Abstract
Glioblastoma multiforme (GBM) is a grade IV glioma with a median survival of 15 months. Recently, bone morphogenetic protein (BMP) signaling has been shown to promote survival in xenograft murine models. To gain a better understanding of the role of BMP signaling in human GBMs, we examined the genomic alterations of 90 genes associated with BMP signaling in GBM patient samples. We completed this analysis using publically available datasets compiled through The Cancer Genome Atlas and the Glioma Molecular Diagnostic Initiative. Here we show how mRNA expression is altered in GBM samples and how that is associated with patient survival, highlighting both known and novel associations between BMP signaling and GBM biology.
Keywords: Glioblastoma multiforme, bone morphogenetic protein, genomics, The Cancer Genome Atlas, REMBRANDT
Introduction
Tumors in the brain known as gliomas are comprised of cells that resemble mature glial cells. These tumors include astrocytomas, oligodendrogliomas, ependymomas, and mixed gliomas.1 Gliomas account for approximately 30% of all central nervous system (CNS) tumors and 80% of all malignant CNS tumors.2 Te World Health Organization (WHO) has classified astrocytomas into four grades based on histological features that correlate with patient outcomes.3 Grade I and II tumors (or low-grade gliomas) are relatively slow-growing tumors that show low proliferative activity. Grade III and IV (or high-grade astrocytomas) are rapidly growing, highly infiltrative tumors that show anaplasia and mitotic activity. Grade IV astrocytomas show, in addition, microvascular proliferation and/or necrosis.3 Approximately 17,000 malignant gliomas are diagnosed each year.4 Glioblastoma multiforme (GBM), a grade IV astrocytoma, is the most common malignant CNS tumor, accounting for approximately 80% of malignant gliomas.2,4 Te survival rate for patients with GBM is extremely low, with less than 5% of patients surviving 5 years after diagnosis and a median survival of only 15 months despite surgical resection, radiation, and chemotherapy.2,5 One of the primary challenges in finding effective therapies is due to the high level of heterogeneity found within GBMs. Although GBMs are diagnosed histopathologically as one disease, recently it has become clear that GBMs are heterogeneous at the molecular level.
Genetic Heterogeneity
GBMs are well known for their histologic heterogeneity. Recently, with the rapid advancement of sequencing technology and accumulation of genomic data, emphasis has been placed on understanding these tumors at the molecular genetic level. Such studies suggest that, while many GBMs share certain genetic and epigenetic alterations, tumors segregate into subclasses based on gene expression analysis. In 2006, Phillips et al established three subclasses of high-grade glioma (HGG) based on large-scale genomic expression profiles of survival-related genes. Tese subgroups are termed proneural (PN), proliferative, and mesenchymal (MES), reflecting the dominant gene expression patterns in each group.6 In 2010, Verhaak et al classified HGGs into four similar subclasses using data compiled by The Cancer Genome Atlas (TCGA).7 These subgroups show gene expression patterns similar to those found within neural lineages.6 Phillips et al likened the gene expression signatures to various stages of neurogenesis in the adult forebrain. Thus, gene expression patterns of MES tumors are akin to that of undifferentiated neural stem cells, PN gene expression is similar to neuroblasts or immature neurons, and proliferative gene expression is reminiscent of transit-amplifying cells.6 Phillips et al further provided evidence of GBM subclass switching, exclusively to the MES pattern, upon tumor recurrence. These data suggest that a better understanding of the pathways regulating neural development is crucial to understand the underlying signaling pathways and networks that give rise to GBM subtypes and may regulate the phenomenon of subclass switching.
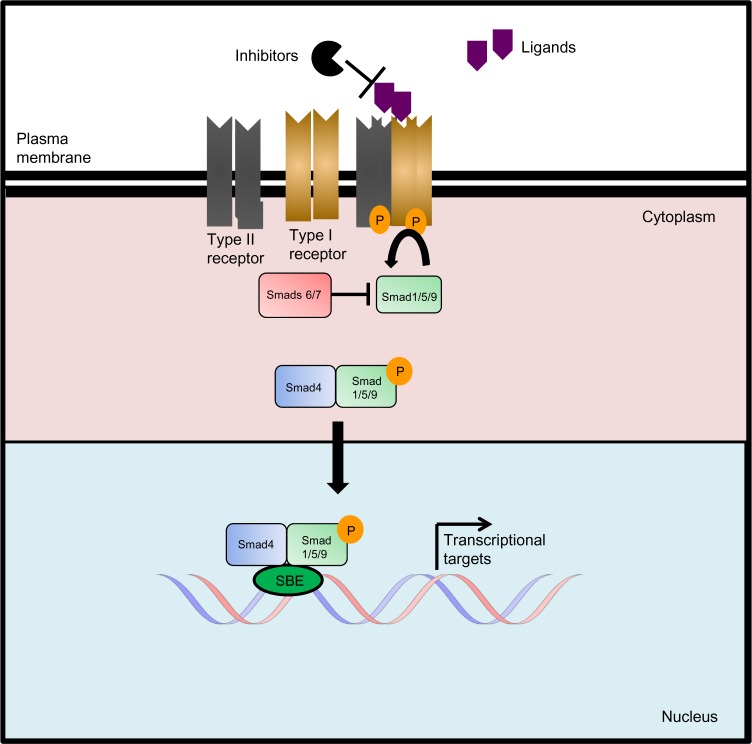
Bone Morphogenetic Protein Pathway
One of the most important signaling pathways regulating neural development is the bone morphogenetic protein (BMP) pathway. Te BMP signaling pathway is part of the transforming growth factor beta family (TGFβ). The TGFβ family has more than 30 members divided into families of activins, nodals, growth differentiation factors (GDFs), and BMPs.8 As with other members of the TGFβ family, during canonical signaling, BMP ligands bind to type I and type II serine-threonine kinase receptors, which transduce the signal downstream through phosphorylation of specific regulatory Smad proteins. During canonical BMP signaling, BMP ligands bind to the BMP type I and II receptors, which phosphorylate SMAD 1, 5, and 9 (human).9 The phosphorylated regulatory Smads then bind to the co-Smad, SMAD4, and the complex is then translocated into the nucleus for transcriptional regulation of specific target genes such as ID1–3, SNAIL, and OASIS.9 BMP signaling is regulated by a large network of extracellular antagonists as well as intracellular modulators such as the inhibitory Smads, SMAD6 and 7. SMAD6 is the primary inhibitor of BMP signaling by inhibiting the regulatory Smads and the co-Smad SMAD4 through a negative feedback mechanism (Fig. 1).BMPs play diverse functional roles in neural development and regulation of neural cell progenitors. During development, BMP signaling plays a major role in the patterning and establishment of early embryonic stem cells. During development, the inhibition of BMP signaling is equally critical for neural development. For example, the presence of a BMP gradient created by the presence of both BMP ligands and antagonists establishes the dorsal–ventral axis during development. During embryonic development, neural precursor cells are systematically driven toward specific neuronal or glial differentiation, and BMPs have been shown to be a key player in this process by regulating proliferation, mitotic arrest, and apoptosis.10 Several reviews have been written detailing these effects.11,12 More recently, it has been discovered that BMP signaling affects the proliferation and differentiation of glioma stem cells (GSC) as well.
