Abstract
The Snail family of transcription factors are core inducers of epithelial-to-mesenchymal transition (EMT). Here we show that the F-box protein FBXO11 recognizes and promotes ubiquitin-mediated degradation of multiple Snail family members including Scratch. The association between FBXO11 and Snai1 in vitro is independent of Snai1 phosphorylation. Overexpression of FBXO11 in mesenchymal cells reduces Snail protein abundance and cellular invasiveness. Conversely, depletion of endogenous FBXO11 in epithelial cancer cells causes Snail protein accumulation, EMT, and tumor invasion, as well as loss of estrogen receptor expression in breast cancer cells. Expression of FBXO11 is downregulated by EMT-inducing signals TGFβ and nickel. In human cancer, high FBXO11 levels correlate with expression of epithelial markers and favorable prognosis. The results suggest that FBXO11 sustains the epithelial state and inhibits cancer progression. Inactivation of FBXO11 in mice leads to neonatal lethality, epidermal thickening, and increased Snail protein levels in epidermis, validating that FBXO11 is a physiological ubiquitin ligase of Snail. Moreover, in C. elegans, the FBXO11 mutant phenotype is attributed to the Snail factors as it is suppressed by inactivation/depletion of Snail homologs. Collectively, these findings suggest that the FBXO11-Snail regulatory axis is evolutionarily conserved and critically governs carcinoma progression and mammalian epidermal development.
Keywords: E-cadherin, Snai1, Scratch, EMT, F-box, epidermis
1. Introduction
As one of the four primary tissues, epithelia cover the external or internal surface of the body as well as its cavities and tubes, and act as protective barriers. This is exemplified by the mammalian skin that protects the body from water loss and infections. Mammalian epidermis has been one of the best-characterized paradigms to study epithelial development and homeostasis [1]. The epidermis develops from a single-layered surface ectoderm during embryogenesis. As the ectodermal cells proliferate, the epidermis progressively stratifies. Only the basal layer is mitotically active. Basal cells destined for terminal differentiation withdraw from the cell cycle, delaminate, and migrate up to become spinous cells, which further differentiate into granular keratinocytes and eventually form the outermost stratum corneum. The vital function of epithelia requires cells to assemble polarized, continuous sheets. Epithelial cells tightly attach to each other through lateral junctional structures, in particular the adherens junctions (AJs) [2]. As a central constituent of epithelial AJs, E-cadherin critically promotes stable cell-cell adhesion and sustains epithelial tissue structure. E-cadherin maintains the quiescence of the cells within the epithelial sheets [3,4]. Ablation of the E-cadherin gene in mouse developing epidermis leads to epidermal hyperproliferation and defective differentiation, in addition to a perturbed water barrier [5–7]. E-cadherin also confines cell motility. In carcinomas, inhibition of E-cadherin facilitates cell detachment from the primary tumor and enhances cell invasiveness [8]. Inactivation of E-cadherin potentiates tumor invasion and metastasis in vivo in a mouse model of carcinogenesis [9]. Indeed, loss of E-cadherin is an indicator of invasive carcinomas and is commonly used for cancer diagnosis and prognosis [8]. Overall, E-cadherin is a crucial suppressor of epithelial cell overproliferation and carcinoma invasion.
The epithelial property is dynamically remodeled during embryonic development, notably via a reprogramming process referred to as epithelial-to-mesenchymal transition (EMT) [10,11]. During EMT, epithelial cells decrease epithelial markers, dissolve cell-cell junctions, and acquire migratory and invasive traits of mesenchymal cells. Downregulation of E-cadherin is a hallmark of EMT. The epithelial plasticity is under strict spatiotemporal regulation during embryonic morphogenesis. In adults, as barriers, epithelia are frequently exposed to a variety of exogenous and endogenous insults including growth factors and inflammatory cytokines, many of which are indeed EMT-inducing signals. Because EMT compromises epithelial integrity, epithelia must resist unscheduled EMT to preserve their functionality. Aberrant activation of EMT may lead to defective barriers and pathological events such as fibrosis and cancer. Indeed, EMT has been implicated in carcinoma invasion, metastatic dissemination, and therapy resistance [10]. Therefore, understanding the regulation of EMT and epithelial maintenance is of crucial importance.
EMT is governed by a cohort of transcription factors, including members of the Snail, Zeb, and Twist families [10–13]. These transcription factors induce extensive gene expression changes and cellular phenotypic switch. As core drivers of EMT, Snail family members Snai1 (also known as Snail) and Snai2 (Slug) are transcriptional repressors that directly inhibit the transcription of E-cadherin and several other intercellular adhesion components [12,14]. The direct repression of E-cadherin by Snai1 has been considered dogma of transcriptional regulation of EMT. Ectopic expression of Snai1/2 induces EMT in various epithelial cells in vitro. Transgenic overexpression of Snai1 in mouse epidermis, albeit insufficient for the induction of EMT [15], decreases E-cadherin expression and causes epidermal hyperplasia [16,17], which phenotypically mimics loss of E-cadherin and is consistent with E-cadherin’s anti-proliferative activity in the developing epithelia. In human cancer, expression of Snai1/2 and E-cadherin is inversely correlated, and elevated levels of Snai1/2 are associated with tumor aggressiveness and metastasis [12,13]. Collectively, the Snail-E-cadherin axis profoundly impacts epithelial cell proliferation and plasticity in normal development and carcinoma progression.
Snail family proteins are highly unstable. Their stability has emerged as a key determinant of the epithelial phenotype [12,13]. Snai1/2 are degraded by the ubiquitin-proteasome pathway. The Skp1-Cul1-F-box-protein (SCF) ubiquitin ligase complexes use diverse F-box proteins as the critical substrate-recognition subunit to selectively bind target proteins for ubiquitination and proteolysis [18]. Three F-box proteins, FBXW1 (βTRCP), FBXL14 (Ppa) and FBXL5, have been shown to promote Snai1/2 degradation [19–25]. Genetic inactivation of FBXW1 and its closely related paralogue FBXW1B (FBXW11, βTRCP2) in mice demonstrates that these factors control Snai1 during spermatogenesis in the testis of adult males, but are dispensable for Snail regulation in embryonic development [26,27]. The physiological roles of mammalian FBXL14 and FBXL5 are unknown. Therefore, physiological regulation of mammalian Snai1/2 remains largely elusive.
Through biochemical purification, we identified a distinct F-box protein, FBXO11, which interacts with Snai1/2 and promotes their ubiquitination and degradation. Depletion of endogenous FBXO11 in carcinoma cells leads to accumulation of Snai1 protein, induction of EMT, and tumor invasion, suggesting that FBXO11 is essential for suppressing EMT and sustaining the epithelial state. Consistently, human cancers with reduced FBXO11 expression are associated with a mesenchymal phenotype and adverse clinical outcomes, suggesting that FBXO11 is an independent prognostic factor. During mouse development, genetic inactivation of FBXO11 causes neonatal lethality. FBXO11-deficient mouse embryos exhibit increased Snai1/2 protein levels, decreased E-cadherin expression, and thickened epidermis. Such phenotypes resemble those of epidermis-specific transgenic overexpression of Snai1 or deletion of E-cadherin [5–7,16,17], indicating that FBXO11 is a physiological ubiquitin ligase of Snai1/2 to inhibit epithelial proliferation during mammalian development. Finally, both FBXO11 and Snail are highly conserved in C. elegans, and epistasis analysis demonstrates that FBXO11-dependent negative regulation of Snail is a phyletically ancient program. Taken together, these data reveal that FBXO11 is a bona fide physiological ubiquitin ligase of Snai1/2, and critically guards epithelial cell proliferation and invasion in mammalian development and cancer.
2. Materials and methods
2.1 C. elegans experiments
C. elegans was grown under standard conditions at 20°C. Strains ST65 ncIs13[ajm-1::gfp] and AA426 dre-1(dh99) were obtained from the CGC, and crossed to obtain IU489 dre-1(dh99) ncIs13[ajm-1::gfp]. For the seam cell fusion assay, gravid adult of ST65 and IU489 worms were allowed to lay eggs on control RNAi (empty vector L4440), dre-1 or ces-1 RNAi plates (from the Ahringer collection) for 3 hours. Seam cell fusion was scored before the L3 molt (around 34 hours after egg lay), developmental age was verified by gonadal outgrowth. L3 worms were anesthetized with 0.7% 1-phenoxy-2-propanol.
2.2 Generation of FBXO11-deficient mice
C57BL/6 mouse ES cells heterozygous for a targeted allele of FBXO11 were obtained from EUCOMM, and microinjected into blastocysts derived from C57BL/6J-Tyrc (albino) female mice. Male chimeras were bred with C57BL/6 female mice to obtain germline transmission of the targeted mutation. FBXO11 homozygous mutants were obtained by breeding heterozygous male and female mice. All mice were maintained in the C57BL/6 background.
2.3 Cell culture and treatments
HEK293, MCF-7, and MDA-MB-231 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% bovine calf serum. H358 and MEF cells were grown in DMEM with 10% fetal bovine serum (FBS). PC3 cells were grown in RPMI-1640 medium with 10% FBS. HCT116 cells were grown in McCoy’s 5A Medium with 10% FBS. Where indicated, the following drugs were used: MG132 (10 μM), cycloheximide (CHX) (20 nM).
