Abstract
The human helicase senataxin (SETX) is implicated in the neurodegenerative diseases amyotrophic lateral sclerosis (ALS4) and ataxia with oculomotor apraxia (AOA2). Here, we reveal a role for SETX in controlling the antiviral response. Cells depleted for SETX and AOA2 patient-derived SETX-deficient cells exhibit increased expression of antiviral mediators in response to infection. Mechanistically, we propose a model whereby SETX attenuates RNA polymerase II (RNAPII) activity at genes stimulated upon viral sensing, thus controlling the magnitude of the host response to pathogens and the biogenesis of numerous RNA viruses (e. g. Influenza A virus and West Nile virus). Our data indicate a potentially causal link between SETX inborn errors, susceptibility to infection and development of neurologic disorders.
The innate immune response to infection represents the first line of cellular defense against invading pathogens. This response is orchestrated by a complex cohort of genes that are specifically induced upon infection1. Despite their protective role, prolonged or aberrant expression of such genes can be deleterious to the host2, 3, 4. Indeed, dysregulated expression of many of these pro- and anti-inflammatory mediators can contribute to deaths caused by septic shock2 and pandemic influenza virus infection3, and has been linked to the aetiology of several life-threatening diseases4. The expression of antiviral mediators is thus strictly regulated, with subsets of genes expressed in distinct temporal patterns during infection5.
A classic example of the temporal regulation of infection-induced gene expression is the induction of type I interferons (IFN) and interferon stimulated genes (ISGs) following viral infection. Upon viral entry, dedicated cytosolic receptors (including RIG-I, LGP2 and Mda5) recognize virus-derived nucleic acids, and initiate a cascade of events that ultimately lead to the activation and nuclear translocation of interferon regulatory factor 3 (IRF3)6. Activated IRF3 is then recruited to the promoters of early response genes, including interferon β (IFNB1), and induces their transcription6. IFN-β subsequently induces the expression of ISGs via a second, IFN-dependent wave of transcription that relies on JAK-STAT signaling5. Importantly, besides IFN-β, the early interferon-independent wave of transcription also allows for the concomitant expression of other important transcription factors (e.g. IRF7) and antiviral mediators (e.g. IFIT1, RIG-I), establishing a feed-forward loop that is likely to potentiate the antiviral response during the course of infection. Light has been shed upon the cellular signaling pathways that culminate in the early induction of antiviral genes, but detailed mechanistic insight into how chromatin regulatory factors control this response is lacking 7.
Stimulus-induced transcription is a tightly regulated, multi-step process that is controlled by a plethora of protein complexes and through multiple mechanisms acting at the level of RNA polymerase II (RNAPII) initiation, pause-release, elongation and termination8. Studies have highlighted important factors and rate limiting steps9, 10 of these processes that operate on chromatin, which function as a signal integration platform during the cell’s response to various stimuli11. For example, transcription initiation and the progression to elongation- and termination- phases of transcription12 is heavily influenced by the dynamic changes to post-translational modifications occurring on histone N-terminal domains and the RNAPII C-terminal domain (CTD)13. The recognition of these chemical marks by dedicated proteins controls transcriptional activity14, co-transcriptional events15, 16 and ultimately, cell responses17. Thus, as previously shown the ability to target key regulators of the chromatin environment presents an attractive means through which to manipulate the transcriptional response to stimuli18, 19.
Using genomics, cell biology and genetic approaches, we report here that the human ATP-dependent helicase, SETX, exerts an inhibitory effect on the transcriptional response to viral infection. We show that SETX promotes RNAPII early termination at IRF3-dependent antiviral genes. This effect is dependent on SETX RNA binding and ATPase activity and results in the inhibition of IRF3-dependent antiviral gene expression. We observe increased expression of antiviral mediators in SETX-depleted and SETX-deficient cells as well as in vivo in Setx−/− mice. In addition, reduced cellular levels of SETX render cells more resistant to viral infection and suppresses viral biogenesis. Notably, SETX-deficient cells derived from patients with AOA2 displayed increased expression of antiviral mediators during infection and inhibition of viral replication compared to wild type controls. These results describe a novel function for SETX as a negative regulator of the antiviral response and may have relevant implications in neurological disorders characterized by SETX mutations.
RESULTS
SETX inhibits antiviral gene expression
We were interested in identifying novel factors that could affect the transcriptional response to viruses. The putative ATP-dependent helicase SETX and its yeast orthologue Sen1 have been implicated as positive regulators of basal gene transcription20, 21, 22. Sen1 has also been shown to negatively regulate the expression of genes induced by cell wall perturbations23, a system functionally equivalent to the cellular response to stimuli in mammals24. This, along with the intriguing possibility of a connection between viral infection and AOA2 and ALS4, led us to hypothesize that SETX might regulate the transcriptional response of mammalian cells to viral infections 1, 5.
