Abstract
Background
Several transcription factors regulate CCS development and function but the role of each in specifying distinct CCS components remains unclear. GATA-binding factor 6 (GATA6) is a zinc-finger transcription factor that is critical for patterning the cardiovascular system. However the role of GATA6 in the embryonic heart and/or CCS has never been shown.
Methods and Results
We report that Gata6 is expressed abundantly in the proximal CCS during mid-gestation in mice. Myocardial-specific deletion of the carboxyl zinc-finger of Gata6 induces loss of HCN4-staining in the compact atrioventricular (AV) node with some retention of HCN4-staining in the AV bundle, but has no significant effect on the connexin40-positive bundle branches. Furthermore, myocardial-specific deletion of the carboxyl zinc-finger of Gata6 alters AV conduction in postnatal life as assessed by surface and invasive electrophysiological evaluation, as well as decreasing the number of ventricular myocytes and inducing compensatory myocyte hypertrophy. Myocardial-specific deletion of the carboxyl zinc-finger of Gata6 is also associated with down-regulation of the transcriptional repressor ID2 and the sodium-calcium exchanger NCX1 in the proximal CCS, where GATA6 transactivates both of these factors. Finally, carboxyl zinc-finger deletion of Gata6 reduces cell-cycle exit of TBX3+ myocytes in the developing AV bundle during the period of AV node specification, which results in fewer TBX3+ cells in the proximal CCS of mature mutant mice.
Conclusions
GATA6 contributes to development and postnatal function of the murine AV node by promoting cell-cycle exit of specified cardiomyocytes towards a conduction system lineage.
Keywords: GATA6, atrioventricular node, cardiac electrophysiology, cardiac embryology, ID2
Insights into the molecules involved with cardiac conduction system (CCS) development are rapidly being uncovered, but the interactions and networks these molecules participate in are not well understood. Several transcription factors implicated in heart development, including NKX2-5, TBX5, TBX3, TBX2, HOPX, IRX3 and ID2, are more highly expressed within the developing CCS than the working myocardium1-6. Furthermore, unique molecular signatures distinguish some components of the atrioventricular (AV) conduction system including the: 1) inferior nodal extension, 2) compact AV node, 3) lower nodal cells, 4) AV bundle and 5) bundle branches that likely contribute to the specification and development of each of these structures7. In addition, Wnt8 and Notch signaling9,10 have been implicated in fate-determination of the AV conduction system.
Patients with mutations in NKX2-511 and TBX512 exhibit congenital heart defects and CCS disease. Loss of functional Nkx2-5 leads to atresia of the AV node anlage in mice, and murine models with deletion of Nxk2-5 develop CCS hypoplasia and atrioventricular block1,13. Haploinsufficiency of Tbx5 in the mouse recapitulates Holt-Oram syndrome with conduction defects and altered distal CCS patterning14, 15. Furthermore, NKX2-5 and TBX5 cooperatively regulate the expression of the helix-loop-helix-containing transcriptional repressor ID2 in the infranodal CCS, where ID2 is involved with normal infranodal conduction system differentiation and function2.
The family of six GATA transcription factors shares C2-H2 zinc-finger DNA binding domains and restrict the developmental potential of multiple cell lineages. The GATA-4/5/6 subfamily of factors is expressed in the heart during embryogenesis, but only GATA-4/6 expression persists in the postnatal myocardium. Both GATA4 and GATA6 are expressed throughout the adult myocardium, and a recent study reported that GATA-dependent enhancers promote transcription in the AV canal and proximal CCS16. While mutations in GATA6 have not been implicated in human conduction system disease, the loss of GATA-factor targets such as SCN5A17 are associated with clinical heart block18. Therefore, we sought to determine if GATA6 itself is involved with CCS function and development.
We report here that Gata6 is abundantly expressed in the proximal CCS during mid-stage murine embryogenesis. Mice engineered with myocardial-specific deletion of the carboxyl zinc-finger domain of Gata6 have fewer TBX3-positive myocytes that fail to exit the cell-cycle during the period of AV node specification. In addition, mice with myocardial-specific deletion of the Gata6 carboxyl zinc-finger develop AV node hypoplasia. Moreover, deletion of the carboxyl zinc-finger of Gata6 induces postnatal AV conduction defects without affecting infranodal conduction. Myocardial-specific deletion of the carboxyl zinc-finger of Gata6 leads to down-regulation of Id2 and Ncx1 in the proximal CCS. At a molecular level GATA6 directly binds to and activates the genes for Id2 and the cardiac sodium-calcium exchanger Ncx1. These data identify a GATA6-dependent program required for embryonic development and functional maintenance of the AV node.
Experimental Methods
Animals
All procedures were performed in accordance with the intuitional guidelines as mandated and approved by the University of Pennsylvania.
Histology, immunohistochemistry and in situ hybridization
Histology, immunohistochemistry and in situ hybridization analyses were performed as previously described3,19 and discussed in detail in the supplemental material.
Electrophysiology studies
Surface ECG recordings were obtained using electrodes placed under each limb, and in vivo electrophysiological studies were performed using an octapolar 1.1-French electrode catheter (Millar) positioned in the right atrium and ventricle. Surface ECG and intracardiac electrograms were displayed on a multi-channel oscilloscope recorder (Bard Electrophysiology) and stored on optical media for off-line analysis.
Chromatin Immunoprecipitation
Whole hearts were excised from euthanized wild-type and Gata6 mutant mice, homogenized in cold PBS plus protease inhibitors and then cross-linked in 1% formaldehyde (Fisher Scientific). The homogenate was washed 2X with PBS and resuspended in lysis buffer containing protease inhibitor cocktail. Samples were sonicated to obtain chromatin fragments between 200 to 500-bp and then incubated overnight with goat anti-GATA6 antibodies (sc-7244X, Santa Cruz) or goat IgG (sc-34665, Santa Cruz). Antibody/histone complexes were collected using protein A agarose beads. The IP sample was washed 5X in wash buffer and DNA was recovered through reverse cross-linking in 5M NaCl. DNA was purified using phenol/chloroform extraction and analyzed by qRT-PCR using SYBR Green (Life Technologies) with primer sequences listed in Supplemental Table 8.
Plasmids and transient co-transfection analyses
The Id2-LUC reporter plasmid containing a 5.2-kb genomic fragment from the mouse inhibitor of DNA 2 gene promoter from basepairs –5992 through –732, and the Ncx1-LUC reporter containing a 5.1-kb promoter fragment of the mouse sodium-calcium exchanger 1 gene from basepairs −4795 through +359 were cloned upstream of firefly luciferase into the KpnI and XhoI sites of the pGL3-promoter vector (Promega). HL-1 immortalized murine cardiomyocytes (1×105) were co-transfected with 100 ng of either Id2-LUC or Ncx1-LUC reporter plasmid; with 0.1-0.5 μg of pcDNA3–GATA6, pcDNA3-GATA4 or pcDNA3–GATA6Δexon4 and 100 ng of pRL-CMV reference plasmid (Promega) using the FuGENE 6 system (Roche Diagnostics).
