODUCTION
Heart diseases remain the primary cause of death in developed countries, and are expected to overtake infection as the primary cause of death in the developing world [1]. Over the past 30 years, the age-adjusted death rate due to heart diseases has steadily decreased in the United States, from its peak of around 900 deaths per 100 000 men a year to roughly 350 deaths per 100 000 [1,2]. This stunning decrease can be attributed roughly equally to changes in public health attitudes, such as the decline in cigarette use over this time frame, and new medical interventions, such as statins and portable defibrillators. Despite these decreases in the overall mortality and morbidity of heart disease, heart failure (HF) has resisted this trend. Indeed, between 2000 and 2010, the number of HF-related hospitalizations in the United States has not decreased [3] and HF remains the leading cause of hospitalization in people over the age of 65 [4]. The incidence of HF is predicted to rise over the next 15 years by roughly 25% [1], a fact that emphasizes the importance and urgency of research that seeks to understand the genetic basis of the disease.
The genetic basis for HF has been well established for many familial cardiomyopathies whose inheritance can be identified as one of the classic ‘simple’ Mendelian inheritance patterns (such as X-linked dilated cardiomyopathy). In contrast, the complexity and heterogeneity of common non-familial forms of HF has made the identification of genes difficult.
The purpose of this review is to examine the difficulties involved in the analysis of human HF using genome-wide association studies (GWAS) (Fig. 1), highlight the current progress in this field, and examine means by which researchers may be able to achieve greater success in the future.
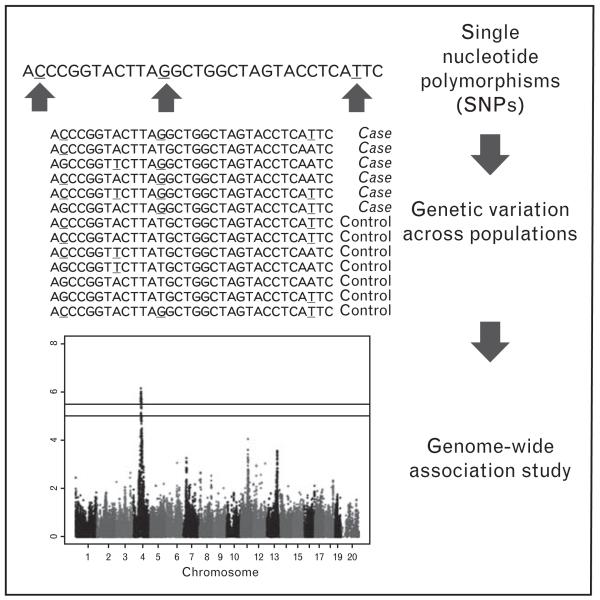
FIGURE 1.
Genome-wide association study overview. Genome-wide association studies harness natural single nucleotide polymorphisms (SNPs) present within a population in order to associate regions of the genome called loci with a phenotype of interest.
THE CHALLENGES OF PERFORMING HEART FAILURE GENOME-WIDE ASSOCIATION STUDIES IN HUMANS
GWAS is a powerful approach to identify genes and genetic basis of complex traits. However, there has been very limited success of GWAS to identify genes for sporadic HF in humans. The limited number of studies are either underpowered or not specific for HF [5–8]. Consequently, human HF GWAS (see Fig. 1) have returned only a handful of associations that reach the accepted genome-wide significance threshold of 5 × 10−8 [9]. By comparison, a single meta-analysis of blood lipid traits returned nearly 100 significant loci [10]. The reasons for this disparity are numerous, but can be roughly categorized as follows (Fig. 2).
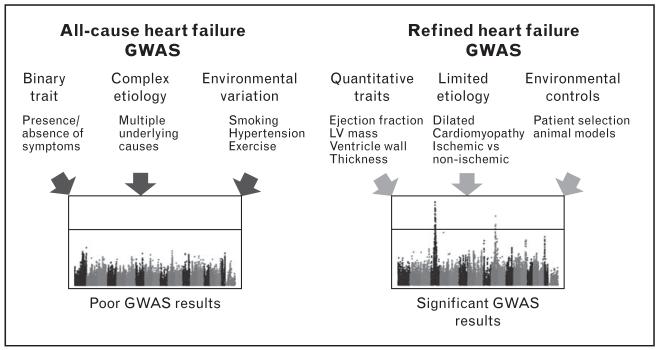
FIGURE 2.