We first examined SETX subcellular localization in lung epithelial cell line A549 prior to and after infection with the influenza A virus A/Puerto Rico/8/1934(ΔNS1) (hereafter PR8ΔNS1) virus. SETX-sufficient wild-type and SETX-deficient human fibroblasts25 were used as controls for antibody staining. Immunohistochemistry showed a nuclear distribution of SETX in infected A549 and wild-type control fibroblasts, indicating that the subcellular localization of SETX was unaffected by infection (Supplementary Fig. 1a). SETX co-eluted with high molecular weight nuclear complexes positive RNAPII in glycerol gradient fractionation (Supplementary Fig. 1b). The association with the transcriptional machinery was further supported by the fact that SETX co-immunoprecipitated with RNAPII (Supplementary Fig. 1c), similar to what has been observed in other cell types21. Altogether, these results are consistent with a potential role for SETX in transcription regulation upon infection.
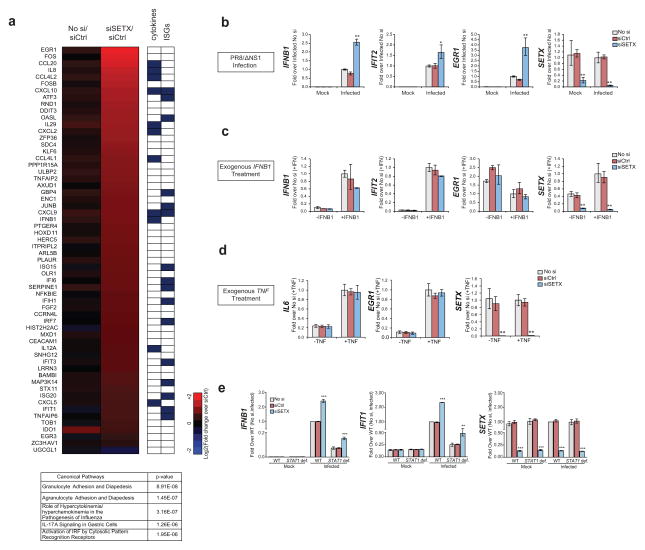
To substantiate this, we profiled the global mRNA expression patterns of SETX siRNA (siSETX)-treated cells and control (siCtrl) cells pre- and 4 hours post-infection with PR8ΔNS1 virus. SETX depletion was specific and resulted in increased upregulation of antiviral genes (hypergeometric test P: 1.72×10−23), (Fig. 1a, Supplementary Figs. 1d and Supplementary Table 1) with negligible effects on housekeeping gene expression (Supplementary Fig. 1e). Conversely, depletion of the exonuclease XRN2, which has been implicated along with SETX in positively controlling the basal expression of two housekeeping genes20 did not result in a similar increased upregulation of antiviral gene expression upon infection (Supplementary Figs. 1f–1h and Supplementary Table 2). This result further corroborates the specificity of SETX during the cellular response to infection and suggests that its negative regulation at antiviral genes is exerted via an unknown mechanism.
Figure 1.
SETX suppresses antiviral gene expression during infection (a) Heatmap of gene expression fold changes in un-transfected (No si) or SETX siRNA (siSETX) transfected A549 cells compared to control siRNA transfected cells (siCtrl) (p<0.01; ANOVA with post-hoc TUKEY HSD test, n=3 per condition) at an early (4 hours) time point post-infection. Known cytokines and interferon-stimulated genes (ISGs) are indicated in the right panel. The top five significant canonical pathways enriched within siSETX-affected genes are shown in the table underneath. (b–d). Expression of indicated genes in siSETX transfected, siCtrl transfected and untransfected A549 cells upon PR8 NS1 infection (b), exogenous IFNB1 treatment (c) or exogenous TNF treatment (d). (e) Expression of the indicated genes in uninfected or PR8ΔNS1 infected (4 hours post-infection) human STAT1-deficient (STAT1 def.) and STAT1-proficient (WT) human fibroblast cells left untreated, or treated with siCtrl or siSETX. For b–e, the means and standard deviation of three independent experiments are shown. Asterisks (*, ** and ***) indicate t-test p-values of <0.05, <0.005 and <0.0005 respectively.
Phosphorylation of the transcription factor IRF3, which is a terminal event in the signaling cascade subsequent to viral sensing6, was unchanged in infected SETX silenced (siSETX) cells as compared to control cells (Supplementary Fig. 1i) demonstrating that the increased expression of early antiviral genes in SETX-depleted cells is not caused by dysregulated signaling. Initial levels of viral genomic and messenger RNA in SETX-depleted and control cells were also equal (Supplementary Figs. 1j and 1k), ruling out indirect effects resulting from differences in infection efficiency. Altogether, these data suggest that upon infection, SETX negatively regulates antiviral genes induced immediately after viral sensing, when transcription of these genes is predominantly driven by the transcription factor IRF3.