Statistical analysis
Continuous variables, ECG intervals and cardiac conduction properties were compared using 2-tailed, unpaired Student's t-test. Datasets with smaller sample sizes (n=5 or less) were compared using the Wilcoxon rank sum test. All values are reported as the mean ± 1 standard deviation, unless otherwise noted. A p-value <0.05 was considered significant.
Results
Myocardial deletion of the Gata6 carboxyl zinc-finger does not affect gross myocardial structure or function
Initially, we examined myocardial-specific deletion of the carboxyl zinc-finger of the Gata6 gene to determine if GATA6 contributes to heart development and function. Gata6 conditional mutant mice (Gata6f/f) were employed where exon 4, which encodes the carboxyl zinc-finger of GATA6 is flanked by loxP sites, is removed but leaves the amino zinc-finger and linker regions intact after Cre-mediated recombination19. Gata6f/f mice were intercrossed with knock-in mice expressing Cre-recombinase under transcriptional control of the myosin light chain 2v (Mlc2v) promoter20 to generate mice with myocardial-specific deletion of the carboxyl zinc-finger domain of Gata6 (Mlc2vCre-Gata6f/f). In the developing and mature rodent heart MLC2V is expressed in the ventricular myocardium, right ventricular outflow tract and AV annulus20. Therefore, MLC2V is expressed in both the proximal and distal elements of the murine CCS. Despite abundant expression of GATA6 throughout the ventricular myocardium, morphometric and histological analyses of 12-month old Mlc2vCre-Gata6f/f hearts showed no gross cardiac abnormalities (Supplemental Figure 1A-B) with no evidence of increased myocardial interstitial fibrosis or myofibril disarray (Supplemental Figure 1C-D). In addition, the hearts of adult Mlc2vCre-Gata6f/f mice were structurally and functionally indistinguishable from control littermates when assessed by echocardiography, as were heart and LV mass corrected for body weight when assessed by necropsy (Supplemental Figure 1E-F & Supplemental Table 1). However, analysis of isolated ventricular myocytes from Mlc2vCre-Gata6f/f and Gata6f/f mice reveals Gata6 mutant myocytes are ~40% larger than those isolated from control mice (Figure 1), and there are correspondingly fewer myocytes per unit area in the mature and developing hearts of the mutant mice (Figure 1). These data suggest that deletion of the carboxyl zinc-finger domain of the Gata6 gene reduces Gata6 function in the working myocardium of our model, since these effects are similar to those reported by van Berlo et al. in their Gata6-null model21. In addition, the fact that Mlc2vCre-Gata6f/f pups are born in the expected Mendelian frequencies also suggests expression of the carboxyl zinc-finger domain of the Gata6 gene is not obligate for myocardial development after mid to late gestation.
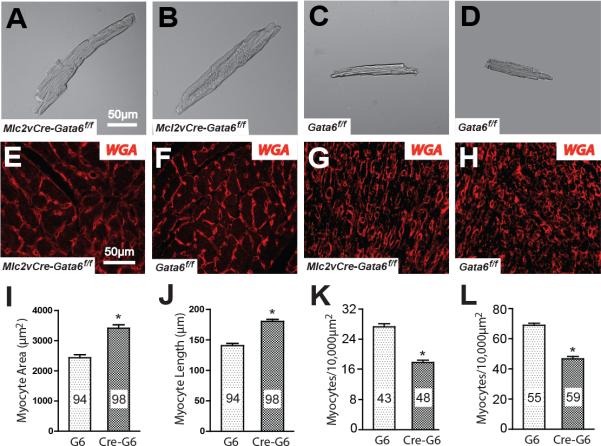
Figure 1.
Gata6 carboxyl zinc-finger deletion increases ventricular myocyte size. (A-D) Bright field images of isolated ventricular myocytes from two mature Mlc2vCre-Gata6f/f mice (A,B) and Gata6f/f mice (C,D). Ventricular myocytes without the Gata6 carboxyl zinc-finger are significantly larger. Scale bar in panel A applies to panels B-D. (E-H) Wheat germ agglutinin (WGA) staining of ventricular sections of mature Gata6f/f and mutant mice (E,F) and E16 Gata6f/f and mutant mice (G,H). Ventricular myocyte cross-sections are larger in the mutant ventricles at both ages. Scale bar in panel E applies to panels F-H. The plots show that mature ventricular myocyte area (I) and length (J) are increased in the Gata6 mutant hearts. Myocytes were isolated from 3 different mice of each genotype. The number of myocytes in each group is shown within the bars. Panels K and L show the number of ventricular myocyte cross-sections in each 10,000μm2 region. The number of regions analyzed from three different hearts in each group is shown within the bars. G6=Gata6f/f; Cre-G6f/f=Mlc2vCre-Gata6f/f. *p<0.01.
Carboxyl zinc-finger deletion of Gata6 induces proximal CCS conduction defects
Although Mlc2vCre-Gata6f/f mice do not display overt defects in myocardial structure or mechanical function, we performed surface ECG analysis in mature animals (54-60 weeks-old) to determine if the carboxyl zinc-finger of the Gata6 gene is necessary for proper CCS development and function. These experiments revealed Mlc2vCre-Gata6f/f mutant mice had prolonged PR-intervals (58.8±3.4 ms vs. 47.6±3.7 ms, p<0.05) compared to Gata6f/f mice (Figure 2A-B, Supplemental Table 2). However, the resting heart rate, P-wave duration, QRS-complex width and QT-interval duration were not different between the two groups. Similar findings were observed using ambulatory ECG recordings in non-sedated mice (Supplemental Table 2) with no episodes of higher-degree heart block in either group. Intracardiac electrophysiologic analysis revealed a longer AH-interval in Mlc2vCre-Gata6f/f mice compared to Gata6f/f controls (37.2±3.1 ms vs. 28.1±2.9 ms, p<0.05), while the HV-interval and His-bundle duration were similar between the two groups (Figure 2A-B, Supplemental Table 3). Furthermore, the Wenckebach cycle length was prolonged in Mlc2vCre-Gata6f/f mice compared to Gata6f/f controls (106±5.0 ms vs. 94.6±4.7 ms, p<0.05). Taken together these findings are consistent with functional defects at the level of the AV node in carboxyl zinc-finger deletion Gata6 mutant mice.