2.4 Lentiviral shRNA depletion and RT-qPCR
To deplete various F-box proteins, cells were infected with lentiviral vector or shRNAs. After puromycin selection, cells were harvested in Trizol (Invitrogen), followed by RNA extraction. Reverse transcription of RNA was conducted using Moloney murine leukemia virus (MuLV) reverse transcriptase with random hexamers. The expression levels of selected genes were determined by real-time PCR. Data were normalized against β-actin. Primer and shRNA sequences are listed in Supporting Information.
2.5 Snail protein degradation assay
HEK293 cells were seeded in 6-well plates for 24 h and transfected with 0.5μg of pCDNA3-Snail1-Flag or Snail mutant (pCDNA3-Snail-6SA-Flag) and 0, 0.4, 0.8, 1.2μg of pCDNA3-FBXO11, the truncated FBXO11 (aa1-335) or the missense mutant FBXO11 (aa1-335, E281K/I282F/V301F). When indicated, 100ng GFP was used as internal transfection control. Cells were harvested after 24 h, and total extracts were obtained using denaturing lysis buffer (50mM Tris(pH7.5) ,1mM EDTA and 1%SDS), boiled for 10min and analyzed by Western blot using the Flag or Snai1 antibody.
2.6 Measurement of Snail half life
HEK293 cells were seeded in 6-well plates for 24 h and transfected with 500 ng of pcDNA3-Snail1-Flag and 100ng GFP was used as internal transfection control. Twenty-four hours after transfection, cells were treated with cycloheximide (CHX) for 0, 0.5, 1, 2, 3, 4 hours and samples were collected in lysis buffer at the indicated time points for immunoblotting.
2.7 In vivo ubiquitination
HEK293T cells were transfected with Flag-tagged Snail or Snail-6SA and either empty vector or Myc-tagged FBXO11. After 18h, transfected cells were treated with 10μM proteasome inhibitor MG132 for 6h and extracts were prepared in denaturing lysis buffer including 50mM Tris (pH7.5),1mM EDTA and 1%SDS. Lysates (100μl volume) were denatured by boiling for 10 min . Lysates were then diluted 10-fold with cold co-immunoprecipitation buffer (20mM Tris PH7.5,150mM NaCl, 0.5%NP-40), followed by centrifuge at 13,200 rpm for 10 min in 4°C. The supernatants were subjected to immunoprecipitation with anti-Flag M2 antibody and Protein A/G+ beads. Western blotting was carried out with mouse anti-ubiquitin monoclonal antibody (Cell Signaling, Billerica, MA).
2.8 Transwell invasion assay
The membranes of Transwell chambers (24-well insert; pore size, 8 μm; BD Biosciences) were coated with 20μg/ml fibronectin at 37°C overnight. Cells (4×104) were resuspended with serum-free medium and were plated in the top chamber. Inserts were placed in complete medium supplemented with serum. After 24h, cells that invaded through the pores and migrated to the lower surface of the membrane were fixed and stained with 1% crystal violet. Pictures were taken under microscope and the numbers of cells were counted.
2.9 Wound-healing migration assay
Cells were plated on 6-well plate and cultured to confluence. A wound was created by manually scraping the cell monolayer with a 200 μl tip and the cells were washed with PBS and maintained in serum-free medium for the duration of the experiment. Closing of the wounds was photographed at 0, 24 and 48 hr following wounding.
2.10 Immunostaining
Paraffin embedded 4 μm thick lung sections from wild type and FBXO11 mutant littermates were stained for H&E. Fresh frozen wild type and FBXO11 mutant littermates at E18.5 were stained for below mentioned antibodies with acetone fixation. Antibodies against Snai2, N-cadherin, β-catenin, p63 (Cell Signaling, Billerica, MA) and Snai1 (Abcam, Cambridge, MA) were used at 1:100 dilution; anti-E-cadherin (Cell Signaling, Billerica, MA) antibody was used at 1:400 dilution.
2.11 Analysis of human microarray data
The genes expression microarray data were downloaded from The Cancer Genome Atlas-Cancer Genome-TCGA (http://cancergenome.nih.gov/), and further studied by clustering analysis of five genes (mesenchymal markers Fibronectin, Vimentin, and MMP2; and epithelial markers E-Cadherin and KRT8) that are positively or inversely coexpressed with FBXO11 in lung cancer cohort samples (n = 286). We next carried out a multi-gene score according to the following algorithm and software: Normalization of probeset log2 values was performed with median values from all samples. These data were uploaded into Cluster 3.0 to hierarchically cluster the genes using “centered correlation” and “complete” as the distance metric and method. The output data were then loaded into Java Tree View software to generate heatmap.
Overall survival of lung cancer patients was analyzed by a Web-based Kaplan-Meier plotter (http://kmplot.com/analysis/index.php?p=service&cancer=lung). Lung cancer cohorts consisted of GSE14814, GSE19188, GSE29013, GSE30219, GSE31210, GSE3141, GSE31908, GSE37745, GSE43580, GSE4573, GSE50081, and GSE8894.
3. Results
3.1 BXO11 interacts with Snai1 and Snai2
To better understand the regulation of Snai1 and its biochemical activity, we employed an affinity purification-mass spectrometry strategy to isolate Snai1-associated proteins [28]. We identified multiple peptides derived from FBXO11 [29] (Fig. S1). FBXO11 is distinct from current F-box ubiquitin ligases of Snai1, especially in regard to the substrate-recognition domains. The potential association between FBXO11 and Snai1 raised the possibility that FBXO11 might act as a new ubiquitin ligase of Snai1. To test this idea, we first verified the interaction between the two proteins. Immunoprecipitation (IP) of FBXO11 from mammalian cellular extracts pulled down both Snai1 and Snai2 (Fig. 1A). In a reciprocal assay, IP of Snai1 co-purified FBXO11 (Fig. 1A). The result suggests that FBXO11 forms complexes with Snai1/2 in cells.
Fig. 1. FBXO11 interacts with Snai1/2 (A).
FBXO11 interacts with Snai1/2 in co-immunoprecipitation (co-IP) assay. HEK293 cells were transfected with Flag-tagged FBXO11 (F-FBXO11) and/or untagged Snai1/2, and treated with MG132 for 6 hours before lysis. Whole-cell extracts were subjected to IP with anti-Flag or anti-Snai1 antibody, and immunoblotting with indicated antibodies.
(B) FBXO11 associates with Snai1/2 in vitro. A schematic of the human FBXO11 protein shows various fragments that are fused to GST. The fusion proteins were purified from bacteria and incubated with lysates prepared from HEK293 cells expressing exogenous Snai1, Snai1-6SA, or Snai2. GST resin-bound proteins were immunoblotted with anti-Snai1 or −Snai2 antibody. Wild type Snai1 protein exhibits a higher molecular weight than Snai1-6SA due to phosphorylation. Coomassie blue gel staining images are shown. Asterisks indicate expected fusion proteins.
(C) FBXO11 binds to Snai1 independently of its phosphorylation in vitro. His-tagged Snai1 and MOF (negative control) proteins were expressed in bacteria, purified, and incubated with HEK293 cell extract expressing Flag-FBXO11. Nickel bead-bound proteins were examined by immunoblotting with a Flag antibody. Asterisks indicate His-tagged proteins.
(D) Domains of Snai1 implicated in FBXO11 association. A schematic of human Snai1 protein shows various fragments that were fused to His tag, which were tested for FBXO11 binding in vitro.
(E) Co-localization of FBXO11 and Snai1 in cells. Myc-tagged FBXO11 and Flag-tagged Snai1 were co-transfected into mouse fibroblast cells, followed by treatment with MG132 prior to immunostaining with anti-Myc and anti-Flag antibodies. Scale bar: 50μm
We next validated the protein interactions and mapped interacting domains by in vitro Glutathione S-transferase (GST) pulldown assay. The FBXO11 protein consists of an amino-terminal F-box, followed by a small Cysteine (C)-rich domain, a central 19 tandem repeats that may adopt a β-helix fold [30], and a carboxyl-terminal UBR motif that is involved in recognition of N-end rule substrates [31] (Fig. 1B). An amino terminus-truncated FBXO11 fragment (ΔN) and the UBR domain were each fused to GST (Fig. 1B), and incubated with lysates prepared from HEK293 cells expressing exogenous Snai1/2. We found that both Snai1 and Snai2 proteins associated specifically with the fusion protein containing FBXO11ΔN, but not with GST alone or the GST-UBR fusion (Fig. 1B). Further dissection of FBXO11ΔN revealed that the C-rich domain was primarily responsible for Snai1 binding (Fig. 1B).
Interaction of Snai1 with FBXW1 requires GSK3β-mediated phosphorylation [19], we thus tested whether its interaction with FBXO11 might have a similar requirement. A mutant form of Snai1, Snai1-6SA, is resistant to phosphorylation by GSK3β and is not recognized by FBXW1 [19]. However, FBXO11 bound to Snai1-6SA as efficiently as to wild type Snai1 (Fig. 1B). Therefore, the FBXO11-Snai1 interaction is independent of GSK3β-mediated phosphorylation, and thus differs from the FBXW1-Snai1 association.
