Abstract
Inflammasomes are critical for mounting host defense against pathogens. The molecular mechanisms controlling activation of the AIM2 inflammasome in response to different cytosolic pathogens is unclear. Here, we show that the transcription factor IRF1 is the upstream molecule leading to AIM2 inflammasome activation during Francisella novicida infection, whereas engagement of the AIM2 inflammasome by mouse cytomegalovirus or transfected dsDNA did not require IRF1. F. novicida infection detected by the cGAS-STING pathway induces type I interferon-dependent expression of IRF1, which drives the expression of guanylate-binding proteins (GBPs) leading to intracellular bacterial killing and DNA release. These results reveal a specific requirement for IRF1 and GBPs in the liberation of DNA for AIM2 sensing depending on the pathogen encountered by the cell.
Inflammasomes are cytosolic multimeric protein complexes which provide host defense against microbial pathogens and play a role in the development of autoinflammatory and metabolic diseases 1. Formation of the inflammasome is initiated by the innate immune sensors nucleotide-binding oligomerization domain-like receptors (NLRs) or AIM2-like receptors (ALRs) following detection of microbial–associated molecular patterns (MAMPs) or damage–associated molecular patterns (DAMPs) in the cytoplasm of a cell 2. In most cases, NLRs and ALRs engage the adaptor protein ASC, wherein its CARD domain is critical for subsequent interaction with the CARD domain of procaspase-1 3. Activation of caspase-1 induces proteolytic cleavage and release of mature interleukin (IL)-1β and IL-18 as well as the induction of a type of pro-inflammatory cell death termed pyroptosis 2. Therefore, inflammasomes are capable of mounting innate immune effector functions in response to a range of stimuli encountered by the cell.
AIM2 is a member of the ALR family which contributes to the host defense against bacterial and viral pathogens, such as Francisella tularensis and cytomegalovirus, respectively 4, 5, 6, 7, 8, 9. AIM2 contains a HIN-200 domain which directly binds double-stranded DNA (dsDNA) released into the cytosol of a cell during a microbial infection4, 5, 6, 7. Similarly, self DNA released into and accumulated in keratinocytes also activates the AIM2 inflammasome to drive the release of IL-1β in lesions of patients with psoriasis 10, indicating that AIM2 also responds to endogenous dsDNA released during cellular damage. Structural analysis demonstrated that the positively charged HIN domain of AIM2 embraces the dsDNA, whereas the pyrin domain of AIM2 facilitates the recruitment of ASC 11.
Compared with the NLRP3 inflammasome, the molecular mechanism governing AIM2 inflammasome assembly is less clear. NLRP3 is activated by a plethora of MAMPs and DAMPs. Activation of the NLRP3 inflammasome requires two signals 3. The first signal, or priming, is provided by engagement of the receptors TLRs, NOD2 or TNFR to activate NF-κB and induce NLRP3 expression. The second signal is provided by an NLRP3 activator, including ATP, uric acid crystals, silica, asbestos, bacterial messenger RNA, bacterial DNA:RNA hybrids, and muramyldipeptide (MDP) 3. In addition, type I interferon signaling is critical for activation of the so-called non-canonical NLRP3 inflammasome 12,13. In this context, extracellular lipopolysaccharide (LPS) from Gram-negative bacteria activates TLR4 and its adapter TRIF to induce type I interferon and pro-caspase-11 expression 12, 13, 14, 15. LPS released into the cytosol by means of vacuolar lysis binds caspase-11, leading to caspase-11-dependent cell death and activation of the so-called non-canonical NLRP3 inflammasome 16, 17, 18, 19, 20. In contrast, AIM2 is constitutively expressed and does not require NF-κB-mediated priming for its activation 6. Instead, type I interferon signaling contributes to AIM2 inflammasome activation in response to certain pathogens, such as Francisella novicida, but not mouse cytomegalovirus (mCMV) 8, 21. Therefore, the downstream mechanism leading to AIM2 inflammasome activation in response to different pathogens is likely to be different and remains to be resolved 3.
Here, we identified two distinct pathways leading to the activation of the AIM2 inflammasome. We find that engagement of the AIM2 inflammasome by F. novicida infection requires interferon regulatory factor 1 (IRF1), but not TRIF or caspase-11. IRF1 is required for robust guanylate-binding protein (GBPs) expression, which kills cytosolic F. novicida and mediates DNA release for access by AIM2. In addition, Irf1−/− mice fail to control F. novicida infection. In contrast, engagement of AIM2 using mCMV or transfected dsDNA does not require IRF1. Therefore, we have unraveled a specific requirement for IRF1 and GBPs in F. novicida-induced activation of the AIM2 inflammasome.
RESULTS
IRF1 mediates AIM2 inflammasome activation by F. novicida
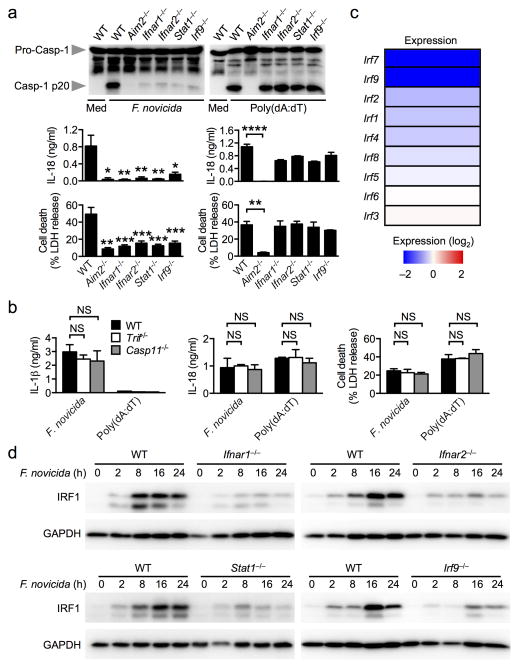
Francisella tularensis, a pathogen which causes a rapid and lethal infection in human and mouse, is recognized by the AIM2 inflammasome 8, 9, 22. In unprimed primary mouse bone marrow-derived macrophages (BMDMs), F. tularensis subspecies novicida (F. novicida) induced AIM2-dependent caspase-1 activation, IL-1β and IL-18 release and cell death in a manner which required type I interferon receptor 1 and 2, and components of the interferon-stimulated gene factor 3 (ISGF3), and STAT1 and IRF9 transcription factors (Fig. 1a and Supplementary Fig. 1a) 8, 9, 21, 22. In marked contrast, activation of the AIM2 inflammasome by transfection of the dsDNA ligand, poly(deoxyadenylic-deoxythymidylic) acid or poly(dA:dT), occurred independently of type I interferon signaling (Fig. 1a). The type I interferon signature leading to activation of the AIM2 inflammasome in response to F. novicida infection is unknown. Type I interferon production induced by the Toll-like receptor adaptor protein, TRIF, is required for robust activation of the Caspase-11-NLRP3 inflammasome in macrophages infected with Citrobacter rodentium, Escherichia coli and Vibrio cholerae 12, 13, 15, however, we found that TRIF and caspase-11 were dispensable for activation of the AIM2 inflammasome by F. novicida (Fig. 1b). Indeed, F. novicida synthesizes a tetra-acylated lipid A, which differs to the hexa-acylated species of many enteric bacteria that normally activates caspase-11, and explains the ability of F. novicida to evade caspase-11-mediated detection 18. These findings suggest that a yet undefined type I interferon signature independent of TRIF and caspase-11 is required to engage the AIM2 inflammasome by F. novicida infection, but not in response to transfected dsDNA.