Figure 2.
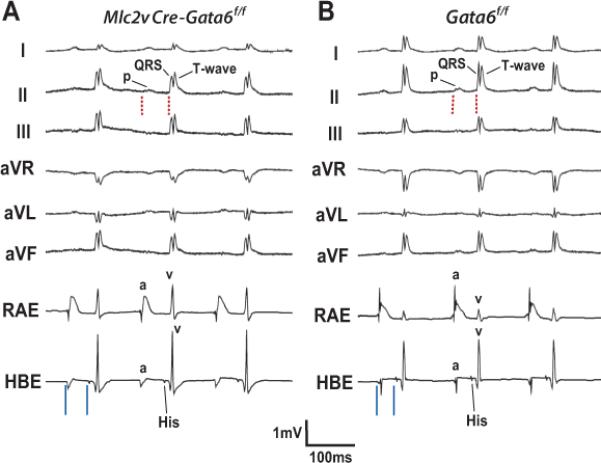
Conduction defects in mature mice without the Gata6 carboxyl zinc-finger. (A, B) Surface ECG leads I, II, III. aVR, aVL & aVF along with intracardiac recordings of the right atrial electrogram (RAE) and His-bundle electrogram (HBE) from an adult Mlc2vCre-Gata6f/f (A) and Gata6f/f (B) mouse. Note the PR-interval is longer (red dashed lines) in the mutant mouse, with no gross defects in overall QRS-complex morphology or other intervals on the surface ECG leads. The Mlc2vCre-Gata6f/f mouse has a PR-interval of 57 ms vs. 46 ms for the Gata6f/f mouse. Recordings from the HBE show atriohisian (AH) interval prolongation (solid blue lines) without functional Gata6. The Mlc2v-CreGata6f/f mouse has an AH-interval of 42 ms vs. 31 ms for the Gata6f/f mouse. P=P-wave; QRS=QRS-complex; T=T-wave; a=atrial electrogram; v=ventricular electrogram.
Gata6 is highly expressed in the embryonic proximal CCS
To determine if the Gata6 gene is expressed within the CCS during embryonic development, we performed in situ hybridization analyses of serial sections prepared from mid-gestation mouse embryos (n=4-5 at each stage). These studies showed Gata6 mRNA (red signal) is most abundant near the AV annulus at embryonic day 12.5 (E12.5) with progressively lower expression in this region at E14.5 and E16.5 (Figure 3A-C). Of note, the region expressing Gata6 mRNA overlaps with those expressing Tbx3 mRNA (red signal), a specific marker of the AV conduction system in mice4, which is most clearly seen at E12.5 (Figure 3D-F). These data confirm Gata6 is expressed in the murine proximal CCS throughout the mid-gestational period. Furthermore, Gata6 gene expression is more abundant in the proximal CCS than in the surrounding myocardium at E12.5 and E14.5 (Figure 3A-B). However, by E16.5 Gata6 gene expression is lower in the proximal CCS than the surrounding Tbx3-negative myocardium (Figure 3C). ID2 is expressed within the distal CCS and has been shown to affect AV conduction in ID2-null mice2. Interestingly, we found Id2 mRNA (red signal) overlaps with Gata6 and Tbx3 in the proximal CCS at E12.5 and E14.5 (Figure 3A, D & G). By E16.5 the expression of Gata6 mRNA is lower in the developing AV bundle, but the expression of Tbx3 and Id2 mRNA is now more robust in this region and overlap (Figure 3C, F & I).
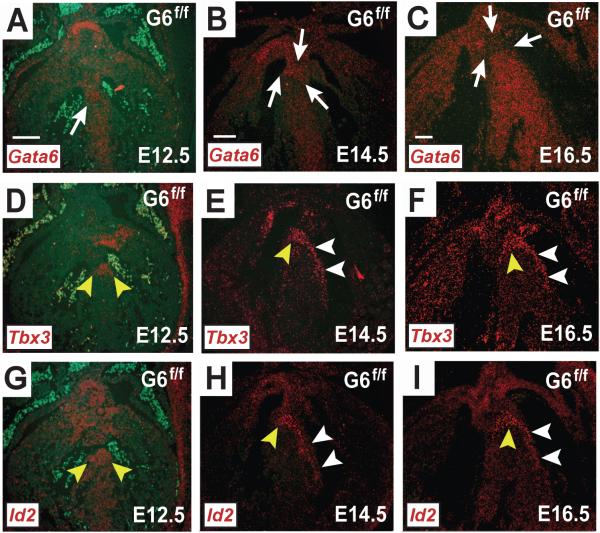
Figure 3.
Gata6 expression in the developing murine AV conduction system. In situ hybridization with probes against Gata6-exon4, Tbx3 and Id2 mRNA in frontal sections from Gata6f/f mouse embryos are shown. At E12.5 with functional Gata6 (A), Gata6 expression overlaps with Tbx3 (D) and Id2 (G). At E14.5 Gata6 transcript expression (B) is weaker in the regions that overlap with Tbx3 (E) and Id2 (H). By E16.5 Gata6 expression (C) is lower in the regions where it overlaps with Tbx3 (F) and Id2 (I), but Gata6 is more robustly expressed in the Tbx3-negative ventricular myocardium. The white arrows point to regions of developing CCS that are either marked or devoid of Gata6+ staining. The yellow arrowheads point to regions of positive signal within the developing AV node and AV bundle. The white arrowheads point to regions of positive signal within the developing bundle branches. Scale bar=100 μm. Scale bar in (A) applies to all E12.5 images; scale bar in (B) applies to all E14.5 images; scale bar in (C) applies to all E16.5 images.
Carboxyl zinc-finger deletion of Gata6 affects proximal CCS development
To determine if Gata6 with deletion of the carboxyl zinc-finger contributes to proximal CCS development, we performed in situ hybridization analyses with Gata6 and Tbx3 riboprobes in serial sections prepared from staged Mlc2vCre-Gata6f/f embryos (n=4-5 at each stage). These experiments showed the region in the proximal CCS containing Tbx3-positive cells was significantly smaller in E12.5-E16.5 conditional mutant embryos compared to similarly staged Gata6f/f control embryos (Figure 4A-F). Moreover, loss of the Gata6 carboxyl zinc-finger was associated with lower Id2 mRNA expression (Figure 4G-I) within the proximal CCS near the crest of the intraventricular septum.