Challenges and solutions for heart failure genome-wide association study (GWAS). On the left, several problems with traditional heart failure classification as well as complex etiological and environmental confounders impede fruitful GWAS analyses. On the right, possible workarounds for these problems are proposed. LV, left ventricle.
First is the difficulty in obtaining quantitative features of HF. As a clinical diagnosis, the current assessment of HF severity relies on largely non-invasive measurement of ventricular function parameters such as ejection fraction or 6-minute walking distance, or New York Heart Association functional classifications, which are highly variable and primarily qualitative in nature. The lack of mechanism-based and disease-specific quantitative measurements for HF results in difficulty to accurately quantifying the disease severity. By comparison, traits such as plasma lipids and blood pressure are easy to measure parameters which can be accurately and quantitatively determined. Unfortunately, most measurements that could be used to quantify ventricular remodeling and function, such as hemodynamic pressure development, tissue weight, tissue fibrosis or gene expression, require invasive procedures that are not feasible in large-scale clinical settings. Echocardiographic parameters provide one possible avenue for HF GWAS success, and indeed several groups [11,12■] have reported loci using these phenotypes.
The second is the extremely complex cause of HF. It is increasingly clear that HF is a disease with many etiological roots and may in fact encompass several mechanistically distinct diseases. Therefore, using a common parameter, such as ejection fraction or mortality, may not be able to discover unique genetic contributions to specific diseases.
The third category is the significant contributions of environmental variation to human HF. HF results in part from the impact of myocardial infarction and valvular disorders that are themselves highly complex and heterogeneous. Nonfamilial HF is typically a disease of the elderly, and is the result not only of genetic factors that act to either protect or predispose individuals, but also of environmental factors, such as smoking, diet and sedentary life style. This heterogeneity complicates attempts to determine the genetic factors underlying HF and even confounds the determination of just how heritable HF is, with estimates ranging from 24% [13] to 69% [14]. Consequently, HF GWAS may require very large cohorts of human cases in order to overcome the noise introduced by environmental factors.
CURRENT HEART FAILURE GENOME-WIDE ASSOCIATION STUDIES RESULTS IN HUMAN POPULATIONS
Current HF GWAS in humans can be divided into three general categories: all-cause genome-wide studies, genome-wide studies on a narrowly defined phenotype, and studies limited by either population or genotype selection (Table 1).
Table 1.
All significant loci for heart failure in human studies
| Phenotype | Study | SNP ID | Chromosome | Peak base pair |
P value | Gene |
|---|---|---|---|---|---|---|
| All-cause HF | ||||||
| HF | Smith et al. [15] | rs10519210 | 15 | 61524978 | 1.4E-8 | USP3 |
| Narrower Characterization of HF | ||||||
| Dilated cardiomyopathy | Villard et al. [6] | rs2234962 | 10 | 121419623 | 1.1E-13 | BAG3 |
| Dilated cardiomyopathy | Meder et al. [16■■] | rs9262636 | 6 | 31025848 | 4.0E-9 | HGC22 |
| LV internal dimension | Vasan et al. [11] | rs89107 | 6 | 118578043 | 1.2E-9 | SLC35F1 |
| LV internal dimension | Vasan et al. [11] | rs11153768 | 6 | 118988152 | 1.7E-8 | PLN |
| Aortic root size | Vasan et al. [11] | rs10852932 | 17 | 2143460 | 2.3E-11 | SMG6 |
| Aortic root size | Vasan et al. [11] | rs4523957 | 17 | 2208899 | 2.3E-11 | SRR |
| Aortic root size | Vasan et al. [11] | rs17470137 | 5 | 123195653 | 1.3E-11 | CCDC100 |
| Aortic root size | Vasan et al. [11] | rs4026608 | 12 | 66000884 | 1.8E-9 | HMGA2 |
| Aortic root size | Vasan et al. [11] | rs10770612 | 12 | 20077705 | 2.4E-8 | PDE3A |
| IVS wall thickness | Fox et al. [12■] | Rs1571099 | 10 | 122256604 | 2.6E-8 | PPAPDC1A |
| Limiting SNPs tested or populations surveyed | ||||||
| HF founder population | Parsa et al. [5] | rs1320448 | 10 | 105836064 | 1.8E-8 | COL17A1 |
| HF limited SNP panel | Cappola et al. [17] | rs1739843 | 1 | 16016759 | 3.9E-6 | HSPB7 |
| HF limited SNP panel | Cappola et al. [17] | rs6787362 | 3 | 69178228 | 6.9E-6 | FRMD4B |
See Supplementary Information for all suggestive loci, http://links.lww.com/HCO/A26, as well as ideograms of GWAS hits across the genome. GWAS, genome-wide association studies; HF, heart failure; IVS, interventricular septum; LV, left ventricle; SNPs, single nucleotide polymorphisms.