To determine whether the activity of SETX in our system is specific to viral infection, we further characterized the involvement of SETX in response to different stimuli. Our analyses confirmed that SETX depletion increased antiviral gene expression in response to influenza PR8ΔNS1 virus infection (Fig. 1b) and showed that neither addition of exogenous IFN-β (which signals through the JAK-STAT signaling cascade6), nor stimulation with tumor necrosis factor (TNF) which acts through different signaling cascades, were able to recapitulate the viral infection-induced gene expression phenotype (Figs. 1c and 1d).
To further corroborate the specificity of SETX activity downstream of viral sensing, we infected immortalized human fibroblasts deficient for STAT1. Since STAT1 protein is activated downstream of IFN-β and is essential for the IFN-β-dependent gene expression program, this system allowed us to uncouple the specific arms of signaling implicated in viral sensing and IFN-β dependent response. Depletion of SETX in STAT1 deficient cells during infection resulted in increased antiviral gene expression compared to control cells (Fig. 1e), further supporting a model in which SETX acts specifically on early IFN-independent antiviral gene expression.
SETX inhibits antiviral gene transcription
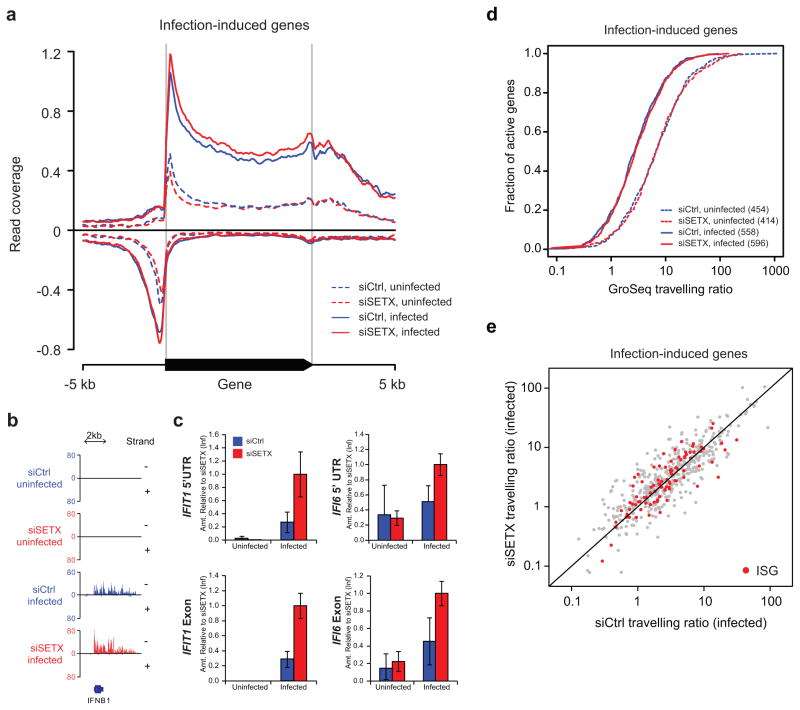
We then investigated the impact that SETX depletion had on viral infection-induced transcription by performing genome-wide analysis of nascent (elongating) RNA via Global Run On sequencing (GRO-seq)18, 26 in control non-targeting siRNA (siCtrl) and SETX siRNA (siSETX)-treated A549 cells prior to, and 4 hours after infection (Fig. 2 and Supplementary Fig. 2) with PR8ΔNS1 virus. Our GRO-Seq analyses show that SETX depletion slightly reduces basal transcription in non-infected cells (dotted lines in Fig. 2a and Supplementary Fig. 2a–d) confirming previous findings of a limited effect of SETX deficiency on housekeeping gene transcription20. In contrast, upon infection, SETX functions as a negative regulator of antiviral transcription: the average profiles of engaged RNAPII indicate that SETX depletion specifically increased transcription of genes induced 4 hours post-infection which are highly enriched for antiviral response genes (solid lines in Fig 2a; Supplementary Table 3), (hypergeometric test P: 2.35×10-78). The increase of inducible gene transcription in SETX-depleted cells is illustrated for representative virus-triggered gene IFNB1 (Fig. 2b) and quantified via Run-on qPCR for representative virus-induced genes IFIT1 and IFI6 (Fig. 2c).
Figure 2.
Loss of SETX increases engaged RNAPII at infection-induced genes. (a) Meta-analysis of GRO-seq signals across 872 genes induced in human A549 cells upon infection with influenza PR8ΔNS1, before (dashed) and 4 hours after infection (solid) in siCtrl (blue lines) and siSETX (red lines) treated cells. Y-axis indicates the geometric mean coverage per 5 million reads in a 30 bp sliding window. (b) GRO-seq read coverage in the IFNB1 gene region. Y-axis indicates base coverage per 5 million reads. − and + refer to the DNA strands. (c) Quantification of GRO-seq by using qPCR for the representative affected IFI6 and IFIT1 genes. The means and standard deviations of three experiments are shown. (d) Travelling ratios (TR) of active infection induced genes for the conditions indicated in a. TRs were calculated as the average number of tags per bp in the promoter-proximal region (30 to +300 bp around TSS) divided by that on the gene body (+300 bp to the end of the gene). The number of active genes identified in each condition is indicated between brackets. (e) Scatter plot of TRs of active genes in siSETX and siCtrl treated A549 cells, 4 hours after infection with PR8ΔNS1. Known interferon stimulated genes (ISG) are highlighted in red.
