Abstract
Biallelic mutations in BLM cause Bloom syndrome (BS), a genome instability disorder characterized by growth retardation, sun sensitivity and a predisposition to cancer. As evidence of decreased genome stability, BS cells demonstrate not only elevated levels of spontaneous sister chromatid exchanges (SCE), but also exhibit chromosomal radial formation. The molecular nature and mechanism of radial formation is not known, but they have been thought to be DNA recombination intermediates between homologs that failed to resolve. However, we find that radials in BS cells occur over 95% between non-homologous chromosomes, and occur non-randomly throughout the genome. BLM must be phosphorylated at T99 and T122 for certain cell cycle checkpoints, but it is not known whether these modifications are necessary to suppress radial formation. We find that exogenous BLM constructs preventing phosphorylation at T99 and T122 are not able to suppress radial formation in BS cells, but are able to inhibit SCE formation. These findings indicate that BLM functions in two distinct pathways requiring different modifications. In one pathway—for which the phosphorylation marks appear dispensable—BLM functions to suppress SCE formation. In a second pathway, T99 and T122 phosphorylation are essential for suppression of chromosomal radial formation, both those formed spontaneously and those formed following interstrand crosslink (ICL) damage.
Introduction
The highly conserved human BLM tumor suppressor protein is a RecQ helicase that functions to maintain genome stability (Bachrati and Hickson 2008; Chu and Hickson 2009; Ellis et al. 1995). Biallelic mutations in BLM cause Bloom syndrome, which is characterized by short stature, sun-sensitive skin lesions primarily affecting the face, and predisposition to a wide spectrum of cancers. BLM is a multifunctional protein with distinct roles in somatic and germ cells where it is essential for regulating homologous recombination (HR) for DNA repair and during meiosis, where it localizes to programmed double strand breaks (DSB) to promote holliday junction resolution (Bugreev et al. 2007; German 1969; Hanada et al. 1997; Holloway et al. 2010; Rockmill et al. 2003; Watt et al. 1996). BLM regulates recombination by acting to ensure dissolution of crossover intermediates, promoting primarily non-crossover resolution (Chu and Hickson 2009; Schvarzstein et al. 2014). Thus, BS cells manifest genomic instability as increased spontaneous and damage-induced sister chromatid exchanges (SCE) (German et al. 1974; Ray and German 1984; Rosin and German 1985). SCE formation appears to reflect instances of HR-dependent DSB repair (Manthei and Keck 2013). Normally, double holliday junction intermediates are resolved via two pathways: BLM/TOP3A-mediated dissolution, which results in non-crossovers, or by HR resolvases, which result in crossovers. In the absence of BLM, repair is skewed towards the crossover pathway of resolution, which can be observed as SCEs at mitosis (Ip et al. 2008; Mankouri and Hickson 2007; Manthei and Keck 2013). More recently, BLM has been localized to ultrafine DNA bridges during anaphase, along with ERCC6L and members of the Fanconi anemia (FA) pathway, where it is proposed to have a role in bridge resolution, ensuring faithful chromosome segregation (Biebricher et al. 2013; Chan and Hickson 2011; Chan et al. 2007; Rouzeau et al. 2012).
BS cells manifest chromosomal radial formation following DNA damage—a sign of genome instability most commonly associated with defects in the FA pathway (German et al. 1974; Hemphill et al. 2009; Schroeder and German 1974). FA [reviewed in (Kee and D'Andrea 2012; Kim and D'Andrea 2012)], is a recessive disorder resulting from a defect in any of sixteen or more proteins acting to maintain genome stability during replication and during interstrand crosslink (ICL) damage repair (Thompson and Hinz 2009; Wang 2007). BLM participates in the FA pathway, interacting specifically with the FA helicase, FANCJ (Suhasini and Brosh 2012). Additionally, several members of the FA core complex associate with BLM in the BRAFT supercomplex (BLM, RPA, TOP3A, and associated proteins), as defined by co-immunoprecipitation (Deans and West 2009; Meetei et al. 2003; Wang 2007). The BLM-TOP3A complex, a conserved association, acts epistatically with the FA pathway during ICL repair, and is necessary for SCE suppression (Hemphill et al. 2009). The BLM-FA interaction appears to be required for phosphorylation of BLM (Naim and Rosselli 2009; Pichierri et al. 2004). Phosphorylation of BLM at T99 and T122 is required for normal checkpoint response to stalled replication forks, and BS cells arrest in G2/M if phosphorylation does not occur following replication stress (Davies et al. 2004). Thus, the functions of BLM and the FA pathway proteins are intricately intertwined, and are required for maintaining genome stability.
Although spontaneous chromosomal radials are a feature of both BS and FA cells, most research has focused on FA radials since they are the basis of diagnostic testing for the disorder (Kuhn 1978; Schroeder et al. 1964; Schroeder and German 1974). Both BS and FA cells show increased radials after exposure to ICL-inducing agents, such as mitomycin C (MMC) or diepoxybutane (DEB). Though spontaneous radials are unique to BS and FA cells, induced radials are not and can be found after ICL damage in cells depleted of proteins that function in DNA repair or maintenance of genome stability, including BRCA1 or BRCA2, non-homologous end joining (NHEJ) proteins Ku70 (now known as XRCC6) and LIG4, and HR proteins RAD51 and RAD52 (Hanlon Newell et al. 2008). However, unlike BS cells, FA cells generally do not exhibit elevated spontaneous SCEs (Gravells et al. 2013; Hemphill et al. 2009). Studies characterizing BS radials have previously classified them as homologous, or occurring between homologous chromosomes (German et al. 1974; Schroeder and German 1974). However, radials are non-homologous in FA cells and occur between various chromosome combinations (Newell et al. 2004). Thus, despite their similarity, BS and FA-derived radials have been documented as different types of chromosome abnormalities and have been thought to arise from different DNA repair defects.