Snai1 protein can be phosphorylated at multiple sites by a variety of kinases other than GSK3β [13]. We prepared bacterial expressed His-tagged full-length Snai1 protein that lacks phosphorylation. We found that FBXO11 strongly bound to bacterial-expressed Snai1 protein (Fig. 1C), suggesting that the interaction between the two proteins in vitro is independent of Snai1 phosphorylation. Snai1 contains an amino-terminal SNAG domain and four carboxyl zinc fingers (ZFs) (Fig. 1D). We further generated two truncated Snai1 constructs: one lacks the SNAG domain and the other consists of only the ZFs. The SNAG domain was dispensable for FBXO11 association, whereas the ZFs retained interaction with FBXO11 albeit at a decreased affinity compared to full-length Snai1 (Fig. 1D).
Phosphorylation of Snai1 controls its subcellular localization, and FBXW1 binds to phosphorylated Snai1 in the cytoplasm [19]. We hence examined the subcellular localization of the FBXO11-Snai1 complex. When Snai1 and FBXO11 were cotransfected into cells, Snai1 displayed a mostly nuclear staining pattern as expected, FBXO11 was predominantly nuclear as well (Fig. 1E), suggesting that the two proteins mainly interact in the nucleus. This is also consistent with FBXO11’s ability to bind to Snail-6SA, which resides exclusively in the nucleus [19].
3.2 FBXO11 promotes ubiquitin-dependent degradation of Snai1 and Snai2
Based on the association of FBXO11 with Snai1/2, we reasoned that FBXO11 might be a ubiquitin ligase for their degradation. When Snai1 and GFP were transfected into HEK293 cells, co-transfection of FBXO11 abolished Snai1 protein expression, but had no effect on GFP (Fig. 2A), suggesting FBXO11 specifically degrades Snai1. FBXW1 only targets phosphorylated Snai1, but not Snai1-6SA [19]. By contrast, FBXO11 efficiently degraded Snai1-6SA (Fig. 2A), suggesting that degradation of Snai1 by FBXO11 does not require GSK3β-mediated phosphorylation. As all Snail family members contain the conserved ZFs that exhibit association with FBXO11 (Fig. 1D), FBXO11 also promoted degradation of Snai2 (Fig. 2A). The Scratch (Scrt) proteins constitute a divergent subgroup of the Snail superfamily [32]. Scrt1 was degraded by FBXO11 as well (Fig. 2A). In all cases, FBXO11-promoted degradation of Snail family proteins was blocked by the proteasome inhibitor MG132. Collectively, these results suggest that FBXO11 is capable of inducing proteasomal degradation of probably all Snail superfamily members.
Fig. 2. FBXO11 promotes degradation and polyubiquitination of Snai1/2 and Scrt1.
(A) FBXO11 promotes proteasomal degradation of Snai1/2 and Scrt1. HEK293 cells were transfected with Flag-tagged Snai1, Snai1-6SA, Scrt1, or untagged Snai2, together with GFP as well as empty vector, full-length or truncated FBXO11, and treated with DMSO or MG132 for 6 hours before harvest. Cellular lysates were immunoblotted with anti-Flag, anti-Snai2, or anti-GFP antibody.
(B) F-box of FBXO11 is required for degrading Snai1. HEK293 cells were transfected with Snai1, GFP, and increasing amounts of full-length FBXO11 or indicated deletion mutants. Cell extracts were subjected to immunoblotting with anti-Snai1 or GFP antibody.
(C) FBXO11 promotes polyubiquitination of Snai1 and Scrt1. HEK293 cells were transfected with Flag-tagged Snai1, Snai1-6SA or Scrt1 together with or without FBXO11, followed by treatment with MG132 for 6 hours prior to harvest. Denatured cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-ubiquitin (top) or anti-Flag (bottom) antibody. Polyubiquitinated Snai1/Scrt1 proteins were indicated as high-molecular-weight species. Immunoglobin heavy chain (from the Flag antibody used in IP) is marked by asterisks.
(D) FBXO11 accelerates Snai1 protein turnover. HEK293 cells were transfected with Snai1 and GFP, in combination with FBXO11 or FBXW1. After 24 hours, cells were treated with cycloheximide (CHX) and harvested at indicated time points for immunoblotting with anti-Snai1 or anti-GFP antibody. The graph shows the quantification of Snai1 protein levels (based on the band intensity from the gels) normalized to GFP over the time course. Snai1 protein level at 0 hour time point of CHX treatment was set as 100%.
We attempted to map the domain of FBXO11 responsible for promoting Snai1/2 degradation. Consistent with its Snai1 binding, a truncated FBXO11 (aa 1-335) that contains the C-rich domain promoted proteasome-dependent degradation of Snai1 (Fig. 2A and S2). When Snai1 was transfected with increasing amounts of FBXO11, reduction of Snai1 protein levels became more pronounced (Fig. 2B). Since the UBR motif is dispensable for Snai1 binding, deletion of UBR did not affect FBXO11’s ability to degrade Snai1 (Fig. 2B and S2). The FBXO11ΔN mutant lacking the F-box failed to decrease Snai1 protein levels (Fig. 2B and S2). F-box is involved in recruiting the ubiquitin-conjugating enzyme [18]. The F-box dependence indicates FBXO11 downregulates Snai1 via a ubiquitin-mediated pathway. Indeed, when Snai1 or Snai1-6SA was cotransfected with FBXO11, FBXO11 stimulated their polyubiquitination (Fig. 2C). Similarly, FBXO11 enhanced polyubiquitination of Scrt1 (Fig. 2C). Consistent with these observations, FBXO11 accelerated protein turnover of Snai1, but not the GFP control protein (Fig. 2D). Taken together, the results suggest that FBXO11 selectively promotes polyubiquitination and degradation of Snail family proteins.
3.3 Overexpression of FBXO11 in mesenchymal cells activates the epithelial phenotype
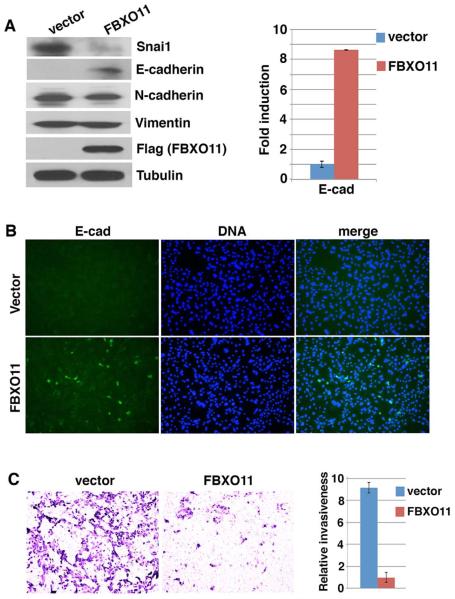
Snai1/2 factors are central drivers of EMT and directly contribute to repression of epithelial genes in mesenchymal cells. The ability of FBXO11 to downregulate Snai1/2 led us to investigate whether overexpression of FBXO11 might de-repress epithelial markers and alter cellular phenotype. When mouse embryonic fibroblast (MEF) cells were transduced with lentivirus expressing FBXO11, endogenous Snai1 protein levels decreased and concomitantly, E-cadherin RNA and protein expression increased (Fig. 3A), whereas mesenchymal markers N-cadherin and Vimentin were not affected. Immunostaining with an E-cadherin antibody revealed that a subset of FBXO11-transduced cells became E-cadherin positive (Fig. 3B). It is likely that degradation of Snail factors by FBXO11 de-represses E-cadherin, but full activation of E-cadherin transcription requires additional recruitment of activators, which appears to be sporadic.
Fig. 3. Overexpression of FBXO11 in MEFs reduces Snai1 protein abundance and activates epithelial features.
MEF cells were infected with lentivirus expressing Flag-tagged FBXO11 or control vector.
(A) FBXO11 reduces endogenous Snai1 protein abundance and activates E-cadherin. Cells were lysed and subjected to immunoblotting with antibodies against Snai1, E-cadherin, N-cadherin, Vimentin, Flag, and tubulin, and RT-qPCR analysis for E-cadherin.
(B) FBXO11-transduced MEF cells express E-cadherin. Cells were analyzed by immunofluorescence staining with anti-E-cadherin antibody and Hoechst (for DNA).
(C) FBXO11 inhibits cell invasion in vitro. Cells were assayed for Transwell invasion. Representative images are shown. The histogram shows the quantification of relative cell numbers migrated through matrix (from 6 random fields).
Upregulation of E-cadherin is indicative of mesenchymal-to-epithelial transition and is expected to decrease cell motility and invasion. In the Transwell invasion assay, MEF cells were highly invasive as they robustly invaded and migrated through the matrix. However, such invasiveness was potently suppressed after transduction with lentiviral FBXO11 (Fig. 3C). FBXO11 did not exert notable effect on cell proliferation, as FBXO11-transduced cells could be regularly cultured and passaged without evident changes in growth rates. Together, these results suggest FBXO11 is able to downregulate endogenous Snai1 protein abundance and activate epithelial features in mesenchymal cells.