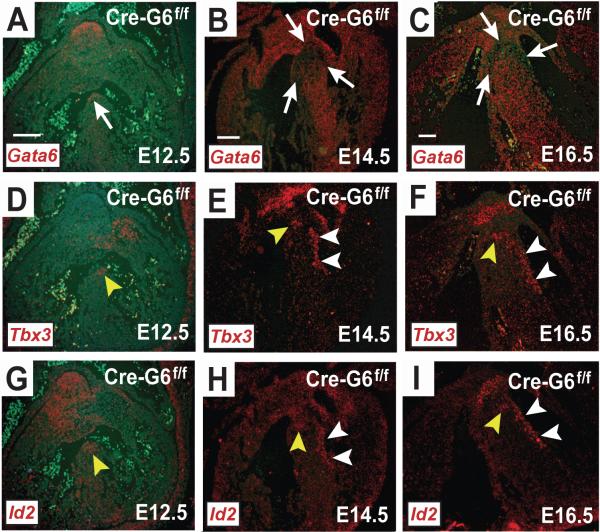
Figure 4.
Effects of Gata6 carboxyl zinc-finger deletion in the developing mouse AV conduction system. In situ hybridization with probes against Gata6-exon4, Tbx3 and Id2 mRNA in frontal sections from Mlc2v-CreGata6f/f mouse embryos are shown. In the absence of Gata6 with a carboxyl zinc-finger, Gata6 expression at E12.5 (A) overlaps with that of Tbx3 (D) and Id2 (G), but the region stained by each of these transcripts is smaller compared to similar anatomic sections from hearts with intact Gata6. A reduced area stained by Gata6 (B), Tbx3 (E) and Id2 (H) persists at E14.5, as well at E16.5 (C, F & I). The white arrows point to regions of the developing CCS that are either marked or devoid of Gata6+ staining. The yellow arrowheads point to regions of positive signal within the developing AV node and AV bundle. The white arrowheads point to regions of positive signal within the developing bundle branches. Scale bar=100 μm. Scale bar in (A) applies to all E12.5 images; scale bar in (B) applies to all E14.5 images; scale bar in (C) applies to all E16.5 images.
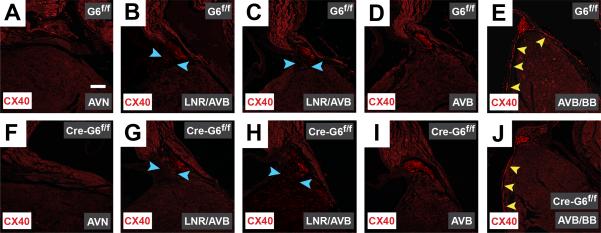
To determine if Gata6 with carboxyl zinc-finger deletion contributes to proper expression of proximal CCS markers during postnatal life, we compared hearts harvested from adult (40-52 week-old) Gata6 conditional mutant mice (Mlc2vCre-Gata6f/f; n=5) to Gata6f/f mice (n=5). We performed immunohistochemical staining of cardiac sections with antibodies that recognize the gap junction protein CX40, a marker of the AV bundle and bundle branches22, and the hyperpolarizing cyclic nucleotide-gated channel, subtype 4 (HCN4) which is expressed in the compact AV node, lower nodal cells and AV bundle of adult mice7. These studies revealed near complete loss of HCN4-positive staining within the compact AV node of Mlc2vCre-Gata6f/f mutant mice (Figure 5A & F). By contrast, HCN4-positive staining was identified in the lower nodal region and AV bundle in hearts from Gata6 conditional mutant mice (Figure 5B-D, G-I). Furthermore, Gata6 with carboxyl zinc-finger deletion did not affect the expression of CX40-positive staining within the AV bundle and bundle branches (Figure 6). Since we saw almost no HCN4 staining in the compact AV node of Gata6 mutant mice, and the compact AV node does not normally express CX40, we examined the expression of TBX3 in both Gata6 mutant and control hearts. These studied showed the region of TBX3 staining in the compact AV node overlaps with that of HCN4 in control mice, while a smaller area of TBX3-postive staining is present in the region of the AV node of Gata6 mutant mice (Supplemental Figure 2). In addition, the expression of Hcn4 and Tbx3 transcripts in the mature AV node are also reduced in Gata6 mutant mice (Supplemental Table 4). These data suggest that GATA6 with an intact carboxyl zinc-finger domain is required for proper expression of compact AV node markers, but not necessarily those of the more distal CCS elements.
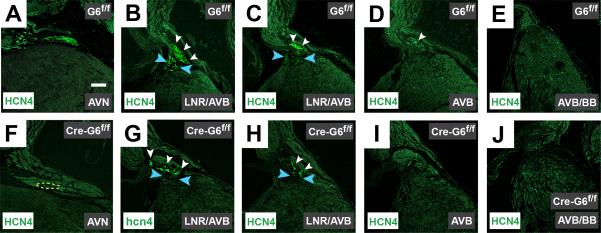
Figure 5.
Gata6 carboxyl zinc-finger deletion reduces HCN4 expression in the AV node. Immunohistochemical staining of frontal sections from mature murine hearts against HCN4 (green) in control hearts (A-E), and Gata6 mutant hearts (F-J), shows HCN4 expression is significantly diminished within the AV node in the absence of Gata6 with a carboxyl zinc-finger (A & F). The dotted yellow line in panel F represents the region of TBX3+ staining in an adjacent section as shown in Supplemental Figure 2. In both the presence and absence of Gata6 with a carboxyl zinc-finger, HCN4-positive cells are expressed in the lower nodal region and portions of the AV bundle (B-D & G-I). The expected lack of HCN4 staining in the bundle branches is seen in the control and mutant hearts (E & J). Blue arrowheads point to the lower nodal region; white arrowheads point to HCN4+ staining in the AV bundle. Scale bar=250 μm and applies to all panels. AVN=atrioventricular node; AVB=atrioventricular bundle; BB=bundle branches; LNR=lower nodal region.
Figure 6.
Gata6 carboxyl zinc-finger deletion does not affect CX40 expression in the mouse CCS. Immunohistochemical staining of frontal sections from mature murine hearts against connexin40 (CX40, red) in Gata6f/f hearts (A-E) and those without the Gata6 carboxyl zinc-finger (F-J) reveals the expected lack of CX40 staining in the AV node, despite the presence of Gata6 (A & F). In the presence and absence of Gata6 with a carboxyl zinc-finger, the expected expression of CX40 within the AV bundle is observed (B-D & G-I). Deletion of the Gata6 carboxyl zinc-finger does not appear to effect the expression of CX40-positive cells within the bundle branches (E & J). The blue arrowheads point to the lower nodal region; the yellow arrowheads point to regions of CX40+ staining in the bundle branches. Scale bar=250 μm and applies to all panels. AVN=atrioventricular node; AVB=atrioventricular bundle; BB=bundle branches; LNR=lower nodal region.