The observation that BS cells show increased SCE and radial formation, while FA cells show only elevated radials, suggests a segregation of BLM activities with one function acting to suppress SCE formation and another to inhibit radial formation. BLM constructs mutated at T99 and T122 to preclude phosphorylation are reported to correct SCE formation to normal levels (Davies et al. 2004). Since BS cells lack functional BLM protein and FA cells are unable to phosphorylate BLM (Pichierri et al. 2004), yet both manifest radials, we questioned whether phosphorylated BLM is required to suppress radial formation after ICL damage.
We find that spontaneous and ICL-induced radials in BS cells are over 95% nonhomologous, appearing to be the same as in FA cells. To determine if phosphorylated BLM is required to suppress radial formation, we used an engineered mutant BLM that cannot be phosphorylated due to substitutions of alanines for native threonines at T99 and T122. We found that such constructs suppress SCEs in BS cells, but fail to suppress radial formation after ICL damage. In fact, exogenous, un-phosphorylatable BLM appears to have a dominant negative effect, increasing spontaneous and clastogen-induced radial formation in normal fibroblasts. These data suggest that BLM functions in two distinct pathways for maintenance of genome stability: one pathway suppresses radial formation and requires the phosphorylation of T99 and T122; the other pathway suppresses SCE formation, independent of those modifications.
Materials and methods
Cell culture and reagents
Primary BS fibroblasts (GM01492) (Coriell) were cultured in α-MEM (Gibco) supplemented with 20% FBS (Gibco), 4mM L-glutamine, and 50μg/ml gentamicin. Cells were grown at 37°C with 5% CO2 and maintained at subconfluency. Transformed fibroblasts were cultured in α-MEM medium (Mediatech) supplemented with 10% fetal bovine serum (Hyclone). Immortalized fibroblast cell lines GM00639 (normal), and GM08505 (BS), were obtained from the NIGMS Human Genetic Cell Repository (Coriell). The functionally complemented control for GM08505 was obtained by transfection with a pMMP vector containing the full-length coding sequence for BLM.
G-banding studies
The GM01492 primary fibroblasts were characterized for the karyotype, as well as for chromosome involvement in radial formation, both spontaneous and clastogen-induced. The treated cells were exposed to 200 ng/ml diepoxybutane (DEB) or 20 ng/ml mitomycin C (MMC) diluted in Hank's balanced salt solution for 48 hr at 37°C with 5% CO2 prior to harvest. Colcemid (0.05 μg/ml) was added overnight to arrest cells at metaphase. Cells were trypsinized, pelleted, and incubated in a hypotonic solution (0.075M KCl, 5% FBS) for 10 min prior to being fixed with 3:1 methanol:acetic acid. Cells were fixed to slides and baked at 90°C for 20 min. After cooling, each slide was trypsinized for 45 sec followed by Wrights stain for 1 min 20 sec, rinsed with diH2O and dried. Radials were imaged using bright field microscopy and analyzed using Cytovision software (Applied Imaging, San Jose, CA). Chromosome breakpoints were identified according to the ISCN (Shaffer 2009).
Chromosome stability
For Sister Chromatid Exchange analysis, cells were allowed to go through two rounds of replication in 25 μg/ml BrdU, 24 hr post-transfection. Clastogen-induced SCEs were elicited by a 4-hr pulse of 20 ng/ml MMC 20 hr prior to harvest. Cells were harvested following a 3 hr exposure to 0.05 μg/ml colcemid (Gibco), treated with a 1:3 solution of 5% fetal calf serum:0.075M KCl, and fixed in 3:1 methanol:acetic acid. Cells were then dropped onto slides and stained for 5 min in 0.01% Acridine Orange. Following staining, slides were rinsed with diH20 and treated with Sorenson Buffer (pH 6.8) (1:1 volume of 0.06M Na2HPO4 and 0.06M KH2PO4). After treatment, slides were placed on a UV transilluminator (UVP model TM-36) for 12 minutes, and then visualized using a FITC filter. Twenty to 25 metaphases from each culture were scored for chromosome counts and numbers of SCEs. The SCE rate was calculated as the number of SCEs per chromosome.
For chromosome breakage studies, 5×104 cells were seeded in T25 flasks, and were treated 24 hr later with 10-20 ng/ml MMC, depending on the cell line and its sensitivity. Following a 48 hr incubation with MMC, cells were harvested as described (Bruun et al. 2003). Slides were stained with Wright stain and 50 metaphases from each culture were scored for radial formation. The results are presented as the percentage of total cells containing at least one radial.