Figure 1. F. novicida infection induces IRF1 expression in a manner that requires type I interferon signaling.
(a) Caspase-1 activation, IL-18 release and cell death in unprimed bone marrow-derived of the indicated strain macrophages (BMDMs) infected with F. novicida (MOI 100) for 20 h or transfected with poly(dA:dT) for 5 h. (b) IL-1β and IL-18 release and cell death in unprimed BMDMs infected with F. novicida for 20 h or transfected with poly(dA:dT) for 5 h. (c) Heat map of microarray analysis showing relative expression of interferon regulatory factors (IRFs) genes in Ifnar1−/− BMDMs compared to wildtype (WT) BMDMs infected with F. novicida for 8 h. (d) Induction of IRF1 expression in unprimed BMDMs infected with F. novicida (MOI 50). Graphs show mean and s.e.m. of two (b) or three (a,d) independent experiments. Microarray analysis was performed using duplicate samples of each genotype (c). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant. One-way ANOVA with a Dunnett’s multiple comparisons test (a,b).
We performed a microarray analysis to identify differentially regulated genes in wild-type and Ifnar1−/− BMDMs that had been infected with F. novicida. Expression levels of a number of interferon regulatory factor (IRF) family members were significantly reduced in the absence of IFNAR1, including IRF1, IRF7 and IRF9 (Fig. 1c). IRF9 is a subunit of the ISGF3 complex and contributed to AIM2 inflammasome activation by F. novicida infection (Fig. 1a and Supplementary Fig. 1a), and we excluded a role for IRF7 in activation of the AIM2 inflammasome upon F. novicida infection (Supplementary Fig. 1b). Validation of the protein expression levels of IRF1 confirmed that it was robustly upregulated by F. novicida infection, through a mechanism that required IFNAR1, IFNAR2, STAT1 and IRF9 (Fig. 1d). Consistently, BMDMs stimulated with recombinant mouse IFN-β also robustly upregulated IRF1 expression, which was critically dependent on type I interferon receptors (Supplementary Fig. 1c).
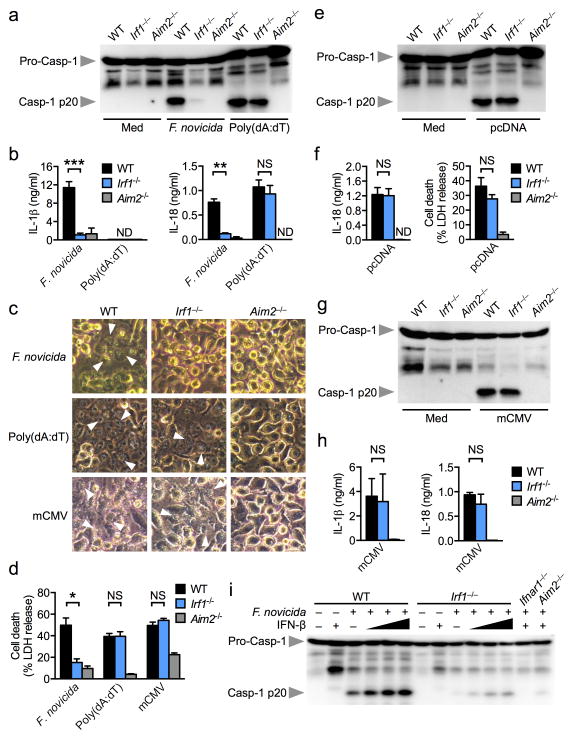
To investigate whether IRF1 is an upstream molecule leading to AIM2 inflammasome activation, we infected unprimed wild-type and Irf1−/− BMDMs with F. novicida and analyzed inflammasome responses 20 h later. Remarkably, unprimed Irf1−/− BMDMs failed to respond to F. novicida-induced AIM2-dependent caspase-1 activation, IL-1β and IL-18 release and cell death, whereas wild-type BMDMs exhibited robust AIM2 inflammasome responses (Fig. 2a–d). In contrast, Irf1−/− BMDMs responded normally to transfected dsDNA ligands, poly(dA:dT) and pcDNA3.1 DNA, and induced AIM2-dependent caspase-1 maturation, IL-18 release and cell death (Fig. 2a–f). Consistently, Irf1−/− BMDMs had an impaired ability to generate an inflammasome speck in response to F. novicida infection, but not in response to transfected poly(dA:dT) (Supplementary Fig. 1d,e). Analysis of Aim2 and Il1β mRNA transcripts using qRT-PCR revealed comparable expression of these genes in wild-type and Irf1−/− BMDMs infected with F. novicida (Supplementary Fig. 2a). The levels of the pro-inflammatory cytokines TNF-α and IL-6 were slightly reduced in Irf1−/− BMDMs (P>0.05), whereas the level of KC was comparable between wild-type and Irf1−/− BMDMs (Supplementary Fig. 2b). Accordingly, the level of IL-12 was lower in Irf1−/− BMDMs compared to wild-type BMDMs infected with F. novicida (Supplementary Fig. 2c).
Figure 2. IRF1 is essential for AIM2 inflammasome activation by F. novicida infection.
(a,b) Caspase-1 activation, IL-1β and IL-18 release in unprimed BMDMs infected with F. novicida (MOI 100) for 20 h or transfected with poly(dA:dT) for 5 h. (c,d) Cell death in unprimed BMDMs infected with F. novicida for 20 h, transfected with poly(dA:dT) for 5 h or infected with mCMV (MOI 10) for 10 h. Arrowheads indicate dead cells. (e,f) Caspase-1 activation, IL-18 release and cell death in unprimed BMDMs transfected with pcDNA for 5 h. (g,h) Caspase-1 activation and IL-1β and IL-18 release in unprimed BMDMs infected with mCMV (MOI 10) for 10 h. (i) Caspase-1 activation in BMDMs infected with F. novicida with or without co-stimulation with recombinant mouse IFN-β (25, 250 and 500 U/ml). Graphs show mean and s.e.m. of two (g,h) or three (a–f, i) independent experiments. *P < 0.01; **P < 0.001; ***P < 0.0001; NS, not significant (two-tailed t-test).
We next investigated whether IRF1 is required for AIM2 inflammasome activation in response to the DNA virus mouse cytomegalovirus (mCMV) 9. Intriguingly, engagement of the AIM2 inflammasome using mCMV did not require IRF1 (Fig. 2c,d,g,h), indicating that detection of bacteria and viruses by AIM2 occurs via distinct pathways governed by the requirement for IRF1.
IRF1 was identified as a transactivator of IFN-β 23, suggesting that defective IFN-β production may lead to impaired AIM2 inflammasome activation. Although we detected reduced Ifnβ gene expression in Irf1−/− BMDMs infected with F. novicida (Supplementary Fig. 2d), priming Irf1−/− BMDMs with recombinant IFN-β failed to rescue AIM2 inflammasome activation by F. novicida, whereas IFN-β stimulation dose-dependently increased AIM2-dependent caspase-1 maturation in wild-type cells (Fig. 2i). These findings suggest that defective AIM2 inflammasome activation in Irf1−/− BMDMs is not solely due to reduced IFN-β production, but largely reflects defects in additional IRF1-mediated processes.