3.4 Depletion of endogenous FBXO11 in epithelial cells stabilizes Snai1 protein and induces EMT and tumor invasion
Because overexpression of exogenous FBXO11 in mesenchymal cells degraded Snai1 protein and activated E-cadherin expression, we decided to determine the function of endogenous FBXO11. We used two independent lentiviral short hairpin RNAs (shRNAs) to deplete FBXO11 in MCF7 human breast carcinoma cells. Depletion of FBXO11 caused marked upregulation of Snai1 protein levels and downregulation of E-cadherin and keratin 8 (KRT8), both of which are epithelial markers and known direct transcriptional targets of Snai1 (Fig. 4A). The mesenchymal marker N-cadherin was increased (Fig. 4A). The E-cadherin to N-cadherin switch is characteristic of EMT. Following depletion of FBXO11, increased Snai1 protein levels and decreased E-cadherin expression were also observed in other carcinoma cells examined, including HCT116 colon cancer (Fig. 4B), H358 lung cancer, and PC3 prostate cancer cells (Fig. S3). The findings suggest that endogenous FBXO11 is required to prevent Snai1 protein accumulation and maintain E-cadherin expression in epithelial cells.
Fig. 4. Depletion of endogenous FBXO11 in carcinoma cells causes EMT and enhances cell migration and invasion.
(A, B) Depletion of endogenous FBXO11 in carcinoma cells stabilizes Snai1 protein and causes gene expression changes characteristic of EMT. MCF7 and HCT116 cells were transduced with lentiviral shRNAs targeting FBXO11 or empty vector pLKO. Cells were collected and subjected to immunoblotting and RT-qPCR analysis for indicated epithelial and mesenchymal markers.
(C) Depletion of FBXO11 induces EMT. Images of control and FBXO11-depleted HCT116 cells show morphological differences.
(D) Depletion of FBXO11 enhances cell migration. Representative images of wound (scratch)-healing assay demonstrate FBXO11-depleted HCT116 cells migrated faster than control cells, as shown 48 hours after scratching.
(E) Depletion of FBXO11 stimulates cell invasiveness in vitro. In the Transwell invasion assay, few control HCT116 cells invade and migrate through the matrix layer. By contrast, FBXO11-depleted HCT116 exhibit dramatically improved invasive potential.
(F) Depletion of FBXO11 enhances tumor invasion in transplant assay. Control and FBXO11-depleted HCT116 cells were injected into immunodeficient mice. Tumors were isolated and sectioned for H&E staining. Dashed line denotes the tumor boundary.
Estrogen receptor ERα is a key regulator of mammary gland development and tumorigenesis [33]. ERα is also a favorable prognostic indicator in breast cancer. Loss of ERα in breast cancer is associated with increased recurrence and a heightened risk of metastasis. ERα was previously identified as a direct target of Snai1 [34]. MCF7 cells are ERα-positive. Consistent with increased Snai1, ERα RNA and protein levels were strikingly reduced in FBXO11-depleted MCF7 cells (Fig. 4A). As the exact mechanisms leading to loss of ERα expression in human breast cancer progression are currently uncertain, this observation implies that loss of FBXO11 might be a contributory factor.
Besides FBXO11, FBXW1 and FBXL14 also regulate Snai1 protein degradation. We thus compared all three F-box factors for their effects on Snai1 protein stability. Each F-box factor was depleted with two different lentiviral shRNAs. Depletion of these F-box factors in MCF7 cells all resulted in accumulation of Snai1 protein to various degrees, and FBXO11 depletion appeared to exhibit the strongest effect on stabilizing Snai1 (Fig. S4), suggesting that multiple ubiquitin ligases can contribute to the regulation of Snai1 stability.
Decreased E-cadherin expression may weaken intercellular adhesion and change cell morphology and behavior. As expected, depletion of FBXO11 in HCT116 carcinoma cells gave rise to a predominance of dispersed and elongated single cells (Fig. 4C). To measure the migratory capacity of cells, the common in vitro wound-healing assay was conducted. Wounding lines (scratches) were introduced into the confluent monolayers of control (vector) and FBXO11-depleted HCT116 cells. Despite slightly reduced cell proliferation (not shown), FBXO11-depleted cells displayed increased motility as they migrated to fill in the wounds (cell-free gaps) faster than control cells (Fig. 4D).
Cells acquire increased invasiveness through EMT. In the Transwell invasion assay, control HCT116 cells rarely invaded, but FBXO11-depleted HCT116 cells were extremely invasive, showing massive trans-matrix migration (Fig. 4E). When transplanted into immunodeficient mice, control HCT116 cells formed encapsulated tumors with a clear boundary. By contrast, FBXO11-depleted tumors showed evident local invasion, exemplified by extensive infiltration of single tumor cells into nearby muscle fibers (Fig. 4F). Collectively, the results suggest that FBXO11 is essential for active suppression of Snai1 protein abundance, EMT, and tumor invasion.
3.5 EMT-inducing signals downregulate FBXO11 expression, and reduced FBXO11 expression in human cancer correlates with mesenchymal markers and adverse clinical outcomes
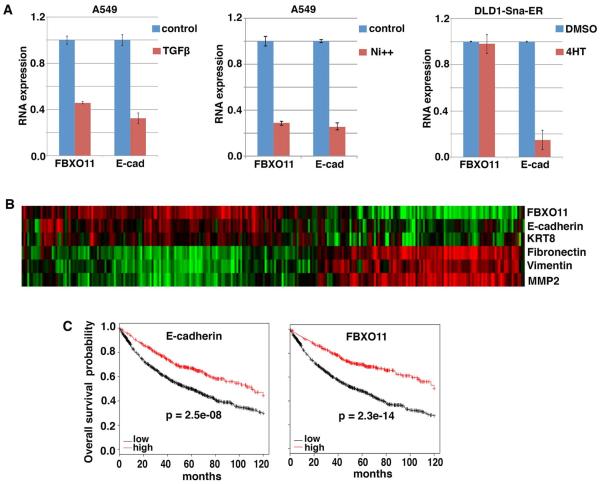
Epithelial cells undergo complete or partial EMT when stimulated with persistent EMT-inducing signals. If FBXO11 prevents Snail protein accumulation in epithelial cells and hence suppresses EMT, how does EMT occur? We investigated whether FBXO11 expression was subjected to regulation during the induction of EMT. At the helm of EMT-inducing signals is TGFβ, which potently induces Snai1/2 expression and EMT [35]. When A549 lung cancer epithelial cells were exposed to TGFβ for two days, cells underwent EMT and E-cadherin was downregulated (Fig. 5A) [28]. We found that FBXO11 RNA levels were reduced too (Fig. 5A). Nickel solution also induces Snai2 and represses E-cadherin expression in A549 cells [28]. We noticed that FBXO11 was markedly downregulated by nickel treatment (Fig. 5A). To test whether FBXO11, like E-cadherin, was a target of Snail or whether its reduced expression was a consequence of EMT, we examined a Snai1-dependent inducible EMT model [28]. Induction of Snai1 led to complete EMT and a strong decrease of E-cadherin expression, however, FBXO11 expression was not affected (Fig. 5A). This result suggests that FBXO11 is not a transcriptional target of Snai1. Collectively, the data suggest that expression of FBXO11 is repressed by EMT-inducing signals TGFβ and nickel, which presumably paves the way for the onset of EMT.
Fig. 5. Regulation of FBXO11 by EMT-inducing signals and expression of FBXO11 in human cancer.
(A) To induce EMT, A549 cells were treated with TGFβ or nickel for 2 days, and DLD1 cells stably expressing inducible Snai1 (Sna-ER) were exposed to 4-hydroxytamoxifen (4HT) for 2 days. Cells were subsequently subjected to RT-qPCR analysis for FBXO11 and E-cadherin.
(B) Expression of FBXO11 correlates with indicated epithelial markers and inversely with mesenchymal markers in a lung cancer cohort (n = 286). Clustering was based on expression levels of EMT markers. Green indicates lower expression, and red indicates higher expression.
(C) FBXO11 is a robust predictor for favorable clinical outcome in cancer. Kaplan-Meier 10-yr overall survival was based on expression levels of E-cadherin or FBXO11 in a large cohort of lung cancer patients (n = 1405). The online survival analysis software was previously reported [36].
We subsequently examined FBXO11 expression in human tumors. Analysis of microarray gene expression datasets revealed that FBXO11 in human cancer overlapped with epithelial markers E-cadherin and KRT8, and inversely correlated with mesenchymal markers Fibronectin, Vimentin, and MMP2 (Fig. 5B). This observation is consistent with FBXO11’s role in suppressing EMT and maintaining the epithelial state. As EMT is an indicator of malignant progression, we thus asked whether reduced FBXO11 expression might lead to adverse prognosis. In a combined large lung cancer cohort [36], like E-cadherin, elevated expression of FBXO11 is strongly correlated with increased recurrence-free survival in patients (Fig. 5C). This association occurs mainly in lung adenocarcinomas but not in squamous cell carcinoma. FBXO11 is thus a robust and independent prognostic indicator. Taken together, the above analyses suggest FBXO11 is associated with epithelial phenotypes and favorable clinical outcome.
