Abstract
The discovery that TNF receptors (TNFR) serve as the binding receptors for progranulin (PGRN) reveals the significant role of PGRN in inflammatory and autoimmune diseases, including inflammatory arthritis. Herein we describe a simple, antibody-free analytical assay, i.e., a biotin-based solid-phase binding assay, to examine the direct interaction of PGRN/TNFR and the PGRN inhibition of TNF/TNFR interactions. Briefly, a 96-well high-binding microplate is first coated with the first protein (protein A), and after blocking, the coated microplate is incubated with the biotin-labeled second protein (protein B) in the absence or presence of the third protein (protein C). Finally the streptavidin conjugated with a detecting enzyme is added, followed by a signal measurement. Also discussed in this chapter are the advantages of the strategy, key elements to obtain reliable results, and discrepancies among various PGRN proteins in view of the binding activity with TNFR.
Keywords: TNF, TNFR, Progranulin, Solid-phase assay, Biotin-labeled protein
1 Introduction
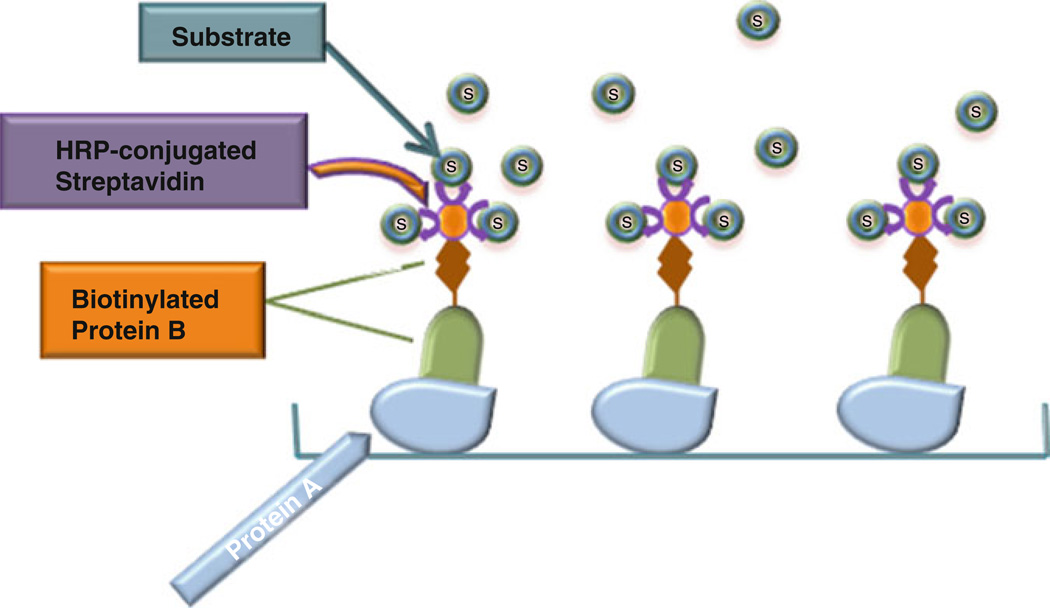
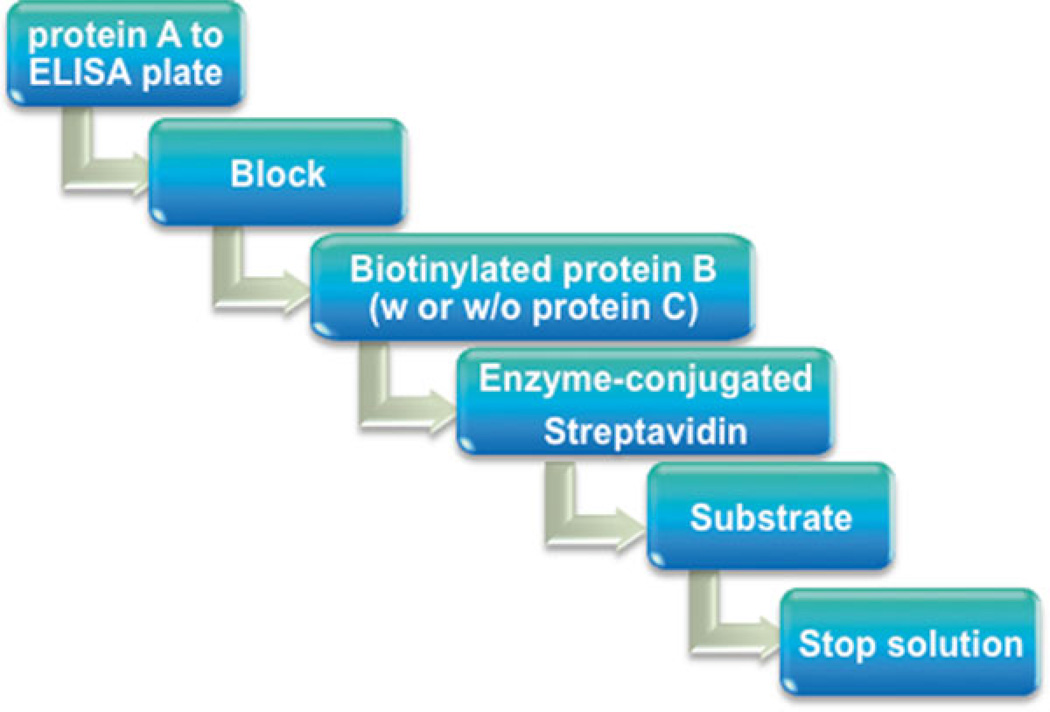
Solid-phase binding assay is a widely used approach to study protein–protein interactions in vitro, especially antibody-based solid- phase assay, in which antigen-specific antibodies are used [1–3]. However, there may be some issues associated with the antibody-based assay, limiting the use of the assay. One common problem is the nonspecific binding of the antibodies, which can result in the high background. Another concern comes from the inconsistency of the antibodies employed, especially when polyclonal antibodies are used. The ideal conditions need to be established for each batch, including antibody concentration, incubation time, dilution buffer, and so on. In addition, some antibodies may not be commercially available. In view of these potential issues, establishing a simple and an antibody-free approach is of importance for routinely detecting direct protein–protein interactions in vitro. Biotin- based solid-phase assay is some sort of a simple and time-saving technique used for protein interaction analysis, and more importantly, it is