IRF1 is dispensable for the NLRP3 and NLRC4 inflammasomes
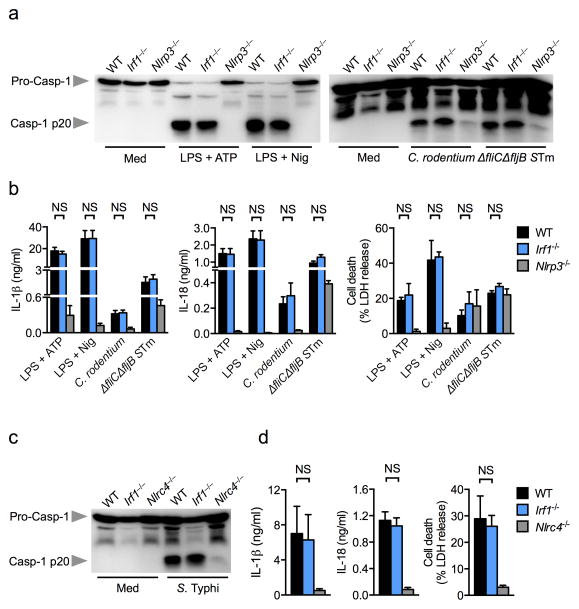
To investigate whether IRF1 is required for activation of additional inflammasomes, we stimulated unprimed wild-type and Irf1−/− BMDMs with the canonical NLRP3 activators ATP and nigericin, and the non-canonical NLRP3 inflammasome activators Citrobacter rodentium and Salmonella enterica serovar Typhimurium (S. Typhimurium) lacking the flagellin subunits fliC and fljB 17. We found that Irf1−/− BMDMs displayed normal caspase-1 activation, IL-1β and IL-18 release and cell death in response to canonical or non-canonical NLRP3 inflammasome activators (Fig. 3a,b). In addition, wild-type and Irf1−/− BMDMs infected with the log-phase grown S. Typhimurium which activates the NLRC4 inflammasome via a SPI-1 Type III secretion system 24, showed comparable levels of caspase-1 activation, IL-1β and IL-18 release and cell death (Fig. 3c,d), indicating that IRF1 does not interfere with activation of the NLRC4 inflammasome.
Figure 3. IRF1 is not required for canonical or non-canonical NLRP3 or NLRC4 inflammasome activation.
(a) Caspase-1 activation, (b) IL-1β and IL-18 release and cell death in LPS-primed BMDMs stimulated with ATP or nigericin or in unprimed BMDMs infected with Citrobacter rodentium (MOI 20) or ΔfliCΔfljB mutant S. Typhimurium (MOI 20) or wild-type S. Typhimurium (STm) for 20 h. (c) Caspase-1 activation, (d) IL-1β and IL-18 release and cell death in unprimed BMDMs infected with log-phase grown S. Typhimurium (MOI 1) for 4 h. Graphs show mean and s.e.m. of three independent experiments. NS, not significant (two-tailed t-test).
IRF1 upregulates GBPs for AIM2 inflammasome activation
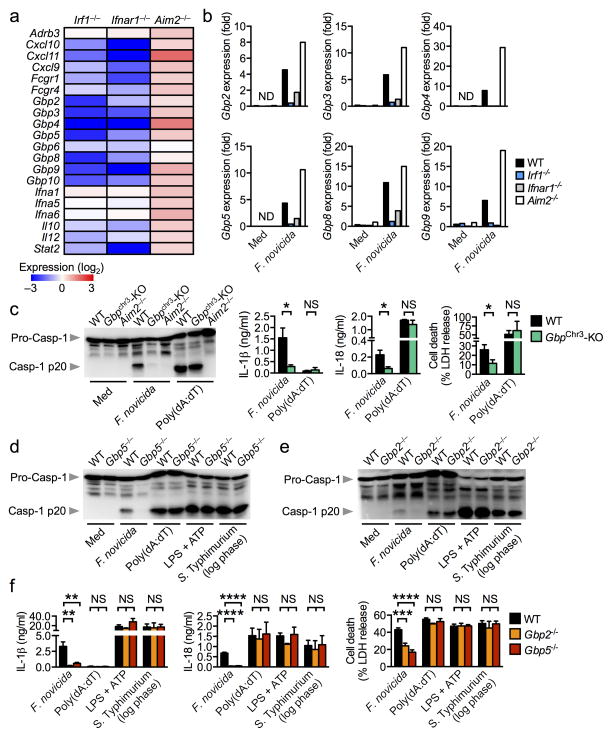
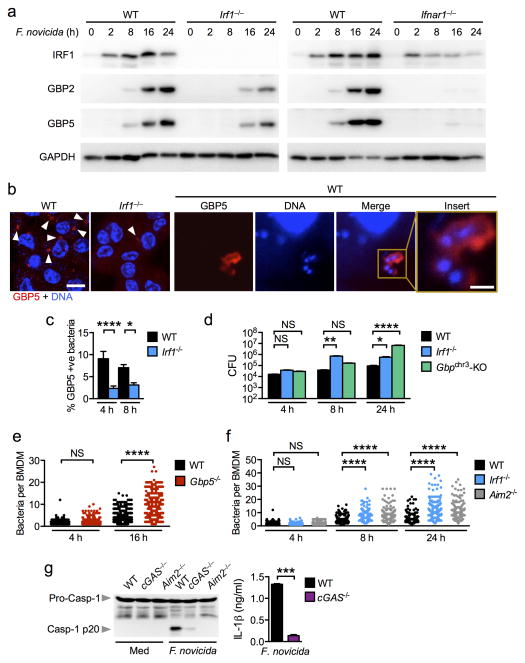
We further investigated the IRF1 signature that leads to AIM2 inflammasome activation by F. novicida. To this end, we performed a microarray analysis to identify differentially expressed genes in F. novicida-infected wild-type, Irf1−/−, Ifnar1−/− and Aim2−/− BMDMs. Analysis of the relative expression of macrophage-mediated immunity genes revealed that the gene set showing reduced expression in both Irf1−/− and Ifnar1−/− BMDMs was dominated by small interferon-inducible GTPases known as guanylate-binding proteins (GBPs), including Gbp2, Gbp3, Gbp4, Gbp5, Gbp6, Gbp8, Gbp9 and Gbp10 (Fig. 4a). Aim2−/− BMDMs, in contrast, showed elevated expression of GBPs and other interferon-inducible genes, an observation supported by the negative regulatory function of AIM2 on interferon responses (Fig. 4a,b) 8, 9. Downregulation of GBPs in the absence of IRF1 and IFNAR1 was further confirmed by real-time qRT-PCR analysis (Fig. 4b).
Figure 4. IRF1 controls expression of GBPs for AIM2 inflammasome activation.
(a) Heat map of microarray data showing relative expression of macrophage-mediated immunity genes found significantly enriched in unprimed Irf1−/−, Ifnar1−/− or Aim2−/− BMDMs compared to unprimed WT BMDMs after 8 h of infection with F. novicida. (b) Gene expression of GBPs relative to β-actin in BMDMs infected with F. novicida for 8 h by real time qRT-PCR. (c–f) Caspase-1 activation, IL-1β and IL-18 release and cell death in unprimed WT, Gbpchr3-deleted (Gbpchr3-KO) or Aim2−/− BMDMs infected with F. novicida (MOI 100, 20 h), log-phase grown S. Typhimurium (MOI 1, 4 h), or transfected with poly(dA:dT) or stimulated with LPS+ATP. Microarray analysis was performed using duplicate samples of each genotype (a). Graphs show mean and s.e.m. of three (b–f) independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant. Two-tailed t-test (c) and One-way ANOVA with a Dunnett’s multiple comparisons test (f).