ALL-CAUSE GENOME-WIDE ASSOCIATION STUDIES FOR HEART FAILURE
To date, there have only been a handful of published all-cause HF GWAS. The earliest published HF GWAS is the Framingham 100k project [8], which used a panel of 71 000 single nucleotide polymorphisms (SNPs) to examine four major cardiovascular diseases (CVDs): atherosclerotic CVDs, myocardial infarction, atrial fibrillation and HF, in a case/control study with 73 cases and 1272 controls wherein HF was diagnosed using standard symptomatic observation. This study was unable to identify any significant (P<5×10−8) associations, and only a single suggestive (P<10−5) association [8], likely due to both an underpowered patient cohort and low SNP density.
The Cohorts for Heart and Aging Research in Genomic Epidemiology consortium published a pair of studies in 2010 [7,15] in which they combined four other studies (including the Framingham study above) to perform a meta-analysis on the incidence of HF in their combined cohorts as well as the survival of those individuals. The combined meta-analysis involved nearly 24 000 individuals, of whom 2500 had HF, and examined 2.5 million imputed SNPs. Despite the improved case number, however, only one locus near the USP3 gene with a low minor allele frequency of 3% passed the accepted significance threshold, although the authors were able to identify 36 other suggestive loci, demonstrating the power of the meta-analysis study to identify genes implicated in complex diseases.
GENOME-WIDE STUDIES OF MORE SPECIFIC PHENOTYPES
The above studies examined all cases of HF without discriminating between individuals based on underlying cause or types of HF. As outlined above, the complex etiologies underlying HF mean that the disease is better appreciated as a set of highly related disorders rather than a single, overarching diagnosis. Subsequent studies have attempted to address these complexities by focusing only on a specific class of HF. Villard et al. [6] focused specifically on sporadic dilated cardiomyopathy. Using 1100 cases and 1100 controls, they were able to identify and replicate a significant locus containing BAG3 as well as three other suggestive loci. The most recent study by Meder et al. [16■■] also chose to examine dilated cardiomyopathy specifically. The authors employed a three-stage strategy in which replication of loci between cohorts was used as positive evidence of association. Using 4100 cases and 7600 controls, they were able to identify novel significant loci, containing the candidate noncoding RNA HGC22, as well as two suggestive loci. The authors also utilized RNA transcriptome arrays to perform an expression quantitative trait analysis in which GWAS techniques are applied to gene expression to suggest that this locus regulates a number of myosin heavy chain genes, whose connection to dilated cardiomyopathy remains unclear.