Because FBXO11 is a favorable prognostic factor, we investigated whether its function might be impaired by somatic mutations in human cancer. At the time of this study, cancer genome sequencing uncovered more than 60 missense, nonsense, and frame-shift somatic mutations in FBXO11 in human cancer samples, mostly carcinomas from various tissues (http://cancer.sanger.ac.uk). There were no evident mutational hotspots in FBXO11. Three missense mutations, namely E281K, I282F, and V301F, are located in the C-rich domain of FBXO11, which is the major determinant of Snai1/2 binding (Fig. 1B). Therefore, we examined the consequence of these mutations on targeting Snai1. In the GST pulldown assay, E281K and V301F mutations abolished the association of FBXO11 with Snai1 (Fig. S5A). Compared to wild type FBXO11, these two mutants were severely impaired in degrading Snai1 (Fig. S5B and S5C). The results suggest that a subset of cancer-derived mutations may compromise FBXO11’s ability to degrade the Snail factors. However, the overall mutation rate of FBXO11 in human cancer was low. Impaired function of FBXO11 may be primarily attributed to its reduced expression (Fig. 5C).
3.6 Inactivation of FBXO11 in mice leads to neonatal lethality, decreased E-cadherin expression, and defective epidermal development
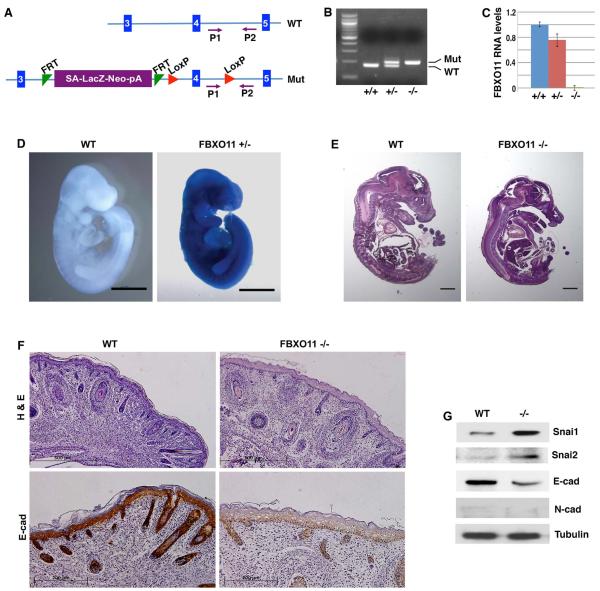
Given its role in cancer, we wondered whether FBXO11 physiologically regulated Snail. Regulation of mammalian Snai1/2 in vivo remained poorly understood. We generated FBXO11 mutant mice through blastocyst microinjection of targeted embryonic stem (ES) cells (Fig. 6A-6C) [37]. The FBXO11 mutant allele carries a knock-in LacZ reporter (Fig. 6A), which is controlled by regulatory elements of the FBXO11 locus. β-Galactosidase staining of FBXO11-heterozygous mouse embryos at embryonic day 10 (E10) (Fig. 6D) and various other stages (not shown) suggested that FBXO11 is ubiquitously expressed throughout mouse embryonic development.
Fig. 6. Generation and characterization of FBXO11 mutant mice.
(A) Schematic representation of FBXO11 inactivation. The mutant allele (Mut) contains a “LacZ-Neo” cassette that also includes the mouse En2 splice acceptor (SA) and the SV40 polyadenylation sequences (pA) [37]. This cassette acts as an exon, which stops the transcription of FBXO11 at the pA and thus creates a null allele. Blue vertical bars represent the exons of FBXO11. The exon 4 encodes the F-box motif. FRT and LoxP are specific DNA elements for potential recombination and conditional deletion (irrelevant in the current study). Primers P1 and P2 are used for genotyping.
(B) Genotyping of mouse FBXO11 mutants. Wild type (WT), FBXO11 heterozygous (+/−) and homozygous (−/−) mutant mice were genotyped by PCR with the P1 and P2 primers (WT: 179 bp; Mut: 204 bp).
(C) Validation of FBXO11 deficiency in the mouse FBXO11 homozygous mutants. Mouse E10.5 embryos with indicated genotypes were analyzed for FBXO11 RNA expression by quantitative RT-PCR (with primers corresponding to the exons 3 and 4). Expression of FBXO11 in the homozygous mutant was undetectable (less than 0.1% of the WT).
(D) Whole mount β-Galactosidase staining of wild type (WT) and FBXO11 heterozygous (het) E10 mouse embryos. FBXO11 mutant allele carries a knock-in LacZ reporter. Scale bar: 1mm.
(E) Sagittal sections of WT and FBXO11 homozygous mutant (KO) E15.5 mouse embryos (H&E, scale bar: 1mm).
(F) FBXO11-deficient mouse epidermis exhibits reduced hair follicles, thickened epidermis and diminished E-cadherin expression. Sagittal sections of newborn wild type (WT) and FBXO11 −/− littermate were stained with H&E (scale bar: 500μm) or immunohistochemically with an anti-E-cadherin antibody (scale bar: 200 μm).
(G) Elevated Snai1 protein levels in FBXO11 mutants. Protein extracts were prepared from the back skin of neonatal WT and FBXO11 KO embryos, and immunoblotted with indicated antibodies.
FBXO11 homozygous mutant embryos survived to term, and did not exhibit gross developmental defects compared to wild type littermates in regard to the size of embryos or organs such as lung, heart, liver, and kidney (Fig. 6E). However, newborn homozygous mutants died shortly after birth. Careful examination of histological sections of newborn pups revealed that the epidermis in FBXO11 homozygous mutants was thicker than that in wild type pups, and the number of hair follicles in FBXO11 mutants was substantially reduced compared to wild type littermates (Fig. 6F). The increased epidermal thickness apparently resulted from keratinocyte hyperplasia. Keratohyalin granules, which are cytoplasmic aggregates characteristic of the normal granular layer [38], were virtually absent in FBXO11 mutant skin (Fig. 6F), implying possible defective epidermal differentiation. These phenotypes, especially the thickening of the epidermis and reduction of hair follicles, were similarly observed in mice with epidermis-specific deletion of E-cadherin [5,7]. We thus performed immunohistochemistry analysis of E-cadherin. Based on the staining intensity, E-cadherin was robustly expressed in all epithelial cells of interfollicular epidermis and hair follicles in newborn wild type mice, however, its expression was strongly reduced in the FBXO11 mutants (Fig. 6F). The result provides in vivo validation that FBXO11 is essential for maintaining E-cadherin expression in epithelial cells.
3.7 FBXO11-deficient mouse mutants exhibit increased Snai1/2 protein abundance in epidermis
Given FBXO11’s role in targeting Snai1/2 for degradation and maintaining E-cadherin expression, we verified whether downregulation of E-cadherin in FBXO11 mutants might be attributed to increased protein abundance of Snai1/2 factors. We examined protein expression in extracts prepared from the back skin (predominantly epidermis) of newborn pups by Western blotting. Snai1 and Snai2 protein levels were markedly increased while E-cadherin was strongly decreased in the FBXO11 mutant compared to wild type littermate (Fig. 6G), suggesting FBXO11 prevents Snai1/2 protein accumulation in vivo. The mesenchymal marker N-cadherin was not detected in either wild type or FBXO11 mutant skin (Fig. 6G).
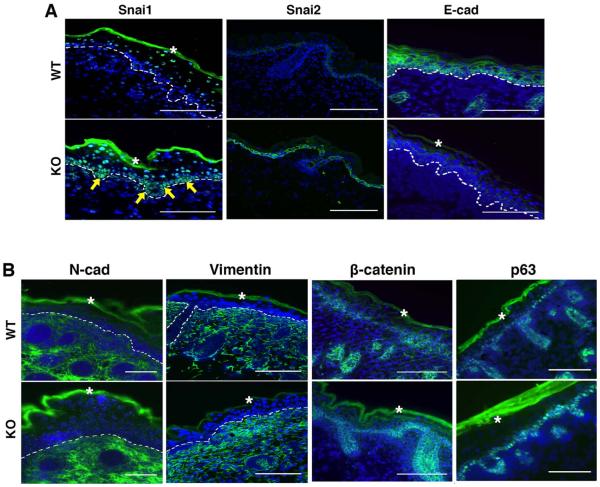
To determine what cells exhibited elevated Snail proteins, we analyzed the expression pattern of Snail factors by immunofluorescence analysis of E18.5 mouse embryos. Snai1 mRNA was reported to be transiently expressed in the developing skin and primarily restricted to the epithelium of nascent hair buds during follicle morphogenesis [16]. The Snai1 antibody detected very weak signals in basal epidermal cells in wild type embryos (Fig. 7A). By contrast, strong staining of Snai1 was observed in the basal epithelial layer of epidermis and hair follicles in the FBXO11 mutant (Fig. 7A). Previous studies have shown that Snai2 RNA was surprisingly primarily confined to the basal layer cells of various stratified and pseudostratified epithelial tissues in late mouse embryonic stages [39–41]. Consistently, Snai2 protein in wild type embryos was weakly detected specifically in the basal cells of interfollicular epidermis and in the outer root sheath of hair follicles, but was absent in the underneath dermal fibroblasts (Fig. 7A). Importantly, fluorescence staining intensity of Snai2 in basal epidermal cells of the FBXO11 mutant was remarkably elevated (Fig. 7A). Unexpectedly, Snai2 protein exhibited cytoplasmic localization (Fig. 7A). We validated the Snai2 antibody on cultured mesenchymal cancer cells, which displayed predominantly nuclear staining (Fig. S6), suggesting that the subcellular localization of Snai2 in basal epidermal cells in vivo is subjected to specific regulation.