Genes encoding GBPs are located on mouse chromosomes 3 and 5 25. Those located on chromosome 3, namely Gbp1, 2, 3, 5 and 7, have been implicated in the control of bacterial and parasitic replication 19, 25. To investigate the role of these GBPs in AIM2 inflammasome activation, BMDMs from mice lacking the GBP locus on chromosome 3 (Gbpchr3)19, 25 were infected with F. novicida. We observed significantly decreased levels of caspase-1 activation, IL-1β and IL-18 release and cell death induction in Gbpchr3-deleted BMDMs relative to infected wild-type BMDMs (Fig. 4c). Gbpchr3-deleted BMDMs, however, displayed normal levels of caspase-1 activation, IL-1β and IL-18 release and cell death induction in response to transfected poly(dA:dT) (Fig. 4c) 19.
Of the GBPs located within chromosome 3, IRF1 controlled upregulation of Gbp2, Gbp3 and Gbp5 expression (Fig. 4a,b). Gbp1 was not identified in our microarray dataset, which is consistent with previous studies showing that Gbp1 is not expressed in the C57BL/6 mouse genetic background (Fig. 4a) 26. Given the importance of IRF1 for AIM2 inflammasome activation, we set out to identify which of the remaining GBPs was responsible for this activity. To this end, we individually knocked down the remaining Gbp found on the chromosome 3 locus (Gbp2, Gbp3, Gbp5 and Gbp7) in primary wild-type BMDMs using an siRNA approach and found that silencing of Gbp2 or Gbp5 led to significant reduction in caspase-1 activation and IL-1β secretion in response to F. novicida infection (Supplementary Fig. 3a–c).
To confirm the role for GBP2 and GBP5 in the activation of the AIM2 inflammasome by F. novicida infection, we stimulated BMDMs from mice lacking GBP2 or GBP5 with F. novicida, poly(dA:dT), LPS+ATP (NLRP3 inflammasome) or log-phase grown S. Typhimurium (NLRC4 inflammasome). Both Gbp2−/− and Gbp5−/− BMDMs showed significantly reduced caspase-1 activation, IL-1β and IL-18 secretion and cell death following F. novicida infection, thus confirming a role for both GBPs in the activation of the AIM2 inflammasome (Fig. 4d–f). In contrast, stimulation of wild-type, Gbp2−/− and Gbp5−/− BMDMs with poly(dA:dT), LPS+ATP or log-phase grown S. Typhimurium resulted in comparable levels of caspase-1 activation, IL-1β and IL-18 release and cell death (Fig. 4d–f) 19. These results indicate that GBP2 and GBP5 play a non-redundant role in the activation of the AIM2 inflammasome in response to F. novicida infection.
GBPs mediate F. novicida killing to induce AIM2 activation
To investigate whether GBP2 and GBP5 expression are entirely under the control of IRF1, we infected wild-type and Irf1−/− BMDMs with F. novicida and monitored the dynamics of GBP2 and GBP5 expression over the course the infection. We found that the levels of GBP2 and GBP5 expression succeeded IRF1 expression and were reduced in the absence of IRF1 (Fig. 5a). However, the residue expression of GBP2 and GBP5 in Irf1−/− BMDMs indicates that an alternative IRF1-independent pathway exists to induce the expression of these GBPs. It is possible that type I interferon signaling may trigger an alternative pathway leading to GBP expression independently of IRF1. To this end, we infected wild-type and Ifnar1−/− BMDMs with F. novicida and found that a lack of type I interferon signaling altogether abolished GBP2 and GBP5 expression (Fig. 5a). Taken together, these findings provide evidence to demonstrate that two different members of the GBP family, GBP2 and GBP5, are under the control of IRF1 and type I interferon signaling, and are expressed to specifically engage the AIM2 inflammasome.
Figure 5. GBPs target bacteria to mediate killing during F. novicida infection.
(a) Induction of IRF1, GBP2 and GBP5 expression in unprimed BMDMs infected with F. novicida (MOI 50). (b) Immunofluorescence staining of GBP5 in unprimed BMDMs infected with F. novicida for 16 h. Scale bar, 10 μm; insert scale bar, 1 μm. (c) The percentage of bacteria colocalized with GBP5 was quantified. n=3,575 bacteria in WT BMDMs and n=3,620 bacteria in Irf1−/− BMDMs. (d) CFU recovered from WT, Irf1−/− and Gbpchr3-deleted BMDMs infected with F. novicida (MOI 100). (e) Quantification of the number of F. novicida bacteria per BMDMs using confocal microscopy. n=758 WT BMDMs and n=679 Gbp5−/− BMDMs. (f) Quantification of the number of F. novicida bacteria per BMDMs using confocal microscopy. n=715 WT BMDMs, n=710 Irf1−/− BMDMs, n=706 Aim2−/− BMDMs. (g) Caspase-1 activation and IL-1β release in unprimed BMDMs infected with F. novicida (MOI 100) for 20 h. Data from one experiment representative of two (d–f) or from three (a–c,g) independent experiments. *P < 0.05; **P < 0.005; ***P < 0.001; ****P < 0.0001; NS, not significant. Two-tailed t-test (c,g) and One-way ANOVA with a Dunnett’s multiple comparisons test (d) or with a Tukey’s multiple comparisons test (e,f).
GBPs can target vacuolar bacteria, such as Salmonella, and induce recruitment of antimicrobial peptides for bacterial killing 19, 27. Whether GBPs mediate killing of cytosolic bacteria such as F. novicida has not been investigated. We therefore used confocal microscopy to observe the spatial distribution of GBP5 relative to F. novicida in infected BMDMs. GBP5 was recruited to and engulfed F. novicida bacteria in wild-type BMDMs (Fig. 5b and Supplementary Fig. 4). The absence of IRF1 led to a significantly reduced prevalence of GBP5-associated bacteria (Fig. 5c). BMDMs lacking IRF1 or Gbpchr3 also failed to control bacterial replication over time (Fig. 5d).
To examine whether GBP5 directly affected bacterial viability in macrophages, we infected wild-type and Gbp5−/− BMDMs with GFP-expressing F. novicida and determined the number of bacteria in these cells over time. Single cell analysis revealed that wild-type BMDMs restricted bacterial replication from 4 to 16 h post-infection, whereas Gbp5−/− BMDMs failed to control bacterial replication at 16 h and harbored a significantly higher number of bacteria compared to the levels observed in wild-type BMDMs (Fig. 5e and Supplementary Fig. 4). In addition, GBP5-associated bacteria tend to lose their GFP expression compared to non-GBP5-associated bacteria, which may suggest a loss of bacterial viability following GBP5 recruitment (Supplementary Fig. 4) 19. To investigate whether IRF1 mediates AIM2-dependent killing of intracellular bacteria, we infected unprimed wild-type, Irf1−/− and Aim2−/− BMDMs with GFP-expressing F. novicida and quantified the number of bacteria in these cells over time. In agreement, Irf1−/− and Aim2−/− BMDMs failed to suppress bacterial replication over 24 h of infection (Fig. 5f).