A number of other studies have focused on even narrower sets of phenotypes, sometimes limiting themselves to a single parameter rather than the disease as a whole in order to attempt to understand a specific pathological feature of HF. Several of these studies [11,12■] have focused on echocardiographic parameters such as left ventricular internal dimension that are routinely obtained in cardiology clinics via noninvasive ultrasonic imaging. Perhaps most importantly, these parameters are quantitative clinical traits rather than qualitative, an important distinction that allows more sophisticated analyses and more reliable and significant results. These two studies examined roughly 23 000 individuals in combination and were able to identify three significant loci for interventricular septal wall thickness with candidates (PPAPDC1A, PLN, SLC35F1) and five significant loci for aortic root size with candidates (SMG6, SPR, CCDC100, HMGA2, PDE3) as well as 16 other suggestive loci. It is striking that, by using a similar number of individuals, studies performed on a more specific phenotype were able to recover eight times as many significant loci as a study that was performed on all-cause HF.
ASSOCIATION STUDIES THAT LIMIT GENETIC DIVERSITY
Another avenue that has been explored is to either limit the number of SNPs tested in order to reduce the significance threshold, or focus explicitly on a small, genetically homogeneous population in order to reduce extraneous factors. Cappola et al. [17] took this first approach, studying 4700 individuals but only examining SNPs near 2000 prioritized candidate CVD genes, and therefore having an adjusted significance threshold of only 5×10−5. They were able to identify two significant loci near the genes HSPB7 and FRMD4B. Parsa et al. [5] examined HF mortality in an Amish founder population of only 850 individuals with follow-up in a cohort of 2000 Caucasians. Using fewer than 3000 individuals, they were able to identify one significant locus near COL17A1 and 18 suggestive loci for left ventricular mass.
ADDRESSING THE DIFFICULTIES OF HUMAN GENOME-WIDE ASSOCIATION STUDIES
As explained above, the striking heterogeneity of human HF has complicated the use of GWAS to uncover the underlying genetic contributors, and the currently identified candidate genes have revealed limited insights in to the underlying mechanism or pathogenic process of the disease. Indeed, beyond USP3 no genome-wide significant GWAS loci for all-cause HF have been identified. Instead, researchers have achieved greater success when studies have been designed to limit potential sources of introduced noise, for instance, by focusing on quantifiable key clinical features, resulting in 13 significant loci. Future genetic studies for common forms of HF will likely benefit from improved diagnosis for disease stratification and quantification, as well as better insights and candidate genes discovered from animal models (Fig. 2).
FINDING QUANTITATIVE MEASURES OF HEART FAILURE
Several papers [11,12■] have already used echocardiographic parameters to successfully query for genes involved in the regulation of left ventricular function and structure, such as chamber dimensions and thickness. Still others [18] have examined the role of certain metabolites in HF. Recent developments in MRI technology [19] suggest that additional critical phenotypes, such as myocardial fibrosis, will soon be available. These better-annotated clinical features are likely to greatly improve the feasibility and success of genome-wide analysis for HF in the coming years.
ADDRESSING VARIABILITY OF CAUSES IN HUMAN HEART FAILURE
Several successful studies [6,16■■] have focused on analyzing subsets of all-cause HF. Since the heterogeneity of HF onset in humans is so varied, these approaches will invariably have more success than studies of all-cause HF. Recent work to focus on further stratified clinical features of HF, such as HF with preserved or reduced ejection fraction and right-sided HF, should also provide better outcomes for GWAS analyses in humans.
ENVIRONMENTAL VARIABILITY
Non-familial HF is a chronic disease and manifests predominantly in the elderly or in people with long-term exposure to other risk factors such as hypertension, metabolic disorder or chronic inflammatory diseases. Furthermore, as the heart is capable of sustained compensatory remodeling, clinicians are rarely able to catch the disease as it progresses, but typically are able to diagnose a patient with HF only after symptoms begin to emerge, further complicating efforts to reveal underlying causes of HF [20,21]. To overcome these challenges, researchers will need to initiate large-scale, decades-long longitudinal studies, which would, themselves, likely address only a portion of the issue.
A more palatable approach to explore the genetics of HF involves the use of animal model systems. There is well-established precedence for the use of animal models to study HF [21–23], however, these studies have primarily been used to investigate the role of individual genes in HF progression. Recently, the development of high-throughput screening technologies, extensive genetic profiling and sophisticated genetic breeding programmes for animal models has allowed researchers to perform similar studies as they would in humans.