Figure 1.
Canonical BMP signaling. The TGFβ family consists of cytokines that signal through serine/threonine kinase receptors. Activated ligands bind to type I and type II serine-threonine kinase receptor complexes, which transduce the signal downstream through the phosphorylation of pathway-specific regulatory Smad proteins (SMADs-1/5/9 are associated with BMP signaling and Smads 2/3 are associated with TGFβ signaling). Generally, the type II receptors phosphorylate the type I receptors, which in turn, phosphorylate the regulatory Smads. Phosphorylated regulatory Smads form a complex with Smad4, the co-Smad, and the complex is translocated to the nucleus for transcriptional regulation. Inhibitory Smads (SMADs 6 and 7) compete with regulatory Smads through both competition for phosphorylation by the type I receptors and binding to Smad4. Signaling antagonists work upstream of the signaling cascade by inhibiting ligands from binding to the receptor complex.
BMPs and GSCs
Glioma stem cells (GSCs) are a subpopulation of neoplastic cells within HGGs that have characteristics similar to those of normal neural stem cells. In 2006, Piccirillo et al published a novel finding in which BMP signaling had a striking tumor-suppressive effect on human GBM GSC. Treating human GSC in vitro and in vivo with BMP4 (a BMP ligand) for 48 hours decreased tumor growth and increased survival in an orthotopic transplant model through a decrease in proliferation and increase in astrocytic differentiation of the GSCs.13 Subsequently, it has been shown that BMP ligands have a similar impact on both human and murine GSCs.14–17 Several reviews have been published detailing the major findings in the field discussing BMP-driven tumor suppressive effects on GSCs and initial developments toward clinical BMP therapies for patients.18,19
Genomic alteration of BMP-related signaling molecules in human GBM
Although many studies have shown the tumor-suppressive effects of BMP signaling on GSC, little has been reported about BMP signaling in the context of GBM genomics. To gain a better understanding of BMP signaling in human GBMs, we queried BMP pathway alterations at the genetic level to assess how the BMP signaling network is altered in patient samples. We used publically available data compiled and analyzed through TCGA to examine gene expression and mutations and the REpository for Molecular BRAin Neoplasia DaTa (REMBRANDT) to analyze associations between gene expression and patient survival. We expanded our search beyond the immediate BMP family and analyzed 90 genes within the TGFβ family including receptors, ligands, inhibitors, and downstream targets known to interact directly with the BMP pathway (Supplementary Table 1). To identify genes within the TGFβ family that show either increased or decreased expression in GBMs, we accessed TCGA data using the cBio portal for cancer genomics maintained through the Memorial Sloan Kettering Cancer Center.20,21 We analyzed the mutational status and mRNA expression of 598 individual GBM samples using the TCGA provisional dataset. mRNA expression is considered to be significantly up or downregulated if the expression is above or below two standard deviations (2σ) of the mean determined from Agilent microarray data. We found that, out of our query of 90 genes, 44 were altered in 5% or more of patients (Table 1). In parallel, we analyzed the association between the mRNA expression of these 90 genes and patient survival using a dataset of 181 GBM patients available using REMBRANDT maintained by the National Cancer Institute (NCI).22 We investigated whether twofold up or downregulation of the mRNA levels analyzed using the U133 2 Plus mRNA expression chips (Affymetrix) is associated with increased or decreased overall survival. mRNA increases or decreases in expression are determined in comparison to nontumor pooled samples.23 Out of our set of 90 genes, 19 genes were significantly associated with either increased or decreased overall survival (Table 2).
Table 1.
Genes altered in ≥5% of human GBMs. All data were collected by the TCGA and analyzed using the cBio Portal for Cancer Genomics. Our dataset consisted of 598 individual GBM samples, and we analyzed 90 genes. For each gene altered in more than 5% of GBMs, we determined in how many tumor samples the gene was mutated and mRNA expression was upregulated or downregulated. mRNA upregulation and downregulation is considered to be >2 standard deviations from the mean expression.