Given the observed changes in the expression of HCN4 and TBX3 within the proximal CCS of Mlc2vCre-Gata6f/f mutant mice, we sought to quantify these changes and determine if deletion of the carboxyl zinc-finger of Gata6 influences CCS structure. First, we examined the expression of acetylcholine esterase (AchE) in the proximal CCS (above the bifurcation of the AV bundle) in adult hearts (aged 46-56 weeks) from Mlc2vCre-Gata6f/f and Gata6f/f mice. AchE marks the compact AV node, lower nodal region, AV bundle and proximal bundle branches of the mature rodent CCS23. These studies showed mutant Gata6 induces a ~50% reduction in the volume of the proximal CCS in mature Mlc2vCre-Gata6f/f mutant mice (n=5, p<0.05; Supplemental Table 5). Next, we examined the effects of myocardial-specific deletion of the carboxyl zinc-finger of Gata6 on HCN4 expression in the compact AV node of mature mouse hearts (aged 46-56 weeks)7. HCN4 is normally restricted to the compact AV node with a few, smaller regions in the lower nodal cells and proximal AV bundle in the mature mouse heart. Remarkably, the volume of HCN4-positive cells in the proximal CCS of adult Mlc2vCre-Gata6f/f mice was reduced by ~80% compared to control littermates (n=4, p<0.05; Supplemental Table 5). Interestingly, almost complete loss of HCN4-staining was observed in the compact AV node versus the lower nodal region and AV bundle of Mlc2vCre-Gata6f/f mice (Figure 5A-C, F-H). Finally, to determine if deletion of the carboxyl zinc-finger of Gata6 affects distal CCS patterning we intercrossed HopxLacz knockin mice with Mlc2vCre-Gata6f/f mice since HOPX marks the distal CCS3. We saw no difference in distal CCS structure of HopxLacz::Mlc2vCre-Gata6f/f mice compared to HopxLacz::Gata6f/f mice (Supplemental Figure 3).
We then crossed MinKCreERT2 BAC transgenic mice with R26R-YFP reporter mice to generate mice that express YFP within the CCS. In addition, we examined Tbx3-GFP BAC transgenic mice that express GFP in the AV node24. MINK is expressed in the AV bundle, bundle branches and Purkinje fibers of the mature murine heart25, so we performed dual immunohistochemical staining with antibodies against GFP and GATA6 in these models to examine GATA6 expression within the mature CCS. These studies showed GATA6 is expressed in all regions of the mature mouse CCS (Supplemental Figure 4A-F). This corresponds with expression of truncated GATA6 mRNA in the heart of mutant mice, whereas only full-length GATA6 mRNA is expressed in control hearts (Supplemental Figure 4G). Together, these data show that deletion of the carboxyl zinc-finger of Gata6 results in loss of HCN4 expression in the AV node of postnatal hearts. By contrast, the expression of HCN4, CX40 and HOPX in the lower nodal cells, AV bundle and bundle branches do not appear to be dependent on expression of Gata6 with a carboxyl zinc-finger, and the infranodal CCS appears to be grossly intact despite reduced numbers of ventricular myocytes with compensated hypertrophy.
Deletion of the carboxyl zinc-finger of Gata6 reduces ID2 expression in the proximal CCS
To identify genes regulated by GATA6 in the CCS, we performed a screen of selected genes with GATA-binding sites in their regulatory elements (Supplemental Table 6). The primer pairs used to detect these genes are listed in Supplementary Table 7. Using mRNA isolated from the region of the adult AV node, quantitative RT-PCR revealed two transcripts that were down-regulated by at least two-fold in Gata6 mutant mice: Ncx1 (0.44; p-value=0.027) and Id2 (0.41; p-value=0.034; Supplemental Table 4). To determine if ID2 is directly regulated by GATA6, we performed immunohistochemical analysis of ID2 in hearts from Mlc2vCre-Gata6f/f mutant and control littermates. These studies showed ID2 is expressed in the proximal CCS of mature mice, but ID2 expression is dramatically reduced in the AV node and lower nodal region in hearts with Gata6 carboxyl zinc-finger deletion (Figure 7A-D). Furthermore, in situ hybridization showed mutation of the Gata6 gene decreased Id2 transcript expression within the proximal CCS from E12.5 through E16.5 (Figure 4G-I). Quantitative RT-PCR analysis revealed Id2 transcript levels in mature Mlc2vCre-Gata6f/f mice are ~60% that of wild-type levels in tissue from the superior intraventricular septum that contains the AV node (Supplemental Table 4). A corresponding 50% decline in ID2 protein level was observed between the two groups (Supplemental Figure 5A). Immunoblot analysis of other GATA-dependent genes from the region of the mature AV node, including CX40, α-myosin heavy chain and cardiac actin were similarly expressed between Mlc2vCre-Gata6f/f and Gata6f/f mice (Supplemental Figure 5B-D).
Figure 7.
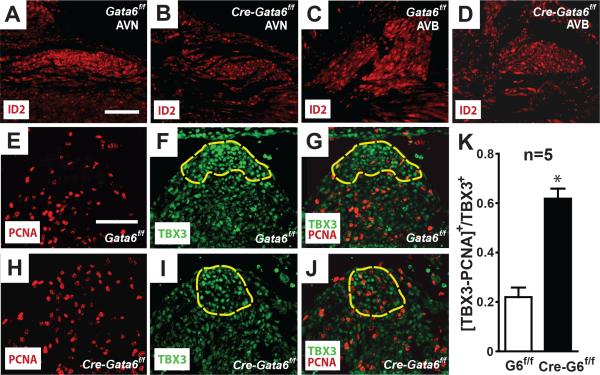
Gata6 carboxyl zinc-finger deletion reduces ID2 expression and cardiomyocyte cell-cycle exit in the developing AV bundle. Immunohistochemical staining in adult murine hearts against ID2 in Gata6f/f hearts (A & C) shows ID2 expression is higher in the proximal CCS compared to the working myocardium. In the absence of Gata6 with a carboxyl zinc-finger (B & D), ID2 expression is reduced in the AV node and AV bundle. Panels E-K show Gata6 with a carboxyl zinc-finger is required for cell-cycle exit of cardiomyocytes in the developing AV bundle. Immunohistochemical staining of E12.5 wild-type hearts (E-G) against PCNA (red) and TBX3 (green), as well as similarly staged Gata6 mutant hearts (H-J). The dashed yellow outline indicates the TBX3-expressing AV bundle. Note there are fewer PCNA/TBX3-positive cells in the Gata6f/f heart compared to the mutant heart. (K) PCNA-positive nuclei in the crest of the septum that overlap with TBX3-expressing cells averaged from five hearts of each genotype. *P<0.01. Scale bar=100 μm. Scale bar in (A) applies to (A-D); scale bar in (E) applies to (E-J). AVN=atrioventricular node; AVB=atrioventricular bundle.