For AIM2 to detect dsDNA, F. novicida or its DNA must be able to escape the vacuole and enter the cytoplasm. Indeed, co-staining of the inflammasome and DNA in F. novicida-infected BMDMs revealed co-localization of DNA with the inflammasome speck (Supplementary Fig. 5a). To investigate whether vacuolar escape by F. novicida engages IRF1- and GBP-dependent AIM2 inflammasome activation, we infected wild-type BMDMs with wild-type F. novicida or an isogenic mutant F. novicida ΔmglA which fails to disrupt the vacuole required for escape into the cytoplasm 28. F. novicida ΔmglA retained the ability to induce IRF1, GBP2 and GBP5 expression, although at a lower capacity relative to wild-type F. novicida (Supplementary Fig. 5b). However, F. novicida ΔmglA failed to activate the AIM2 inflammasome to trigger IL-1β and IL-18 release and cell death (Supplementary Fig. 5c,d). Production of other pro-inflammatory cytokines, TNF-α, IL-6 and KC, were comparable to wild-type F. novicida (Supplementary Fig. 5e), demonstrating that the inability of F. novicida ΔmglA to activate the AIM2 inflammasome was not due to a general defect in its ability to stimulate cytokines in macrophages. These results suggest that cytosolic escape is essential for AIM2 inflammasome activation, and that cytosolic escape enhances, but is not indispensable, for the induction of IRF1-mediated GBP expression. It is possible that additional processes complementing GBP expression are required to fully engage the AIM2 inflammasome. Indeed, deficiency in IRF1 does not completely abolish GBP2 and GBP5 expression (Fig. 5a), which suggests that IRF1 may direct additional processes other than driving GBP expression to activate the AIM2 inflammasome.
We next investigated whether the absence of IRF1 prevents cytosolic escape of F. novicida, which may be an additional mechanism governing IRF1-dependent activation of the AIM2 inflammasome. Cytosolic pathogens, such as F. novicida, escape the vacuole and can be used as a mechanism to deliver ligands into the cytosol of a cell 18. We took advantage of this cytosolic delivery method and infected wild-type, Irf1−/− and Aim2−/− BMDMs with F. novicida in the presence of ultrapure Salmonella LPS to examine whether Salmonella LPS can be efficiently introduced into the cytosol to activate the inflammasome in the absence of IRF1. We found that wild-type, Irf1−/− and Aim2−/− BMDMs all induced caspase-1 activation in response to F. novicida infection in the presence of Salmonella LPS, indicating that LPS was delivered into the cytosol, which likely activated the non-canonical NLRP3 inflammasome (Supplementary Fig. 6a). These findings suggest that in the absence of IRF1 cytosolic escape was not fully compromised at 20 h post-infection.
cGAS and STING contribute to AIM2 inflammasome activation
Although the downstream signaling pathway of type I interferon which engages the AIM2 inflammasome requires IRF1, the upstream pathway leading to type I interferon signaling in response to F. novicida infection is largely unexplored. A major pathway that mediates recognition of cytosolic bacteria to generate IFN-β production is the cGAS and STING signaling axis 22, 29. In agreement, both Mb21d1−/− (the gene encoding cGAS and hereafter written as cGas−/−) and Tmem173gt which harbors a nonfunctional Gt allele in Tmem173 (the gene encoding STING) that renders a lack of detectable protein (hereafter referred to as StingGt/Gt)29 BMDMs had an impaired ability to robustly activate caspase-1, induce IL-1β and IL-18 or cell death in response to F. novicida infection (Fig. 5g and Supplementary Fig. 6b,c). In addition, cGAS−/− BMDMs failed to robustly induce IRF1 expression (Supplementary Fig. 6d). The residue inflammasome responses in cGAS−/− and StingGt/Gt BMDMs infected with F. novicida could be due to a minor source of type I interferon induced by TLRs, RLRs or other PRRs. Collectively, these findings indicate that the cGAS signaling axis induces type I interferon-dependent IRF1 expression necessary to engage GBP-mediated killing of F. novicida, which ultimately results in the release of bacterial DNA to activate the AIM2 inflammasome (Supplementary Fig. 6e).
IRF1 provides protection against F. novicida infection
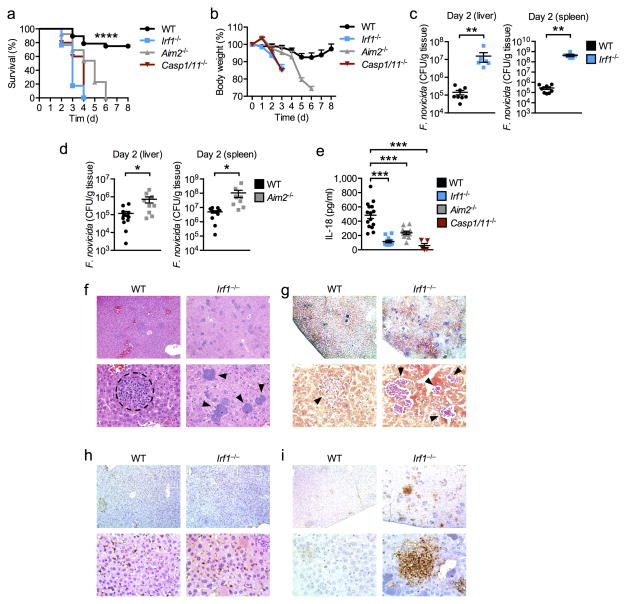
We extended our findings to an in vivo setting and infected wild-type, Irf1−/−, Aim2−/− and Casp1 x Casp11 double-deficient mice (hereafter Casp1−/−/11−/−) mice with F. novicida and monitored their susceptibility to infection. Mice lacking IRF1, AIM2 or caspase-1/11 lost more body weight and all succumbed to infection within 6 days, whereas 75% of the wild-type mice survived beyond day 6 (P<0.0001; Fig. 6a,b). Analysis of bacterial burdens showed that mice lacking IRF1 harbored significantly increased F. novicida colony-forming units (CFU) in the liver and spleen compared to wild-type mice (P<0.001; Fig. 6c). Similarly, Aim2−/− mice were significantly more susceptible to F. novicida infection than wild-type mice (P<0.05; Fig. 6d) 8, 22. The increased susceptibility of Irf1−/− mice to F. novicida infection compared to Aim2−/− mice may suggest that IRF1 is important in multiple antimicrobial defense mechanisms. Analysis of serum IL-18 showed that mice lacking IRF1 had an impaired ability to produce this cytokine following F. novicida infection, an observation phenocopied by mice lacking AIM2 or caspase-1/11 (Fig. 6e). Histopathology analysis of liver tissues showed that Irf1−/− mice failed to control F. novicida dissemination (Fig. 6f,g). In contrast, granulomas that surround infectious particles were found in livers of wild-type mice (Fig. 6g). In addition, mice lacking IRF1 showed increased cell death but normal levels of granulocyte recruitment in the liver, as revealed by terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling (TUNEL) and myeloperoxidase (MPO) staining, respectively (Fig. 6h,i). The increased prevalence of TUNEL staining in Irf1−/− mice is likely the result of increased damage from increased bacterial burden, which leads to different cell types undergoing different types of cell death in the liver. Taken together, these results highlight an important role for IRF1 in the host defense against F. novicida infection by engaging the AIM2 inflammasome.
Figure 6. IRF1 provides host protection against F. novicida infection in vivo.