To explore the mechanism responsible for the increased transcription in response to SETX depletion we compared the RNAPII density at the promoter divided by the density in the gene body between conditions. This parameter, called the RNAPII travelling ratio (TR)27, defines the propensity of RNAPII to be paused or licensed into elongation. The TR of infection-induced genes and ISGs in siCtrl- and siSETX-treated cells were indistinguishable (Fig. 2d and 2e; Kolmogorov–Smirnov test, p-value 0.7093), indicating that the increased antiviral gene transcription in siSETX-treated cells did not involve changes in RNAPII elongation18 or in pause-release27, and is therefore likely achieved at (or shortly after) transcription initiation28. Indeed, treatment of control and SETX-depleted cells with the positive transcription elongation factor b (P-TEFb) inhibitor Flavopiridol, that blocks transcription immediately following initiation, reduced the induction of antiviral genes in infected siCtrl- and siSETX-treated cells to similar levels (Supplementary Fig. 2e). Taken together, our results suggest that during infection, SETX negatively regulates transcription and, in turn, the expression levels of virus-induced genes.
SETX promotes premature termination of antiviral genes
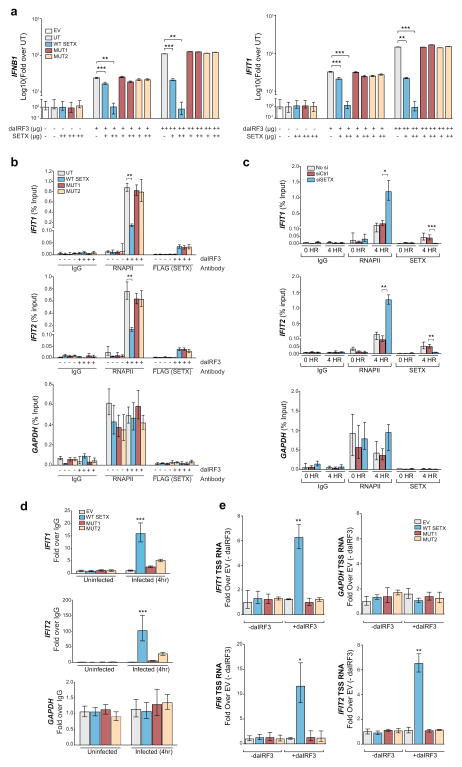
To gain additional insight into the mechanism of action of SETX, we undertook a series of experiments that rely on the transient expression of a constitutively active form of the transcription factor IRF3 (hereafter dominant active IRF3; daIRF3), which is sufficient to induce IFN-β and ISG expression in non-infected cells6. If the effects of SETX were exerted downstream of IRF3, co-expression of SETX should counteract daIRF3-induced activation; this effect was indeed observed (Fig. 3a; blue bars).
Figure 3.
SETX controls IRF3-dependent antiviral gene expression. (a) Expression levels of IFNB and IFIT1 genes in A549 cells upon transient expression of the indicated proteins. The means and standard deviation of three independent experiments are shown. The amount of DNA transfected per condition is indicated as (−) : 0 μg; (+): 1 μg; (++): 5 μg. (b) ChIP-qPCR analysis of RNAPII and Flag-SETX using the experimental system described in (a), and showing the recruitment of RNAPII and wild type and mutant SETX to the transcription start sites (TSS) of antiviral (IFIT1 and IFIT2) and housekeeping (GAPDH) genes. IgG represents the control IP. A representative experiment of three independent ChIP experiments is shown, where the mean and standard deviation of three technical replicates are displayed. (c) ChIP analysis of endogenous RNAPII and SETX at the TSS of the indicated genes upon infection in control or SETX-depleted A549 cells. A representative experiment of three independent experiments is shown. The means and standard deviation of three technical replicates are shown. (d). RNA-IP of uninfected or PR8ΔNS1 infected A549 cells transfected with empty vector (EV) or vectors expressing wild type or mutant SETX. Specific enrichment of SETX binding to 5′ UTR-derived RNAs of IFIT1, IFIT2 and GAPDH was analyzed by qPCR. The means and standard deviation of three independent experiments are shown. (e) qPCR analyses of TSS-associated RNA in A549 cells overexpressing wild type and mutant SETX along with daIRF3. Means and standard deviations of three independent experiments are shown. For a–e, asterisks (*,** and ***) indicate t-test p-values <0.05, 0.005 and <0.0005 respectively. Abbreviations used: UT: untreated; EV: empty vector; WT SETX: wild type SETX; MUT1: K1969Q SETX helicase mutant; MUT2: G2343D SETX RNA-binding mutant; daIRF3: dominant active IRF3