Since ID2 expression appears to be dependent on expression of Gata6 with an intact carboxyl zinc-finger, and ID2 acts as a transcriptional repressor that promotes differentiation of the ventricular conduction system2, we tested the hypothesis that Gata6 is necessary for AV node myocyte differentiation. For these experiments we utilized cell-cycle exit as an early indicator of conduction system lineage specification. Myocytes of the AV node exit the cell-cycle between E10 and E12, which differs from non-conduction embryonic ventricular myocytes that have continuous cell-cycle progression throughout embryogenesis26. We exploited these differences to evaluate AV node cell-cycle exit in Gata6 mutant and control mice. Nuclear PCNA is a marker of cellular proliferation and the lack of PCNA staining suggests exit from the cell-cycle27. When we examined control hearts we found localized absence of proliferating nuclear cell antigen (PCNA) in the developing AV bundle at E12 (Figure 7E). To confirm that the non-dividing cells contribute to the conduction system, we co-labeled the hearts with an antibody against TBX3 (Figure 7F). Control hearts showed few PCNA+/TBX3+ cells while PCNA staining was observed in TBX3- cells at E12 (Figure 7E-G,K). In contrast Gata6 mutant hearts showed uniform PCNA staining throughout the embryonic ventricle, including the developing AV bundle (Figure 7H-J). In the mutant hearts there were more PCNA+/TBX3+ cells in the crest of the interventricular septum, but there are fewer PCNA+/TBX3- cells in the mutant heart compared to control hearts (Figure 7G, J & K). From these findings we conclude GATA6 with an intact carboxyl-zinc finger in specified AV nodal myocardial cells results in cell-cycle exit and adoption of a conduction system fate.
GATA6 regulates transcription of Id2 and Ncx1
Prior studies suggest Id2 and Ncx1 gene expression are regulated by GATA428,29. Since Id2 and Ncx1 mRNA expression in the proximal CCS is decreased by deletion of the carboxyl zinc-finger of Gata6, Id2 and Ncx1 may also be transcriptional targets of GATA6 in the CCS. Comparing the mouse Id2 and human ID2 genes by VISTA analysis (http://genome.lbl.gov/vista/index.shtml) revealed greater than 80% sequence identity between these promoter regions (Figure 8A). Within or adjacent to the regions of sequence homology there are 4 putative GATA-binding sites (Figure 8A, Supplemental Table 8) conserved between the mouse Id2 and human ID2 genes. Each species also contains additional consensus GATA sites in this region (ENSEMBL and NCBI databases, http://www.ensembl.org/indix.html and http://www.ncbi.nih.gov/BLAST, respectively). A similar bioinformatic analysis shows the mouse Ncx1 and human NCX1 genes share ~55% sequence identity within their promoter regions, with 5 shared putative GATA-binding sites in or near the region of sequence homology (Figure 8B, Supplemental Table 8). To determine if Id2 and Ncx1 transcription is activated by GATA6, we performed transient transactivation studies using reporter constructs driven by Id2 and Ncx1 promoter fragments (Figure 8C-D). As shown in Figure 8, co-transfection with increasing concentrations of the pcDNA3–GATA6 expression plasmid results in stepwise activation of both the Id2-LUC and Ncx1-LUC reporter, suggesting that the Id2 and Ncx1 promoters are directly activated by GATA6 (Figure 8E-F). In contrast, co-transfection with pcDNA3–GATA6Δexon4 (encoding GATA6 without the carboxyl zinc-finger) does not increase luciferase activity of either reporter. Co-transfection with the pcDNA3–GATA4 expression plasmid also results in activation of both the Id2-LUC and Ncx1-LUC reporter, which is augmented by the presence of pcDNA3-GATA6 but not pcDNA3-GATA6 Δexon4 (Supplemental Figure 6). These results demonstrate that transcriptional activation of the Id2 and Ncx1 promoters are dependent on carboxyl zinc-finger–mediated binding of GATA6 to DNA and that deletion of the carboxyl zinc-finger of GATA6 does not alter the ability of GATA4 to activate these genes.
Figure 8.
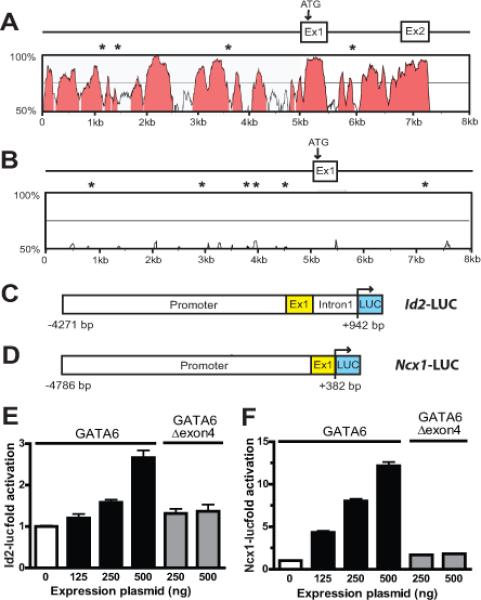
GATA6 transactivates Id2 and Ncx1. (A) VISTA comparison of the murine and human Id2 proximal promoter and exon 1, and (B) of the murine and human Ncx1 proximal promoter and exon 1. In panels A and B the x and y axes indicate sequence length (kb) and percent homology (≥ 75%, pink), respectively. GATA6 binding sites conserved in the mouse and human sequence are indicated by asterisks. (C) Schematic of the Id2-LUC reporter containing the 5.2-kb proximal promoter upstream of firefly luciferase (LUC), and (D) the Ncx1-LUC reporter containing the 5.1-kb proximal promoter that is also upstream of firefly luciferase. (E) Activation of the Id2-LUC reporter by GATA6. HL-1 cells were transiently transfected with Id2-LUC and 125–500 ng of expression plasmid encoding wild-type GATA6 or GATA6 lacking exon 4 (Gata6Δexon4). The reporter is activated by wild-type GATA6 but not by GATA6Δexon4. (F). Activation of the Ncx1-LUC reporter by GATA6. HL-1 cells were similarly transiently transfected with Ncx1-LUC and 125–500 ng of expression plasmid encoding wild-type GATA6 or GATA6Δexon4. As with Id2-LUC, the Ncx1-LUC reporter was activated by the expression of wild-type GATA6 but not by GATA6Δexon4. The transient transection studies in panels E and F show the mean change in report activity from at least 3 different experiments at each concentration of expression plasmid.