antibody-free. Briefly, the first protein (protein A) is immobilized to an ELISA plate (solid phase). After blocking, biotin- labeled second protein (protein B), alone or together with the third protein (protein C), is added to the well. After washing, bound protein is detected by addition of a streptavidin conjugated with an enzyme followed by a detection step and signal measurement (Figs. 1, 2, and 3).
Fig. 1.
Schematic diagram of biotin-based solid-phase binding assay
Fig. 3.
Pipeline of detecting protein–protein interactions using biotin-based solid-phase assay
Progranulin (PGRN), also known as proepithelin (PEPI), PC cell-derived growth factor (PCDGF), and granulin epithelin precursor (GEP), is a 593-amino-acid autocrine growth factor [4, 5]. PGRN is highly expressed in epithelial cells, certain types of neurons, and macrophages [6], and is likewise expressed in a broad range of other tissues and cell types including skeletal muscle, chondrocytes, adipose tissue, hematopoietic cells, and immune cells, including T cells and dendritic cells [7–9]. PGRN has multiple physiological functions, and is involved in many types of disease processes, including autoimmune disorders, cancer, and neurodegenerative diseases. PGRN promotes cell proliferation, and is crucial to the development and generation of fast-growing epithelial, endothelial cells, and cancer cells [10]. PGRN levels are elevated in many types of cancer, including breast cancer, ovarian cancer, and cholangiocarcinoma [11–13]. Mutations of the PGRN gene are known to lead to the development of frontotemporal lobar degeneration (FTLD) [14, 15], and the protein levels of PGRN in serum and cerebral spinal fluid (CSF) were significantly reduced in both unaffected and affected PGRN-mutation carriers in FTLD [16–18]. The importance of PGRN has also been emerging in respect to the immune system [19]. PGRN has been reported to promote CD4 + T cells to differentiate into Foxp3 + regulatory T cells [20] and to enhance antibody diversity in B cells [21]. PGRN also binds with Toll-like receptor 9 (TLR9) and assists in the recruitment of CpG oligonucleotides (CpG-ODNs) in macrophages [22], a process which is critical for the elimination of infection.