| GENES | PERCENT ALTERED IN GBMS (598) | NUMBER OF PATIENTS WITH MUTATED GENE | NUMBER OF PATIENTS WITH mRNA UPREGULATION (>2 STD. DEV) | NUMBER OF PATIENTS WITH mRNA DOWNREGULATION (<2 STD. DEV) |
|---|---|---|---|---|
| BMP Ligands | ||||
| BMP7 | 5.2 | 0 | 15 | 16 |
| BMP8B | 10.2 | 0 | 9 | 52 |
| Receptors | ||||
| BMPR1A | 5.7 | 0 | 1 | 33 |
| TGFBR1 | 7. 7 | 2 | 6 | 38 |
| TGFBR2 | 5 | 2 | 15 | 11 |
| ACVR1 | 6 | 0 | 26 | 10 |
| ACVR1B | 8 | 0 | 7 | 41 |
| ACVRL1 | 7.5 | 1 | 13 | 31 |
| ACVR2B | 6 | 0 | 29 | 6 |
| ACVR2A | 5 | 0 | 26 | 2 |
| AMHR2 | 7 | 1 | 11 | 31 |
| Co-receptor | ||||
| BMPER | 5.5 | 1 | 32 | 0 |
| Intracellular | ||||
| Mediators | ||||
| SMAD1 | 6.5 | 0 | 17 | 22 |
| SMAD2 | 7.2 | 0 | 22 | 21 |
| SMAD3 | 6.2 | 0 | 13 | 24 |
| SMAD4 | 7.7 | 1 | 22 | 23 |
| SMAD6 | 6.2 | 0 | 12 | 25 |
| SMAD9 | 7.2 | 0 | 12 | 31 |
| SMURF2 | 6 | 0 | 15 | 21 |
| IDs | ||||
| ID2 | 6.2 | 0 | 20 | 17 |
| ID3 | 5.2 | 0 | 3 | 28 |
| Modulators | ||||
| CHRD | 8.9 | 1 | 7 | 46 |
| AMN | 8.7 | 0 | 9 | 43 |
| ENG | 6.7 | 1 | 14 | 25 |
| FSTL1 | 6 | 1 | 11 | 24 |
| SOST | 6 | 0 | 19 | 17 |
| TWSG1 | 5.7 | 0 | 10 | 24 |
| GREM2 | 5 | 0 | 16 | 14 |
| TGFβ Ligands | ||||
| TGFβ1 | 5 | 0 | 14 | 16 |
| TGFβ2 | 5 | 0 | 12 | 18 |
| TGFβ3 | 6.4 | 0 | 14 | 24 |
| Activins | ||||
| NODAL | 10.7 | 0 | 62 | 2 |
| AMH | 8.4 | 0 | 8 | 42 |
| GDNF | 7.2 | 2 | 12 | 29 |
| LEFTY2 | 5.9 | 2 | 16 | 17 |
| INHBC | 5.4 | 0 | 16 | 16 |
| INHBE | 5.7 | 1 | 26 | 7 |
| Misc. | ||||
| NRTN | 9.2 | 0 | 11 | 44 |
| BMP1 | 7 | 0 | 15 | 27 |
| ARTN | 6.4 | 0 | 12 | 26 |
| PSPN | 6.2 | 0 | 19 | 18 |
Table 2.
mRNA expression of genes associated with a significant increase or decrease in overall survival. Using the publically available database REMBRANDT, we were able to determine in which genes upregulation or downregulation of mRNA expression is associated with significant (P < 0.05) increased or decreased patient overall survival. Upregulation and downregulation are considered to be more than twofold change from expression in nontumor pooled samples. Log-rank P-values were calculated using the Mantel–Haenszel procedure. For each gene that was associated with overall survival, we determined the number of patients that expressed the alteration and calculated the average overall survival in months. Our dataset consisted of 181 GBM patients with a total overall survival of 19.6 months.
| GENE | mRNA EXPRESSION | LOG RANK P-VALUE | ASSOCIATED WITH INCREASED OR DECREASED SURVIVAL: | AVERAGE OVERALL SURVIVAL (MONTHS) | NUMBER OF PATIENTS WITHmRNA ALTERATION |
|---|---|---|---|---|---|
| GDNF | Up-regulated | 0.01 | Increased | 26.3 | 46 |
| BMP8B | Down-regulated | 0.02 | Increased | 29.1 | 19 |
| GDF5 | Up-regulated | 0.04 | Increased | 24.9 | 36 |
| BMP5 | Up-regulated | 0.05 | Increased | 42.6 | 7 |
| CHRDL1 | Up-regulated | 0.05 | Increased | 39.2 | 6 |
| RGMA | Down-regulated | 0.05 | Increased | 72.9 | 2 |
| BMP6 | Down-regulated | 0.000009 | Decreased | 8.9 | 12 |
| GDF15 | Down-regulated | 0.000065 | Decreased | 2.2 | 1 |
| ACVR1B | Down-regulated | 0.0014 | Decreased | 15.5 | 82 |
| GDNF | Down-regulated | 0.01 | Decreased | 14.0 | 32 |
| NBL1 | Down-regulated | 0.01 | Decreased | 16.6 | 110 |
| NRTN | Down-regulated | 0.01 | Decreased | 13.5 | 33 |
| SMAD6 | Up-regulated | 0.01 | Decreased | 10.4 | 10 |
| SOST | Up-regulated | 0.01 | Decreased | 14.0 | 45 |
| TSKU | Up-regulated | 0.01 | Decreased | 17.8 | 152 |
| FSTL1 | Up-regulated | 0.02 | Decreased | 18.1 | 154 |
| INHBE | Down-regulated | 0.02 | Decreased | 14.7 | 39 |
| SMAD1 | Up-regulated | 0.02 | Decreased | 15.4 | 65 |
| TWSG1 | Up-regulated | 0.02 | Decreased | 17.5 | 121 |
We found four genes, BMP8B, ACVR1B, SMAD1, and NRTN, to be both altered in more than 5% of patients and show an association with survival. Te role of these four genes in relation to GBMs is largely unknown. BMP8B (OP-2), first described by Ozkaynak et al in 1992, was found to be a member of the TGFβ family identified through cDNA library screenings. BMP8B is expressed early in embryogenesis.23 Although little is known about this protein in relation to gliomas, BMP8B treatment has been shown to decrease the proliferation of glioma stem cells.13 Interestingly, the mRNA expression available through the TCGA shows that BMP8B mRNA expression is downregulated in 52 patients and upregulated in only 9 patients. Therefore, downregulation of BMP8B mRNA expression accounts for 85% of the total BMP8B alterations. Within the REMBRANDT GBM dataset, downregulation of BMP8B correlates with increased patient survival (P = 0.02). The 19 GBM patients within this dataset with downregulated BMP8B had an average overall survival of 29.1 months.