(a) % Survival of 8 week-old WT (n=28), Irf1−/− (n=17), Aim2−/− (n=13) and Casp1 x Casp11 double-deficient (Casp1/11−/−) (n=5) mice subcutaneously infected with 7.5×104 CFU of F. novicida. (b) Body weight change of 8 week-old WT (n=16), Irf1−/− (n=9), Aim2−/− (n=9) and Casp1/11−/− (n=5) subcutaneously infected with 7.5×104 CFU of F. novicida. (c) Bacterial burden in the liver and spleen of 8 week-old male WT (n=8) and Irf1−/− (n=6) mice infected with 1×105 CFU of F. novicida. (d) Bacterial burden in the liver and spleen of 8-week old female WT (n=12) and Aim2−/− (n=9) mice infected with 1×105 CFU of F. novicida. (e) Concentrations of IL-18 in the serum of WT (n=15), Irf1−/− (n=11), Aim2−/− (n=11), and Casp1/11−/− (n=5) mice infected with 1×105 CFU of F. novicida for 24 h. (f) H&E and (g) Gram staining of F. novicida-infected livers collected on day 3. (h) Myeloperoxidase (MPO) and (i) TUNEL staining of F. novicida-infected livers collected on day 3. Top panels, 10× magnification; bottom panels 40× magnification. The dashed circle indicates a granuloma. Arrowheads indicate bacterial colonies. Data are from one experiment representative of two independent experiments (b) or pooled from two independent experiments (a,c–e). Error bars indicate s.e.m. *P < 0.05; **P < 0.001; ***P < 0.0001. Log-rank test (a), Two-tailed t-test (c,d) and One-way ANOVA with a Dunnett’s multiple comparisons test (e).
DISCUSSION
Multiple pathways exist for the recognition of DNA to trigger innate immune responses in the cell. TLR9 recognizes CpG DNA in the endosomal compartment to activate transcription of pro-inflammatory cytokine genes 30 and STING responds directly to cyclic dinucleotides or serves as an adaptor for a range of DNA sensors to induce type I interferon responses 31. AIM2 mediates sensing of cytosolic dsDNA to initiate the assembly of the inflammasome. However, the precise mechanisms by which different pathogens trigger AIM2 have remained unclear 3. In addition, the functional role for type I interferon signaling in the activation of the AIM2 inflammasome is enigmatic, given that type I interferon signaling is important for mediating AIM2 inflammasome activation in response to F. novicida infection, but not for mCMV infection 3.
Here, we have identified an IRF1-dependent pathway downstream of type I interferon signaling that is responsible for activation of the AIM2 inflammasome by F. novicida. The existence of this ‘non-canonical’ AIM2 inflammasome pathway defined by its requirement for IRF1, GBPs and type I interferon signaling is analogous to the non-canonical NLRP3 inflammasome and its requirement for type I interferon signaling and caspase-11 12, 13, 15, 17. IRF1 was first shown as a virus-induced transcription factor which interacts with human IFN-β regulatory DNA elements and contributes to the transcription of the Ifnβ gene 32. Later studies revealed that both human and mouse IRF1 are involved in the expression of IFN-α, MHC class I, MX (mouse), 2′ 5′ oligo A synthetase (mouse), and ISG54 (human) 23. IRF1 is not absolutely required for the production of IFN-β and certain subtypes of IFN-α in F. novicida-infected macrophages. Instead, we found cGAS and STING are the likely upstream sensors that initiate type I interferon production.
Notably, IRF1 and type I interferon induce GBPs to kill and lyse cytosolic bacteria to drive bacterial DNA release for access by AIM2. Although our microarray analysis suggests that IRF1 and IFNAR induce a similar expression profile of ISGs, protein expression analysis revealed that GBP2 and GBP5 expression are completely abolished in the absence of IFNAR1, whereas some residue expression of these proteins is observed in the absence of IRF1. This suggests that IRF1 and IFNAR1 are likely to regulate their own set of genes in F. novicida-infected macrophages, some of which may be common.
Herpesviruses, including mCMV, enter the host cell via a mechanism that requires binding and fusion of the viral envelope with the host cell membrane, which results in the release of the viral capsid into the host cell. Under these circumstances, viral DNA may have access to the cytoplasm directly for detection by AIM2. We and others show that mutant F. novicida that fails to escape the vacuole cannot activate the AIM2 inflammasome 33. In addition, we show that phagosomal escape enhances IRF1 and GBP expression, suggesting that this process may provide an additional signal to enhance GBP-mediated killing of the bacteria in the cytosol. F. tularensis LVS escapes the vacuole as early as 1 h post-infection 34, suggesting that vacuolar escape precedes GBP expression which occurs mostly after 16–24 h of infection following type I interferon signaling. However, the precise method by which GBPs kill F. novicida is unknown. Previous studies have found that GBP1 or GBP7 can recruit additional effector proteins to mediate pathogen killing. GBP7 interacts with and recruits p22phox and other components of the NADPH oxidase to the mycobacteria-containing vacuole to mediate bacterial killing, whereas GBP1 facilitates delivery of ubiquitinylated cargo (e.g. bacteria decorated with ubiquitin) to autolysosomes to mediate killing 27. GBP1, GBP2 and GBP5 share a common feature in that they harbor a C-terminal CaaX motif, which is targeted by prenylation – a posttranslational modification process that results in the attachment of a lipid hydrophobic moiety to mediate docking of the protein to cellular membranes 35. It is possible that GBP2 and GBP5 could directly target cytosolic F. novicida to induce recruitment of antimicrobial process to mediate bacterial killing. Indeed, IRF1 controls the expression of inducible nitric oxide synthase (iNOS) 36. The contribution of iNOS and its connection with GBPs in the activation of the AIM2 inflammasome warrants further investigation.
We observed that macrophages lacking either IRF1 or AIM2 failed to restrict F. novicida replication, whereas mice lacking IRF1 are more susceptible to F. novicida infection compared to mice lacking AIM2. These results suggest that IRF1 controls multiple antimicrobial pathways to suppress bacterial burden in vivo, which ultimately leads to lysis of the bacterial pathogen for AIM2 inflammasome activation. IRF1 has been linked to IL-12 production. However, previous studies have shown that mice lacking IL-12 p40 or IL-12 p30 or mice treated with anti-IL-12 antibodies infected with F. tularensis LVS have similar bacterial burden compared to wild-type mice 3 days post-infection 37. In our study, mice lacking IRF1 harbor a significantly higher level of bacteria compared to wild-type mice on day 2, and by day 3 more than 80% of the mice lacking IRF1 succumbed to infection. Taken together, we speculate that IL-12 is unlikely to account for the major difference in bacterial burden between wild-type and Irf1−/− mice.
Additional mechanisms have been identified to control the assembly and degradation of the AIM2 inflammasome. Polyubiquitination of ASC recruits the autophagic marker, p62, to target the AIM2 inflammasome for autophagic degradation 38. The human pyrin-containing protein, POP3, interact with the pyrin domain of AIM2 and competes with ASC to inhibit AIM2 inflammasome activation in response to poly(dA:dT) and mCMV 39. The existence of these pathways indicates that controlling AIM2 inflammasome activation is important for maintaining homeostasis of a cell. Indeed, aberrant AIM2 inflammasome activation in response to dsDNA could lead to important physiological consequences. Increased AIM2 expression is associated with the development of psoriasis, abdominal aortic aneurysm and systemic lupus erythematosus, whereas reduced AIM2 expression is linked to colorectal and prostate cancer 40. Our findings uncovered a new layer of regulation that governs cytoplasmic DNA sensing upon infection by intracellular pathogens. Therapies that modulate IRF1 activities could lead to enhanced protection against rapid and lethal Francisella tularensis infection.