Our recent studies have led to the isolation of PGRN as a binding partner of TNFR in a functional genetic screen [20, 23]. We have confirmed the interactions between PGRN and TNFR using various approaches, including Co-IP, FastStep Kinetic Assay, and antibody-based solid-phase assay [20]. Here we describe a biotin-based solid-phase binding assay using PGRN and TNFR as examples. In addition, a biotin-based assay for examining PGRN inhibition of TNF binding to TNFR is also presented.
2 Materials
-
Recombinant proteins.
Recombinant human progranulin: Purified from progranulin stable cell line-PGRN HEK293 EBNA stable cell line (Lab progranulin) [24], or R&D Systems Inc. (Mouse myeloma cell line NS0-derived, Thr18-Leu593).
Recombinant Human TNF RII (Invitrogen or R&D Systems Inc.), also known as CD120b and p75.
Recombinant Human TNF-α (R&D systems Inc.): E. coli-derived, Val77-Leu233.
EZ-Link Sulfo-NHS-Biotinylation Kit (Thermo scientific) (see Note 1).
Zeba™ Spin Desalting Column: It contains a high-performance resin that offers exceptional desalting or buffer exchange for protein samples.
TBS buffer: Tris-buffered saline, 10 mM Tris–HCl, pH 7.4, 150 mM NaCl.
Binding buffer: TBS buffer containing 0.1 % (w/v) BSA, 1 mM CaCl2. Prepare fresh.
Streptavidin-HRP (Invitrogen): Diluted 1:2,500 in binding buffer.
TMB Solution (Invitrogen): Ready to use, no dilution or further preparation required. In the presence of HRP, TMB will turn to blue.
Stop solution: 2 N Sulfuric Acid.
Blocking solution: 5 % (w/v) bovine serum albumin (BSA) Fraction V, Heat Shock Treated (Fisher scientific), 0.05 % (w/v) sodium azide in TBS. Prepare sodium azide as 20 % stock solution, then add to TBS buffer, dissolve BSA to the buffer, mix gently up and down. Prepare fresh
3 Methods
3.1 Biotinylation of Protein
Sulfo-NHS-Biotin is a popularly used water-soluble biotinylation reagent for labeling antibodies, proteins, and any other macromolecules that contain primary amine. It can be conjugated to many proteins of the primary amino groups to form stable amide bonds without perturbing the function of the molecule due to the small size of biotin, and it has high binding affinity to streptavidin and avidin.
The manufacturer’s protocol is followed to label proteins. Briefly, calculate the amount of reagent needed, and prepare the biotin solution; add to the protein solution for labeling reaction, then perform buffer exchange to remove excess biotin reagent using a desalting column, followed by an HABA assay [2–(4-hydroxyazobenzene) benzoic acid] to estimate biotin label incorporation.
3.2 Purification of Recombinant PGRN Protein
Culture the progranulin stable cell line-PGRN HEK293 EBNA in tissue culture dishes in DMEM supplemented with 10 % of fetal bovine serum (FBS) and 50 µg/mL of G418 (Geneticin). After reaching confluence, add serum-free medium to the cells, and continue incubation for another 2 days, then collect the medium.
To purify recombinant PGRN protein, add nickel-chelating resin to the medium and mix at 4 °C for overnight. After washing, the bound protein was eluted and analyzed by SDS-PAGE and Western blot with anti-PGRN antibody.