ACVR1B (activin A type IB receptor, ALK4) is part of the activin subfamily within the TGFβ family. Activins are members of the TGFβ family known for their role as growth and differentiation factors. ACVR1B was originally discovered using a sequence-based polymerase chain reaction (PCR) approach by Dijke et al in 1993. ACVR1B mRNA is ubiquitously expressed in all tissues, most strongly in the kidneys, pancreas, brain, lung, and liver.24 ACVR1B mutations have been identified and found to have varying effects in several types of cancer. In prostate cancer, Nomura et al showed that cell lines with constitutively active AVCR1B had increased migratory ability aiding in epithelial–mesenchymal transition (EMT). In a neuroblastoma cell line, Suzuki et al showed that ACVR1B-specific activin signaling induced neuronal differentiation.25 Little is known about the role of ACVR1B on gliomas. The TCGA data show that ACVR1B expression is frequently decreased (41 out of 49 alterations, 85%) when altered in GBMs. Furthermore, within the REMBRANDT dataset, downregulation of ACVR1B was associated with decreased survival (P = 0.0014). Te 82 patients with ACVR1B downregulation had an average overall survival of 15.5 months compared to the average overall survival of the 181 patients at 19.6 months.
SMAD1 belongs to the Smad family, a family of proteins that serve as the signal transducers for canonical BMP and TGFβ signaling. While these Smad genes were originally discovered and understood in Drosophila and C. elegans, SMAD1, the human homolog, was first discussed and cloned in 1996.8,26 This protein is activated through BMP receptor phosphorylation, leading to downstream transcriptional regulation. In gliomas, Liu et al showed that phospho-SMAD1 is expressed at lower levels in glioma samples in comparison to normal brain tissue.27 Additionally, it was found that patients with a high ratio of phosphorylated SMAD1/5/9 to SMAD1 had increased survival, demonstrating that increased SMAD1 activation is beneficial to patient survival.28 This suggests that increased BMP signaling and increased SMAD1 phosphorylation provides a survival benefit. Using REMBRANDT, we observed that increased expression of SMAD1, found in 65 patients, is associated with decreased survival (P = 0.02) when compared to the total overall survival of REMBRANDT GBM patients (15.4 months vs 19.6 months). The mRNA expression data available through the TCGA shows that SMAD1 mRNA is equally up and downregulated within SMAD1-altered GBM samples (17 and 22 patients show upregulation and downregulation, respectively). As described above, analyzing the expression of SMAD1 in parallel with the phosphorylation of SMAD1 will be more informative with regard to GBMs and patient survival.
NRTN (neurturin) is a neurotrophic factor serving to promote the survival of various neuronal populations.27 NRTN was first isolated in 1996 by Kotzbauer et al after being identified by its ability to support sympathetic neurons in culture. NRTN is closely related to the glial cell line-derived neurotrophic factor (GDNF), both known as TGF-beta-related neurotrophins (TRNs). TRNs belong to the TGFβ family based on structural similarity and the presence of conserved cysteine residues found within TGFβ signaling molecules; yet TRNs share less than 20% amino acid sequence similarity to other TGFβ family members.27,29 Little is known about the role of NRTN in gliomas; however it has been reported that NRTN promotes pancreatic cell aggressiveness through both proliferation and invasion.30 The mRNA expression data we examined show that NRTN expression is frequently downregulated when altered in GBMs (44 out of 55 alterations). Using the REMBRANDT dataset NRTN downregulation is present in 33 patient samples and is associated with decreased survival with the average survival of 13.5 months (P = 0.01).
Additional genes of interest
In addition to the genes described above, we chose to further investigate five genes: SMAD4, BMPR1A, BMP5, ID1, and GREM1. SMAD4, BMPR1A, and ID1 were primarily selected because of their crucial role in mediating canonical BMP signaling. We selected BMP5 because we found 4% of all GBM patients posessed upregulation of BMP5, which was also found to be associated with increased survival. Finally, GREM1 was chosen because of the recently published finding showing that glioma stem cells secrete GREM1 to promote tumorigenesis through inhibition of BMP signaling.31
SMAD4 is a member of the Smad family, and of particular interest, as it is a central regulator of TGFβ family canonical signaling. SMAD4 was originally discovered as a tumor suppressor in pancreatic cancer in 1996 by Hahn et al after it was found to be homozygously deleted in 25 of 84 pancreatic tumors.32 SMAD4 has been shown to be involved in many other types of cancer primarily through chromosome deletion. SMAD4 has been shown to be inactivated in 48% of pancreatic tumors but is inactivated in less than 10% of tumors in other types of cancer.33 These deletions have been described in colon cancer,34 head and neck squamous cell carcinoma,35 and breast and ovarian cancer.33 In gliomas, He et al showed that SMAD4 expression is reduced in all gliomas in comparison with normal brain tissue with the lowest expression in HGGs. In addition He et al found that the loss of SMAD4 is correlated with poor survival.36 In our analysis, we found SMAD4 to be dysregulated in about 8% of the GBM dataset queried with equally distributed up and downregulation (22 and 23, respectively). Furthermore, our analysis showed no significant association between survival and up or downregulation of SMAD4 mRNA expression (P = 0.68, P = 0.73 respectively).
BMPR1A (bone morphogenetic protein receptor IA) (also known as ALK3), is a type I receptor in the TGFβ family. Using PCR technology, Dijke et al discovered BMPR1A in 1993 based on sequence homology to the human activin receptor type II and a type I-like TGFβ receptor in C. elegans.24 Mutations in BMPR1A have been shown to cause juvenile polyposis in many patients, a condition characterized by benign growth within the gastrointestinal tract.37–42 Guo et al in 2014 showed that the micro-RNA, miR-656, acted as a tumor suppressor in gliomas by specifically repressing expression of BMPR1A.43 Our analysis from TCGA data shows that BMPR1A mRNA expression is altered in approximately 5.7% of GBMs, with the vast majority of those being downregulated (33 out of 34 alterations). Within the REMBRANDT dataset, there were no GBM patient samples available with downregulation of BMPR1A. We found no association between upregulation of mRNA expression of BMPR1A and survival (P = 0.23).