The yeast orthologue of SETX, Sen1, induces transcription termination at small nucleolar RNAs (sno-RNAs)29 by virtue of its RNA binding and ATPase activities30. We thus assessed whether RNA binding and enzymatic activities were similarly required for human SETX function. We first confirmed that purified human SETX possesses helicase activity (Supplementary Fig. 3a and 3b). In addition, we generated a human SETX mutant devoid of its enzymatic (helicase and translocase) activity by altering the conserved Walker A motif shared by many cellular ATPases (K1969Q; MUT1) and an RNA-binding mutant (G2343D; MUT2) by altering SETX motif IV31 (Supplementary Fig. 3a–3e). Our results demonstrated that both activities are essential for inhibiting daIRF3-dependent transcription (Fig. 3a; red and yellow bars). Chromatin immunoprecipitation (ChIP) experiments performed under the same experimental conditions further showed that although wild type and mutant SETX proteins were present in equal amounts at promoter regions of induced antiviral genes, only wild type SETX caused a concomitant reduction of RNAPII at promoters (Fig. 3b). This effect is then dependent upon the ATPase and RNA binding activity of SETX. Consistent with these observations, reduction of endogenous SETX lead to an increase of RNAPII loading at promoters of the viral-induced genes IFIT1 and IFIT2 (Fig. 3c). Our data, describing the amounts of RNAPII at anti-viral gene promoters being inversely correlated with SETX activity and not SETX binding, suggest that SETX acts to promote premature termination of RNAPII at IRF3-dependent genes.
We thought that our results would fit a simple model whereby SETX, via binding to nascent RNA, would translocate (by means of its ATPase activity) toward RNAPII and induce early termination. This model is reminiscent of how the RhoA helicase controls termination in Escherichia coli32 and is analogous to how Sen1, the yeast orthologue of human SETX, controls termination at sno-RNAs30. For this model to be true, wild type SETX, unlike the RNA binding mutant, should bind 5′ RNA of antiviral genes induced by active IRF3. Moreover, in the same experimental setting, wild type SETX expression, unlike the SETX mutants, should lead to the enrichment of prematurely terminated 5′ end derived RNA. To test this model, we first investigated SETX RNA binding by RNA Immunoprecipitation. Consistent with our hypothesis, we observed that the RNA binding mutant (and to a similar extent, the SETX ATPase mutant) is severely impaired in binding 5′ end derived RNA from antiviral genes compared to wild type SETX (Fig. 3d). These results suggest that, although both mutants can be targeted as efficiently as wild type SETX to promoters of IRF3-dependent genes (Fig. 3b), they are both unable to bind such nascent RNAs. These results imply that targeting of SETX to promoters is independent of nascent RNA binding and ATPase activity, but that both activities are required to promote RNAPII termination.
We next investigated the effect of SETX action on the levels of transcription start site (TSS)-associated short RNAs (tssRNA)33, 34. We observed that expression of wild type SETX, but not the ATPase or RNA-binding mutants, increased the amount of tssRNAs at IRF3 dependent genes (Fig. 3e and Supplementary Fig. 3f–3h). Together, these data suggest that SETX promotes premature transcription termination at virus-induced gene promoters, and that its depletion renders the cells amenable to expressing higher levels of antiviral genes.
SETX and TAF4 coordinate the antiviral response
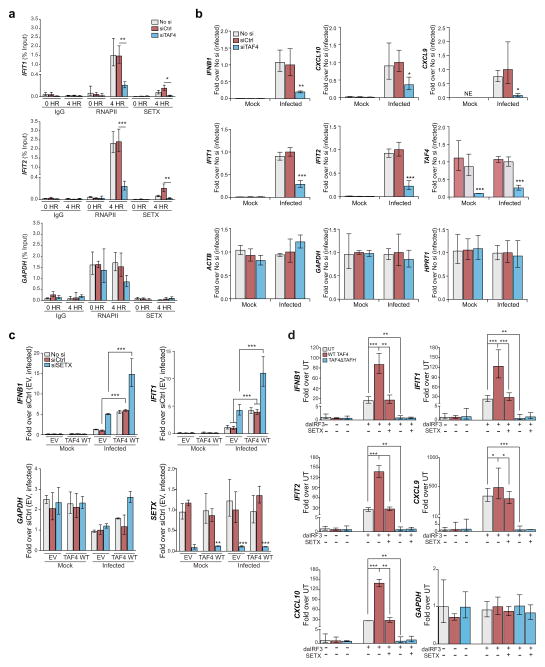
We next sought to determine which factors were responsible for coordinating SETX targeting at antiviral loci. Use of an affinity purification mass spectrometry assay (IP-MS/MS) revealed that Transcription initiation factor TFIID subunit (TAF4) is a primary interacting partner of SETX in infected cells (Supplementary Figs. 4a and 4b and Supplementary Table 4). In line with this, siRNA-mediated depletion of TAF4 in infected A549 cells resulted in reduced amounts of SETX at the target genes IFIT1 and IFIT2 (Fig. 4a). TAF4 has been shown to function as a transcriptional co-activator in inducible and stimulus-dependent gene expression programs (for example Wnt signaling)35, 36. TAF4 depletion led to a decrease in the antiviral gene expression upon infection (Fig. 4b). Conversely, overexpression of TAF4 increased the expression of antiviral genes upon infection (Fig. 4c). These results suggest that SETX expression can inhibit TAF4 dependent transactivation of IRF3 dependent genes. To prove this directly, we overexpressed the dominant active form of IRF3 with TAF4 and SETX. Consistent with our hypothesis, our results show that IRF3 dependent transcription was enhanced by TAF4 coexpression and counteracted by SETX (Fig. 4d).