Constructs
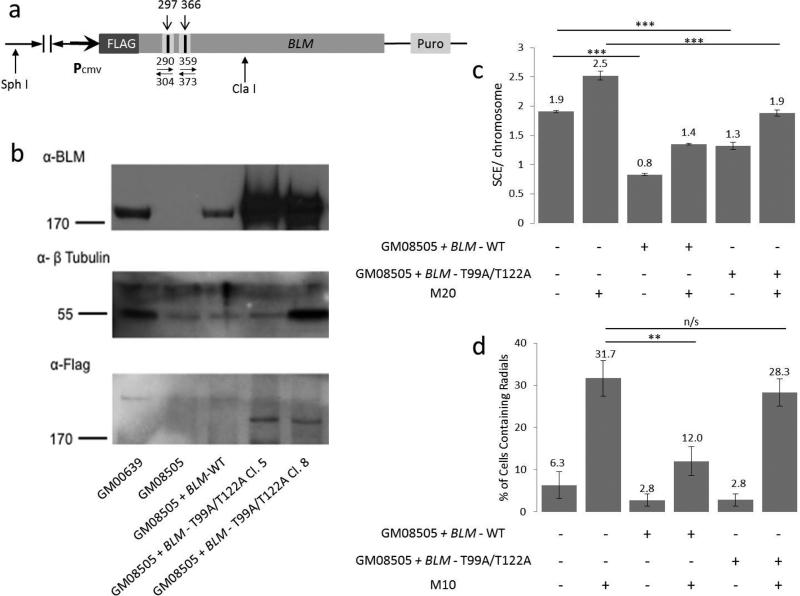
FLAG-tagged full-length BLM coding sequence in a pMMPpuro retroviral vector was digested with ClaI and SphI (NEB) to produce a 3.5 kb pMMP-BLM junction fragment containing the T99 and T122 phosphorylation sites (Fig. 3a). The 3.5 kb ClaI-SphI fragment was ligated into ClaI-SphI digested pBR322 vector and minipreps were sequence-verified. Miniprep DNA was used as the template for site directed mutagenesis (Stratagene). Individual mutant candidate minipreps were sequence-verified. Mutations were then shuttled back into the full-length pMMP-BLM construct via the ClaI-SphI sites. Final midipreps were sequenced prior to transfection of human cells.
Figure 3. BLM Construct and Expression.
(a) BLM Construct. The numbers above the linear construct refer to nucleotide position of targeted sites. The numbers below with opposing arrows indicate PCR primers used for mutagenesis. (b) Immunoblots of representative clones expressing BLM constructs with anti-BLM or anti-FLAG antibody, as indicated. Tubulin was used as a loading control. GM00639 are wild-type cells, GM08505 are BS cells, and lanes 3 through 5 contain GM08505 cells transfected with various constructs. BLM-WT encodes full length BLM protein and BLM - T99A/T122A encodes BLM protein with two threonine→alanine mutations at key phosphorylation sites. (c)SCE formation in BS cells, cells expressing BLM-WT construct, and cells expressing the BLM - T99A/T122A construct. Data represent 25 or more metaphase analyses from each of four independent clones. ‘M20’ is treatment with 20ng/ml MMC. (d) Radial formation in cells from part “c”. M10 is a dose of 10ng/ml of MMC. The error bars in “c” and “d” represent standard error. **p<0.01; ***p<0.001; n/s is “not significant”.
Transfection
Cell lines were transfected with pMMP-BLM constructs using Lipofectamine 2000 (Invitrogen) or TransIT-293 (Mirus) in 6-well plates as directed by the manufacturer. The red fluorescent plasmid pDs-Red2-C1 (ClonTech) was used in a parallel transfection to monitor transfection efficiency. After 24 hours cells were re-plated in 100mm or 150mm plates, based on transfection efficiency, and placed under 0.1-0.4 μg/ml puromycin (Sigma) selection. After 10 days of growth colonies were selected, expanded, and expression was confirmed by immunoblotting.
Immunoblotting
Cells grown in T-75 flasks were washed with PBS, trypsinized, pelleted, and frozen at -80°C. Whole cell extracts were prepared as previously described (Bruun et al. 2003). 50 μg whole cell extract per lane was run on a 7.5% acrylamide gel and transferred to a nitrocellulose membrane (Osmonics). Membranes were blocked overnight in TBST (TBS plus 0.1% Tween) plus 5% dry milk. Membranes were probed with anti-BLM rabbit polyclonal antibody (Abcam) at a 1:1000 dilution in TBST with 5% dry milk. For β-tubulin blots, membranes were probed with a rabbit polyclonal antibody (H-235, Santa Cruz) at a 1:3000 dilution in TBST. All antibodies were visualized with HRP-conjugated secondary antibodies and enhanced chemiluminescence.
Statistics
Hot and cold chromosomes, and hot bands were identified using a Monte Carlo simulation. Multiple iterations were run (300, 500, 1000, and 2500) to compute z-scores for each sample, and then converted to a p-value. P-values below 0.05 were considered “hot” and above 0.95 considered “cold”. For chromosome bands, “hot-spots” were considered those with an observed frequency two standard deviations from the mean (95% confidence interval).
Results
Radials in BS cells are non-homologous
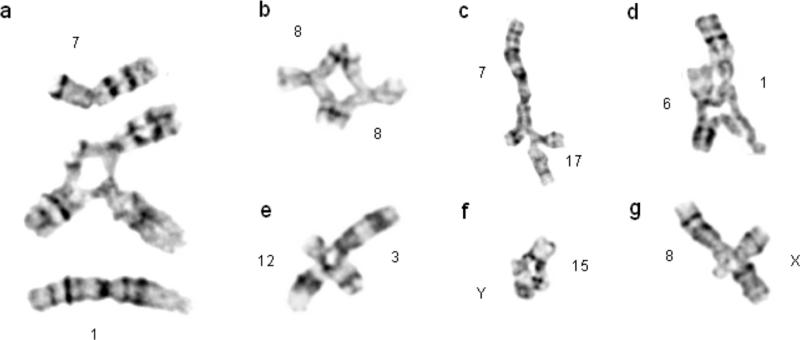
The GM01492 BS primary fibroblast cell line was karyotyped and found to be 46,XY (data not shown). Analysis of rare spontaneous chromosome radials showed them to occur primarily between non-homologous chromosomes (Table 1). To increase the number of radial formations available for characterization, the cells were treated with two different clastogens, DEB or MMC. Representative radials are shown with an example of intact chromosomes juxtaposed in order to orient the chromosomes within a radial (Figure 1a). Again, radials occurred predominantly between non-homologous chromosomes. Homologous radials, such as in Figure 1b, occurring between two homologs of chromosome 8 at breakpoints 8q13, were rare. Radials can also occur between homologous chromosomes, but at non-homologous positions, such as between sites on the short and long arms of chromosome 6 (data not shown). These radials were also uncommon, but were observed in the present study. Despite occurring between homologous chromosomes, these radials do not occur at regions of gross homology—rather, they are akin to non-homologous radials that chanced to form at a non-homologous region on the other homolog present in each cell. Results of radial formation are summarized in Table 1. Spontaneous (n=42), DEB-induced (n=101), and MMC-induced (n=101) radials all primarily (>95%) formed between non-homologous chromosomes.
Table 1.
Summary of radials observed in the present study occurring between various chromosome configurations
| Non-homologous chromosomes | Non-homologous sites on homologous chromosomes | Homologous Chromosomes | |
|---|---|---|---|
| Spontaneous | 95.2% | 2.4% | 2.4% |
| DEB | 96% | 2% | 2% |
| MMC | 93% | 3% | 4% |
Fig. 1. Radials occurring in Bloom Syndrome cells are non-homologous.
(a) Intact chromosomes are superimposed alongside a radial occurring between two non-homologous chromosomes to orient individual chromosomes within a radial. (b) A homologous radial and (c-g) representative non-homologous radials isolated from clastogen-treated BS cells.
Breakpoint hotspots are independent of chromosome size
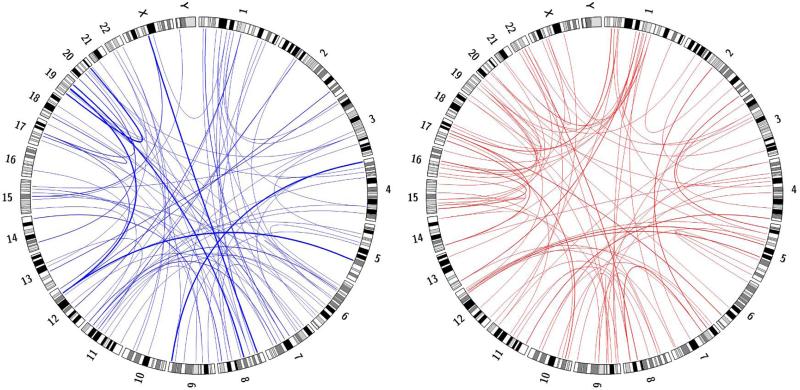
Radials seen in FA cells are not formed with a uniform distribution along the chromosomes, but are concentrated in hotspots (Newell et al. 2004). We evaluated whether the radials seen in BS cells showed similar clustering. We started by plotting the radials observed in a Circos plot (Figure 2), which allowed for the visualization of every chromosome pair within a radial including the specific chromosome bands that were broken. “Hot” and “cold” chromosomes for radial formation could be visualized, as well as the overall non-homologous nature of the radials. For a more detailed analysis of the clustering, a Monte Carlo simulation was done to determine if observed chromosome radial frequencies suggested hot and cold spots. The Monte Carlo simulation randomizes choice based on a discrete distribution. We performed 300, 500, 1,000, and 2,500 iterations. The discrete distribution was defined to be chromosome size, with the assumption that radial frequencies were proportionate to chromosome size. For each sample (observed count and those generated from Monte Carlo simulation), z-scores were computed then converted to a p-value. Table 2 summarizes the results for chromosomes that were determined to contain hot or cold spots for 1,000 iterations of the Monte Carlo simulation.
Fig. 2. Circos plots depicting radial breakpoints and chromosome partners for radials isolated from DEB (blue) or MMC (red)–treated BS cells.
Ideogram depictions of chromosomes are arranged p-arm to q-arm. Blue and red lines connect the breakpoint in the first chromosome to the breakpoint in the partner chromosome for each radial analyzed. The line thickness is quantitative, with the thin lines indicating one observation, medium lines two, and thick lines three.
Table 2.
Summary of Monte Carlo simulation (1,000 iterations) to determine hot and cold spots for chromosome radial formation.
| Chromosome | ICL agent | # Obs. Radials | P-value | Comment |
|---|---|---|---|---|
| 2 | MMC | 6 | 0.996384862 | Cold |
| 2 | DEB | 6 | 0.996264578 | Cold |
| 5 | DEB | 5 | 0.984008375 | Cold |
| 6 | MMC | 5 | 0.978934717 | Cold |
| 14 | DEB | 2 | 0.978817400 | Cold |
| 11 | DEB | 16 | 0.011362183 | Hot |
| 17 | MMC | 10 | 0.024462551 | Hot |
| 19 | DEB | 9 | 0.005868607 | Hot |
| 19 | MMC | 11 | 0.000203494 | Hot |
| 20 | DEB | 8 | 0.036558558 | Hot |
| 22 | MMC | 8 | 0.006393191 | Hot |
Chromosomes 2, 5, 6, and 14 were determined to have low radial formation and were designated as “cold”, while chromosomes 11, 17, 19, 20, and 22 were determined to have high radial formation and were designated as “hot”. Chromosome 19 was the only chromosome to be “hot” when exposed to both MMC and DEB as a DNA damaging agent. Conversely, chromosome 2 was “cold” when exposed to either damaging agent. This can be visualized by the densely clustered lines at chromosome 19, and the lack of chromosome 2 involvement given its size, shown in Figure 2.
To determine hot spots by chromosome band, we assumed a uniform distribution across the genome and set a threshold of observed radial frequency to be greater than two standard deviations from the mean, representing >95% confidence interval. Table 3 summarizes these results. Chromosome 22 was the only “hot” chromosome to not contain any “hot” bands. Chromosome 14 was determined to be “cold” for radial formation, yet band 14q16 was determined to be a “hot” band. Chromosome 1 had three “hot” bands, but was not determined to be a “hot” chromosome overall. The remaining “hot” bands occurred alone in unique chromosomes. Our results indicate that there are hotspots for radial formation, but the occurrence of hotspots is independent of the chromosome average. Seven of the twelve hot bands coincided with recognized fragile sites indicating a possible link between fragile site stability and radial formation (Schwartz et al. 2006).
Table 3.
Summary of radial hot-spots by chromosome band
| Observed Radials | Hot Spot | |||||
|---|---|---|---|---|---|---|
| Bands | DEB | MMC | Total | DEB | MMC | Total |
| 1p13 | 2 | 4 | 6 | YES | YES | |
| 1p12 | 2 | 6 | 8 | YES | YES | |
| 1q21 | 4 | 4 | 5 | YES | YES | |
| 8q24.1 | 1 | 4 | 5 | YES | YES | |
| 14p11.2 | 0 | 5 | 5 | YES | YES | |
| 19q13.3 | 2 | 6 | 8 | YES | YES | |
| 20q11.2 | 1 | 4 | 5 | YES | YES | |
| 1q12 | 4 | 2 | 6 | YES | YES | |
| 4p16 | 4 | 2 | 6 | YES | YES | |
| 11q23 | 6 | 2 | 8 | YES | YES | |
| 12q24.1 | 5 | 2 | 7 | YES | YES | |
| 17q21 | 3 | 3 | 6 | YES | ||
BLM T99A/T122A suppresses SCEs, but not radials
In order for BLM to act in DNA replication recovery, it must be phosphorylated at specific sites—T99 and T122 (Davies et al. 2004). To determine whether phosphorylation of BLM is also required to suppress chromosomal radial formation after ICL damage, a BLM construct was made encoding BLM with amino acids T99 and T122 converted to alanine, preventing phosphorylation of those sites (BLM-T99A/T122A) (Figure 3a). Expression from the modified BLM constructs was monitored using anti-BLM or anti-FLAG detection by immunoblotting to verify expression and size (Figure 3b). BS cells transfected with wildtype BLM (BLM-WT) showed a decrease in SCEs as expected (Figure 3c). BS cells transfected with BLM-T99A/T122A suppressed SCE formation both with and without ICL damage as well, as previously reported (Davies et al. 2004). This result indicates that BLM protein is required for regulation of SCE formation, but phosphorylation of these two sites is not needed. Similarly, BS cells transfected with BLM-WT had a significant decrease in radials following MMC treatment (Figure 3d). However, BS cells transfected with the BLM-T99A/T122A construct failed to suppress chromosome radial formation following ICL damage. This indicates that T99 and T122 phosphorylation are required for BLM to perform its function in preventing radial formation, but not for it to suppress SCEs.
BLM T99A/T122A acts as a dominant negative
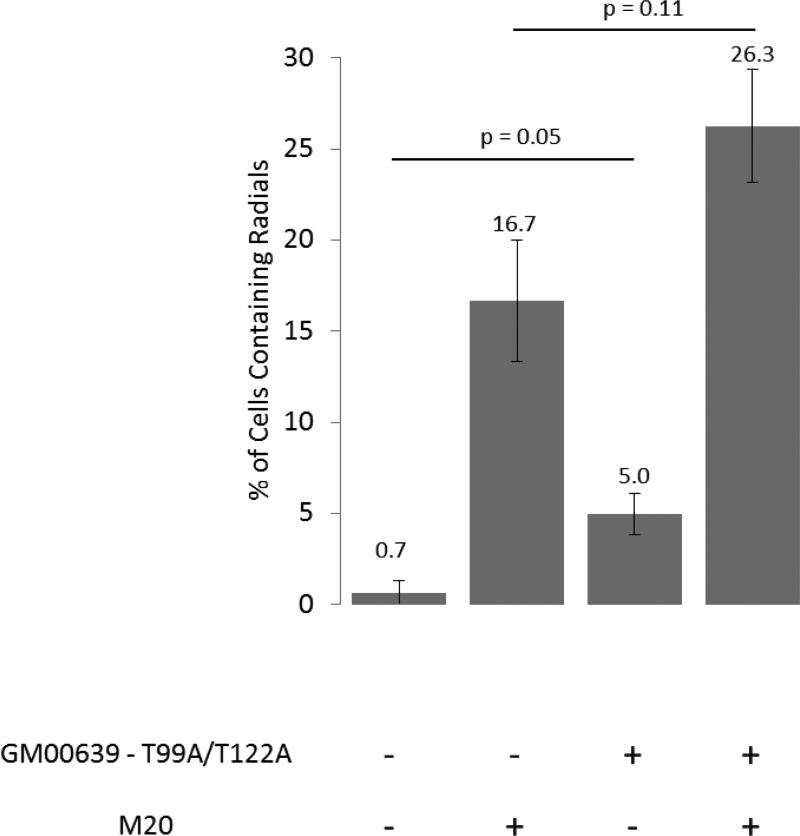
Normal transformed human fibroblasts (GM00639) were transfected with BLMT99A/T122A in an effort to determine whether dysfunctional BLM protein interfered with functional BLM. Without ICL damage, the rate of spontaneous radial formation increased with the addition of BLM-T99A/T122A (Figure 4). Additionally, an increase (approximately 65%) in radial formation was observed with ICL damage. Thus, expression of a BLM that cannot be phosphorylated appears to hinder the function of normal phosphorylated BLM present in the cells, suggesting a possible dominant negative effect on suppression of radial formation.
Figure 4. Radial formation in normal transformed human fibroblasts (GM00639) expressing the BLM - T99A/T122A construct.
Data are the average of combined results from two trials each of four independent clones. ‘M20’ represents treatment with 20ng/ml MMC. The error bars represent standard error.
Discussion
Radials in both BS and FA cells form preferentially between non-homologous chromosomes. This is contrary to previous reports that had classified BS radials as occurring between primarily homologous chromosomes. Previous studies utilized a solid staining method, such as Giemsa or Wright stain, which does not allow specific identification of the chromosomes involved. The homologous interpretation was largely based on symmetry, which we now appreciate can also be the presentation for non-homologous radials. For example, the radial formation between chromosomes 1 and 7 in Figure 1a would appear to be symmetrical and, without identification by G-banding, assumed to involve homologous chromosomes.
The chromosome pairing with respect to radial formation provides valuable information about the underlying repair defect that causes them. Homologous radials were thought to result from a failure to resolve recombination intermediates, and thus represented a defect in homologous recombination. However, non-homologous radials suggest that a non-homologous end-joining pathway—either canonical or alternative—is more likely responsible for radial formation in both BS and FA cells. The physical similarity between both spontaneous and induced radials in BS and FA cells indicates that they most likely arise from a shared DNA repair defect. Similar to FA radials, BS radials also exhibit “hot” and “cold” chromosomes and bands, suggesting preferential spots in the genome for radial formation. The origin of these “hotspots” requires further investigation, but may reflect differences in chromatin compaction or preference of clastogens to target GC-rich regions (Kumar et al. 1992). Either of these situations may explain the trend of gene-rich chromosomes being “hotspots” for radial formation. For instance, chromosomes 1 and 19 are similar in size to 2 and 18, respectively. However, chromosomes 2 and 18 are both gene-poor and “colder” for radial formation, while chromosomes 1 and 19 are “hotter” and much more gene-rich. It is also possible that specific fragile sites play a role in radial formation. Interestingly, some of the bands that are considered “hot” are also documented chromosome fragile sites (Schwartz et al. 2006). BLM has been localized to ultrafine bridges that form between chromosome fragile sites, especially after replication stress, and is required for their resolution (Chan et al. 2009). In addition, the FA pathway has been shown to be involved in maintaining fragile site stability and localize to these sites regardless of whether they are broken at metaphase (Chan et al. 2009; Howlett et al. 2005). Given this, fragile site instability may have an unexplored role in radial formation.
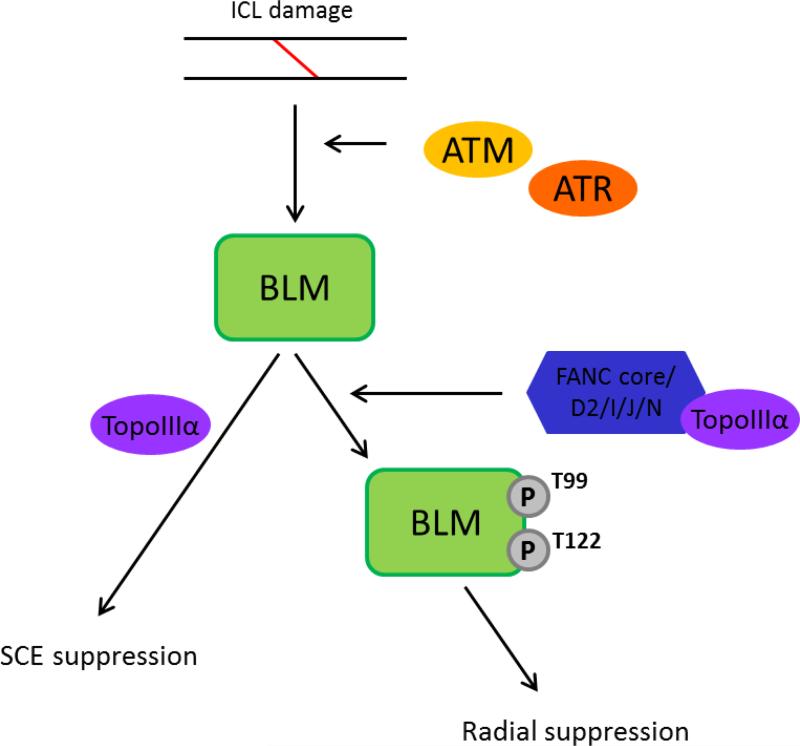
The findings presented show that phosphorylation of BLM at T99 and T122 acts as a molecular switch inhibiting chromosomal radial formation, but that these marks are dispensable for the suppression of SCEs, separating the regulation of these two events. Thus, BLM acts in different modes to limit these two kinds of genomic instability (Figure 5). When phosphorylated, BLM prevents radial formation and is known to interact with FA pathway elements. Suppression of SCEs does not require BLM phosphorylation and evidence is lacking to support interaction with the core FA pathway.