BMP5 (Bone morphogenetic protein 5) BMP ligands were originally identified by Urist in 1965 because of their ability to induce endochondral osteogenesis in vivo at an extraskeletal site.44 BMP5, based on sequence homology, is in a subgroup with BMP 6, 7, and 8b.45 In adrenocortical carcinoma and pancreatic cancer, expression of BMP5 was found to be downregulated.46,47 BMP5 was shown to inhibit cell proliferation yet increase migration and invasion in pancreatic cancer cell lines.47 BMP5 has been shown to decrease proliferation of glioma stem cells.13 Our mRNA expression data show that BMP5 is altered in 4.8% of GBMs. Eighty-six percent of those alterations were due to upregulation of BMP5 (25 out of 29 alterations), and we found upregulation of BMP5 to be associated with increased survival of GBM patients (P = 0.05). The seven patients available within the REMBRANDT database with upregulation of BMP5 had an average overall survival of 42.6 months, more than twice the average survival.
ID1 (inhibitor of DNA-binding 1) is a key transcriptional regulator that is a specific downstream target of active BMP signaling. ID proteins inhibit the binding of DNA to other transcriptional factors by binding to the helix–loop–helix motif of transcriptional factors. ID1 was isolated from human fibroblasts in 1994 by Hara et al.48 ID1 has been shown to regulate the cell cycle and differentiation of cells in a wide variety of cell types48–51 including normal neural and glioma cells. In 2009 Nam and Benezra showed that ID1 and ID3 are required to maintain self-renewal of the type B adult neural stem cells. In addition, it was shown that ID1 can be used to identify type B neural stem cells within the stem cell niches of the brain and that ID1 protein levels decrease during the process of cell differentiation.52 However, the role of ID1 in GBM biology has proven to be very complex. ID1 expression has been shown to be upregulated in human gliomas and murine experimental models of glioma.53,54 In 2013, Soroceanu et al showed that ID1 levels correlate with tumor histopathologic grades and tumor cell invasiveness in vitro and that knockdown of ID1 increased survival in an orthotopic model of GBM.55 Contrastingly, Barrett et al showed, using a murine model of HGG, that glioma cells with both high and low levels of ID1 are tumorigenic and, surprisingly, the low ID1 expressing cells formed tumors more rapidly and with higher penetrance.56 According to our analyses, ID1 expression is altered in approximately 4.5% of GBMs, equally up and downregulated in the TCGA sample population (13 patients each). In the patient samples available through REMBRANDT, the five GBM patients with low ID1 mRNA expression had significantly decreased survival with an average overall survival of 8.5 months (P = 0.01). ID1 plays a prominent role in regulating both normal and tumor cells and warrants further investigation.
GREM1 (Gremlin 1) is an antagonist of BMP signaling. GREM1 is a secreted molecule that binds to BMP ligands to prevent them from binding to their receptors. GREM1 was isolated in 1998 by Hsu et al by cloning the human homolog of the gremlin gene in Xenopus.57 GREM1 is a crucial mediator of development by restricting BMP signaling.58 We chose to specifically investigate the expression of GREM1 due to the recent publication: “Glioma cancer stem cells secrete Gremlin1 to promote their maintenance within the tumor hierarchy” by Yan et al in 2014. This publication shows that GREM1 is endogenously expressed by GSCs to protect their self-renewal ability and stem-like state from the pro-differentiation effects of BMP signaling. GREM1 secretion is thought to contribute to treatment resistance through maintaining cellular proliferation and cellular hierarchies within the tumor, as well as increasing resistance to differentiation therapy.31 Our analysis shows that GREM1 is altered in approximately 3.8% of GBMs (23 patients), all of which are mRNA upregulations. Interestingly, using the REMBRANDT dataset, mRNA upregulation was present in 12 of the 181 patients showing enhanced survival with an average survival of 40 months, which is more than twice the average survival (P = 0.06).
Conclusions and Future Outlook
The BMP signaling pathway is a complex network of receptors, ligands, and antagonists, all of which may be capable of dynamically impacting GBM growth, maintenance, and progression, both positively and negatively. As with TGFβ signaling, it appears as though BMP signaling can modulate tumor growth and maintenance in various ways and most likely plays a context-dependent role in GBM tumor growth. Our analysis shows that upregulation and downregulation of ligands, receptors, and intracellular modulators are associated with both increased and decreased survival. Similarly, previous reports have shown conflicting data regarding the expression of various BMP signaling molecules and survival in human GBM.59–61 To better understand this pathway and how we may be able to exploit the signaling for novel drug treatments, we examined patient samples at the genetic level to explore the BMP pathway in GBMs. BMP signaling is required for development and the regulation of neural cells. As reviewed by Guang-Quan Zhao et al in 2002, targeted mutagenesis of BMP ligands, receptors, and other pathway modulators has shown that BMP signaling is involved and critical to almost all aspects of development.62 Given the high importance of BMP signaling, especially with a focus on neural cells, we hypothesize that BMP signaling is likely necessary for the survival of tumor cells and is very tightly regulated within the tumor environment, which may explain why the pathway is attenuated and not genetically deleted within GBMs. In support of our hypothesis, we observed that within the 598 tumors surveyed using the TCGA GBM provisional dataset, none of the genes queried are altered in more than 15% of the tumors, and mutations are exceedingly rare. The majority of the genes examined are not altered in more than 5% of GBMs. The alterations seen primarily occur in nonoverlapping tumors, indicating that compensation or redundancy in the pathway is possible and perhaps necessary for survival of tumor cells in GBMs. However, we may be able to exploit this importance to tumor cell survival in a way to benefit patients. As shown in xenograft models, there have been several forms of BMP treatments that have shown increased survival and decreased tumor growth, which should be considered for development in the clinic.13,17,63,64 More studies need to be completed to show that increased levels of BMP signaling in vivo at high doses do not act as tumor promoters. Understanding the larger impact of BMP treatment on the bulk of the tumor and the microenvironment is crucial prior to the development of BMPs into the clinic. As our analysis shows, increased expression of molecules involved in BMP signaling is not always associated with increased survival. The nature of genomic studies allows for a network view of individual pathways and uncovers genes within the pathway that may have remained unnoticed. Our analysis primarily serves as a guide for future research, emphasizing genes that are altered in a significant number of patients and are associated with survival. Here we highlighted several genes largely unstudied in the context of GBMs, which are both altered in a high percentage of patients and have associations with survival. Our analysis of 90 genes related to the BMP pathway provides a network view of the alterations occurring within the pathway to complement the many single-gene research studies that have been done. However, there are several factors that should be considered when interpreting this data. GBMs are known for their intratumoral heterogeneity; therefore it is likely that the gene expression and gene alterations differ throughout the tumor. Additionally, it is impossible to know when these mutations and alterations arose during the progression of the tumor. It is unknown which of the alterations reported were present in the initial tumor-promoting cells and which have been acquired in response to the selective pressures of the tumor microenvironment, treatments, and tumor resections. Johnson et al demonstrated this phenomena showing vast differences in the genomic alterations and mutations present in initial and recurrent tumors.32 For example, Johnson et al showed that SMADs 4,6,7, and 9 were mutated in recurrent tumors and not initial tumors, suggesting that alterations in Smads may not be involved in driving the initial tumor formation.32 Finally, our analysis is based on limited patient data with diverse genetic and treatment variables, which are important components to consider when determining appropriate therapies.