ONLINE METHODS
Mice
Irf1−/−,41 Irf7−/−,42 Irf9−/− (also known as p48),43 Ifnar1−/−,44 Ifnar2−/−,44 Stat1−/−,45 Aim2−/−,22 Nlrp3−/−,46 Nlrc4−/−,47 Casp1−/− (Casp11−/−),17 Casp11−/−,17 Gbpchr3-deleted,25 Gbp5−/−,19 Trif−/−48 and StingGt/Gt 29 mice have been described previously. Mice were bred at the St. Jude Children’s Research Hospital. Animal studies were conducted under protocols approved by the St. Jude Children’s Research Hospital on the Use and Care of Animals.
Cell culture and stimulation
Primary bone marrow–derived macrophages (BMDMs) were grown for 5–6 days in IMDM (Gibco) supplemented with 1% non-essential amino acids (Gibco), 10% FBS (Atlanta Biologicals), 30% L929 conditioned media and 1% penicillin and streptomycin (Sigma). BMDMs were seeded in antibiotic–free media at a concentration of 1 × 106 cells onto 12-well plates and incubated overnight. Francisella novicida strain U112 or its isogenic mutant F. novicida ΔmglA were grown in BBL™ Trypticase™ Soy Broth (TSB) (211768, BD) supplemented with 0.2% L-cysteine (Fisher) overnight under aerobic conditions at 37°C. Bacteria were subcultured (1:10) in fresh TSB supplemented with 0.2% L-cysteine for 4 h and resuspended in PBS. S. Typhimurium SL1344, an isogenic mutant lacking fliC and fljB (ΔfliCΔfljB STm) and Citrobacter rodentium (ATCC 51459) were inoculated into LB broth and incubated overnight under aerobic conditions at 37°C. S. Typhimurium SL1344 was subcultured (1:10) into fresh LB broth for 3 h at 37°C to generate log-phase grown bacteria. The following conditions were used: F. novicida or F. novicida ΔmglA (MOI 100 and 20 h for caspase-1 activation; MOI 50 for 2, 8, 16 and 24 h for IRF1 or GBP expression), S. Typhimurium (MOI 1, 4 h), ΔfliCΔfljB STm (MOI 20 for 20 h) and C. rodentium (MOI 20 for 20 h). Gentamicin (50 μg/ml, Gibco) was added after 2 h (S. Typhimurium), 4 h (C. rodentium and ΔfliCΔfljB STm), and 8 h (F. novicida) post-infection to kill extracellular bacteria. The mouse CMV Smith MSGV strain (ATCC® VR-1399™) was obtained from P.G. Thomas (St. Jude). Virus was added to unprimed BMDMs at an MOI of 10 for 10 h to activate the AIM2 inflammasome.
To activate the canonical NLRP3 inflammasome, BMDMs were primed using 500 ng/ml ultrapure LPS from Salmonella minnesota R595 (InvivoGen) for 4 h and stimulated with 5 mM ATP or 10 μM nigericin (Sigma) for 45 min. For DNA transfection, each reaction consisted of 2.5 μg of poly(dA:dT) (InvivoGen) resuspended in PBS and mixed with 0.6 μl of Xfect polymer in Xfect reaction buffer (Clontech Laboratories, Inc.). After 10 min, DNA complexes were added to BMDMs in Opti-MEM (Gibco) and incubated for 5 h.
For CFU analysis, supernatant from BMDMs infected with F. novicida for 3 or 7 h was replaced with media containing 50 μg/ml gentamicin (Gibco). Cells were incubated for an additional 1 h, washed twice with PBS and scraped before plating onto TSB agar. For 24 h infection, media containing 50 μg/ml gentamicin (1 h incubation) was replaced with 10 μg/ml gentamicin and cells were further incubated for 16 h.
Levels of lactate dehydrogenase released by cells were determined using the CytoTox 96 Non-Radioactive Cytotoxicity Assay according to the manufacturer’s instructions (Promega). Cell culture supernatants were collected for ELISA.
siRNAs knockdown
BMDMs were transfected with siRNA from siGENOME smart pools (Dharmacon) for 48 h using GenMute siRNA Transfection Reagent and the according to the manufacturer’s instructions (Fisher Scientific). The siGENOME SMARTpool siRNA to mouse Gbp2 (M-040199-00-0005), Gbp3 (M-063076-01-0005), Gbp5 (M-054703-01-0005), Gbp7 (M-061204-01-0005) were used in the study. A control siRNA pool was used. Transfected cells were infected with F. novicida as described above. See Supplementary Table 1 for sequences.
Immunoblotting analysis
For caspase-1 immunoblotting, BMDMs and supernatant were lysed in RIPA buffer and sample loading buffer containing SDS and 100 mM DTT. For IRF1 or GBP signaling immunoblotting, supernatant was removed and BMDMs washed once with PBS, followed by lysis in RIPA buffer and sample loading buffer containing SDS and 100 mM DTT. Proteins were separated on 8–12% polyacrylamide gels. Following electrophoretic transfer of protein onto PVDF membranes, membranes were blocked in 5% skim milk and incubated with primary antibodies against caspase-1 (clone Casper-1, 1:3,000 dilution, AG-20B-0042, Adipogen), IRF1 (clone D5E4, 1:1,000 dilution, #8478, Cell Signaling Technologies), GBP2 (1:1,000 dilution, 11854-1-AP, Proteintech), GBP5 (1:1,000 dilution, 13220-1-AP, Proteintech) or GAPDH (clone D16H11, 1:10,000 dilution, #5174, Cell Signaling Technologies) followed by secondary anti-rabbit or anti-mouse HRP antibodies (1:1,000 dilution; Jackson Immuno Research Laboratories).
Immunofluorescence staining
F. novicida strain U112 or a strain expressing GFP was used for infection. For visualization of inflammasomes, BMDMs were infected for 20 h and further incubated in fresh media containing 1× FAM FLICA active caspase-1 for 1 h at 37°C (ImmunoChemistry Technologies). Cells were washed 3 times with PBS and fixed in 4% paraformaldehyde for 15 min at room temperature, followed by blocking in 10% normal goat serum (Dako) supplemented with 0.1% saponin (Sigma) for 1 h. Cells were incubated with a rabbit anti-ASC antibody (clone AL177, 1:500 dilution, AG-25B-0006-C100, AdipoGen) for 50 min at 37°C. For GBP5 staining, BMDMs were infected for the indicated times and washed 3 times with PBS. Cells were fixed and blocked as described above and stained using a rabbit anti-GBP5 antibody (1:500 dilution, 13220-1-AP, Proteintech) overnight at 4°C. The secondary antibody used was an Alexa Fluor 568 anti-rabbit IgG (Life technologies). Cells were counterstained in DAPI mounting medium (Vecta Labs). Bacteria, inflammasomes and BMDMs were visualized, counted, and imaged using a Nikon C2 confocal microscope.
Real time qRT-PCR analysis
RNA was extracted using TRIzol according to the manufacturer’s instructions (Life Technologies). Isolated RNA was reverse transcribed into cDNA using the First-Strand cDNA Synthesis Kit (Life Technologies). Real-time qPCR was performed on an ABI 7500 real-time PCR instrument with 2× SYBR Green (Applied Biosystems). See Supplementary Table 2 for sequences.
Cytokine analysis
Cytokine levels were determined using multiplex ELISA (Millipore) or IL-18 ELISA (MBL international) according to the manufacturers’ instructions.