GENOMICS OF HEART FAILURE IN ANIMAL MODELS
Animal models have several key advantages over human studies, most notably in the ability for researchers to carefully control the onset and progression of a disease while simultaneously limiting both environmental and genetic variation. By eliminating most sources of biological noise, researchers are able to obtain results comparable with large human studies using only a few dozen to a few hundred genetic strains of animals.
LINKAGE ANALYSIS
Traditional genetics studies in animal models have focused on using linkage analysis to uncover genes modulating sensitivity or resistance to HF [24,25]. In mice, careful work has been done by the Rockman and Marchuk groups to identify seven genome-wide significant loci influencing the progression of HF in a calsequestrin knockout model [25–27]. In other model organisms, McDermott-Roe et al. were able to identify the soluble epoxide hydrolase and mitochondrial endonuclease G (Endog) as a HF locus in rats [28], whereas in canines multiple groups have identified several cardiomyopathy loci [29–31] and a single locus around the sarcoglycan delta gene has been identified in hamsters [32].
Despite these successes, the identified loci linked to HF are typically quite large, on the order of tens to hundreds of megabases, which significantly complicates efforts to identify and validate candidate genes. For example, despite identifying seven HF loci in mice, only one [24] has had its gene identified.
GENOME-WIDE ASSOCIATION STUDIES
Recently, driven by advances in computational techniques that deal with the complex interrelatedness of most types of animal models [33,34], several GWAS have emerged in both canines [35,36] and mice [37■,38]. These animal models have returned a number of highly significant results. For instance, using only 800 mice, Rau et al. [37■] were able to identify seven significant and 18 suggestive loci for HF-related conditions, whereas Hersch et al. [38] were able to identify a large number of both significant and suggestive results using only 23 strains of mice. As a result of the reduced genetic complexity and environmental influences, the outcome of animal-based GWAs for HF has begun to reveal interesting insights into the underlying disease mechanisms. For instance, in the study by Rau et al. [37■] genetic variants of calcineurin and sarcoglycan delta were identified to be significantly associated with isoproterenol-induced hypertrophy, whereas Abcc6 was associated with fibrosis in mice. In addition, Hersch et al. [38] were able to identify mir-21 and Meurs et al. [36] were able to associate PDK4 with HF in canines. These animal studies have potential to integrate with other omics approaches and lead to additional insights into HF.
CONCLUSIONS
Human HF is a complicated disease that has resisted efforts to understand it using traditional GWAS approaches. Despite these setbacks in the analysis of all-cause HF, researchers have made progress toward identifying the underlying pathways, genes and mechanisms of HF through analysis of subphenotypes, such as echocardiographic parameters, the segregation of HF into a number of component disorders, such as dilated cardiomyopathy or heart failure with preserved ejection fraction, and through the use of animal models. Further research and GWAS using these approaches will undoubtedly lead to a deeper understanding of HF and progression toward more sophisticated and advanced diagnosis and treatment options.
Supplementary Material
KEY POINTS.
The genetics underlying common forms of human HF are poorly understood, likely due to a combination of imprecise categorization and environmental factors.
All-cause HF GWAS have proven unable to identify more than a handful of potential candidate genes.
Narrowly focused analyses on specific quantitative phenotypes or limited populations as well as animal models have been able to reveal a number of novel and interesting genetic factors contributing to HF.
Future research should continue to focus on well-defined, quantitative measures of HF when conducting GWAS.
Acknowledgements
The authors would like to thank Dr James Weiss for discussion and encouragement.
Financial support and sponsorship
This work was supported in part by grants from National Institutes of Health_HL123295 and HL114437.
Footnotes
Conflicts of Interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■ of outstanding interest
- 1.Go AS, Mozaffarian D, Roger VL, et al. Heart disease and stroke statistics: 2013 update – a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ford ES, Capewell S. Proportion of the decline in cardiovascular mortality disease due to prevention versus treatment: public health versus clinical care. Annu. Rev. Public Health. 2011;32:5–22. doi: 10.1146/annurev-publhealth-031210-101211. [DOI] [PubMed] [Google Scholar]
- 3.Hall MJ, Levant S, DeFrances CJ. Hospitalization for congestive heart failure: United States, 2000-2010. NCHS Data Brief. 2012:1–8. [PubMed] [Google Scholar]
- 4.Krumholz HM, Chen YT, Wang Y, et al. Predictors of readmission among elderly survivors of admission with heart failure. Am Heart J. 2000;139:72–77. doi: 10.1016/s0002-8703(00)90311-9. [DOI] [PubMed] [Google Scholar]
- 5.Parsa A, Chang Y-PC, Kelly RJ, et al. Hypertrophy-associated polymorphisms ascertained in a founder cohort applied to heart failure risk and mortality. Clin Transl Sci. 2011;4:17–23. doi: 10.1111/j.1752-8062.2010.00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villard E, Perret C, Gary F, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32:1065–1076. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrison AC, Felix JF, Cupples LA, et al. Genomic variation associated with mortality among adults of European and African ancestry with heart failure: the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet. 2010;3:248–255. doi: 10.1161/CIRCGENETICS.109.895995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larson MG, Atwood LD, Benjamin EJ, et al. Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet. 2007;8(Suppl 1):S5. doi: 10.1186/1471-2350-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasan RS, Glazer NL, Felix JF, et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–178. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12■.Fox ER, Musani SK, Barbalic M, et al. Genome-wide association study of cardiac structure and systolic function in African Americans: the Candidate Gene Association Resource (CARe) study. Circ Cardiovasc Genet. 2013;6:37–46. doi: 10.1161/CIRCGENETICS.111.962365. This study used echocardiography and MRI to study intermediate phenotypes of HF using GWAS.
- 13.Post WS, Larson MG, Myers RH, et al. Heritability of left ventricular mass: the Framingham Heart Study. Hypertension. 1997;30:1025–1028. doi: 10.1161/01.hyp.30.5.1025. [DOI] [PubMed] [Google Scholar]
- 14.Swan L, Birnie DH, Padmanabhan S, et al. The genetic determination of left ventricular mass in healthy adults. Eur Heart J. 2003;24:577–582. doi: 10.1016/s0195-668x(02)00524-9. [DOI] [PubMed] [Google Scholar]
- 15.Smith NL, Felix JF, Morrison AC, et al. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry: a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2010;3:256–266. doi: 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16■■.Meder B, Rühle F, Weis T, et al. A genome-wide association study identifies 6p21 as novel risk locus for dilated cardiomyopathy. Eur Heart J. 2014;35:1069–1077. doi: 10.1093/eurheartj/eht251. This study analyzed a subset of HF and combined analysis with transcriptomics to convincingly identify a very strong locus for dilated cardiomyopathy and implicate several genes in the locus.
- 17.Cappola TP, Li M, He J, et al. Common variants in HSPB7 and FRMD4B associated with advanced heart failure. Circ Cardiovasc Genet. 2010;3:147–154. doi: 10.1161/CIRCGENETICS.109.898395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu B, Zheng Y, Alexander D, et al. Genome-wide association study of a heart failure related metabolomic profile among African Americans in the Atherosclerosis Risk in Communities (ARIC) study. Genet Epidemiol. 2013;37:840–845. doi: 10.1002/gepi.21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Jong S, Zwanenburg JJ, Visser F, et al. Direct detection of myocardial fibrosis by MRI. J Mol Cell Cardiol. 2011;51:974–979. doi: 10.1016/j.yjmcc.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 20.Creemers EE, Wilde AA, Pinto YM. Heart failure: advances through genomics. Nat Rev Genet. 2011;12:357–362. doi: 10.1038/nrg2983. [DOI] [PubMed] [Google Scholar]