Given these caveats, our ultimate goal in sharing these findings is that our analysis will guide future studies to a novel and more complete understanding of the BMP pathway in relation to GBM pathology. As more genomic information is acquired, and as diverse samples are rapidly added to these publically available datasets, we will be able to generate more distinct and conclusive data on the genetic alterations and mutations that are critical regulators in GBM development and progression. In future studies with the addition of patient data, we will be able to stratify the population based on factors such as tumor subtype, initial or recurrent tumors, or previous treatments to take a more hypothesis-driven approach to genomic analysis. Recently, GBM genomic studies have begun to explore the extent of intratumoral heterogeneity when stratified by tumor stage or at the single-cell level enforcing the need for larger, increased depth of publically available GBM datasets.32,65
As we begin to combine the rapidly growing knowledge of epigenetic and proteomic information with genomic studies, our understanding of GBM tumor biology will vastly increase. Our current understanding of these tumors is clearly not proficient, given the dismal, almost uniformly fatal, outcome of this disease; however, genomic analyses can lead to new diagnostic tests, classifications, and treatment combinations and will direct both basic scientists and clinicians toward a future of successful individualized treatments. With the arrival of personalized medicine, it is more imperative than ever to gain a further understanding of these heterogeneous tumors at the genetic level for the optimization of new therapies.
Materials and Methods
TCGA analysis: cBioPortal for cancer genomics
Gene mutation status and mRNA expression were analyzed using publically available data obtained through the cBio Cancer Genomics Portal ((http://www.cbioportal.org/public-portal/ (accessed May 2014)). We selected the GBM (TCGA, Provisional) dataset from the Brain CNS Cancer Study category. Within the Genomic Profiles options, we selected mutations and mRNA expression data from Agilent microarray data using a z-score threshold of 2.0. Z-scores were determined by comparing the mRNA expression of each tumor sample to the mean expression value of all tumors that are diploid for the gene of interest. At the time of access, there were 598 patients available within the “All Tumors” GBM TCGA provisional dataset.
Survival analysis: REMBRANDT
Microarray gene expression and survival data were acquired from the publically available NCI REMBRANDT database (https://caintegrator.nci.nih.gov/rembrandt/home.do (accessed May 2014)). To analyze associations with survival, we selected the graph format: Kaplan-Meier survival plot for Gene Expression Data. We restricted the analysis to GBM patient samples. At the time of access, there were 181 GBM samples. Gene expression was determined from U133 2 Plus mRNA expression chips (Affymetrix). Upregulation and downregulation were determined as twofold or greater difference than pooled nontumor samples. Log-rank P-values were calculated using the Mantel–Haenszel procedure to determine the significance between groups of samples stratified by levels of gene expression.66 Gene associations with overall survival were compared to the overall survival for all 181 GBM patients. Te average overall survival for all 181 patients was 19.6 months. To determine the average overall survival, we reviewed the clinical reports of patients segregated by gene expression and calculated the mean overall survival.
Acknowledgments
We would like to thank the members of the Harold L. Moses and Ty Abel laboratories for critically reading the manuscript. Laura Hover has been supported by the VUMC Department of Pathology, Microbiology and Immunology. We thank Dr. Samuel Santoro for the support and encouragement.
Footnotes
ACADEMIC EDITOR: Todd W. Miller, Editor in Chief
FUNDING: Dr. Philip Owens has been previously supported by DoD BCRP postdoctoral fellowship grant number W81XWH-09–1-0421. He is also supported by NIH grants CA085492, CA102162, the Robert J. and Helen C. Kleberg Foundation, and the T.J. Martell Foundation. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the committee on Publication Ethics (COPE).
Author contributions
Conceived and designed the experiments: LDH, PO. Analyzed the data: LDH, PO. Wrote the first draft of the manuscript: LDH. Contributed to the writing of the manuscript: LDH, TWA, PO. Agree with the manuscript results and conclusions: LDH, TWA, PO. Jointly developed the structure and arguments for the paper: LDH, TWA, PO. Made critical revisions and approved final version: LDH, TWA, PO. All authors reviewed and approved of the final manuscript.
Supplementary Data
Supplementary Table 1. List of 90 genes related to TGFβ family that was analyzed at the genomic level. Genes were analyzed at the mRNA level for both alterations and any associations with survival in GBM patients.