3.3 Solid-Phase Binding Procedure for Detecting the Binding of PGRN to TNFR
Coat various doses (0–0.5 µM) of PGRN (protein A) or BSA (serving as control) in TBS buffer containing 0.5 % BSA to ELISA plate (100 µL/well) (see Note 2). Perform the assay using triplicates for each set of samples (see Note 3). Cover the plate and store at room temperature overnight. The plate can be stored at 4 °C for several days.
Discard the solution in the well and add 100 µL of blocking solution to each well using a multichannel pipette. After soaking for 1 min, invert the plate over a sink and strike it onto adsorbent paper toweling to remove the solution.
Add 200 µL of blocking solution to each well using a multichannel pipette. Leave at room temperature for 1–3 h (see Note 4).
Discard the blocking solution, and wash the wells three times with binding buffer.
Flick out the final wash. Invert the plate and tap it hard onto adsorbent paper towel to remove residual liquid (see Note 5).
Dilute the biotin-labeled TNFR2 (protein B) in binding buffer to a concentration of 1 ng/µL. Add 100 µL of this solution to each experimental well. Cover the plate and incubate at 37 °C for 2–3 h.
Aspirate the wells to remove unbound protein. Wash the wells three times with binding buffer. Remove residual buffer as in step 5.
Dilute streptavidin–HRP reagent 1:2,500 in binding buffer. Add 100 µL of this reagent to each well using a multichannel pipette. Incubate the plate for 15 min at room temperature (see Note 6).
Aspirate the wells to remove unbound streptavidin–HRP. Wash the wells seven to ten times with binding buffer. Remove residual buffer as in step 5.
Add 100 µL of TMB to each well using a multichannel pipette. Keep the plate in the dark until a blue color is obtained (typically 5–30 min) (see Note 7).
Stop the reaction by adding 100 µL of 2 N H2 SO4 solution to each well using a multichannel pipette.
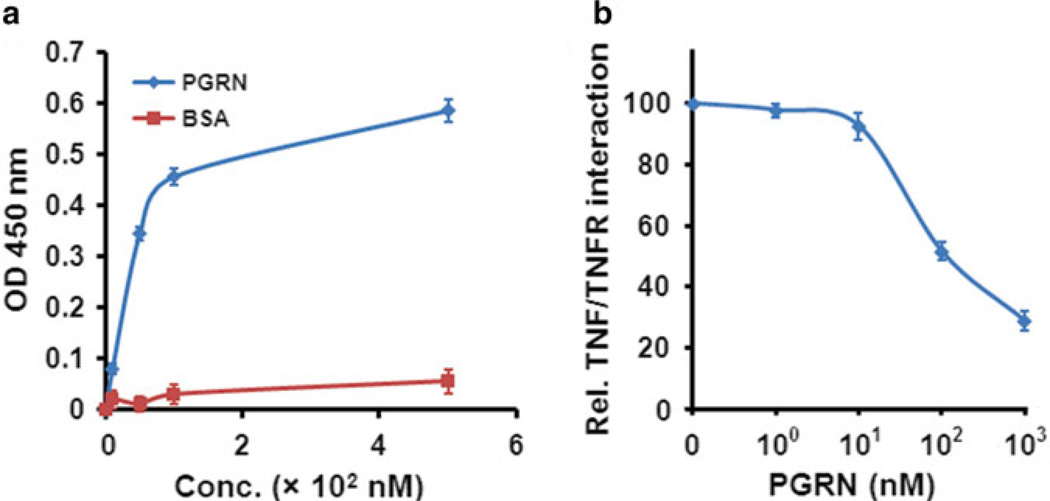
Read the plate using an automatic plate reader at 450 nm (see Notes 8– 11) and analyze the data (see Fig. 4a).
Fig. 4.
Reprehensive examples of solid-phase assay. (a) Binding of PGRN to TNFR2. Various doses of PGRN or BSA, as indicated, were absorbed to an ELISA plate. After blocking biotin-labeled TNFR2 was added, and the bound protein from the liquid phase was then detected by streptavidin conjugated with horseradish peroxidase. (b) PGRN inhibition of TNFα binding to TNFR2. After blocking, plates precoated with TNFR2 were incubated with various amounts of PGRN, as indicated, and biotin-labeled TNFα, and the bound biotin-labeled TNFα was detected as described in (a)
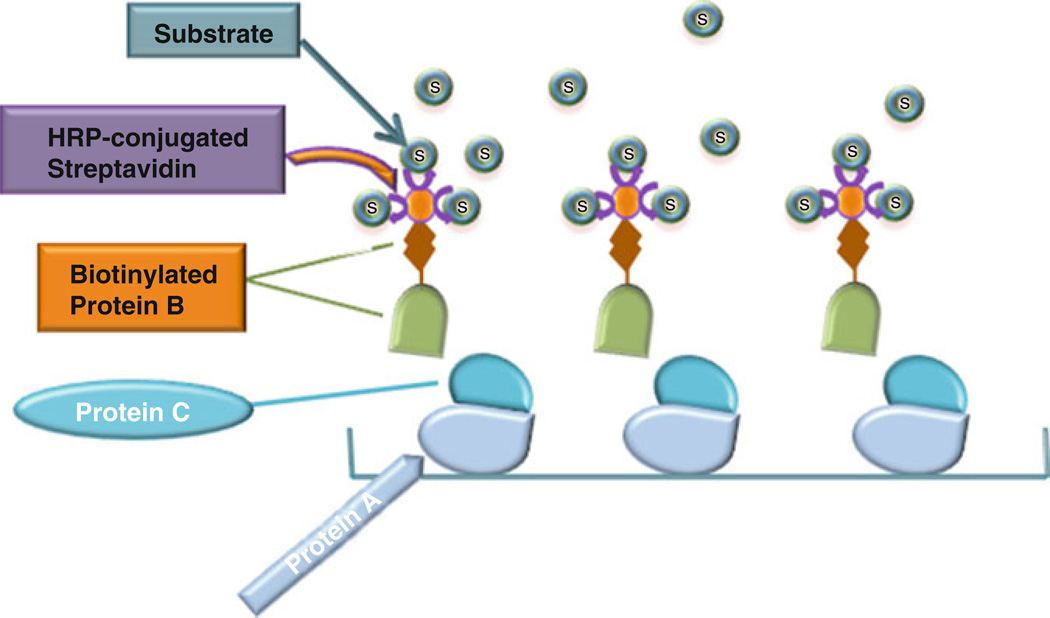
3.4 Solid-Phase Binding Procedure for Detecting the Inhibition of TNF/TNFR Interactions by PGRN
Coat ELISA plate with TNFR2 (100 µL/well) in TBS buffer containing 0.2 % BSA. The well without TNFR2 serves as a blank. Perform the assay using triplicates for each set of samples. Cover the plate with plastic film and store at room temperature overnight or 4 °C for up to 1 week.
Follow the steps 2–5 in Subheading 3.3.
Add various doses (0–1 µM) of PGRN (protein C) or BSA (serving as control) in binding buffer to each experimental well. Cover the plate with plastic film and incubate at 37 °C for 1 h. Then add biotin-labeled TNFα (protein B) to each experimental well and incubate at 37 °C for another 2 h.
Follow the steps 7–12 in Subheading 3.3 (Fig. 4b).
Fig. 2.
Schematic diagram of biotin-based solid-phase inhibition assay
Acknowledgements
This work was supported partly by NIH research grants R01AR062207, R01AR061484, R56AI100901, and a Disease Targeted Research Grant from Rheumatology Research Foundation. Researches in the laboratory of C. J. Liu were also supported by Atreaon, Inc.
Abbreviations
- BSA
Bovine serum albumin
- CpG-ODNs
CpG oligonucleotides
- CSF
Cerebral spinal fluid
- EBNA
Epstein-Barr nuclear antigen
- FGF
Fibroblast growth factor
- FTLD
Frontotemporal lobar degeneration
- GEP
Granulin–epithelin precursor
- HEK
Human embryonic kidney
- HRP
Horseradish peroxidase
- PCDGF
GP88/PC-cell derived growth factor
- PEPI
Proepithelin
- PGRN
Progranulin
- TLR9
Toll-like receptor
- TMB
3,3′,5,5″-Tetramethylbenzidine
- TNFR
Tumor necrosis factor receptor
- TNF-α
Tumor necrosis factor-α
Footnotes
EZ-Link Sulfo-NHS-Biotin is moisture-sensitive. Prepare the biotin reagent immediately before use.