To verify that GATA6 directly binds to consensus GATA sequences in the Id2 and Ncx1 genes we performed chromatin immunoprecipitation using chromatin isolated from wild-type and Gata6 mutant hearts. These studies showed chromatin fragments containing two consensus GATA sequences in the Id2 gene (−1561 to −1566; +668 to +673), and four consensus sequences in the Ncx1 gene (−4115 to −4120; −3097 to −3102; − 1271 to 1276; −1037 to −1042) were specifically immunoprecipitated from wild-type, but not from mutant hearts, compared to control regions (Supplemental Tables 9 & 10). These results suggest GATA6 directly binds to the Id2 and Ncx1 genes via the carboxyl zinc-finger domain at consensus sequences in the heart.
Discussion
We report here that Gata6 is required for normal development and function of the AV node. In the compact AV node, HCN4-expression is dependent on Gata6 while this does not appear to be the case for more distal CCS elements. Mechanistically, we show the transcriptional repressor ID2 is expressed within the proximal murine CCS and Gata6 directly regulates ID2 expression. In addition, deletion of the carboxyl zinc-finger of Gata6 leads to down-regulation of ID2 with AV node hypoplasia, reduced cell-cycle exit of TBX3+ myocytes in the proximal CCS and AV nodal conduction defects in mature mice. Finally, myocardial-specific deletion of the carboxyl zinc-finger of Gata6 does not significantly affect gross structure or function of the infranodal CCS despite decreasing proliferation of ventricular myocytes.
Several transcription factors regulate the size and function of the proximal and distal CCS1,13,14. Other transcription factors such as Hopx appear to be more specific for maintenance of the distal CCS in postnatal life3. Interestingly, we found that deletion of the carboxyl zinc-finger of Gata6 affects the structure and function of the AV node without significant affects on the infranodal CCS. The fact that infranodal conduction and gross infranodal CCS morphology are unaltered in the absence of the Gata6 carboxyl zinc-finger makes it unlikely the CCS is structurally abnormal below the His-bundle. These findings support the existence of a modular, CCS molecular program involving GATA6 that targets ID2 and NCX1 within HCN4-positive cell of the proximal CCS.
While ID2 is highly expressed in the murine AV node2, ID2 expression is lower in the human AV node during embryogenesis30. Although our data supports Id2 as a transcriptional target of GATA6, the fact that Id2 is not ‘robustly’ transcribed by GATA6 and ID2 is not highly expressed in the human AV node suggests GATA6 may recruit other transcriptional repressors to maintain the AV node.
It is interesting that AV node hypoplasia is observed in the setting of myocardial-specific deletion of the Gata6 carboxyl zinc-finger with preserved Gata4 transcript expression (Supplemental Table 4), and we found that carboxyl zinc-finger deletion of Gata6 induces proximal CCS defects with no gross effect on infranodal CCS structure or function despite changes in ventricular myocyte proliferation and size. These results could be explained by the fact that GATA4 and GATA6 can compensate for one another by interacting with the same regulatory regions of common target genes31. Our findings suggest similar non-redundant, functional overlap may exist with regards to Id2 and Ncx1, where preserved GATA4 function is unlikely to maintain functional levels of these genes in the proximal CCS without GATA6 with an intact carboxyl zinc-finger. With regards to the CCS, GATA6 is highly expressed in the AV canal and proximal CCS during mid-embryogenesis when AV node myocytes are specified. Therefore, carboxyl zinc-finger deletion of GATA6 appears to affect the number and specification of AV nodal myocytes compared to those in the infranodal CCS. In addition, we need to consider the fact that we only removed the Gata6 carboxyl zinc-finger from Mlc2v-positive myocytes and not necessarily from atrial myocytes or other cells populations that may contribute to the proximal CCS. Therefore, further studies examining CCS-specific deletion of GATA4 and GATA6 will be useful to assess the relative contributions of these factors to regionalized CCS structure and development.
In summary, the findings presented here support the existence of a molecular hierarchy where GATA6 contributes to AV node structure and function through the transcriptional repressor ID2. This is the first evidence GATA6 is required for development and maintenance of the AV node or the embryonic heart. A recent report described a role for GATA4 in proximal CCS function where haploinsufficiency of GATA4 enhanced AV node conduction. In this report GATA4 increased conduction by down-regulating the low-conductance gap junction protein connexin30.232. This is another example where GATA4 and GATA6 have non-redundant overlapping functions, since we found the loss of Gata6 does not affect connexin30.2 mRNA levels (Supplemental Table 4). The findings presented in this report demonstrate that Gata6 plays a critical role in proximal CCS function, and supports a paradigm where upstream transcription factors contribute to CCS development by suppressing cell-cycle exit and differentiation of cardiomyocytes through the transcriptional repressor ID2.
Supplementary Material
Acknowledgments
We thank Mike Parmacek for reading the manuscript and providing suggestions. We also thank Ivan Moskowitz (Univ. of Chicago) for the Id2 in situ probe and MinKCreERT2 BAC transgenic mice, Mike Parmacek (Univ. of Pennsylvania) for the GATA expression vectors and Vincent Christoffels (Academic Medical Center, Univ. of Amsterdam) for the Tbx3-GFP BAC transgenic mice.
Funding Sources: This work was supported by grants from the NIH to V.V.P (R01 HL105734), the Commonwealth of Pennsylvania and the Gunther Cardiology Research Fund. V.V.P. is an Innovative Researcher of the American Heart Association supported by grant 11IRG4930008.