Fig. 7. Molecular characterization of epidermis from FBXO11-deficient mouse embryos.
Cryosections of FBXO11 mutant and control littermate embryos at E18.5 were stained for immunofluorescence analysis with antibodies specific for Snai1, Snai2, and E-cadherin (A) as well as N-cadherin, Vimentin, β-catenin, and p63 (B). Dashed lines demarcate the basement membrane that separates the epidermis from dermis. Arrows indicate basal cells with increased Snai1 expression. The asterisks denote autofluorescence of the cornified layer. Scale bar: 100μm.
As the FBXO11-Snai1/2 pathway modulates epithelial phenotype, we further examined expression of EMT markers in E18.5 mouse embryos. Epidermal basal and suprabasal cells as well as hair follicle epithelial cells stained strongly positive for E-cadherin in wild type embryos (Fig. 7B). In FBXO11 mutant, E-cadherin staining was diminished in all epidermal epithelial cells (Fig. 7B). This observation is similar to reduced E-cadherin expression due to epidermis-specific transgenic Snai1 [16,17]. The mesenchymal markers N-cadherin and Vimentin specifically label the dermal compartment (Fig. 7B). There was no evident ectopic expression of the mesenchymal markers in the FBXO11 mutant epidermis in which E-cadherin was downregulated. This is consistent with the absence of mesenchymal marker induction in Snai1 transgenic epidermis [15]. While downregulation of E-cadherin and upregulation of N-cadherin often seem coupled in vitro (for instance, Fig. 4A and 4B), these two gene regulatory programs appear to be separable in vivo. β-catenin marks cell-cell contacts and was detected in all epidermal epithelial cells, and there was no obvious difference in the distribution or abundance of β-catenin in wild-type and FBXO11 mutant (Fig. 7B). Expression of p63, a specific marker for the basal epidermal cells [42], generally remained to be confined to a single layer in the epidermis of FBXO11 mutant (Fig. 7B), suggesting epidermal thickening in the FBXO11 mutant mainly results from increased suprabasal cells. Overall, inactivation of FBXO11 in mice leads to increased epidermal Snai1/2 protein abundance, decreased E-cadherin expression, and thickened epidermis. These phenotypes resemble at least in part those of aforementioned epidermal deletion of E-cadherin or transgenic overexpression of Snai1. The results validate that FBXO11 is a physiological ubiquitin ligase of Snai1/2, and provide a molecular basis for the phenotypic similarities between FBXO11 deficiency and transgenic overexpression of Snai1 in the basal layer of epidermis [16,17].
3.8 The FBXO11-Snail regulation is conserved in evolution and the FBXO11 deficient phenotype is attributed to Snail factors in C. elegans
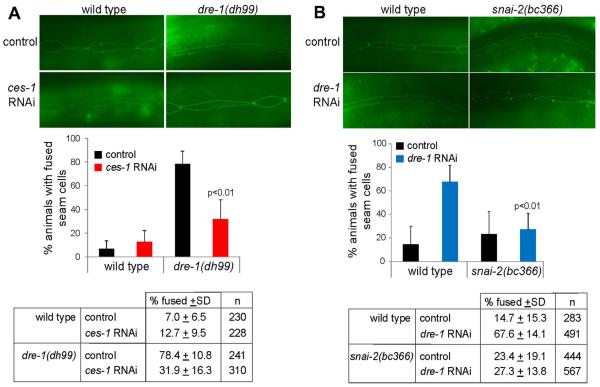
The genetic study in mice demonstrates that FBXO11 physiologically suppresses Snails in epidermis, however, it does not prove that the FBXO11 deficient epidermal phenotype, which is albeit similar to that of transgenic overexpression of Snai1, is caused by abnormally accumulated Snail factors. To address this point, we took advantage of the nematode C. elegans model organism. Both FBXO11 and Snail have homologs in C. elegans. We were interested in verifying whether the FBXO11-Snail regulatory axis was evolutionarily conserved. In C. elegans, differentiation of the hypodermis starts as epidermal seam cells exit the cell cycle at the L4 larval stage and fuse to form a continuous cuticular structure [43]. Null mutations in dre-1, the C. elegans counterpart of FBXO11, cause embryonic lethality; but hypomorphic loss-of-function mutants, such as dre-1(dh99), as well as dre-1 depletion by RNAi starting from the L1 stage, display specific precocious seam cell fusion at the L3 stage [43] (Fig. 8A and 8B). If the dre-1 phenotype is attributed to aberrant accumulation of Snail homologs, one may expect that removal of Snail factors should at least in part rescue the dre-1 epidermal defects.
Fig. 8. ces-1 depletion or snai-2 mutation suppresses the seam cell fusion phenotype of reduced dre-1 in C. elegans.
(A) Precocious seam cell fusion in dre-1(dh99) L3 larvae is suppressed by ces-1 RNAi. (top) Images of seam cell adherence junctions in the L3 molt as visualized by the ajm-1::gfp reporter. Wild type seam cells were unfused on control RNAi (empty vector L4440) or ces-1 RNAi. dre-1(dh99) seam cells were precociously fused on control RNAi but showed rescue of this phenotype on ces-1 RNAi. Worms were scored fused if more than 80% of cells were fused. Significance was determined using the Student-T-test. Data are the mean + the standard deviation (SD) from 5 independent experiments.
(B) Precocious seam cell fusion caused by dre-1 RNAi in L3 larvae is suppressed by the snai-2(bc366) allele. (top) Images of seam cell adherence junctions in the L3 molt as visualized by the ajm-1::gfp reporter. Seam cells are mostly unfused in wild type or K02D7.2/snai-2(bc366) on control RNAi (empty vector L4440). dre-1 RNAi causes precocious seam cell fusion at the L3 molt in the wild type strain but not in the snai-2(bc366) mutant, indicating a suppression by snai-2. Data in histogram are the mean + SD from 3 independent experiments.
The C. elegans genome encodes three Snail superfamily members: CES-1, SCRT-1 and K02D7.2 [44]. CES-1 and SCRT-1 are homologs of human Scrt proteins, whereas K02D7.2 is most closely related to human Snai2 [32] and is hence renamed SNAI-2 in this study. CES-1 regulates cell cycle progression, polarity and apoptosis, and functions as a transcriptional repressor of the target gene egl-1 [45–47]. As FBXO11 regulates Scrt in human cells (Fig. 2A and 2C), we indeed found that RNAi depletion of ces-1 significantly reduced the precocious seam fusion in the dre-1(dh99) mutants from approximately 80% to 30% (Fig. 8A). The C. elegans SCRT-1 and SNAI-2 remain poorly characterized and have no described functions. According to the WormBase, there exists a mutant allele of snai-2, snai-2(bc366), which causes a premature stop codon in the coding region due to deletion, and the resulting truncated mutant protein lacks most zinc fingers and is expected to be nonfunctional. We found that this mutation in snai-2 could also rescue the precocious phenotype of depletion of dre-1 by RNAi (Fig. 8B). Therefore, depletion/inactivation of either ces-1 or snai-2 suppresses the dre-1 seam cell phenotype caused by a hypomorphic mutation or RNAi, suggesting that the dre-1 phenotype is at least in part attributed to Snail homologs. These results validate the in vivo role of FBXO11/DRE-1 in negatively regulating Snail-like proteins and demonstrate that such regulation is evolutionarily conserved.
4. Discussion
E-cadherin-based epithelial intercellular adhesion confines cell proliferation and motility. During epithelial morphogenesis, dynamic adhesive remodeling ensures coordinated cell proliferation, differentiation, and migration. In cancer, deregulated epithelial plasticity contributes to malignant progression. However, the molecular mechanisms regulating epithelial cohesion in development and cancer are complex and remain poorly understood. In this study, we discovered that the F-box protein FBXO11 functions as a ubiquitin ligase for the EMT-driving transcription factors Snai1/2. FBXO11 interacts with Snai1/2 proteins and promotes their ubiquitination and proteasome-mediated degradation. Overexpression of FBXO11 reduces Snai1 protein levels and activates epithelial features in mesenchymal cells. Conversely, depletion of endogenous FBXO11 in epithelial cancer cells leads to increased Snai1 protein abundance, decreased E-cadherin expression, EMT, and tumor invasion, demonstrating that FBXO11 actively maintains the epithelial state to suppress carcinoma progression. FBXO11 is downregulated by EMT-inducing signals. Consistently, human cancers with low FBXO11 expression are associated with a mesenchymal phenotype and adverse clinical outcome. Genetic inactivation of FBXO11 in mice causes aberrant accumulation of Snai1/2 proteins in epidermis, demonstrating mammalian FBXO11 physiologically regulates Sna1/2. Diminished epidermal E-cadherin expression and thickened epidermis in FBXO11 mutants partly phenocopy transgenic overexpression of Snai1 [5–7,16,17], suggesting the FBXO11-deficient phenotype is potentially attributed to Snai1/2 accumulation. In support of this notion, depletion or inactivation of Snail homologs in C. elegans rescued the FBXO11 mutant worms, suggesting that the FBXO11 mutant phenotype is indeed at least in part caused by the Snail family members and the FBXO11-Snail regulatory module is an evolutionarily conserved program. Taken together, these findings indicate FBXO11 is an indispensable physiological ubiquitin ligase of the Snail factors, and highlight the importance of the FBXO11-Snail pathway in the control of epithelial proliferation, homeostasis and plasticity during mammalian epidermal development and carcinoma progression.