Figure 5.
Schematic displaying the segregated actions of BLM. The proposed scheme depicts the separate roles BLM plays in suppression of excess SCE and radial formation, each differentially dependent on the phosphorylation status of BLM.
Blocking phosphorylation of BLM at T99 and T122 has a dominant negative effect on radial suppression that is compatible with models suggesting that BLM acts in a complex with FA proteins, TOP3A, RPA and other proteins functioning in response to ICL formation. Overexpression of an unphosphorylatable BLM may hinder complex formation, which is likely dependent on those phosphorylation marks, and thus compromises the ability of cells to prevent radial formation.
These results raise the question as to whether the radials characteristically seen in FA cells are the result of defective BLM phosphorylation. If FA cells are unable to phosphorylate BLM protein as reported (Pichierri et al. 2004), then radials could ultimately be the result of impaired BLM protein function. This defect would not hinder the ability of BLM to suppress SCEs in FA cells—as we have shown this process to be independent of BLM phosphorylation—but only affect the ability to suppress radials. If this is in fact so, then the interaction of BLM and FA in a single epistatic pathway to process ICLs might reflect a requirement for FA protein function to promote ATM or ATR activation of BLM protein via phosphorylation. The structural similarities observed between FA and BS radials—as both occurring between non-homologous chromosomes—also supports a single pathway of radial formation dependent on both FA and BLM protein function.
Acknowledgments
The authors thank C. Lopez and M. Dai for helpful technical contributions. We thank Dr. Stephen Moore for useful discussions. This work was supported by NHLBI grant PO1HL048546 and NIH training grant 5T32HL007781.
References
- Bachrati CZ, Hickson ID. Recq helicases: Guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- Biebricher A, Hirano S, Enzlin JH, Wiechens N, Streicher WW, Huttner D, Wang LH, Nigg EA, Owen-Hughes T, Liu Y, Peterman E, Wuite GJ, Hickson ID. Pich: A DNA translocase specially adapted for processing anaphase bridge DNA. Molecular cell. 2013;51:691–701. doi: 10.1016/j.molcel.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun D, Folias A, Akkari Y, Cox Y, Olson S, Moses R. Sirna depletion of brca1, but not brca2, causes increased genome instability in fanconi anemia cells. DNA Repair (Amst) 2003;2:1007–1013. doi: 10.1016/s1568-7864(03)00112-5. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Hickson ID. New insights into the formation and resolution of ultra-fine anaphase bridges. Seminars in cell & developmental biology. 2011;22:906–912. doi: 10.1016/j.semcdb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Chan KL, North PS, Hickson ID. Blm is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. The EMBO journal. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nature cell biology. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Chu WK, Hickson ID. Recq helicases: Multifunctional genome caretakers. Nature reviews Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the bloom's syndrome helicase and its role in recovery from s-phase arrest. Molecular and cellular biology. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AJ, West SC. Fancm connects the genome instability disorders bloom's syndrome and fanconi anemia. Molecular cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The bloom's syndrome gene product is homologous to recq helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- German J. Bloom's syndrome. I. Genetical and clinical observations in the first twenty-seven patients. American journal of human genetics. 1969;21:196–227. [PMC free article] [PubMed] [Google Scholar]
- German J, Crippa LP, Bloom D. Bloom's syndrome. Iii. Analysis of the chromosome aberration characteristic of this disorder. Chromosoma. 1974;48:361–366. doi: 10.1007/BF00290993. [DOI] [PubMed] [Google Scholar]
- Gravells P, Hoh L, Solovieva S, Patil A, Dudziec E, Rennie IG, Sisley K, Bryant HE. Reduced fancd2 influences spontaneous sce and rad51 foci formation in uveal melanoma and fanconi anaemia. Oncogene. 2013;32:5338–5346. doi: 10.1038/onc.2012.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. Recq DNA helicase is a suppressor of illegitimate recombination in escherichia coli. Proc Natl Acad Sci U S A. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon Newell AE, Hemphill A, Akkari YM, Hejna J, Moses RE, Olson SB. Loss of homologous recombination or non-homologous end-joining leads to radial formation following DNA interstrand crosslink damage. Cytogenetic and genome research. 2008;121:174–180. doi: 10.1159/000138882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill AW, Akkari Y, Newell AH, Schultz RA, Grompe M, North PS, Hickson ID, Jakobs PM, Rennie S, Pauw D, Hejna J, Olson SB, Moses RE. Topo iiialpha and blm act within the fanconi anemia pathway in response to DNA-crosslinking agents. Cytogenetic and genome research. 2009;125:165–175. doi: 10.1159/000230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Morelli MA, Borst PL, Cohen PE. Mammalian blm helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol. 2010;188:779–789. doi: 10.1083/jcb.200909048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW. The fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Human molecular genetics. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Kee Y, D'Andrea AD. Molecular pathogenesis and clinical management of fanconi anemia. J Clin Invest. 2012;122:3799–3806. doi: 10.1172/JCI58321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the fanconi anemia/brca pathway. Genes Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn EM. Mitotic chiasmata and other quadriradials in mitomycin c-treated bloom's syndrome lymphocytes. Chromosoma. 1978;66:287–297. doi: 10.1007/BF00330557. [DOI] [PubMed] [Google Scholar]
- Kumar S, Lipman R, Tomasz M. Recognition of specific DNA sequences by mitomycin c for alkylation. Biochemistry. 1992;31:1399–1407. doi: 10.1021/bi00120a016. [DOI] [PubMed] [Google Scholar]
- Levitt NC, Hickson ID. Caretaker tumour suppressor genes that defend genome integrity. Trends Mol Med. 2002;8:179–186. doi: 10.1016/s1471-4914(02)02298-0. [DOI] [PubMed] [Google Scholar]
- Mankouri HW, Hickson ID. The recq helicase-topoisomerase iii-rmi1 complex: A DNA structure-specific 'dissolvasome'? Trends in biochemical sciences. 2007;32:538–546. doi: 10.1016/j.tibs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Manthei KA, Keck JL. The blm dissolvasome in DNA replication and repair. Cellular and molecular life sciences : CMLS. 2013;70:4067–4084. doi: 10.1007/s00018-013-1325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. A multiprotein nuclear complex connects fanconi anemia and bloom syndrome. Molecular and cellular biology. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V, Rosselli F. The fanc pathway and blm collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nature cell biology. 2009;11:761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- Newell AE, Akkari YM, Torimaru Y, Rosenthal A, Reifsteck CA, Cox B, Grompe M, Olson SB. Interstrand crosslink-induced radials form between nonhomologous chromosomes, but are absent in sex chromosomes. DNA repair. 2004;3:535–542. doi: 10.1016/j.dnarep.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Rosselli F. Blm and the fanc proteins collaborate in a common pathway in response to stalled replication forks. The EMBO journal. 2004;23:3154–3163. doi: 10.1038/sj.emboj.7600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray JH, German J. Bloom's syndrome and em9 cells in brdu-containing medium exhibit similarly elevated frequencies of sister chromatid exchange but dissimilar amounts of cellular proliferation and chromosome disruption. Chromosoma. 1984;90:383–388. doi: 10.1007/BF00294165. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Fung JC, Branda SS, Roeder GS. The sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Current biology : CB. 2003;13:1954–1962. doi: 10.1016/j.cub.2003.10.059. [DOI] [PubMed] [Google Scholar]
- Rosin MP, German J. Evidence for chromosome instability in vivo in bloom syndrome: Increased numbers of micronuclei in exfoliated cells. Hum Genet. 1985;71:187–191. doi: 10.1007/BF00284570. [DOI] [PubMed] [Google Scholar]
- Rouzeau S, Cordelieres FP, Buhagiar-Labarchede G, Hurbain I, Onclercq-Delic R, Gemble S, Magnaghi-Jaulin L, Jaulin C, Amor-Gueret M. Bloom's syndrome and pich helicases cooperate with topoisomerase iialpha in centromere disjunction before anaphase. PloS one. 2012;7:e33905. doi: 10.1371/journal.pone.0033905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TM, Anschutz F, Knopp A. [spontaneous chromosome aberrations in familial panmyelopathy]. Humangenetik. 1964;1:194–196. doi: 10.1007/BF00389636. [DOI] [PubMed] [Google Scholar]
- Schroeder TM, German J. Bloom's syndrome and fanconi's anemia: Demonstration of two distinctive patterns of chromosome disruption and rearrangement. Humangenetik. 1974;25:299–306. doi: 10.1007/BF00336905. [DOI] [PubMed] [Google Scholar]
- Schvarzstein M, Pattabiraman D, Libuda DE, Ramadugu A, Tam A, Martinez-Perez E, Roelens B, Zawadzki KA, Yokoo R, Rosu S, Severson AF, Meyer BJ, Nabeshima K, Villeneuve AM. DNA helicase him-6/blm both promotes mutsgamma-dependent crossovers and antagonizes mutsgamma-independent inter-homolog associations during caenorhabditis elegans meiosis. Genetics. 2014 doi: 10.1534/genetics.114.161513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M, Zlotorynski E, Kerem B. The molecular basis of common and rare fragile sites. Cancer letters. 2006;232:13–26. doi: 10.1016/j.canlet.2005.07.039. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Slovak ML, Campbell LJ. Iscn: An international system for human cytogenetic nomenclature. Basel; Karger: 2009. 2009. [Google Scholar]
- Suhasini AN, Brosh RM., Jr Fanconi anemia and bloom's syndrome crosstalk through fancj-blm helicase interaction. Trends in genetics : TIG. 2012;28:7–13. doi: 10.1016/j.tig.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Hinz JM. Cellular and molecular consequences of defective fanconi anemia proteins in replication-coupled DNA repair: Mechanistic insights. Mutation research. 2009;668:54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of fanconi anaemia and brca proteins. Nature reviews Genetics. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Watt PM, Hickson ID, Borts RH, Louis EJ. Sgs1, a homologue of the bloom's and werner's syndrome genes, is required for maintenance of genome stability in saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]