- 21.Breckenridge R. Heart failure and mouse models. Dis Model Mech. 2010;3:138–143. doi: 10.1242/dmm.005017. [DOI] [PubMed] [Google Scholar]
- 22.Patten RD, Hall-Porter MR. Small animal models of heart failure: development of novel therapies, past and present. Circ Heart Fail. 2009;2:138–144. doi: 10.1161/CIRCHEARTFAILURE.108.839761. [DOI] [PubMed] [Google Scholar]
- 23.Zaragoza C, Gomez-Guerrero C, Martin-Ventura JL, et al. Animal models of cardiovascular diseases. J Biomed Biotechnol. 2011;2011:497841. doi: 10.1155/2011/497841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheeler FC, Tang H, Marks OA, et al. Tnni3k modifies disease progression in murine models of cardiomyopathy. PLoS Genet. 2009;5:e1000647. doi: 10.1371/journal.pgen.1000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Corvoisier P, Park H-Y, Carlson KM, et al. Multiple quantitative trait loci modify the heart failure phenotype in murine cardiomyopathy. Hum Mol Genet. 2003;12:3097–3107. doi: 10.1093/hmg/ddg333. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki M. Genetic modifier loci affecting survival and cardiac function in murine dilated cardiomyopathy. Circulation. 2002;105:1824–1829. doi: 10.1161/01.cir.0000014926.32463.89. [DOI] [PubMed] [Google Scholar]
- 27.Wheeler FC, Fernandez L, Carlson KM, et al. QTL mapping in a mouse model of cardiomyopathy reveals an ancestral modifier allele affecting heart function and survival. Mamm Genome. 2005;16:414–423. doi: 10.1007/s00335-005-2468-7. [DOI] [PubMed] [Google Scholar]
- 28.McDermott-Roe C, Ye J, Ahmed R, et al. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;7367:114–118. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philipp U, Broschk C, Vollmar A, Distl O. Evaluation of tafazzin as candidate for dilated cardiomyopathy in Irish wolfhounds. J Hered. 2007;98:506–509. doi: 10.1093/jhered/esm045. [DOI] [PubMed] [Google Scholar]
- 30.Meurs KM, Fox PR, Norgard M, et al. A prospective genetic evaluation of familial dilated cardiomyopathy in the Doberman pinscher. J Vet Intern Med. 2007;21:1016–1020. doi: 10.1892/0891-6640(2007)21[1016:apgeof]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 31.Werner P, Raducha MG, Prociuk U, et al. A novel locus for dilated cardiomyopathy maps to canine chromosome 8. Genomics. 2008;91:517–521. doi: 10.1016/j.ygeno.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okazaki Y, Okuizumi H, Ohsumi T, et al. A genetic linkage map of the Syrian hamster and localization of cardiomyopathy locus on chromosome 9qa2.1-b1 using RLGS spot-mapping. Nat Genet. 1996;13:87–90. doi: 10.1038/ng0596-87. [DOI] [PubMed] [Google Scholar]
- 33.Kang HM, Zaitlen NA, Wade CM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lippert C, Listgarten J, Liu Y, et al. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833–835. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- 35.Lanfear DE, Yang JJ, Mishra S, Sabbah HN. Genome-wide approach to identify novel candidate genes for beta blocker response in heart failure using an experimental model. Discov Med. 2011;11:359–366. [PMC free article] [PubMed] [Google Scholar]
- 36.Meurs KM, Lahmers S, Keene BW, et al. A splice site mutation in a gene encoding for PDK4, a mitochondrial protein, is associated with the development of dilated cardiomyopathy in the Doberman pinscher. Hum Genet. 2012;131:1319–1325. doi: 10.1007/s00439-012-1158-2. [DOI] [PubMed] [Google Scholar]
- 37■.Rau CD, Wang J, Avetisyan R, et al. Mapping genetic contributions to cardiac pathology induced by beta-adrenergic stimulation in mice. Circ Cardiovasc Genet. 2015;8:40–49. doi: 10.1161/CIRCGENETICS.113.000732. This study performed GWAS in a mouse model of HF. It demonstrates the power of animal models with regard to environmental controls, stressor selection and population management.
- 38.Hersch M, Peter B, Kang HM, et al. Mapping genetic variants associated with beta-adrenergic responses in inbred mice. PLoS One. 2012;7:e41032. doi: 10.1371/journal.pone.0041032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.