Microarray
Transcript profiling was performed using two biological replicate samples of unprimed BMDMs obtained from WT and knockout mice. Total RNA (100 ng) was converted to biotin-labeled cRNA using the Ambion WT expression kit (Life Technologies) and hybridized to a Mouse Gene 2.0 ST GeneChip (Affymetrix, Inc). After staining and washing, array signals were normalized and transformed into log2 transcript expression values using the Robust Multi-array Average algorithm (Partek Genomics Suite v6.6) 49. Differential expression was defined by applying a 0.5 log2(signal) difference between conditions. Lists of differentially expressed transcripts were analyzed for functional enrichment using the DAVID bioinformatics databases (http://david.abcc.ncifcrf.gov/)50 and Ingenuity Pathways Analysis software (www.qiagen.com/ingenuity).
Animal infection
Francisella novicida strain U112 was grown as described above. For survival and weight change analyses, mice were injected subcutaneously with 7.5×104 colony-forming units (CFU) of F. novicida in 200 μl PBS. For CFU analysis, mice were injected subcutaneously with 1×105 CFU of F. novicida in 200 μl PBS. After 2 days, liver and spleen were harvested and homogenized in PBS with metal beads for 2 min using the TissueLyser II apparatus (Qiagen). CFU were determined by plating lysates onto TSB agar supplemented with 0.2% L-cysteine and incubated overnight. No randomization or blinding was performed.
Histopathology
Formalin-preserved livers were processed and embedded in paraffin according to standard procedures. Sections (5 μm) were stained with hematoxylin and eosin (H&E) and examined by a pathologist blinded to the experimental groups. For immunohistochemistry, formalin-fixed paraffin-embedded livers were cut into 4 μm sections. To stain for granulocytes, we used an anti-MPO antibody (1:500 dilution for 30 min, A0398, Dako) followed by rabbit on rodent polymer-HRP (RMR622L, BioCare Medical) for 30 min. TUNEL staining was performed using the Dead End kit (#PRG7130) according to the manufacturer’s instructions (Promega).
Statistical analysis
GraphPad Prism 6.0 software was used for data analysis. Data are shown as mean +/− s.e.m. Statistical significance was determined by t tests (two-tailed) for two groups or One-way ANOVA (with Dunnett’s or Tukey’s multiple comparisons tests) for three or more groups. Survival curves were compared using the log-rank test. P<0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank V. M. Dixit, N. Kayagaki (Genentech) and R. Flavell (Yale) for the mutant mouse strains. We thank K.A. Fitzgerald (University of Massachusetts Medical School), P. Broz (University of Basel) and K. Pfeffer (Heinrich-Heine-University Duesseldorf) for the cGAS−/−, Gbpchr3-deleted and Gbp2−/− femurs, respectively. We would like to acknowledge P.G. Thomas (St. Jude) for mCMV and X. Qi and M. Barr (St. Jude) for technical assistance. This work was supported by grants from the National Institutes of Health (grants AR056296, CA163507 and AI101935), the American Lebanese Syrian Associated Charities (to T.-D.K), European Research Council (grant 281600) and the Fund for Scientific Research-Flanders (grant G030212N) (to M.L.). S.M.M. is a recipient of the Neoma Boadway Endowed Fellowship funded by the St. Jude Children’s Research Hospital and the NHMRC R.G. Menzies Early Career Fellowship funded by the National Health & Medical Research Council, Australia.
Footnotes
Author Contributions
S.M.M., R.K. and T.-D.K. designed the study. S.M.M., M.L. and T.-D.K. wrote the manuscript. S.M.M., R.K., R.K.S.M., G.N., and P.V. performed the experiments and analyzed the data; M.Y. contributed reagents.
COMPETING INTERESTS STATEMENT
The authors declare no conflict of interest.
References
- 1.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481(7381):278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 2.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Rathinam VAK, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nature immunology. 2012;14(4):333–342. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature immunology. 2009;10(3):266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 5.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458(7237):509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458(7237):514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323(5917):1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 8.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nature immunology. 2010;11(5):385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature immunology. 2010;11(5):395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dombrowski Y, Peric M, Koglin S, Kammerbauer C, Goss C, Anz D, et al. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med. 2011;3(82):82ra38. doi: 10.1126/scitranslmed.3002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36(4):561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rathinam VA, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, et al. TRIF Licenses Caspase-11-Dependent NLRP3 Inflammasome Activation by Gram-Negative Bacteria. Cell. 2012;150(3):606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490(7419):288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474(7351):385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurung P, Malireddi RK, Anand PK, Demon D, Walle LV, Liu Z, et al. Toll or Interleukin-1 Receptor (TIR) Domain-containing Adaptor Inducing Interferon-beta (TRIF)-mediated Caspase-11 Protease Production Integrates Toll-like Receptor 4 (TLR4) Protein- and Nlrp3 Inflammasome-mediated Host Defense against Enteropathogens. The Journal of biological chemistry. 2012;287(41):34474–34483. doi: 10.1074/jbc.M112.401406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341(6151):1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 17.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479(7371):117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 18.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341(6151):1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509(7500):366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 20.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014 doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 21.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. The Journal of experimental medicine. 2007;204(5):987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O’Rourke K, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(21):9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, et al. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell. 1988;54(6):903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 24.Man SM, Hopkins LJ, Nugent E, Cox S, Gluck IM, Tourlomousis P, et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(20):7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, et al. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37(2):302–313. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Staeheli P, Prochazka M, Steigmeier PA, Haller O. Genetic control of interferon action: mouse strain distribution and inheritance of an induced protein with guanylate-binding property. Virology. 1984;137(1):135–142. doi: 10.1016/0042-6822(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 27.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332(6030):717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 28.Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cellular microbiology. 2005;7(7):969–979. doi: 10.1111/j.1462-5822.2005.00526.x. [DOI] [PubMed] [Google Scholar]
- 29.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infection and immunity. 2011;79(2):688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 31.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita T, Sakakibara J, Sudo Y, Miyamoto M, Kimura Y, Taniguchi T. Evidence for a nuclear factor(s), IRF-1, mediating induction and silencing properties to human IFN-beta gene regulatory elements. The EMBO journal. 1988;7(11):3397–3405. doi: 10.1002/j.1460-2075.1988.tb03213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. The Journal of experimental medicine. 2005;202(8):1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(39):14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Britzen-Laurent N, Bauer M, Berton V, Fischer N, Syguda A, Reipschlager S, et al. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PloS one. 2010;5(12):e14246. doi: 10.1371/journal.pone.0014246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamijo R, Harada H, Matsuyama T, Bosland M, Gerecitano J, Shapiro D, et al. Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science. 1994;263(5153):1612–1615. doi: 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- 37.Elkins KL, Cooper A, Colombini SM, Cowley SC, Kieffer TL. In vivo clearance of an intracellular bacterium, Francisella tularensis LVS, is dependent on the p40 subunit of interleukin-12 (IL-12) but not on IL-12 p70. Infection and immunity. 2002;70(4):1936–1948. doi: 10.1128/IAI.70.4.1936-1948.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nature immunology. 2012;13(3):255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khare S, Ratsimandresy RA, de Almeida L, Cuda CM, Rellick SL, Misharin AV, et al. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nature immunology. 2014;15(4):343–353. doi: 10.1038/ni.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connolly DJ, Bowie AG. The emerging role of human PYHIN proteins in innate immunity: Implications for health and disease. Biochemical pharmacology. 2014 doi: 10.1016/j.bcp.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 41.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75(1):83–97. [PubMed] [Google Scholar]
- 42.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 43.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, et al. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes to cells : devoted to molecular & cellular mechanisms. 1996;1(1):115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 44.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 45.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84 (3):443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 46.Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440(7081):233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 47.Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430(6996):213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301(5633):640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 49.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 50.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.