Figure 4.
TAF4 controls antiviral gene expression. (a) ChIP analyses of IgG control, RNAPII and SETX in PR8ΔNS1 infected or control cells treated with control non-targeting siRNA (siCtrl) or siRNAs against TAF4 (siTAF4). Untreated cells (No Si) are included as controls. Analyses were performed at the transcriptional start (TSS) of the indicated genes. (b) Expression levels of the indicated genes upon infection in siTAF4 treated or control cells (siCtrl or No si). (c) Expression levels of the indicated genes in siCtrl and siSETX treated cells overexpressing wild type TAF4 or its empty vector control (EV). (d) Expression levels of the indicated genes in upon transient expression of the indicated proteins. Abbreviations: UT: untreated; SETX: wild type SETX; daIRF3: dominant active IRF3; TAF4: wild type TAF4; TAF4 Δ TAFH: TAFH deletion mutant. For a–d, A549 cells were used and the means and standard deviation of three independent experiments are shown; Asterisks (*,**,***) indicate p-values <0.05, < 0.005 and <0.0005 as calculated by t-tests against siCtrl treated cells (a–c) or the corresponding daIRF3 overexpressing cells (d).
TAF4 contains a homology domain (TAFH) previously shown to be required for activator-dependent gene expression35 via direct recruitment of positive and negative regulators of transcription37. As expected, overexpression of the TAFH deletion mutant (TAF4ΔTAFH) abolished TAF4 transactivation of antiviral gene expression (Fig. 4d). These data, along with protein interaction studies showing TAFH and SETX binding (Supplementary Fig. 4c and Table 4), suggest that TAF4 is a co-activator for antiviral genes, and that its effect is modulated by SETX. Our data show that a SETX-TAF4 axis could thus represent a pivotal arm of the cellular response to pathogens, whereby loss of function in either protein results in dysregulated innate immune responses to pathogens.
SETX-deficient human cells are hyper-responsive to infection
Finally, to determine the physiological consequence of SETX loss-of-function on viral replication, we first profiled viral growth kinetics in human cells depleted for SETX. Both wild type influenza virus PR8 and PR8ΔNS1 influenza viral growth were repressed in the absence of SETX (Supplementary Fig. 5a and 5b), consistent with a role of SETX in controlling the expression of cytokines and antiviral mediators. Consistent with the role of SETX in controlling the early antiviral response to viral sensing, its effect on viral growth depended on the presence of the cellular sensor for influenza virus, RIGI (Supplementary Fig. 5c). Overall, these results strongly suggest that the increased expression of antiviral genes in SETX deficient cells inhibits viral growth, directly implicating SETX as an attenuator of the antiviral response and representing a key player in a cellular buffering system that controls the magnitude of the host response to viruses.
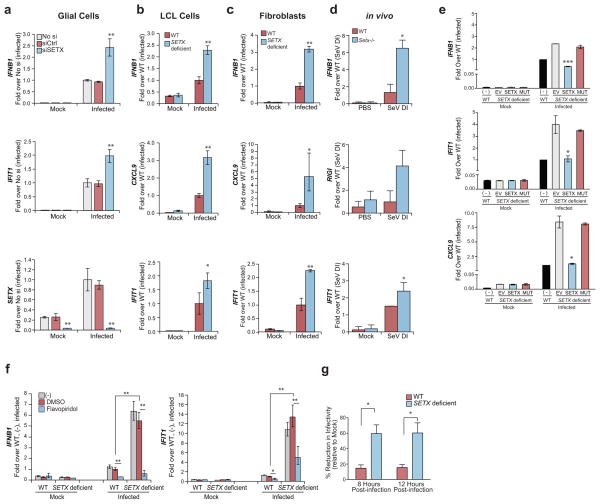
Loss of SETX function has been implicated in the neurodegenerative diseases ALS438 and ataxia with AOA239. We therefore performed ex vivo experiments to determine whether AOA2 patient-derived SETX-deficient cells could recapitulate the increased antiviral response observed in SETX-depleted A549 cells (Fig. 1c–1d) and siSETX treated immortalized glial cells (Fig. 5a). In line with our initial observations, SETX-deficient lymphoblastoid cells (LCL) (Fig. 5b) and primary human fibroblasts (Fig. 5c) derived from AOA2 patients25 displayed increased antiviral gene expression upon infection. We were also able to recapitulate these observations in an in vivo setting by intranasal administration of Setx−/− mice40 with immunostimulatory Sendai virus defective-interfering RNA. We observed increased expression of antiviral genes in Setx−/− lungs compared to wild type controls, further reinforcing the physiological role of SETX in regulating antiviral gene expression (Fig. 5d).