The addition of BSA is not always required when preparing protein samples. And when it is used, the concentration in dilution buffer should be optimized from various trials aiming at the particular target protein and reagents. An appropriate selection of the buffer helps to block nonspecific binding sites to the upmost and retain the specific binding affinity at the same time.
Replicating samples for each set is preferred to get reliable results. Usually, the samples should be performed using at least triplicates, if possible, six to eight replicates, the more the better. The experiments are also expected to be repeated for several times, to ensure that the results are reproducible.
Blocking is the principal procedure to reduce nonspecific binding. A preferable blocking effect results from the proper choice of blocking reagents and the proper dosage of it. Different reagents may bring about dramatically varied background. The same blocker may perform distinctly different in different tests. Nonfat dry milk, for example, is regarded as an effective blocking buffer in certain kinds of solid-phase assay, like antibody-based assay, which, however, is not the same situation as in this biotin-based solid-phase assay. In this study we used BSA as the blocker. To determine the optimal BSA concentration, preliminary experiments should be performed with different concentrations of BSA in blocking buffer. Meanwhile, a longer blocking time should be expected in case of high background binding.
Another basic way to get rid of high background is to present enough washes. Insufficient washing will lead to reagent residual in the wells, which may attenuate the difference between control and experimental groups. It is critical to guarantee plenty of washing times and complete elimination of the washing buffer each time.
To avoid excessive HRP signal, it is important to perform multiple dilutions of streptavidin-HRP before use. A feasible concentration of HRP can be figured out by performing experiments with different dilutions.
The signal development is time-dependent in the assay. After adding the substrate to start coloration, the plate should be kept in the dark for maintaining the stability of TMB. But it is important to check the plate every few minutes. Stop solution should be added whenever the coloration is obvious. Overtime incubation might result in overreactions. A typical time period is 5–30 min, which relies on different properties, concentrations, and binding affinities of reagents.
A highly reliable OD450 value is conventionally considered as ranging between 0.2 and 1.5. The values exceeding this range may be not reliable. High OD value may occur because of improper and inefficient washing or high concentration of reagents. Low OD value is probably due to insufficient incubation time, improper sample dilutions, expiration of reagents, or incorrect operation on ELISA readers (such as using wrong wavelength).
Another reason that leads to low OD value is probably due to the use of sodium azide. Sodium azide is an inorganic compound; it has an inhibitory effect on the activity of HRP and can quench peroxidase activity and suppress the growth of bacteria during incubations. It may be added for its bacteriostatic properties, but should be avoided by the step of incubation with HRP reagents.
In summary, biotin-based solid-phase assay is a simple, costeffective analytical assay for studying protein interactions. For instance, it is antibody free and significantly reduces the workload, as there is no need to optimize antibody conditions. More importantly, it avoids the inconsistency and nonspecific binding resulted from the application of various antibodies. Although the biotin-based solid-phase assay offers numerous advantages over other assays, one potential disadvantage associated with this assay is that the biotinylation process may alter the structure and properties of the proteins of interest, and the labeled protein may thus exhibit less or no binding activities to its binding partners.
The fact that PGRN is composed of 593 amino acids but displays an apparent molecular weight of approximately 90 kDa indicates that PGRN is modified, including glycosylation [25]. It appears that the various glycosylation and conformation are probably critical for the binding of PGRN to TNFR, since recombinant PGRN produced from a HEK-EBAN stable line, but not purchased from R&D System (data not shown), bound to TNFR (Fig. 4a). These findings are in line with the binding activities of perlecan, a PGRN-binding glycoprotein [26]. Perlecans purified from different cell types or same cell type with different expression systems have been shown to have the potential for significant variation in glycosylation and function, and can vary dramatically in FGF receptor-based assays or in growth factor binding assays [27–29]. It is worthwhile to note that the PGRN-binding activities of TNFR2 obtained from several commercial sources also significantly varied. For instance, TNFR2 purchased from Invitrogen or Sigma, but not from R&D System, showed reproducible binding with PGRN (data not shown), indicating that proper conformation of TNFR is also important for their ligand binding properties.
References
- 1.Aroca-Aguilar JD, Sanchez-Sanchez F, Ghosh S, Fernandez-Navarro A, Coca-Prados M, Escribano J. Interaction of recombinant myocilin with the matricellular protein SPARC: functional implications. Invest Ophthalmol Vis Sci. 2011;52:179–189. doi: 10.1167/iovs.09-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham NA, Pope MD, Rimchala T, Huang BK, Asthagiri AR. A microtiter assay for quantifying protein-protein interactions associated with cell-cell adhesion. J Biomol Screen. 2007;12:683–693. doi: 10.1177/1087057107301941. [DOI] [PubMed] [Google Scholar]
- 3.Arlinghaus FT, Eble JA. The collagen-binding integrin alpha2beta1 is a novel interaction partner of the T. flavoviridis venom protein flavocetin-A. J Biol Chem. 2012;288:947–955. doi: 10.1074/jbc.M112.399618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu CJ. Progranulin: a promising therapeutic target for rheumatoid arthritis. FEBS Lett. 2011;585:3675–3680. doi: 10.1016/j.febslet.2011.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu CJ, Bosch X. Progranulin: a growth factor, a novel TNFR ligand and a drug target. Pharmacol Ther. 2012;133:124–132. doi: 10.1016/j.pharmthera.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniel R, He Z, Carmichael KP, Halper J, Bateman A. Cellular localization of gene expression for progranulin. J Histochem Cytochem. 2000;48:999–1009. doi: 10.1177/002215540004800713. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Bai X, Tian Q, Lai Y, Lin EA, Shi Y, et al. GEP constitutes a negative feedback loop with MyoD and acts as a novel mediator in controlling skeletal muscle differentiation. Cell Mol Life Sci. 2012;69:1855–1873. doi: 10.1007/s00018-011-0901-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsubara T, Mita A, Minami K, Hosooka T, Kitazawa S, Takahashi K, et al. PGRN is a key adipokine mediating high fat diet-induced insulin resistance and obesity through IL-6 in adipose tissue. Cell Metab. 2012;15:38–50. doi: 10.1016/j.cmet.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Elkabets M, Gifford AM, Scheel C, Nilsson B, Reinhardt F, Bray MA, et al. Human tumors instigate granulin-expressing hematopoietic cells that promote malignancy by activating stromal fibroblasts in mice. J Clin Invest. 2011;121:784–799. doi: 10.1172/JCI43757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Z, Bateman A. Progranulin gene expression regulates epithelial cell growth and promotes tumor growth in vivo. Cancer Res. 1999;59:3222–3229. [PubMed] [Google Scholar]
- 11.Frampton G, Invernizzi P, Bernuzzi F, Pae HY, Quinn M, Horvat D, et al. Interleukin-6-driven progranulin expression increases cholangiocarcinoma growth by an Akt-dependent mechanism. Gut. 2012;61:268–277. doi: 10.1136/gutjnl-2011-300643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serrero G. Autocrine growth factor revisited: PC-cell-derived growth factor (progranulin), a critical player in breast cancer tumorigenesis. Biochem Biophys Res Commun. 2003;308:409–413. doi: 10.1016/s0006-291x(03)01452-9. [DOI] [PubMed] [Google Scholar]
- 13.Diaz-Cueto L, Arechavaleta-Velasco F, Diaz-Arizaga A, Dominguez-Lopez P, Robles-Flores M. PKC signaling is involved in the regulation of progranulin (acrogranin/PC-cell-derived growth factor/granulin-epithelin precursor) protein expression in human ovarian cancer cell lines. Int J Gynecol Cancer. 2012;22:945–950. doi: 10.1097/IGC.0b013e318253499c. [DOI] [PubMed] [Google Scholar]
- 14.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 15.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, et al. Null mutations in progranulin cause ubiquitinpositive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 16.Ghidoni R, Benussi L, Glionna M, Franzoni M, Binetti G. Low plasma progranulin levels predict progranulin mutations in frontotemporal lobar degeneration. Neurology. 2008;71:1235–1239. doi: 10.1212/01.wnl.0000325058.10218.fc. [DOI] [PubMed] [Google Scholar]
- 17.Sleegers K, Brouwers N, Van Damme P, Engelborghs S, Gijselinck I, van der Zee J, et al. Serum biomarker for progranulin-associated frontotemporal lobar degeneration. Ann Neurol. 2009;65:603–609. doi: 10.1002/ana.21621. [DOI] [PubMed] [Google Scholar]
- 18.Finch N, Baker M, Crook R, Swanson K, Kuntz K, Surtees R, et al. Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain. 2009;132:583–591. doi: 10.1093/brain/awn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jian J, Konopka J, Liu C. Insights into the role of progranulin in immunity, infection, and inflammation. J Leukoc Biol. 2012;93:199–208. doi: 10.1189/jlb.0812429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang W, Lu Y, Tian QY, Zhang Y, Guo FJ, Liu GY, et al. The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science. 2011;332:478–484. doi: 10.1126/science.1199214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puga I, Cols M, Barra CM, He B, Cassis L, Gentile M, et al. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat Immunol. 2012;13:170–180. doi: 10.1038/ni.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park B, Buti L, Lee S, Matsuwaki T, Spooner E, Brinkmann MM, et al. Granulin is a soluble cofactor for toll-like receptor 9 signaling. Immunity. 2011;34:505–513. doi: 10.1016/j.immuni.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 23.Tian QY, Zhao YP, Liu CJ. Modified yeast-two-hybrid system to identify proteins interacting with the growth factor progranulin. J Vis Exp. 2012;59:e3562. doi: 10.3791/3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng JQ, Guo FJ, Jiang BC, Zhang Y, Frenkel S, Wang DW, et al. Granulin epithelin precursor: a bone morphogenic protein 2-inducible growth factor that activates Erk1/2 signaling and JunB transcription factor in chondrogenesis. FASEB J. 2010;24:1879–1892. doi: 10.1096/fj.09-144659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Songsrirote K, Li Z, Ashford D, Bateman A, Thomas-Oates J. Development and application of mass spectrometric methods for the analysis of progranulin N-glycosylation. J Proteomics. 2010;73:1479–1490. doi: 10.1016/j.jprot.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez EM, Mongiat M, Slater SJ, Baffa R, Iozzo RV. A novel interaction between perlecan protein core and progranulin: potential effects on tumor growth. J Biol Chem. 2003;278:38113–38116. doi: 10.1074/jbc.C300310200. [DOI] [PubMed] [Google Scholar]
- 27.Whitelock JM, Graham LD, Melrose J, Murdoch AD, Iozzo RV, Underwood PA. Human perlecan immunopurified from different endothelial cell sources has different adhesive properties for vascular cells. Matrix Biol. 1999;18:163–178. doi: 10.1016/s0945-053x(99)00014-1. [DOI] [PubMed] [Google Scholar]
- 28.Knox S, Merry C, Stringer S, Melrose J, Whitelock J. Not all perlecans are created equal: interactions with fibroblast growth factor (FGF) 2 and FGF receptors. J Biol Chem. 2002;277:14657–14665. doi: 10.1074/jbc.M111826200. [DOI] [PubMed] [Google Scholar]
- 29.Knox S, Fosang AJ, Last K, Melrose J, Whitelock J. Perlecan from human epithelial cells is a hybrid heparan/chondroitin/ keratan sulfate proteoglycan. FEBS Lett. 2005;579:5019–5023. doi: 10.1016/j.febslet.2005.07.090. [DOI] [PubMed] [Google Scholar]