REFERENCES
- 1.Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2(9):494–503. doi: 10.1038/ncpneuro0289. [DOI] [PubMed] [Google Scholar]
- 2.CBTRUS . CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2004–8. Central Brain Tumor Registry of the United States; Hinsdale, IL: 2012. pp. 1–52. [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, et al. Te 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–50. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 5.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 6.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 7.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riggins GJ, Tiagalingam S, Rozenblum E, et al. Mad-related genes in the human. Nat Genet. 1996;13:347–9. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 9.Miyazono K, Kamiya Y, Morikawa M. Bone morphogenetic protein receptors and signal transduction. J Biochem. 2010;147(1):35–51. doi: 10.1093/jb/mvp148. [DOI] [PubMed] [Google Scholar]
- 10.Panchision DM, Pickel JM, Studer L, et al. Sequential actions of BMP receptors control neural precursor cell production and fate. Genes Dev. 2001;15:2094–110. doi: 10.1101/gad.894701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H-L, Panchision DM. Concise review: bone morphogenetic protein pleiotropism in neural stem cells and their derivatives – alternative pathways, convergent signals. Stem Cells. 2007;25(1):63–8. doi: 10.1634/stemcells.2006-0339. [DOI] [PubMed] [Google Scholar]
- 12.Bond AM, Bhalala OG, Kessler JA. Te dynamic role of bone morphogenetic proteins in neural stem cell fate and maturation. Dev Neurobiol. 2012;72(7):1068–84. doi: 10.1002/dneu.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piccirillo SG, Reynolds BA, Zanetti N, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444(7120):761–5. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 14.Chirasani SR, Sternjak A, Wend P, et al. Bone morphogenetic protein-7 release from endogenous neural precursor cells suppresses the tumourigenicity of stemlike glioblastoma cells. Brain. 2010;133:1961–72. doi: 10.1093/brain/awq128. [DOI] [PubMed] [Google Scholar]
- 15.Klose A, Waerzeggers Y, Monfared P, et al. Imaging bone morphogenetic protein 7 induced cell cycle arrest in experimental gliomas. Neoplasia. 2011;13(3):276–85. doi: 10.1593/neo.101540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Z, Sun L, Wang Y, et al. Bone morphogenetic protein 4 inhibits cell proliferation and Induces apoptosis in glioma stem cell. Cancer Biother Radiopharm. 2011;26:77–83. doi: 10.1089/cbr.2010.0857. [DOI] [PubMed] [Google Scholar]
- 17.Lee J, Son MJ, Woolard K, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein developmental pathway inhibits differentiation of human glioblastoma tumor initiating cells. Cancer Cell. 2008;13(1):69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reguera-Nuñez E, Roca C, Hardy E, de la Fuente M, Csaba N, Garcia-Fuentes M. Implantable controlled release devices for BMP-7 delivery and suppression of glioblastoma initiating cells. Biomaterials. 2014;35(9):2859–67. doi: 10.1016/j.biomaterials.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Nakano I, Saigusa K, Kornblum HI. BMPing of glioma stem cells. Cancer Cell. 2008;13(1):3–4. doi: 10.1016/j.ccr.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, et al. Te cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):1–19. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Cancer Institute REMBRANDT [Internet] 2005. Available at: http://rembrandt.nci.nih.gov.
- 23.Ozkaynak E, Schnegelsberg PN, Jin DF, et al. Osteogenic protein-2. J Biol Chem. 1992;267(35):25220–7. [PubMed] [Google Scholar]
- 24.Ten Dijke P, Ichijo H, Franzen P, et al. Activin receptor-like kinases: a novel subclass of cell-surface receptors with predicted serine/threonine kinase activity. Oncogene. 1993;8(10):2879–87. [PubMed] [Google Scholar]
- 25.Suzuki K, Kobayashi T, Funatsu O, Morita A, Ikekita M. Activin A induces neuronal differentiation and survival via ALK4 in a SMAD-independent manner in a subpopulation of human neuroblastomas. Biochem Biophys Res Commun. 2010;394(3):639–45. doi: 10.1016/j.bbrc.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 26.Hoodless PA, Haerry T, Abdollah S, et al. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell. 1996;85(4):489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 27.Kotzbauer PT, Lampe PA, Heuckeroth RO, et al. Neurturin, a relative of glial-cell-line-derived neurotrophic factor. Nature. 1996;384:467–70. doi: 10.1038/384467a0. [DOI] [PubMed] [Google Scholar]
- 28.Liu S, Tian Z, Yin F, et al. Expression and functional roles of Smad1 and BMPR-IB in glioma development. Cancer Invest. 2009;27(7):734–40. doi: 10.1080/07357900802620786. [DOI] [PubMed] [Google Scholar]
- 29.Airaksinen MS, Saarma M. Te GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3(5):383–94. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- 30.Wang K, Demir IE, D’Haese JG, et al. Te neurotrophic factor neurturin contributes toward an aggressive cancer cell phenotype, neuropathic pain and neuronal plasticity in pancreatic cancer. Carcinogenesis. 2014;35(1):103–13. doi: 10.1093/carcin/bgt312. [DOI] [PubMed] [Google Scholar]
- 31.Yan K, Wu Q, Yan DH, et al. Glioma cancer stem cells secrete Gremlin1 to promote their maintenance within the tumor hierarchy. Genes Dev. 2014;28(10):1085–100. doi: 10.1101/gad.235515.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–93. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schutte M, Hruban RH, Hedrick L, et al. DPC4 gene in various tumor types. Cancer Res. 1996;56:2527–30. [PubMed] [Google Scholar]
- 34.Tiagalingam S, Lengauer C, Leach FS, et al. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13:343–6. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 35.Kim SK, Fan Y, Papadimitrakopoulou V, et al. DPC4, a candidate tumor suppressor gene, is altered infrequently in head and neck squamous cell carcinoma. Cancer Res. 1996;56:2519–21. [PubMed] [Google Scholar]
- 36.He S, Zhao Z, Wang Y, et al. Reduced expression of SMAD4 in gliomas correlates with progression and survival of patients. J Exp Clin Cancer Res. 2011;30(1):70. doi: 10.1186/1756-9966-30-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou X, Woodford-richens K, Lehtonen R, et al. Germline mutations in BMPR1 A/ALK3 cause a subset of cases of juvenile polyposis syndrome and of Cowden and Bannayan-Riley-Ruvalcaba syndromes. Am J Hum Genet. 2001;69:704–11. doi: 10.1086/323703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim I-J, Park J-H, Kang HC, et al. Identification of a novel BMPR1A germline mutation in a Korean juvenile polyposis patient without SMAD4 mutation. Clin Genet. 2003;63(2):126–30. doi: 10.1034/j.1399-0004.2003.00008.x. [DOI] [PubMed] [Google Scholar]