Since prior submissions of this manuscript, it has recently been reported that FBXO11 promotes tumor metastasis by acting as a ubiquitin ligase of Snai1, in particular, FBXO11 specifically recognizes Serine 11 (S11)-phosphorylated Snai1 [48]. In the present study, FBXO11 was able to bind to unphosphorylated Snai1 in vitro (Fig. 1C). Truncated Snai1 lacking residues 1-11 still strongly associated with FBXO11 (Fig. 1D). Furthermore, the S11 phosphorylation site of Snai1 is not conserved in Snai2 and SCRT1 as well as their C. elegans homologs, all of which are expected substrates of FBXO11. Despite this discrepancy, both studies revealed that FBXO11 degrades Snail and suppresses tumor progression. The present study further demonstrates the physiological regulation of Snail family members by FBXO11 in mice and C. elegans in vivo.
F-box proteins confer substrate specificity. In general, a single F-box protein targets multiple substrates [49], and its function probably depends on the distribution pattern of diverse substrates. It is often challenging to identify specific substrate that is responsible for a particular phenotype. While Snail factors were hereby identified as physiological substrates of FBXO11, FBXO11 was shown to promote the degradation of BCL6 and BLIMP1, both of which are zinc finger transcription factors critical for B cell development, and CDT2, a cell cycle regulator [50–53]. In addition, FBXO11 was previously reported to regulate TGFβ signaling, although the underlying biochemical mechanism remains unknown [54]. However, little was known about the physiological function of mammalian FBXO11 and the corresponding substrates [29]. In this study, we generated FBXO11-deficient mice. FBXO11 mutant embryos displayed decreased E-cadherin and epidermal hyperplasia. Because the skin phenotype mirrors transgenic overexpression of Snai1 or deletion of E-cadherin [5–7,16,17], and Snai1/2 protein levels are in fact elevated in FBXO11 mutant epidermis, it is conceivable that Snail factors may contribute to the epidermal defects. Nevertheless, contributions from other FBXO11 substrates cannot be excluded. Different FBXO11 substrates may function in different cell types and/or developmental stages. The neonatal lethality of FBXO11 mutant mice is possibly due to pulmonary defects, however, we did not detect Snai1/2 proteins in wild type and FBXO11 mutant lungs with current antibodies (not shown). Therefore, the FBXO11 substrate(s) involved in lung development remain to be identified. There are likely additional defects in the FBXO11 mutants. Based on FBXO11’s regulation of BCL6 and BLIMP1, FBXO11 mutants are expected to have hematopoietic abnormalities. In C. elegans, the FBXO11/DRE-1 mutant phenotype is suppressed by depletion of CDT2, BLIMP1, or Snail homologs, suggesting all these substrates are essential contributors.
Given their critical function in development and pathogenesis, Snail transcription factors are tightly regulated by transcriptional, translational, and post-translational mechanisms [12,13]. Control of their protein stability represents an important mechanism in epithelial regulation. It is not unusual that a single substrate protein may be controlled by multiple ubiquitin ligases. For example, mammalian cell cycle regulator Cyclin D1 is ubiquitinated by at least five distinct F-box factors [49]. Different F-box regulators may function under diverse cellular contexts. Snail factors are vital for early developmental EMT processes, in particular mesoderm and neural crest formation [14]. Inactivation/depletion of Snai1/2 severely impairs early embryonic morphogenesis. Conversely, overexpression of Snai2 in the neural tube of the chick embryo increases neural crest production [14]. Transgenic overexpression of Snai1 in developing epidermis of mouse embryos represses E-cadherin, and causes epidermal hyperproliferation and thickening [16,17], although the EMT phenotype is incomplete as there is no ectopic induction of mesenchymal marker expression in the epidermal cells [15,16]. Multiple F-box proteins have now been identified to degrade Snails. Genetic studies in mice show that FBXW1 and related FBXW1B are critical for Snai1 regulation during spermatogenesis in adult male mice, but dispensable for embryonic development [26,27]. In the present study, FBXO11 deficiency leads to epidermal thickening and accumulation of Snai1/2 primarily in the basal layer of developing epidermis in late stage mouse embryos, suggesting FBXO11 principally controls Snai1/2 during mammalian epidermal development. It remains to be investigated how other Snai1/2-mediated developmental EMT events (e.g. gastrulation, neural crest) may be regulated by F-box proteins.
Supplementary Material
Highlights.
FBXO11 interacts with and degrades the EMT-driving Snail transcription factors
FBXO11 inhibits EMT and tumor invasion and is a favorable prognostic factor
FBXO11 deficient mice show increased Snail protein levels and epidermal hyperplasia
Epistasis analysis in C. elegans validates the FBXO11-Snail regulatory axis
Acknowledgements
C. elegans strains ST65 and AA426 were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The snai-2(bc366) allele was kindly provided by Dr. Barbara Conradt. We would like to thank Gena V. Topper for the construction of the IU489 dre-1(dh99) ajm-1::gfp strain, and Dr. Mien-Chie Hung for providing the Snail-6SA cDNA. This work was in part supported by grants to J.L. from Florida Department of Health (4KB07) and NIH (R01CA137021).
Footnotes
Conflict of interest statement The authors confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- [1].Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 2009;10:207–17. doi: 10.1038/nrm2636. doi:10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Harris TJC, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 2010;11:502–514. doi: 10.1038/nrm2927. doi:10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- [3].McClatchey AI, Yap AS. Contact inhibition (of proliferation) redux. Curr. Opin. Cell Biol. 2012;24:685–94. doi: 10.1016/j.ceb.2012.06.009. doi:10.1016/j.ceb.2012.06.009. [DOI] [PubMed] [Google Scholar]
- [4].Müller EJ, Williamson L, Kolly C, Suter MM. Outside-in signaling through integrins and cadherins: a central mechanism to control epidermal growth and differentiation? J. Invest. Dermatol. 2008;128:501–16. doi: 10.1038/sj.jid.5701248. doi:10.1038/sj.jid.5701248. [DOI] [PubMed] [Google Scholar]
- [5].Tinkle CL, Lechler T, Pasolli HA, Fuchs E. Conditional targeting of E-cadherin in skin: insights into hyperproliferative and degenerative responses. Proc. Natl. Acad. Sci. U. S. A. 2004;101:552–7. doi: 10.1073/pnas.0307437100. doi:10.1073/pnas.0307437100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tunggal JA, Helfrich I, Schmitz A, Schwarz H, Günzel D, Fromm M, et al. E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J. 2005;24:1146–56. doi: 10.1038/sj.emboj.7600605. doi:10.1038/sj.emboj.7600605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Young P, Boussadia O, Halfter H, Grose R, Berger P, Leone DP, et al. E-cadherin controls adherens junctions in the epidermis and the renewal of hair follicles. EMBO J. 2003;22:5723–33. doi: 10.1093/emboj/cdg560. doi:10.1093/emboj/cdg560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Paredes J, Figueiredo J, Albergaria A, Oliveira P, Carvalho J, Ribeiro AS, et al. Epithelial E- and P-cadherins: role and clinical significance in cancer. Biochim. Biophys. Acta. 2012;1826:297–311. doi: 10.1016/j.bbcan.2012.05.002. doi:10.1016/j.bbcan.2012.05.002. [DOI] [PubMed] [Google Scholar]
- [9].Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–3. doi: 10.1038/32433. doi:10.1038/32433. [DOI] [PubMed] [Google Scholar]
- [10].Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. doi:10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- [11].Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. doi:10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Peinado H, Olmeda D, Cano A. Snail. Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype?, Nat. Rev. Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. doi:10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- [13].De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. doi:10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- [14].Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–66. doi: 10.1038/nrm757. doi:10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- [15].Du F, Nakamura Y, Tan T-L, Lee P, Lee R, Yu B, et al. Expression of snail in epidermal keratinocytes promotes cutaneous inflammation and hyperplasia conducive to tumor formation. Cancer Res. 2010;70:10080–9. doi: 10.1158/0008-5472.CAN-10-0324. doi:10.1158/0008-5472.CAN-10-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jamora C, Lee P, Kocieniewski P, Azhar M, Hosokawa R, Chai Y, et al. A signaling pathway involving TGF-beta2 and snail in hair follicle morphogenesis. PLoS Biol. 2005;3:e11. doi: 10.1371/journal.pbio.0030011. doi:10.1371/journal.pbio.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].De Craene B, Denecker G, Vermassen P, Taminau J, Mauch C, Derore A, et al. Epidermal Snail expression drives skin cancer initiation and progression through enhanced cytoprotection, epidermal stem/progenitor cell expansion and enhanced metastatic potential. Cell Death Differ. 2014;21:310–20. doi: 10.1038/cdd.2013.148. doi:10.1038/cdd.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 2013;14:369–81. doi: 10.1038/nrm3582. doi:10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial mesenchymal transition. Nat. Cell Biol. 2004;6:931–40. doi: 10.1038/ncb1173. doi:10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- [20].Yook JI, Li X-Y, Ota I, Fearon ER, Weiss SJ. Wnt-dependent regulation of the E-cadherin repressor snail. J. Biol. Chem. 2005;280:11740–8. doi: 10.1074/jbc.M413878200. doi:10.1074/jbc.M413878200. [DOI] [PubMed] [Google Scholar]