Figure 5.
Dysregulated antiviral response in SETX deficient human cells. (a–c) mRNA expression in PR8ΔNS1 infected (4 hours post-infection) SETX depleted and control human glial (a) or lymphoblastoid (LCL; b) and fibroblast (c) cells isolated from AOA2 patients derived SETX deficient and wild type human cells (WT). (d) qPCR expression analyses of the indicated genes in the lungs of WT or Setx−/− mice treated for 6 hours with 4 μg of in vitro transcribed Sendai virus defective interfering RNA (SeV DI; n = 3 per group) or PBS (n = 4 per group) intranasally. (e) qPCR analyses of representative antiviral genes in uninfected or PR8ΔNS1 infected SETX deficient LCL reconstituted with empty vector (EV), wild type (SETX) and ATPase mutant (MUT) SETX. (f) Expression of IFNB1 and IFIT1 in SETX deficient and WT fibroblasts in the presence of flavopiridol, DMSO vehicle control or untreated control cells. (g) WT and SETX deficient LCL cells were infected with PR8ΔNS1 at a high MOI and supernatants collected at 8 and 12 hours post-infection were used to pretreat A549 cells for 12h prior to infection with a PR8-GLuc reporter virus at an MOI 1. Luciferase signal was quantified 8 hours post-infection. The mean and standard error of 3 independent experiments are shown. For a–c, e and f the means and standard deviation of three independent experiments are shown. For a–g, asterisks (*,** and ***) indicate t-test p-values < 0.05, < 0.005 and 0.0005 respectively.
We were also able to confirm that SETX activity was dependent on its ATPase activity in primary human fibroblasts wild type SETX expression in human SETX-deficient fibroblasts, unlike the expression of the SETX enzymatic mutant, could reconstitute a normal host response to infection (Fig. 5e and Supplementary Fig. 5d). Notably, similar to epithelial cells (Supplementary Fig. 2e), the up-regulation of antiviral genes in SETX deficient cells was reduced to wild type levels by the pTEFB inhibitor Flavopiridol (Fig. 5f).
The consequences of dysregulated antiviral gene expression in SETX deficient cells were also apparent at the protein level in infected cells; secreted cytokines and chemokines were higher in SETX-deficient fibroblasts compared to control wild type fibroblasts (Supplementary Fig. 5e). Naive cells pretreated with the supernatants of infected SETX-deficient fibroblasts were also more resistant to a reporter virus infection than cells treated with supernatants derived from infected wild type cells (Fig. 5g). Similar to siSETX treated A549 cells, SETX-deficient fibroblasts suppressed viral growth of wild type influenza PR8 (Supplementary Fig. 5f). Finally, increased levels of antiviral gene expression and concomitant inhibition of viral biogenesis in SETX-deficient LCL cells were also observed upon infection with the neurotropic Kunjin virus (KUNV), a West Nile Virus subtype (Supplementary Fig. 5g–5h). Although SETX absence or depletion rendered the epithelial, fibroblast and glial cells we tested hyper-responsive to viral infection, the overall transcriptional regulation of the SETX protein itself was different amongst the three cell types (Fig. 5a), and suggests an additional layer of cell-type and stimulus specificity in the control of SETX-dependent transcription activities. Of note, we found that infection of glial cells resulted in increased SETX expression at RNA and protein levels (Fig. 5a; Supplementary Fig. 5i). It is possible that SETX up-regulation could reflect a cell type-specific built-in system to downregulate antiviral genes via a negative feedback loop controlling the magnitude of the antiviral response. Overall, these results indicate that SETX-mediated transcriptional control is disrupted in cells isolated from patients with SETX deficiency and corroborates, across a broad range of cell types and viruses, the role of SETX in regulating the initial steps of antiviral gene transcription (Supplementary Fig. 5j).