- 39.Howe JR, Bair JL, Sayed MG, et al. Germline mutations of the gene encoding bone morphogenetic protein receptor 1 A in juvenile polyposis. Nat Genet. 2001;28(2):184–7. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 40.Howe JR, Sayed MG, Ahmed AF, et al. Te prevalence of MADH4 and BMPR1 A mutations in juvenile polyposis and absence of BMPR2, BMPR1B, and ACVR1 mutations. J Med Genet. 2004;41:484–91. doi: 10.1136/jmg.2004.018598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao X, Eu KW, Kumarasinghe MP, Li HH, Loi C, Cheah PY. Mapping of hereditary mixed polyposis syndrome (HMPS) to chromosome 10q23 by genomewide high-density single nucleotide polymorphism (SNP) scan and identification of BMPR1A loss of function. J Med Genet. 2006;43:e13. doi: 10.1136/jmg.2005.034827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delnatte C, Sanlaville D, Mougenot J-F, et al. Contiguous gene deletion within chromosome arm 10q is associated with juvenile polyposis of infancy, reflecting cooperation between the BMPR1A and PTEN tumor-suppressor genes. Am J Hum Genet. 2006;78:1066–74. doi: 10.1086/504301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo M, Jiang Z, Zhang X, et al. miR-656 inhibits glioma tumorigenesis through repression of BMPR1A. Carcinogenesis. 2014;00(00):1–9. doi: 10.1093/carcin/bgu030. [DOI] [PubMed] [Google Scholar]
- 44.Urist MR. Bone: formation by autoinduction. Science. 1965;150(3698):893–9. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- 45.Kawabata M, Imamura T, Miyazono K. Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev. 1998;9(1):49–61. doi: 10.1016/s1359-6101(97)00036-1. [DOI] [PubMed] [Google Scholar]
- 46.Johnsen IK, Kappler R, Auernhammer CJ, Beuschlein F. Bone morphogenetic proteins 2 and 5 are down-regulated in adrenocortical carcinoma and modulate adrenal cell proliferation and steroidogenesis. Cancer Res. 2009;69(14):5784–92. doi: 10.1158/0008-5472.CAN-08-4428. [DOI] [PubMed] [Google Scholar]
- 47.Virtanen S, Alarmo E-L, Sandström S, Ampuja M, Kallioniemi A. Bone morphogenetic protein -4 and -5 in pancreatic cancer – novel bidirectional players. Exp Cell Res. 2011;317(15):2136–46. doi: 10.1016/j.yexcr.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Hara E, Yamaguchi T, Nojima H, et al. Id-related genes encoding helix-loop-helix proteins are required for G1 progression and are repressed in senescent human fibroblasts. J Biol Chem. 1994;269(3):2139–45. [PubMed] [Google Scholar]
- 49.Ohtani N, Zebedee Z, Huot TJ, et al. Opposing effects of Ets and Id proteins on p16 INK4a expression during cellular senescence. Nature. 2001;409:1067–70. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 50.Singh J, Itahana Y, Parrinello S, Murata K, Desprez PY. Molecular cloning and characterization of a zinc finger protein involved in Id-1-stimulated mammary epithelial cell growth. J Biol Chem. 2001;276(15):11852–8. doi: 10.1074/jbc.M006931200. [DOI] [PubMed] [Google Scholar]
- 51.Lyden D, Young AZ, Zagzag D, et al. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401(6754):670–7. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 52.Nam H, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5(5):515–26. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anido J, Sáez-Borderías A, Gonzàlez-Juncà A, et al. TGF-β Receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18(6):655–68. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 54.Vandeputte DA, Troost D, Leenstra S, et al. Expression and distribution of Id helix-loop-helix proteins in human astrocytic tumors. Glia. 2002;38:329–38. doi: 10.1002/glia.10076. [DOI] [PubMed] [Google Scholar]
- 55.Soroceanu L, Murase R, Limbad C, et al. Id-1 is a key transcriptional regulator of glioblastoma aggressiveness and a novel therapeutic target. Cancer Res. 2013;73(5):1559–69. doi: 10.1158/0008-5472.CAN-12-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrett LE, Granot Z, Coker C, et al. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell. 2012;21(1):11–24. doi: 10.1016/j.ccr.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 57.Hsu DR, Economides AN, Wang X, Eimon PM, Harland RM. Te Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol Cell. 1998;1:673–83. doi: 10.1016/s1097-2765(00)80067-2. [DOI] [PubMed] [Google Scholar]
- 58.Zúñiga A, Haramis AP, McMahon AP, Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- 59.Bao Z, Zhang C, Yan W, et al. BMP4, a strong better prognosis predictor, has a subtype preference and cell development association in gliomas. J Transl Med. 2013;11(100):1–7. doi: 10.1186/1479-5876-11-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu C, Tian G, Tu Y, Fu J, Lan C, Wu N. Expression pattern and clinical prognostic relevance of bone morphogenetic protein-2 in human gliomas. Jpn J Clin Oncol. 2009;39(10):625–31. doi: 10.1093/jjco/hyp094. [DOI] [PubMed] [Google Scholar]
- 61.Yamada N, Kato M, Dijkel P, et al. Bone morphogenetic protein type IB receptor is progressively expressed in malignant glioma tumours. Br J Cancer. 1996;73:624–9. doi: 10.1038/bjc.1996.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao GQ. Consequences of knocking out BMP signaling in the mouse. Genesis. 2003;35(1):43–56. doi: 10.1002/gene.10167. [DOI] [PubMed] [Google Scholar]
- 63.Tate CM, Pallini R, Ricci-Vitiani L, et al. A BMP7 variant inhibits the tumorigenic potential of glioblastoma stem-like cells. Cell Death Differ. 2012;19:1–11. doi: 10.1038/cdd.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duggal R, Geissinger U, Zhang Q, et al. Vaccinia virus expressing bone morphogenetic protein-4 in novel glioblastoma orthotopic models facilitates enhanced tumor regression and long-term survival. J Transl Med. 2013;11(155):1–14. doi: 10.1186/1479-5876-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–67. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. List of 90 genes related to TGFβ family that was analyzed at the genomic level. Genes were analyzed at the mRNA level for both alterations and any associations with survival in GBM patients.