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, et al. Nkx2-5 pathways and congenital heart disease: loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:378–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 2.Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, et al. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 3.Ismat FA, Zhang M, Kook H, Huang B, Zhou R, Ferrari VA, et al. Homeobox protein Hop functions in the adult cardiac conduction system. Circ Res. 2005;96:898–903. doi: 10.1161/01.RES.0000163108.47258.f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakker ML, Boukens BJ, Mommersteeg MT, Brons JF, Wakker V, Moorman AF, et al. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- 5.Aanhaanen WT, Brons JF, Domínguez JN, Rana MS, Norden J, Airik R, et al. The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ Res. 2009;104:1264–1274. doi: 10.1161/CIRCRESAHA.108.192450. [DOI] [PubMed] [Google Scholar]
- 6.Zhanga S-S, Kime K-H, Rosene A, Smythh JW, Sakumac R, Delgado-Olguína P, et al. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His–Purkinje network. Proc Natl Acad Sci U.S.A. 2011;108:13576–13581. doi: 10.1073/pnas.1106911108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aanhaanen WT, Mommersteeg MT, Norden J, Wakker V, de Gier-de Vries C, Anderson RH, et al. Developmental origin, growth, and three-dimensional architecture of the atrioventricular conduction axis of the mouse heart. Circ Res. 2010;107:728–736. doi: 10.1161/CIRCRESAHA.110.222992. [DOI] [PubMed] [Google Scholar]
- 8.Bond J, Sedmera D, Jourdan J, Zhang Y, Eisenberg CA, Eisenberg LM, et al. Wnt11 and Wnt7a are up-regulated in association with differentiation of cardiac conduction cells in vitro and in vivo. Dev Dyn. 2003;227:536–543. doi: 10.1002/dvdy.10333. [DOI] [PubMed] [Google Scholar]
- 9.Milan DJ, Giokas AC, Serluca FC, Peterson RT, MacRae CA. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development. 2006;133:1125–1132. doi: 10.1242/dev.02279. [DOI] [PubMed] [Google Scholar]
- 10.Rentschler S, Harris BS, Kuznekoff L, Jain R, Manderfield L, Lu MM, et al. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J Clin Invest. 2011;121:525–533. doi: 10.1172/JCI44470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 12.Basson CT, Huang T, Lin RC, Bachinsky DR, Weremowicz S, Vaglio A, et al. Different TBX5 interactions in heart and limb defined by Holt-Oram syndrome mutations. Proc Natl Acad Sci U.S.A. 1999;96:2919–2924. doi: 10.1073/pnas.96.6.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, et al. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113:1130–1137. doi: 10.1172/JCI19846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moskowitz IPG, Pizard A, Patel VV, Bruneau BG, Kim JB, Kupershmidt S, et al. The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development. 2004;131:4107–4116. doi: 10.1242/dev.01265. [DOI] [PubMed] [Google Scholar]
- 15.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 16.Stevanovic S, Barnett P, van Duijvenboden K, Weber D, Gessler M, Christoffels VM. GATA-dependent regulatory switches establish atrioventricular canal specificity during heart development. Nat Comm. 2014;5:3680–3690. doi: 10.1038/ncomms4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang P, Kupershmidt S, Roden DM. Cloning and initial characterization of the human cardiac sodium channel (SCN5A) promoter. Cardiovasc Res. 2004;61:56–65. doi: 10.1016/j.cardiores.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 18.Viswanathan PC, Benson DW, Balser JR. A common SCN5A polymorphism modulates the biophysical effects of an SCN5A mutation. J Clin Invest. 2003;111:341–346. doi: 10.1172/JCI16879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lepore JJ, Mericko PA, Cheng L, Lu MM, Morrisey EE, Parmacek MS. GATA-6 regulates semaphorin 3C and is required in cardiac neural crest for cardiovascular morphogenesis. J Clin Invest. 2006;116:929–939. doi: 10.1172/JCI27363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Kubalak SW, Minamisawa S, Price RL, Becker KD, Hickey R, et al. Selective requirement of myosin light chain 2v in embryonic heart function. J Biol Chem. 1998;273:1252–1256. doi: 10.1074/jbc.273.2.1252. [DOI] [PubMed] [Google Scholar]
- 21.van Berlo JH, Elrod JW, van den Hoogenhof MM, York AJ, Aronow BJ, Duncan SA, et al. The transcription factor GATA-6 regulates pathological cardiac hypertrophy. Circ Res. 2010;107:1032–1040. doi: 10.1161/CIRCRESAHA.110.220764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delorme B, Dahl E, Jarry-Guichard T, Marics I, Briand JP, Willecke K, et al. Developmental regulation of connexin 40 gene expression in mouse heart correlates with the differentiation of the conduction system. Dev Dyn. 1995;204:358–371. doi: 10.1002/aja.1002040403. [DOI] [PubMed] [Google Scholar]
- 23.Hegab ES, Ferrans VJ. A histochemical study of the esterases of the rat heart. Am J Anat. 1966;119:235–261. doi: 10.1002/aja.1001190204. [DOI] [PubMed] [Google Scholar]
- 24.Horsthuis T, Buermans HP, Brons JF, Verkerk AO, Bakker ML, Wakker V, et al. Gene expression profiling of the forming atrioventricular node using a novel tbx3-based node-specific transgenic reporter. Circ Res. 2009;105:61–69. doi: 10.1161/CIRCRESAHA.108.192443. [DOI] [PubMed] [Google Scholar]
- 25.Arnolds DE, Moskowitz IP. Inducible recombination in the cardiac conduction system of minK: CreERT2 BAC transgenic mice. Genesis. 2011;49:878–884. doi: 10.1002/dvg.20759. [DOI] [PubMed] [Google Scholar]
- 26.Sedmera D, Reckova M, DeAlmeida A, Coppen SR, Kubalak SW, Gourdie RG, et al. Spatiotemporal pattern of commitment to slowed proliferation in the embryonic mouse heart indicates progressive differentiation of the cardiac conduction system. Anat Rec. 2003;274:773–777. doi: 10.1002/ar.a.10085. [DOI] [PubMed] [Google Scholar]
- 27.Matthew MB, Bernstein RM, Franza BR, Jr, Garrels JI. Identity of the proliferating cell nuclear antigen and cyclin. Nature. 1984;309:374–376. doi: 10.1038/309374a0. [DOI] [PubMed] [Google Scholar]
- 28.Lim JY, Kim WH, Kim J, Park SI. Induction of Id2 expression by cardiac transcription factors GATA4 and NKX2.5. J Cell Biochem. 2008;103:182–194. doi: 10.1002/jcb.21396. [DOI] [PubMed] [Google Scholar]
- 29.Nicholas SB, Philipson KD. Cardiac expression of the Na(+)/Ca(2+) exchanger NCX1 is GATA factor dependent. Am J Physiol. 1999;277:H324–330. doi: 10.1152/ajpheart.1999.277.1.H324. [DOI] [PubMed] [Google Scholar]
- 30.Sizarov A, Ya J, de Boer BA, Lamers WH, Christoffels VM, Moorman AF. Formation of the building plan of the human heart: morphogenesis, growth, and differentiation. Circulation. 2011;123:1125–1135. doi: 10.1161/CIRCULATIONAHA.110.980607. [DOI] [PubMed] [Google Scholar]
- 31.Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol Cell Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munshi NV, McAnally J, Bezprozvannaya S, Berry JM, Richardson JA, Hill JA, et al. Cx30.2 enhancer analysis identifies Gata4 as a novel regulator of atrioventricular delay. Development. 2009;136:2665–2674. doi: 10.1242/dev.038562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.