DISCUSSION
The cellular transcriptional response to infection is regulated by numerous cellular factors that ensure temporal and dynamic expression of specific antiviral genes. We have described here several lines of evidence that identify SETX as a novel factor controlling the antiviral response. First, depletion of SETX resulted in increased expression of several early infection-induced genes, including key cytokines (e.g. IFN-β) and other antiviral mediators (e.g. IFI6, IFIT1) important for the resolution of the infection. Second, overexpression of SETX was able to counteract IRF3-dependent activation of antiviral genes at endogenous loci. Third, human cells (lymphoblastoid and fibroblast) derived from patients that carry loss-of function mutations in the SETX gene displayed upregulation of virally induced genes upon infection compared to wild type cells. Fourth, SETX-deficient human cells, like SETX-depleted cells, inhibit viral biogenesis. Finally, we were also able to recapitulate in vivo the effects seen with SETX depletion where Setx−/− mice expressed higher amounts of inflammatory mediators upon immune stimulation compared to wild type mice.
Our findings indicate that SETX negatively regulates antiviral genes by promoting RNAPII early termination. The mechanism employed by SETX to inhibit antiviral genes parallels the mechanism described for Sen1, the yeast homologue of SETX, in controlling basal transcription at non-coding RNA loci. In fact, based on elegant experiments using mutant yeast strains with altered RNAPII processivities, it has been shown that Sen1 tracks along nascent RNA and induces RNAPII termination29 in a manner that is very similar to Rho dependent attenuation of transcription in bacteria. In vitro experiments have supported this view and revealed novel features of Sen1 activity30 suggesting that such a mechanism could be in place for regulating promoter activity.
Several of our observations support promoter-proximal termination events during RNAPII-mediated transcription as the mechanism by which SETX limits transcription of antiviral genes. Our experimental data show that SETX depletion causes a specific increase in transcriptionally active RNAPII across viral induced genes, with a concomitant increase in their mRNA levels, in a manner that is independent of RNAPII pause-release, elongation and termination. The ability of SETX to counteract transcriptional activity depends on its ATPase activity. In line with this, wild type SETX, unlike the SETX enzymatic and RNA binding mutants, binds to the 5′ end of nascent RNA at antiviral genes and increases the level of prematurely terminated RNA at the TSS of IRF3-dependent antiviral genes. Finally, overexpression of SETX modulates IRF3-dependent gene expression in a dose-dependent manner, suggesting that SETX levels can dynamically compete with IRF3-mediated transcription. Overall, our data reveal a novel function of SETX in negatively regulating the magnitude of antiviral response at both the cellular and organismal level. We thus suggest that SETX recruitment to its target genes drives early termination of transcription at the antiviral genes and limits their overall expression levels. SETX-mediated termination may depend directly on sequence-specific recognition of the target RNA itself through its RNA binding domain, and require SETX translocase activity. Alternatively, SETX-mediated termination may be driven by its interaction with structural elements, such as R-loops, formed at nascent transcript. The latter would require SETX helicase activity and it is a likelier possibility, given that (i) SETX can resolve RNA/DNA hybrids, (ii) such structures can be formed at promoters of inflammatory genes41, (iii) free 5′-end RNA at promoters can increase R loop formation and avoid the requirement for RNA cleavage to promote transcriptional termination.
Mechanistically, SETX depletion leads to diminished transcription at some housekeeping genes at basal state, while during infection it increases the activation of antiviral genes. Such positive and negative regulation could be simply dependent on differential binding partners targeting SETX activity to different gene regions (3′ end region for housekeeping genes versus the promoter region for antiviral genes). This would then result in a differential effect on transcription at specific gene subsets. At the housekeeping genes, interaction of SETX with 3′ end cleavage and termination complexes is supposed to coordinate proper termination16. In contrast, at the antiviral genes, control of premature termination by SETX at promoters is dependent on IRF3, a master transcription factor activated by the incoming virus, and requires the cofactor TAF4. Indeed, upon infection, IRF3-dependent gene expression is potentiated by TAF4 and subject to negative regulation by SETX. Such multilevel regulation is key during the cell response to viruses due to the detrimental effect of high/prolonged antiviral gene expression (see below) and it is reminiscent of how E protein and Wnt signaling coordinate recruitment of transcription factors and other cofactors to regulate stimulus dependent gene expression35, 36. Strikingly, in these studies, as in ours, TAF4 appears to play a pivotal role as a binding platform for the recruitment of positive and negative effectors that coordinate transcriptional activity during cell response.
In summary, we have revealed a new mechanism of regulating antiviral gene expression by an ATP-dependent helicase that controls the expression levels of IFN-β and other key antiviral genes. Our results suggest that, during viral infection, SETX functions as a surveillance factor to ensure that an appropriate physiological response to viruses is elicited. We hypothesize that SETX has evolved a role in controlling transcriptional responses to viruses, and as such, may be an amenable target for treatment of infections and inflammatory disorders. In the conditions studied here, SETX activity and regulation were specific for signaling downstream of viral sensing; however, we do not discount the possibility that loss of SETX disrupts other transcriptional networks.
Dysregulation of transcription has been linked to disease development42, as shown for TAF4 deficiency conferring susceptibility to Huntington disease43. