SUMMARY
During embryogenesis, ectodermal stem cells adopt different fates and form diverse ectodermal organs, such as teeth, hair follicles, mammary glands and salivary glands. Interestingly, these ectodermal organs differ in their tissue homeostasis, which leads to differential abilities for continuous growth postnatally. Mouse molars lose the ability to grow continuously whereas incisors retain this ability. In this study, we found that a BMP-Smad4-SHH-Gli1 signaling network may provide a niche supporting transient Sox2+ dental epithelial stem cells in mouse molars. This mechanism also plays a role in continuously growing mouse incisors. The differential fate of epithelial stem cells in mouse molars and incisors is controlled by this BMP/SHH signaling network, which partially accounts for the different postnatal growth potential of molars and incisors. Collectively, our study highlights the importance of crosstalk between two signaling pathways, BMP and SHH, in regulating the fate of epithelial stem cells during organogenesis.
Keywords: BMP, SHH signaling, postnatal organogenesis, stem cell niche, tooth
INTRODUCTION
Ectoderm-derived organs include a large variety of highly specialized biological structures such as teeth, hair follicles, mammary glands and salivary glands. Despite varying in number, shape and function, all these ectodermal organs develop through continuous and reciprocal epithelial–mesenchymal interactions, sharing common morphological and molecular features especially during their embryonic development. The first morphological sign of their development is a thickening of the epithelium that forms the placode. In most cases, the placode invaginates into the mesenchyme and proliferates to give rise to a bud, while the surrounding mesenchyme starts to condense. During later stages of embryogenesis, the putative ectodermal stem cells adopt different fates and consequently generate a variety of tissue-specific stem cells, which are the sources for the various cell lineages that form the diverse ectodermal organs (Pispa and Thesleff, 2003).
Most ectodermal organs undergo terminal differentiation and become functional postnatally. After birth, ectodermal organs are exposed to a high risk of damage and it is critical that they retain the ability to repair and regenerate throughout the lifespan of the organism. In a number of ectodermal organs, epithelial adult stem cells have already been identified and shown to support tissue homeostasis and injury repair, such as in mouse incisors (Biehs et al., 2013; Harada et al., 1999; Harada et al., 2002; Juuri et al., 2012), hair follicles (Blanpain, 2010; Blanpain and Fuchs, 2006; Blanpain et al., 2004; Tumbar et al., 2004), mammary glands (Visvader and Smith, 2011) and salivary glands (Lombaert, 2008). These epithelial adult stem cells reside in niches that provide them with the proper signals to regulate their function and maintenance. In contrast, some ectodermal organs, such as human teeth and mouse molars, lack adult epithelial stem cells and lose the capacity for renewal and regeneration after development.
In mice, incisors maintain their epithelial stem cells along with their mesenchymal stem cells, allowing them to grow continuously throughout life (Biehs et al., 2013; Feng et al., 2011; Harada et al., 1999; Harada et al., 2002; Zhao et al., 2014). These epithelial stem cells reside in their niche, the cervical loop at the proximal end of the labial side of the incisor, and eventually differentiate into ameloblasts that deposit enamel along the labial side of the incisor from the proximal to the distal end (Harada et al., 1999; Harada et al., 2002). In contrast, the cervical loop structure is lost after crown formation in mouse molars, and a double layer of root sheath epithelium forms to direct limited root growth postnatally, similar to human tooth development (Tummers and Thesleff, 2003). Therefore, mouse teeth provide an ideal model system for the comparison of epithelial stem cell fate regulation in molars versus continuously growing incisors.
Previous studies have shown that the BMP/TGFβ signaling pathways serve as key regulators that individually or coordinately control epithelial stem cell fate. BMP signaling appears to influence the activation of different types of epithelial stem cells, because inhibition of BMP signaling by overexpression of Noggin results in induction of hair placode formation as well as de novo formation of intestinal crypts (Botchkarev VA et al., 2001; Haramis et al., 2004; He et al., 2004). Mutations that affect BMP signaling account for almost half of the cases of juvenile polyposis syndrome, a condition that can lead to intestinal cancer (Sancho et al., 2004). Similarly, conditional ablation of the Bmpr1a gene, encoding a BMP receptor protein, results in the continued activation of hair follicle stem cells and the eventual formation of follicular tumors (Andl et al., 2004). BMP signaling is also required for proper stem cell differentiation, as targeted ablation of Bmpr1a results in the accretion of undifferentiated hair follicle and intestinal epithelial cells (Andl et al., 2004; Kobielak et al., 2007; Sancho et al., 2004). In contrast, epithelial stem cell populations in hair follicles show differential responses to TGFβ signaling, including proliferation, differentiation and apoptosis (Lin and Yang, 2013). Additionally, activated TGFβ/phospho-Smad target genes and/or Smad interacting proteins are found in hair follicle bulge stem cells (Fuchs et al., 2004; Tumbar et al., 2004). Blocking TGFβ signaling in bulge stem cell culture abolishes the colony-forming ability of these stem cells, suggesting that TGFβ signaling is required for their maintenance (Lin and Yang, 2013). Moreover, paracrine TGFβ signaling counterbalances BMP-mediated inhibition of hair follicle stem cell activation (Oshimori and Fuchs, 2012). Smad4 serves as a central intracellular mediator for both BMP and TGFβ signaling transduction and plays a crucial role in regulating BMP/TGFβ signaling during organogenesis (Ko et al., 2007; Massague, 2000; Xu et al., 2008; Yang et al., 1998).
In this study, we investigate (i) the effects of BMP/TGFβ signaling on dental epithelial stem cells during development and (ii) whether BMP/TGFβ signaling contributes to the differential fate of epithelial stem cells in postnatal tooth development. In addition, we identify BMP/TGFβ downstream target genes to elucidate the molecular mechanism that regulates epithelial stem cells during postnatal tooth development and test whether the BMP/TGFβ signaling network discovered here is also utilized in regulating epithelial stem cells during the formation of other organs.
RESULTS
Transient Sox2+ stem cells contribute to all epithelial cell lineages during molar development
Sox2 was recently identified as an epithelial stem cell marker in mouse incisors (Juuri et al., 2012). Sox2 is expressed in the oral epithelium, dental cord and dental epithelium during initiation and morphogenesis of both mouse molars and incisors (Juuri et al., 2013; Juuri et al., 2012). Sox2+ cells in the mouse incisor contribute to all epithelial cell lineages and are dental epithelial stem cells (Juuri et al., 2012). To determine whether Sox2+ cells in the mouse molar are also epithelial stem cells, we performed inducible genetic cell fate mapping in Sox2-CreER;R26R mice, in which tamoxifen transiently induces Cre-recombinase, leading to permanent expression of lacZ in Sox2+ cells and their progeny (Arnold et al., 2011). We genetically labeled Sox2+ cells by administering tamoxifen at E11.5 and traced their descendants by detecting lacZ expression after 48 hours and 1 week (Figure 1A). After 48 hours, a small number of lacZ+ cells were detectable in the epithelial compartments of the lower first molar, including the oral epithelium, dental cord and dental epithelium (Figure 1B), corresponding to the areas of Sox2 expression (Juuri et al., 2013). One week after induction, the number of lacZ+ cells had increased, covering almost the entire enamel organ, including the ameloblasts (AM), outer enamel epithelium (OEE), stellate reticulum (SR) and stratum intermedium (SI) (Figure1C, D), demonstrating the self-renewing capacity of Sox2+ cells. In control R26R mice, no lacZ+ cells were detectable in the epithelial compartment (data not shown). These results demonstrate that Sox2+ cells in the mouse molar are epithelial stem cells, contributing to all epithelial cell lineages.
Figure 1. Transient Sox2+ stem cells contribute to all epithelial cell lineages during molar development.
(A) Timing of tamoxifen induction and sample harvest in Sox2-CreER;R26R mice. (B-D) LacZ expression assayed by X-gal staining (blue) in sagittal sections of the lower first molar 48 hr (B) and 1 week (C,D) after tamoxifen induction. Broken line in (B) indicates basement membrane. Boxed area in (C) is shown magnified in (D). X-gal staining is detectable in dental epithelial cells, but not in the dental mesenchyme. Note that all lacZ+ epithelial cell types in the enamel organ (D), including AM, OEE, SR, and SI, are derived from Sox2+ cells. (E-J) Immunofluorescence of Sox2 (green) in sagittal sections of the lower first molar and incisor at E16.5 (E-G) and PN0.5 (H-J). Boxed areas in (E) and (H) are shown magnified in (F/G) and (I/J), respectively. Broken lines indicate cervical loop areas. Note the absence of Sox2 expression (white arrow, I) in mouse molars at PN0.5. AM: Ameloblast; DE: Dental epithelium; DM: Dental mesenchyme; DP: Dental papilla; IN: Incisor; M1: Lower first molar; M2: Lower second molar; OEE: Outer enamel epithelium; SI: Stratum intermedium; SR: Stellate reticulum. Scale bars (B, D, F, G, I, J): 50μm. Scale bars (C, E, H): 200μm. See also Figure S1.
Using a time-course study of Sox2 expression in the mouse molar, we found that Sox2 expression was largely restricted to both sides of the cervical loop, a putative stem cell niche in the dental epithelium, at E16.5, similar to the mouse incisor (Figure 1E-G). Sox2 expression was undetectable after birth in mouse molars (Figure 1H, I). In contrast, Sox2 expression was maintained postnatally in the cervical loop of mouse incisors (Figure 1H, J). These data were further supported by our detection of Sox2 expression using Sox2-EGFP mice (Figure S1) and are consistent with the findings of a previous study (Juuri et al., 2012). Thus, Sox2+ dental epithelial stem cells transiently reside in developing mouse molars.
Loss of Smad4 in the dental epithelium prolongs the maintenance of the cervical loop and molar crown development
BMP/TGFβ signaling is a key regulator for stem cell fate determination in many epithelial tissues, such as hair (Oshimori and Fuchs, 2012) and intestine (He et al., 2004), so we investigated whether Sox2+ epithelial stem cell fate in the developing mouse molar is also controlled by this pathway. We generated KRT14-rtTA;tetO-Cre;Smad4fl/fl mice in which deletion of the gene encoding Smad4, the common mediator of BMP/TGFβ signaling, can be induced by doxycycline. In order to explore the effect of BMP/TGFβ signaling on the regulation of dental epithelial stem cell fate during tooth development, we decided to initiate the deletion of Smad4 at E16.5 (Figure 2A), based on the following reasons: (1) at this stage, Sox2+ stem cells become restricted to the cervical loop but have yet to disappear; (2) also, at this stage, dental epithelial cells remain in an undifferentiated state; and (3) earlier ablation of Smad4 in the epithelium results in embryonic lethality (Xu et al., 2008).
Figure 2. Loss of Smad4 in the dental epithelium prolongs the maintenance of the cervical loop and molar crown development.
(A) Timing of doxycycline induction and sample harvest in control and KRT14-rtTA;tetO-Cre;Smad4fl/fl (K;T;S) mice. (B-E) Macroscopic views (B), quantitation of the crown length (C) and H&E staining (D, E) of PN21.5 control and KRT14-rtTA;tetO-Cre;Smad4fl/fl lower first molars. Arrows indicate crown length. N=3. *, p < 0.05. Error bars indicate standard deviation (SD). (F-Q) Time-course analysis of H&E staining (F, G, J, K, N, O) and Keratin 14 (K14; green) immunofluorescence (H, I, L, M, P, Q) of control and KRT14-rtTA;tetO-Cre;Smad4fl/fl lower first molars at PN7.5 (F-I), PN12.5 (J-M), and PN21.5 (N-Q). Arrows indicate the HERS in control lower first molars. Broken lines indicate the cervical loop in KRT14-rtTA;tetO-Cre;Smad4fl/fl lower first molars. M1: Lower first molar; M2: Lower second molar; R: Root. Scale bars (D, E): 500μm. Scale bars (F-Q): 50μm.
Loss of Smad4 in the dental epithelium prolonged molar crown formation and resulted in a longer crown in KRT14-rtTA;tetO-Cre;Smad4fl/fl mice (Figure 2B-E). Using time-course histological analysis, we found that the multi-layered cervical loop structure, including the SR core, disappeared after crown formation and the bi-layered epithelial structure termed the HERS (Hertwig's Epithelial Root Sheath) formed at postnatal (PN) day 7.5 in control mice (Figure 2F). Afterwards, the HERS grew downwards, dissociated and guided root formation (Figure 2J, N). In contrast, the absence of Smad4 in the dental epithelium affected HERS formation and resulted in maintenance of the cervical loop structure including the SR core as late as PN21.5 (Figure 2G, K, O). We analyzed Keratin 14 expression to examine the dental epithelial structure (Figure 2H-I, L-M, P-Q) and found that Smad4 mutant dental epithelium grew continuously and failed to dissociate (Figure 2M, Q). Taken together, these data indicate that ablation of Smad4 in the dental epithelium prolongs the maintenance of the cervical loop and molar crown development.
Ablation of Smad4 affects dental epithelial cell fate and Sox2+ dental epithelial stem cell maintenance during molar development
The cervical loop area has been proposed to be the putative epithelial stem cell niche compartment in the mouse incisor (Harada et al., 1999) and vole molar (Tummers and Thesleff, 2003), which grow continuously throughout life. The maintenance of the cervical loop in KRT14-rtTA;tetO-Cre;Smad4fl/fl molars suggests there may be a change in dental epithelial cell fate in these mice. Proliferation was increased by almost 3-fold in PN7.5 Smad4 mutant dental epithelial cells compared to control dental epithelial cells in the cervical loop area, as indicated by Ki67 expression (Figure 3A-C). We detected more proliferative activity in the cervical loop region of Smad4 mutant mice than in controls as late as PN21.5 (Figure S2). In addition, we evaluated the differentiation ability of dental epithelial cells in KRT14-rtTA;tetO-Cre;Smad4fl/fl mice compared to controls. To examine the status of ameloblast differentiation in the dental epithelium, we assayed the expression of Amelogenin, an ameloblast differentiation marker, using in situ hybridization. In control mice, we detected strong Amelogenin expression in the ameloblast layer (Figure 3D). In KRT14-rtTA;tetO-Cre;Smad4fl/fl mice, however, Amelogenin was not detectable (Figure 3E). Our data therefore suggest that lack of Smad4 in the dental epithelium affects cell proliferation and differentiation.
Figure 3. Ablation of Smad4 affects dental epithelial cell fate and Sox2+ dental epithelial stem cell maintenance in vivo and in vitro during molar development.
(A-C) Immunofluorescence of Ki67 (red) in control and KRT14-rtTA;tetO-Cre;Smad4fl/fl (K;T;S) lower first molars at PN7.5. Quantitation of the percentage of Ki67-labeled nuclei in the dental epithelium (indicated by broken lines) of control and KRT14-rtTA;tetO-Cre;Smad4fl/fl lower first molars. N=3. *, p < 0.05. Error bars, SD. (D, E) In situ hybridization of Amelogenin in control and KRT14-rtTA;tetO-Cre;Smad4fl/fl lower first molars at PN7.5. Black arrow indicates Amelogenin expression in control ameloblasts. Black arrowhead indicates lack of expression of Amelogenin in KRT14-rtTA;tetO-Cre;Smad4fl/fl mice. (F-I) In situ hybridization of dental epithelial stem cell niche markers Notch1 or Lfng in control (F, H) and KRT14-rtTA;tetO-Cre;Smad4fl/fl (G, I) lower first molars at PN7.5. Black arrows indicate expression, whereas black arrowheads indicate lack of detectable expression. (J-M) Immunofluorescence of Sox2 (green) in control and KRT14-rtTA;tetO-Cre;Smad4fl/fl lower first molars at PN7.5 (J, K) and PN21.5 (L, M). White arrows indicate expression, whereas white arrowheads indicate absence of expression. (N) Timing of doxycycline induction and sample harvest for cell culture in vitro in control and KRT14-rtTA;tetO-Cre;Smad4fl/fl (K;T;S) mice. (O) Schematic diagram depicting dissection of PN3.5 lower first molar, separation of the epithelium and mesenchyme, dissociation of the epithelium into single cells, and culture of dental epithelial cells. Four days after plating, single stem cells in KRT14-rtTA;tetO-Cre;Smad4fl/fl molars proliferated to produce colonies. Colonies were not detectable in controls. (P, Q) Colony-forming assay after 7 days of culture in control versus KRT14-rtTA;tetO-Cre;Smad4fl/fl molars. Total number of colonies was quantified. N=3. *, p < 0.05. Error bars, SD. (R) Immunofluorescence of Sox2 (red) and Keratin 14 (K14; green) in epithelial cell colony from KRT14-rtTA;tetO-Cre;Smad4fl/fl molars after 7 days culture. (S) Immunofluorescence of Bmi1 (red) in epithelial cell colony from KRT14-rtTA;tetO-Cre;Smad4fl/fl molars after 7 days culture. Scale bars (A, B, D-M): 50μm. Scale bars (O, R, S): 100μm. See also Figure S2.
Based on these results, we hypothesized that a population of cells in the cervical loop might remain in an undifferentiated, progenitor status due to the absence of Smad4 in the dental epithelium. Previous studies have demonstrated that Notch signaling pathway genes, such as Notch1 and Lunatic fringe (Lfng), can serve as molecular markers of the epithelial stem cell niche in self-renewing tooth types, such as the mouse incisor and vole molar (Harada et al., 1999; Tummers and Thesleff, 2003, 2008). We examined the expression of Notch1 and Lfng using in situ hybridization. In control mouse molars, we did not detect Notch1 or Lfng expression in the dental epithelium following HERS formation at PN7.5 (Figure 3F, H). In contrast, Notch1 expression was detectable in the SR and SI cells of the cervical loop in Smad4 mutant mice, and Lfng expression was detectable in the transit-amplifying (TA) epithelial cells of the inner enamel epithelium (IEE) (Figure 3G, I), consistent with their expression patterns in the continuously growing mouse incisor and vole molar (Harada et al., 1999; Tummers and Thesleff, 2003, 2008). Similarly, we found that Sox2+ cells persisted in the cervical loop region of Smad4 mutant molars at PN7.5 (Figure 3K) and were detectable as late as PN21.5 (Figure 3M). In contrast, Sox2+ cells were not detectable in control molars (Figure 3J, L). To rule out the possibility that the persistence of Sox2+ cells in dental epithelium is due to the delayed development of the HERS in Smad4 mutant molars, we harvested samples as late as 1 month after birth. In contrast to controls, continuous growth of the epithelium and maintenance of Sox2+ cells were also detectable in Smad4 mutant molars at PN31.5 (Figure S2), indicating that the phenotype of Smad4 mutant molars is not simply due to the delayed development of the HERS.
Next, we examined whether the Sox2+ cells that persist in Smad4 mutant molars are functional stem cells, rather than merely the result of upregulated Sox2 expression due to deletion of Smad4. We used a dental epithelial stem cell (DESC) culture system (Chavez et al., 2014) to analyze molar DESCs in vitro. Dental epithelia from PN3.5 control or Smad4 mutant molars were dissociated to create single-cell suspensions (Figure 3N, O). Seven days after plating the cells, we detected a significant increase in the total number of colonies formed in Smad4 mutant molar samples (Figure 3P, Q), with strongly positive expression of typical incisor DESC markers, Sox2 and Bmi1 (Biehs et al., 2013; Juuri et al., 2012) (Figure 3R, S). In contrast, few colonies were detectable in control molar samples (Figure 3P, Q). Taken together, our results suggest that loss of Smad4 affects dental epithelial cell fate and maintenance of Sox2+ dental epithelial stem cells.
The BMP-Smad4 signaling cascade regulates Sox2+ dental epithelial stem cell maintenance during molar development
Previous studies have shown that BMP/TGFβ signaling pathways individually or coordinately regulate epithelial stem cell fate determination (Andl et al., 2004; Botchkarev VA et al., 2001; Fuchs et al., 2004; Haramis et al., 2004; He et al., 2004; Kobielak et al., 2007; Lin and Yang, 2013; Oshimori and Fuchs, 2012; Sancho et al., 2004; Tumbar et al., 2004). In order to determine whether the BMP-Smad4 or TGFβ-Smad4 signaling cascade is responsible for the phenotypes in Smad4 mutant mice, we generated both KRT14-rtTA;tetO-Cre;Bmpr1afl/fl and KRT14-rtTA;tetO-Cre;Tgfbr2fl/fl mice in which BMP or TGFβ signaling, respectively, is specifically blocked in the dental epithelium (Figure 4A-O).
Figure 4. The BMP-Smad4, not TGFβ-Smad4, signaling cascade regulates Sox2+ dental epithelial stem cell maintenance during molar development.
(A) Timing of doxycycline induction and sample harvest in control, KRT14-rtTA;tetO-Cre;Bmpr1afl/fl (K;T;B) and KRT14-rtTA;tetO-Cre;Tgfbr2fl/fl (K;T;T) mice. (B, C) Macroscopic views of control compared to KRT14-rtTA;tetO-Cre;Bmpr1afl/fl lower first molars (B) and control compared to KRT14-rtTA;tetO-Cre;Tgfbr2fl/fl lower first molars (C) at PN48.5. (D-G) H&E staining of control compared to KRT14-rtTA;tetO-Cre;Bmpr1afl/fl lower first molars (D, F) and control compared to KRT14-rtTA;tetO-Cre;Tgfbr2fl/fl lower first molars (E, G) at PN7.5. Red arrows indicate the HERS. Red broken line in (F) indicates the cervical loop. (H-K) In situ hybridization of Amelogenin in control compared to KRT14-rtTA;tetO-Cre;Bmpr1afl/fl lower first molars (H, I) and control compared to KRT14-rtTA;tetO-Cre;Tgfbr2fl/fl lower first molars (J, K) at PN7.5. Black arrows indicate Amelogenin expression, whereas arrowhead indicates lack of expression. (L-O) Immunofluorescence of Sox2 (green) in control compared to KRT14-rtTA;tetO-Cre;Bmpr1afl/fl lower first molars (L, M) and control compared to KRT14-rtTA;tetO-Cre;Tgfbr2fl/fl lower first molars (N, O) at PN7.5. White arrow indicates expression, whereas white arrowheads indicate absence of expression. Scale bars: 50μm.
We found that KRT14-rtTA;tetO-Cre;Bmpr1afl/fl mice, with specific disruption of BMP signaling in the dental epithelium, phenocopied KRT14-rtTA;tetO-Cre;Smad4fl/fl mice, with prolonged molar crown formation, visible cervical loop structure, disturbed ameloblast differentiation and persistence of Sox2+ stem cells (Figure 4B, D/F, H/I, L/M). In contrast, disruption of Tgfbr2 in the dental epithelium did not affect molar crown formation or Sox2+ stem cell disappearance (Figure 4C, E/G, J/K, N/O). Taken together, our data indicate that the BMP-Smad4, not TGFβ-Smad4, signaling cascade regulates Sox2+ epithelial stem cell maintenance during molar development.
The BMP-Smad4 signaling cascade inhibits SHH-Gli1 signaling activity to control Sox2+ epithelial stem cell maintenance during molar development
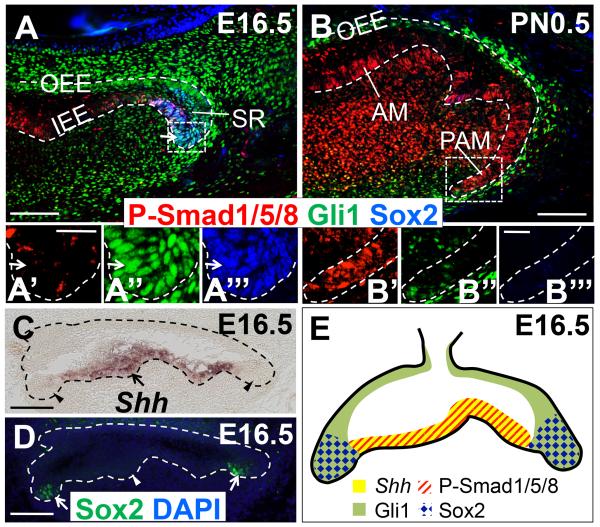
To determine whether BMP-Smad4 regulation of Sox2+ stem cells is direct or indirect, we analyzed the expression pattern of pSmad1/5/8 in developing molars. At E16.5, pSmad1/5/8 was strongly expressed in the IEE and weakly expressed in SI and SR cells in the dental epithelium, except the cervical loop region (Figure 5A, A’). The majority of pSmad1/5/8+ cells did not overlap with Sox2+ stem cells in the cervical loop region (Figure 5A, A’, A’’’), indicating that BMP-Smad4 signaling might not directly regulate Sox2+ dental epithelial stem cells. Therefore, we infer that some other factor(s) must be released from pSmad1/5/8+ cells to regulate Sox2+ dental epithelial stem cell maintenance in the cervical loop region.
Figure 5. Temporal and spatial relationship of BMP-Smad4, SHH-Gli1 signaling pathway and Sox2+ dental epithelial stem cells during molar development.
(A, B) Immunofluorescence of P-Smad1/5/8 (red), Gli1 (green), and Sox2 (blue) in lower first molars of Gli1-lacZ mice (control) at E16.5 (A) and PN0.5 (B). Detection of Gli1 expression was performed using anti-beta galactosidase antibody. Boxed areas in (A) and (B) are shown magnified in (A’-A’’’) and (B’-B’’’), respectively. Arrows indicate co-expression of Gli1 and Sox2 (light blue). Broken lines indicate dental epithelium. (C, D) In situ hybridization of Shh (C) and immunofluorescence of Sox2 (green) (D) in adjacent sections of E16.5 lower first molars. Arrows indicate expression, whereas arrowheads indicate lack of detectable expression. Broken lines indicate dental epithelium. (E) Schematic diagram of the expression patterns of Shh (yellow), P-Smad1/5/8 (red), Gli1 (green), and Sox2 (blue) in the dental epithelium (indicated by black line) of lower first molars at E16.5. AM: Ameloblast; IEE: Inner enamel epithelium; OEE: Outer enamel epithelium; PAM: Preameloblast; SR: Stellate reticulum. Scale bars (A-D): 100μm. Scale bars (A’-A’’’, B’-B’’’): 25μm.
SHH, a member of the hedgehog signaling family, is expressed in the dental epithelium and plays an essential role during tooth development (Dassule et al., 2000; Nakatomi et al., 2006). Previous studies have shown that Shh is induced or inhibited by BMP-Smad4 signaling during development (Bastida et al., 2009; Huang et al., 2010). We found that Shh is mainly expressed in IEE and some SI and SR cells at E16.5, but not in dental epithelial cells in the cervical loop region (Figure 5C), similar to the pSmad1/5/8 expression pattern (Figure 5A). Gli1, a transcription factor activated by SHH, is specifically expressed in dental epithelial cells in the cervical loop region and OEE at E16.5, complementary to the Shh and pSmad1/5/8 expression patterns (Figure 5A, C). Moreover, all Sox2+ dental epithelial stem cells in the cervical loop region overlapped with Gli1+ cells (Figure 5A, A’’, A’’’) and were surrounded by SHH-secreting cells at E16.5 (Figure 5C, D). Therefore, we hypothesized that SHH may be one of the key factors controlled by BMP-Smad4 signaling and may provide a niche to regulate Sox2+ dental epithelial stem cell maintenance during molar development (Figure 5E).
To test our hypothesis, we analyzed the expression patterns of pSmad1/5/8, Gli1 and Sox2 in the dental epithelium of KRT14-rtTA;tetO-Cre;Smad4fl/fl and control mice at PN0.5. We found that pSmad1/5/8 expression in the dental epithelium expanded to the entire cervical loop region at PN0.5, whereas Gli1 expression was dramatically reduced in the cervical loop region and Sox2+ dental epithelial stem cells were not detectable (Figure 5B-B’’’ and 6A-A’’). In contrast, more Gli1+ cells and Sox2+ dental epithelial stem cells were present and overlapped with each other in the cervical loop region at PN0.5 following inhibition of the BMP-Smad4 signaling cascade in Smad4 mutant mice (Figure 6B-B’’). Postnatally, SHH-secreting cells persisted in the dental epithelium adjacent to the cervical loop in Smad4 mutant mice (Figure 6D, F). Activated Gli1 expression was also detectable in the cervical loop region at PN21.5 in Smad4 mutant mice combined with a Gli1-LacZ reporter line (Figure 6H). In contrast, Shh expression and Gli1 activity were significantly decreased after birth in the dental epithelium in control mice (Figure 6C, E, G). To determine whether persistent Sox2+ dental epithelial stem cells might be the result of extended activation of SHH signaling in Smad4 mutant dental epithelium, we generated KRT14-rtTA;tetO-Cre;Smad4fl/fl;Shhfl/fl mice in which the gene encoding Shh was deleted in Smad4 mutant dental epithelium (Figure 6I-Q). MicroCT analysis showed that the abnormal crown and root morphology in Smad4 mutant mice were partially rescued after loss of Shh in the dental epithelium (Figure 6J, K). Histological analysis further revealed that loss of Shh in Smad4 mutant dental epithelium restored the root and periodontal tissue formation (Figure 6L-N). More importantly, Sox2+ epithelial stem cells were no longer detectable in the dental epithelium of KRT14-rtTA;tetO-Cre;Smad4fl/fl;Shhfl/fl mice (Figure 6O-Q). These data suggest that maintenance of SHH-Gli1 signaling activity in the dental epithelium contributed to Sox2+ dental epithelial stem cell persistence in KRT14-rtTA;tetO-Cre;Smad4fl/fl mice.
Figure 6. The BMP-Smad4 signaling cascade inhibits SHH-Gli1 signaling activity to control Sox2+ epithelial stem cell maintenance during molar development.
(A, B) Immunofluorescence of Gli1 (green) and Sox2 (blue) in tetO-Cre;Smad4fl/fl;Gli1-LacZ (T;S;Gli1-LacZ; control) and KRT14-rtTA;tetO-Cre;Smad4fl/fl;Gli1-LacZ (K;T;S;Gli1-LacZ) lower first molars at PN0.5. Detection of Gli1 expression was performed using anti-beta galactosidase antibody. Boxed areas in (A) and (B) are shown magnified in (A’-A’’) and (B’-B’’), respectively. Arrows indicate co-expression of Gli1 and Sox2 (light blue). Broken lines indicate the cervical loop. (C-F) In situ hybridization of Shh in control (C, E) and KRT14-rtTA;tetO-Cre;Smad4fl/fl (K;T;S) (D, F) lower first molars at PN7.5 and PN12.5. Arrows indicate expression of Shh. Broken lines indicate the HERS (C, E) or the cervical loop (D, F). (G, H) LacZ expression assayed by X-gal staining (blue) in tetO-Cre;Smad4fl/fl;Gli1-LacZ (control) and KRT14-rtTA;tetO-Cre;Smad4fl/fl;Gli1-LacZ lower first molars at PN21.5. Arrows indicate activated Gli1 expression in the dental epithelium. (I) Timing of doxycycline induction and sample harvest in control, KRT14-rtTA;tetO-Cre;Smad4fl/fl (K;T;S) and KRT14-rtTA;tetO-Cre;Smad4fl/fl;Shhfl/fl (K;T;S;Shhfl/fl) mice. (J, K) MicroCT 3D reconstructions (J) and sagittal images (K) of the lower first molars in PN21.5 control, KRT14-rtTA;tetO-Cre;Smad4fl/fl and KRT14-rtTA;tetO-Cre;Smad4fl/fl;Shhfl/fl mice. (L-N) H&E staining of control, KRT14-rtTA;tetO-Cre;Smad4fl/fl and KRT14-rtTA;tetO-Cre;Smad4fl/fl;Shhfl/fl lower first molars at PN21.5. (O-Q) Immunofluorescence of Sox2 (red) in control (O), KRT14-rtTA;tetO-Cre;Smad4fl/fl (P) and KRT14-rtTA;tetO-Cre;Smad4fl/fl;Shhfl/fl (Q) lower first molars at PN21.5. Boxed insert in (Q) shows positive Sox2 expression in the oral epithelium of the same section of KRT14-rtTA;tetO-Cre;Smad4fl/fl;Shhfl/fl lower first molar. White arrows indicate expression, whereas white arrowheads indicate absence of expression. DE: Dental epithelium; M1: Lower first molar; P: Pulp; PDL: Periodontal ligament; R: Root. Scale bars (A-F, O-Q): 50μm. Scale bars (A’-A’’, B’-B’’): 25μm. Scale bars (G, H, L-N): 100μm.
Deletion of Smad4 in the dental epithelium results in ectopic activation of SHH-Gli1 signaling and ectopic Sox2+ epithelial stem cells during postnatal incisor growth
The maintenance of Sox2+ dental epithelial stem cells in the cervical loop allows mouse incisors to grow continuously throughout life (Harada et al., 1999; Harada et al., 2002; Juuri et al., 2012). Previous studies have reported that SHH, which is produced by the differentiating progeny of Gli1+ dental epithelial stem cells, regulates the generation of ameloblast progenitors from Gli1+ dental epithelial stem cells in mouse incisors (Seidel et al., 2010). We analyzed the expression patterns of pSmad1/5/8, Gli1 and Sox2 in the dental epithelium during incisor development. pSmad1/5/8 has a similar expression pattern to that of SHH in the differentiating progeny of the epithelial stem cells (Figure S3). Sox2 and Gli1 co-localization is only detectable in the cervical loop (Figure S3). These expression patterns persist at postnatal stages in mouse incisors (Figure S3), unlike in mouse molars (Figure 5). To investigate whether the mechanism regulating dental epithelial stem cell fate in the mouse molar is shared with the incisor, we specifically ablated Smad4 in the dental epithelium of mouse incisors and molars at the same stage. Loss of Smad4 in the dental epithelium resulted in significantly reduced enamel formation in Smad4 mutant incisors, indicated by microCT analysis (Figure 7A, A’, B, B’). We found ectopic epithelial outgrowths in the TA cell region in Smad4 mutant incisors (Figure 7C, D). We also detected increased proliferation in these ectopic epithelial outgrowths, based on Ki67 expression (Figure 7E, F). The ectopic outgrowths are located in the same region as pSmad1/5/8+ and SHH- secreting cells, suggesting that BMP-SHH interaction might also be involved in this phenotype in KRT14-rtTA;tetO-Cre;Smad4fl/fl mice. To test our hypothesis, we analyzed the activated SHH signaling pathway in incisors of Smad4 mutant mice combined with a Gli1-LacZ reporter line. We detected ectopic Gli1 expression in the TA cell region including the ectopic outgrowth sites in KRT14-rtTA;tetO-Cre;Smad4fl/fl mice (Figure 7H). In contrast, Gli1 expression was not detectable in the TA cell region in control mice (Figure 7G). Surprisingly, Sox2+ dental epithelial stem cells were detectable in the TA cell region of Smad4 mutant mice, outside the cervical loop region, consistent with the location of ectopic Gli1+ cells (Figure 7I, J). These results suggest that loss of the BMP-Smad4 signaling cascade during mouse incisor development results in ectopic Sox2+ dental epithelial stem cells via activation of the SHH-Gli1 signaling cascade.
Figure 7. Deletion of Smad4 in the dental epithelium results in ectopic activation of SHH-Gli1 signaling and ectopic Sox2+ epithelial stem cells during postnatal incisor growth.
(A, B) Sagittal micro-CT images of PN21.5 control and KRT14-rtTA;tetO-Cre;Smad4fl/fl (K;T;S) lower incisors. Coronal sections (A’ and B’) were sampled at comparable positions, indicated by broken lines in (A) and (B). Arrowheads indicate enamel of the incisors. (C, D) H&E staining of PN21.5 control and KRT14-rtTA;tetO-Cre;Smad4fl/fl (K;T;S) lower incisors. Arrow indicates ectopic epithelial outgrowth in the TA cell region. (E, F) Immunofluorescence of Ki67 (green) in the TA cell region of PN7.5 control and KRT14-rtTA;tetO-Cre;Smad4fl/fl lower incisors. Broken lines indicate basement membrane. (G, H) LacZ expression assayed by X-gal staining (blue) in tetO-Cre;Smad4fl/fl;Gli1-LacZ (T;S;Gli1-LacZ; control) and KRT14-rtTA;tetO-Cre;Smad4fl/fl;Gli1-LacZ (K;T;S;Gli1-LacZ) lower incisors at PN14.5. Arrow indicates ectopic Gli1 expression, whereas arrowhead indicates absence of Gli1 expression. (I, J) Immunofluorescence of Sox2 (green) in control and KRT14-rtTA;tetO-Cre;Smad4fl/fl lower incisors at PN14.5. Arrows indicate Sox2 expression, whereas arrowhead indicates absence of Sox2 expression. Scale bars (A-F): 100μm. Scale bars (G, H): 200μm. See also Figure S3.
DISCUSSION
Ectodermal organs share many common features during development, with some important differences emerging postnatally. The mouse dentition provides an ideal model to study the difference in postnatal epithelial stem cells, because molars lose their stem cells whereas incisors maintain them. In this study, we utilized Sox2 as a dental epithelial stem cell marker to demonstrate the transient presence of dental epithelial stem cells during mouse molar development. Our in vivo and in vitro studies demonstrate that Sox2+ stem cells exist transiently during molar crown development, contribute to all epithelial cell lineages of the molar and disappear prior to root formation. Sox2 expression appears to be primarily restricted to the cervical loop, the putative stem cell niche, in the developing molar, as it is in the developing incisor (Juuri et al., 2012). However, Sox2+ dental epithelial stem cells in the developing molar adopt a different fate than those in the incisor, disappearing during postnatal development. Therefore, our results suggest a mechanism at the cellular level for the loss of renewal and regeneration ability in mouse molars postnatally.
In this study, loss of Smad4 in the dental epithelium of developing molars results in maintenance of the cervical loop structure and Sox2+ dental epithelial stem cells postnatally. Alternatively, loss of Smad4 might have de-repressed Sox2 expression in a small subpopulation of epithelial stem cells. These findings suggest that BMP/TGFβ signaling regulates dental epithelial stem cell fate determination during molar development. In self-renewing organs, such as the hair follicle, BMP signaling appears to influence the activation of epithelial stem cells and is required for proper stem cell differentiation (Andl et al., 2004; Kobielak et al., 2007). Paracrine TGFβ signaling counterbalances BMP-mediated repression of hair follicle stem cell activation (Oshimori and Fuchs, 2012). In non-self-renewing mouse molars, we have found that BMP-Smad4, not TGFβ-Smad4, signaling is required for the disappearance of Sox2+ dental epithelial stem cells in the cervical loop after birth.
Communication between stem cells and their niche enables adult stem cells to receive and respond to environmental changes, balancing their growth and regenerative potential or initiating terminal differentiation programs. The latter may provide a fail-safe mechanism to avoid dysplastic cell growth or amplification of the stem cell pool, while maintaining appropriate tissue homeostasis (Scadden, 2006). In the mouse dentition, molars and incisors start with similar developmental processes but differ in tissue homeostasis due to the differing fates of their dental epithelial stem cells during development. In continuously growing mouse incisors, dental epithelial stem cells and their surrounding niche are maintained throughout life, resulting in the continuous differentiation of ameloblasts and deposition of enamel on the labial side of the incisor, analogous to the crown of a molar (Harada et al., 1999; Harada et al., 2002). In contrast, in non-continuously growing mouse molars, the cervical loop niche including putative epithelial stem cells disappears after crown formation and a double layer of root sheath epithelium directs limited root growth postnatally.
BMP activity negatively regulates Shh transcription during limb development (Bastida et al., 2009). In this study, our time-course analyses show the dynamic and complementary expression patterns of pSmad1/5/8 and Gli1, and the dynamic and colocalized expression patterns of Gli1 and Sox2 in the dental epithelium of developing molars. Blockage of the BMP-Smad4 signaling cascade in the dental epithelium results in persistence of SHH-Gli1 signaling activity and Sox2+ epithelial stem cells. Moreover, ablation of Shh in Smad4 mutant mice results in the disappearance of Sox2+ dental epithelial stem cells and the restoration of crown and root morphology. Our findings suggest that the BMP-Smad4-regulated SHH-Gli1 signaling pathway likely regulates Sox2+ epithelial stem cell maintenance during molar development. Our results are consistent with the role of the SHH-Gli1 pathway in stem cell niche maintenance in the developing neocortex and dental mesenchyme (Palma and Altaba, 2004; Zhao et al., 2014). Interestingly, our previous study has shown that Smad4-mediated BMP signaling is required for SHH expression during early tooth development and postnatal root development (Huang et al., 2010; Xu et al., 2008). This difference in regulatory outcomes of BMP/Smad4 and SHH signaling at different stages of tooth development highlights the complex and context-dependent nature that each signaling pathway may play within its network, varying at different developmental time points during organogenesis.
In incisors, loss of Smad4 in the dental epithelium releases the BMP inhibitory effect on the SHH pathway and expands the SHH-Gli1 signaling activity from the cervical loop to the TA cell region. This altered spatial distribution of SHH-Gli1 signaling activity seems to expand the stem cell niche in continuously growing mouse incisors, supported by the expansion of Sox2+ dental epithelial stem cells, consistent with our findings in developing molars in Smad4 mutant mice. Thus, the interplay between the BMP/Smad4 and SHH/Gli1 signaling pathways also regulates the fate of dental epithelial stem cells during mouse incisor development. Taken together, these findings suggest the BMP/Smad4/SHH/Gli1 signaling cascade is a well-conserved mechanism regulating epithelial stem cells during tooth morphogenesis.
In addition, activation of WNT/β-catenin signaling in the dental epithelium of postnatal mouse incisors resulted in similar phenotypes to those of Smad4 mutant incisors (Liu et al., 2010; Wang et al., 2009). Constitutive activation of β–catenin decreased BMP signaling activity in incisor dental epithelium (Liu et al., 2010). These findings suggest that the WNT-BMP signaling cascade might also participate in the regulation of dental epithelial cell fate. However, specific ablation of Bmpr1a in the dental epithelium using KRT5-rtTA;tetO-Cre;Bmpr1afl/fl mice induced from E14.5 resulted in switched differentiation of crown epithelia into the root lineage and led to formation of ectopic cementum-like structures, which is related to the upregulation of Wnt/β-catenin signaling and epithelial-mesenchymal transition (Yang et al., 2013). The different findings from our observations might be due to the utilization of a different transgenic mouse model induced at a different time point.
From an evolutionary-developmental biology perspective, the regulation of the epithelial stem cell niche seems to control the switch between crown and root during tooth development, allowing for the existence of different tooth types in nature, such as brachydont (lower crowned), hypsodont (higher crowned) and hypselodont (continuously growing) teeth (Tummers and Thesleff, 2003, 2008). Our study suggests that a BMP/Smad4/SHH signaling network might control the crown/root switch via the regulation of dental epithelial stem cells and their niche during tooth development. Moreover, our study highlights the importance of crosstalk between two major pathways, BMP and SHH, in the regulation of epithelial stem cell fate during tooth development. This discovery has significant implications for our understanding of the regulatory mechanisms of epithelial stem cell maintenance and suggests novel molecular approaches for tooth regeneration.
EXPERIMENTAL PROCEDURES
Animals
All animal models (10 different transgenic lines), their source of origin (e.g., Jackson Laboratory ID#), and original references describing each of these 10 transgenic lines are listed in the Supplemental Experimental Procedures. All mouse experiments were conducted in accordance with protocols approved by the Department of Animal Resources and the Institutional Animal Care and Use Committee of the University of Southern California.
Tamoxifen and Doxycycline Administration
Tamoxifen (Sigma, T5648) was dissolved in corn oil (Sigma, C8267) at 20 mg/ml and injected intraperitoneally (single injection, 2mg/10g body weight). Doxycycline rodent diet (Harlan, TD.01306) was administered every day.
Immunofluorescence Staining, X-gal Staining and In Situ Hybridization
The following primary antibodies were used in our study: Beta galactosidase (Abcam ab9361, 1:50) (used for detecting Gli1 expression in Gli1-lacZ mice), Bmi1 (Abcam ab14389, 1:200), Keratin 14 (Abcam ab7800, 1:200), Ki67 (Abcam ab16667, 1:100), pSmad1/5/8 (Cell Signaling 9511S, 1:500), Sox2 (Abcam ab97959, 1:2000 for in vivo, 1:500 for in vitro). Mouse cDNA clones were kindly provided by several laboratories: Amelogenin, Malcolm Snead; Lfng and Notch1, Thomas Gridley; Shh, Andrew McMahon. Detailed protocols are provided in the Supplemental Experimental Procedures.
Isolation and Culture of Molar Epithelial Stem Cells
Lower first molars were carefully dissected at PN3.5 with fine forceps. Dissected lower first molars were then placed in 500 μl 4 mg/mL Dispase II (Roche, 04942078001) in a 12-well plate for 20-30 min at 37°C. The molars were removed from Dispase II and placed in cold DMEM/F12 media (Invitrogen, 11320). The dental epithelium and mesenchyme were gently separated using two syringes (0.5 ml, 28 G 1/2) and the dental epithelial tissues were immediately placed in cold DMEM/F12 in a 1.5 ml centrifuge tube on ice. The tubes were spun down and the medium was removed, then replaced with 500 μl cell detachment solution (Sigma, A7089). After incubating the tissues in cell detachment solution for 30-40 min at 37 °C, the tissues were dissociated by gently pipetting up and down using a 1,000 μl low-adhesion pipette tip (USA Scientific, 1182-1830). A single cell suspension was further produced through a 70 μm sterile cell strainer (Fisher Scientific, 08-771-2). After counting cells using an Automated Cell Counter (Bio-Rad, TC10), 6 × 104 cells were seeded in a 6-well plate in 1.5 ml DESC media, which consists of DMEM/F12 supplemented with mouse EGF recombinant protein (R&D, 2028-EG-200) at a concentration of 20 ng/ml, FGF recombinant protein (R&D, 233-FB-025) at a concentration of 25 ng/ml, 1X B27 supplement (Invitrogen, 10889-038), and 1% antibiotic solution (penicillin, 100 U/ml, streptomycin, 50 μg/ml) (Invitrogen, 15140-122). Initial cultures were allowed to rest undisturbed for 4 days after plating to allow cell adhesion and colony formation. After 4 days culture in 5% CO2 at 37°C, colonies were checked under a microscope and half of the volume (750 μl) of the old medium was replaced with an equal volume (750 μl) of new medium. After the first medium change, additional changes of 2 ml were performed every other day and colonies were checked under a microscope each time.
Colony-forming Assay
Independent dental epithelial cells (6 × 104) isolated from PN3.5 molars were seeded into 6-well culture plates. After 7 days, the culture plates were stained with a mixture of 0.1% toluidine blue and 2% paraformaldehyde solution. Colonies containing more than 50 cells were counted as single colony clusters.
MicroCT Analysis
MicroCT analysis was performed using a SCANCO μCT50 device at the University of Southern California Molecular Imaging Center. The microCT images were acquired with the x-ray source at 70 kVp and 114 μA. The data were collected at a resolution of 10 μm. The three-dimensional (3D) reconstruction was done using AVIZO 7.1 (Visualization Sciences Group).
Statistics
A two-tailed Student’s t test was applied for statistical analysis. A p value of less than .05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Julie Mayo and Bridget Samuels for critical reading of the manuscript. This study was supported by a grant from the NIDCR, NIH (R01 DE022503) to Yang Chai.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, Croft NJ, Cebra-Thomas JA, Metzger D, Chambon P, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram Mary A., Polo Jose M., Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. Sox2+ Adult Stem and Progenitor Cells Are Important for Tissue Regeneration and Survival of Mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastida MF, Sheth R, Ros MA. A BMP-Shh negative-feedback loop restricts Shh expression during limb development. Development. 2009;136:3779–3789. doi: 10.1242/dev.036418. [DOI] [PubMed] [Google Scholar]
- Biehs B, Hu JK-H, Strauli NB, Sangiorgi E, Jung H, Heber R-P, Ho S, Goodwin AF, Dasen JS, Capecchi MR, et al. BMI1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat Cell Biol. 2013;15:846–852. doi: 10.1038/ncb2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C. Stem cells: Skin regeneration and repair. Nature. 2010;464:686–687. doi: 10.1038/464686a. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annual Review of Cell and Developmental Biology (Palo Alto: Annual Reviews) 2006:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-Renewal, Multipotency, and the Existence of Two Cell Populations within an Epithelial Stem Cell Niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Botchkarev VA, Botchkareva NV, Nakamura M, Huber O, Funa K, Lauster R, Paus R, BA. G. Noggin is required for induction of the hair follicle growth phase in postnatal skin. The FASEB Journal. 2001;15:2205–2214. doi: 10.1096/fj.01-0207com. [DOI] [PubMed] [Google Scholar]
- Chavez MG, Hu J, Seidel K, Li C, Jheon A, Naveau A, Horst O, Klein OD. Isolation and culture of dental epithelial stem cells from the adult mouse incisor. J Vis Exp. 2014 doi: 10.3791/51266. doi: 10.3791/51266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- Feng J, Mantesso A, De Bari C, Nishiyama A, Sharpe PT. Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proceedings of the National Academy of Sciences. 2011;108:6503–6508. doi: 10.1073/pnas.1015449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Tumbar T, Guasch G. Socializing with the Neighbors: Stem Cells and Their Niche. Cell. 2004;116:769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung H-S, Mustonen T, Wang YA, Thesleff I. Localization of Putative Stem Cells in Dental Epithelium and Their Association with Notch and Fgf Signaling. The Journal of Cell Biology. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Toyono T, Toyoshima K, Yamasaki M, Itoh N, Kato S, Sekine K, Ohuchi H. FGF10 maintains stem cell compartment in developing mouse incisors. Development. 2002;129:1533–1541. doi: 10.1242/dev.129.6.1533. [DOI] [PubMed] [Google Scholar]
- Haramis A-PG, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJA, Clevers H. De Novo Crypt Formation and Juvenile Polyposis on BMP Inhibition in Mouse Intestine. Science. 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- He XC, Zhang J, Tong W-G, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-[beta]-catenin signaling. Nat Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- Huang X, Xu X, Bringas P, Hung YP, Chai Y. Smad4-Shh-Nfic signaling cascade–mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. Journal of Bone and Mineral Research. 2010;25:1167–1178. doi: 10.1359/jbmr.091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juuri E, Jussila M, Seidel K, Holmes S, Wu P, Richman J, Heikinheimo K, Chuong C-M, Arnold K, Hochedlinger K, et al. Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development. 2013;140:1424–1432. doi: 10.1242/dev.089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, Klein Ophir D., Thesleff I, Michon F. Sox2+ Stem Cells Contribute to All Epithelial Lineages of the Tooth via Sfrp5+ Progenitors. Developmental Cell. 2012;23:317–328. doi: 10.1016/j.devcel.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SO, Chung IH, Xu X, Oka S, Zhao H, Cho ES, Deng C, Chai Y. Smad4 is required to regulate the fate of cranial neural crest cells. Developmental Biology. 2007;312:435–447. doi: 10.1016/j.ydbio.2007.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proceedings of the National Academy of Sciences. 2007;104:10063–10068. doi: 10.1073/pnas.0703004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-Y, Yang L-T. Differential response of epithelial stem cell populations in hair follicles to TGF-β signaling. Developmental Biology. 2013;373:394–406. doi: 10.1016/j.ydbio.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Liu F, Dangaria S, Andl T, Zhang Y, Wright AC, Damek-Poprawa M, Piccolo S, Nagy A, Taketo MM, Diekwisch TGH, et al. β-catenin Initiates Tooth Neogenesis in Adult Rodent Incisors. Journal of Dental Research. 2010;89:909–914. doi: 10.1177/0022034510370090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert IMA, Brunsting JF, Wierenga PK, Faber H, Stokman MA, Kok T, Visser WH, Kampinga HH, de Haan G, Coppes RP. Rescue of Salivary Gland Function after Stem Cell Transplantation in Irradiated Glands. PLoS ONE. 2008;3:e2063. doi: 10.1371/journal.pone.0002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. How cells read TGF-[beta] signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- Nakatomi M, Morita I, Eto K, Ota MS. Sonic Hedgehog Signaling is Important in Tooth Root Development. Journal of Dental Research. 2006;85:427–431. doi: 10.1177/154405910608500506. [DOI] [PubMed] [Google Scholar]
- Oshimori N, Fuchs E. Paracrine TGF-β Signaling Counterbalances BMP-Mediated Repression in Hair Follicle Stem Cell Activation. Cell Stem Cell. 2012;10:63–75. doi: 10.1016/j.stem.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma V, Altaba A.R.i. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 2004;131:337–345. doi: 10.1242/dev.00930. [DOI] [PubMed] [Google Scholar]
- Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Developmental Biology. 2003;262:195–205. doi: 10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Sancho E, Batlle E, Clevers H. SIGNALING PATHWAYS IN INTESTINAL DEVELOPMENT AND CANCER. Annual Review of Cell and Developmental Biology. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, Cao T, Carano RAD, Curran T, Schober M, et al. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development. 2010;137:3753–3761. doi: 10.1242/dev.056358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the Epithelial Stem Cell Niche in Skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummers M, Thesleff I. Root or crown: a developmental choice orchestrated by the differential regulation of the epithelial stem cell niche in the tooth of two rodent species. Development. 2003;130:1049–1057. doi: 10.1242/dev.00332. [DOI] [PubMed] [Google Scholar]
- Tummers M, Thesleff I. Observations on continuously growing roots of the sloth and the K14-Eda transgenic mice indicate that epithelial stem cells can give rise to both the ameloblast and root epithelium cell lineage creating distinct tooth patterns. Evolution & Development. 2008;10:187–195. doi: 10.1111/j.1525-142X.2008.00226.x. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Smith GH. Murine Mammary Epithelial Stem Cells: Discovery, Function, and Current Status. Cold Spring Harbor Perspectives in Biology. 2011;3 doi: 10.1101/cshperspect.a004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-P, O'Connell DJ, Lund JJ, Saadi I, Kuraguchi M, Turbe-Doan A, Cavallesco R, Kim H, Park PJ, Harada H, et al. Apc inhibition of Wnt signaling regulates supernumerary tooth formation during embryogenesis and throughout adulthood. Development. 2009;136:1939–1949. doi: 10.1242/dev.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Jr, Deng C, Chai Y. Ectodermal Smad4 and p38 MAPK Are Functionally Redundant in Mediating TGF-β/BMP Signaling during Tooth and Palate Development. Developmental Cell. 2008;15:322–329. doi: 10.1016/j.devcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li C, Xu X, Deng C. The tumor suppressor SMAD4/DPC4 is essential for epiblast proliferation and mesoderm induction in mice. Proceedings of the National Academy of Sciences. 1998;95:3667–3672. doi: 10.1073/pnas.95.7.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Hai B, Qin L, Ti X, Shangguan L, Zhao Y, Wiggins L, Liu Y, Feng JQ, Chang JYF, et al. Cessation of Epithelial Bmp Signaling Switches the Differentiation of Crown Epithelia to the Root Lineage in a β-Catenin-Dependent Manner. Molecular and Cellular Biology. 2013;33:4732–4744. doi: 10.1128/MCB.00456-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Feng J, Seidel K, Shi S, Klein O, Sharpe P, Chai Y. Secretion of Shh by a Neurovascular Bundle Niche Supports Mesenchymal Stem Cell Homeostasis in the Adult Mouse Incisor. Cell Stem Cell. 2014;14:160–173. doi: 10.1016/j.stem.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.