- [21].Yook JI, Li X-Y, Ota I, Hu C, Kim HS, Kim NH, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat. Cell Biol. 2006;8:1398–406. doi: 10.1038/ncb1508. doi:10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- [22].Vernon AE, LaBonne C. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development. 2006;133:3359–70. doi: 10.1242/dev.02504. doi:10.1242/dev.02504. [DOI] [PubMed] [Google Scholar]
- [23].Viñas-Castells R, Beltran M, Valls G, Gómez I, García JM, Montserrat-Sentís B, et al. The hypoxia-controlled FBXL14 ubiquitin ligase targets SNAIL1 for proteasome degradation. J. Biol. Chem. 2010;285:3794–805. doi: 10.1074/jbc.M109.065995. doi:10.1074/jbc.M109.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wu Z-Q, Li X-Y, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16654–9. doi: 10.1073/pnas.1205822109. doi:10.1073/pnas.1205822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Viñas-Castells R, Frías Á, Robles-Lanuza E, Zhang K, Longmore GD, García De Herreros A, et al. Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic Acids Res. 2014;42:1079–1094. doi: 10.1093/nar/gkt935. doi:10.1093/nar/gkt935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer. 2008;8:438–49. doi: 10.1038/nrc2396. doi:10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kanarek N, Horwitz E, Mayan I, Leshets M, Cojocaru G, Davis M, et al. Spermatogenesis rescue in a mouse deficient for the ubiquitin ligase SCF{beta}-TrCP by single substrate depletion. Genes Dev. 2010;24:470–7. doi: 10.1101/gad.551610. doi:10.1101/gad.551610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tang M, Shen H, Jin Y, Lin T, Cai Q, Pinard M. a, et al. The Malignant Brain Tumor (MBT) domain protein SFMBT1 is an integral histone reader subunit of the LSD1 demethylase complex for chromatin association and epithelial-to mesenchymal transition. J. Biol. Chem. 2013:0–25. doi: 10.1074/jbc.M113.482349. doi:10.1074/jbc.M113.482349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hardisty-Hughes RE, Tateossian H, Morse SA, Romero MR, Middleton A, Tymowska-Lalanne Z, et al. A mutation in the F-box gene, Fbxo11, causes otitis media in the Jeff mouse. Hum. Mol. Genet. 2006;15:3273–9. doi: 10.1093/hmg/ddl403. doi:10.1093/hmg/ddl403. [DOI] [PubMed] [Google Scholar]
- [30].Ponting CP, Mott R, Bork P, Copley RR. Novel protein domains and repeats in Drosophila melanogaster: insights into structure, function, and evolution. Genome Res. 2001;11:1996–2008. doi: 10.1101/gr.198701. doi:10.1101/gr.198701. [DOI] [PubMed] [Google Scholar]
- [31].Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu. Rev. Biochem. 2012;81:261–89. doi: 10.1146/annurev-biochem-051710-093308. doi:10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–61. doi: 10.1242/dev.01907. doi:10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- [33].Nilsson S, Mäkelä S, Treuter E, Tujague M, Thomsen J, Andersson G, et al. Mechanisms of estrogen action. Physiol. Rev. 2001;81:1535–65. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- [34].Dhasarathy A, Kajita M, Wade PA. The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol. Endocrinol. 2007;21:2907–18. doi: 10.1210/me.2007-0293. doi:10.1210/me.2007-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–30. doi: 10.1038/nrm3434. doi:10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. doi:10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–42. doi: 10.1038/nature10163. doi:10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Manabe M, O’Guin WM. Keratohyalin, trichohyalin and keratohyalin-trichohyalin hybrid granules: an overview. J. Dermatol. 1992;19:749–55. doi: 10.1111/j.1346-8138.1992.tb03774.x. [DOI] [PubMed] [Google Scholar]
- [39].Parent AE, Choi C, Caudy K, Gridley T, Kusewitt DF. The developmental transcription factor slug is widely expressed in tissues of adult mice. J. Histochem. Cytochem. 2004;52:959–65. doi: 10.1369/jhc.4A6277.2004. doi:10.1369/jhc.4A6277.2004. [DOI] [PubMed] [Google Scholar]
- [40].Parent AE, Newkirk KM, Kusewitt DF. Slug (Snai2) expression during skin and hair follicle development. J. Invest. Dermatol. 2010;130:1737–9. doi: 10.1038/jid.2010.22. doi:10.1038/jid.2010.22. [DOI] [PubMed] [Google Scholar]
- [41].Shirley SH, Hudson LG, He J, Kusewitt DF. The skinny on Slug. Mol. Carcinog. 2010;49:851–61. doi: 10.1002/mc.20674. doi:10.1002/mc.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Blanpain C, Fuchs E. p63: revving up epithelial stem-cell potential. Nat. Cell Biol. 2007;9:731–3. doi: 10.1038/ncb0707-731. doi:10.1038/ncb0707-731. [DOI] [PubMed] [Google Scholar]
- [43].Fielenbach N, Guardavaccaro D, Neubert K, Chan T, Li D, Feng Q, et al. DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev. Cell. 2007;12:443–55. doi: 10.1016/j.devcel.2007.01.018. doi:10.1016/j.devcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- [44].Reece-Hoyes JS, Deplancke B, Barrasa MI, Hatzold J, Smit RB, Arda HE, et al. The C. elegans Snail homolog CES-1 can activate gene expression in vivo and share targets with bHLH transcription factors. Nucleic Acids Res. 2009;37:3689–98. doi: 10.1093/nar/gkp232. doi:10.1093/nar/gkp232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Metzstein MM, Horvitz HR. The C. elegans cell death specification gene ces-1 encodes a snail family zinc finger protein. Mol. Cell. 1999;4:309–19. doi: 10.1016/s1097-2765(00)80333-0. [DOI] [PubMed] [Google Scholar]
- [46].Thellmann M, Hatzold J, Conradt B. The Snail-like CES-1 protein of C. elegans can block the expression of the BH3-only cell-death activator gene egl-1 by antagonizing the function of bHLH proteins. Development. 2003;130:4057–71. doi: 10.1242/dev.00597. [DOI] [PubMed] [Google Scholar]
- [47].Yan B, Memar N, Gallinger J, Conradt B. Coordination of cell proliferation and cell fate determination by CES-1 snail. PLoS Genet. 2013;9:e1003884. doi: 10.1371/journal.pgen.1003884. doi:10.1371/journal.pgen.1003884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zheng H, Shen M, Zha Y-L, Li W, Wei Y, Blanco MA, et al. PKD1 Phosphorylation-Dependent Degradation of SNAIL by SCF-FBXO11 Regulates Epithelial-Mesenchymal Transition and Metastasis. Cancer Cell. 2014;26:358–73. doi: 10.1016/j.ccr.2014.07.022. doi:10.1016/j.ccr.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Skaar JR, D’Angiolella V, Pagan JK, Pagano M. SnapShot: F Box Proteins II. Cell. 2009;137:1358, 1358.e1. doi: 10.1016/j.cell.2009.05.040. doi:10.1016/j.cell.2009.05.040. [DOI] [PubMed] [Google Scholar]
- [50].Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–3. doi: 10.1038/nature10688. doi:10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Abbas T, Mueller AC, Shibata E, Keaton M, Rossi M, Dutta A. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Mol. Cell. 2013;49:1147–58. doi: 10.1016/j.molcel.2013.02.003. doi:10.1016/j.molcel.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rossi M, Duan S, Jeong Y-T, Horn M, Saraf A, Florens L, et al. Regulation of the CRL4(Cdt2) ubiquitin ligase and cell-cycle exit by the SCF(Fbxo11) ubiquitin ligase. Mol. Cell. 2013;49:1159–66. doi: 10.1016/j.molcel.2013.02.004. doi:10.1016/j.molcel.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Horn M, Geisen C, Cermak L, Becker B, Nakamura S, Klein C, et al. DRE-1/FBXO11-Dependent Degradation of BLMP-1/BLIMP-1 Governs C. elegans Developmental Timing and Maturation. Dev. Cell. 2014 doi: 10.1016/j.devcel.2014.01.028. doi:10.1016/j.devcel.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tateossian H, Hardisty-Hughes RE, Morse S, Romero MR, Hilton H, Dean C, et al. Regulation of TGF-beta signalling by Fbxo11, the gene mutated in the Jeff otitis media mouse mutant. Pathogenetics. 2009;2:5. doi: 10.1186/1755-8417-2-5. doi:10.1186/1755-8417-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.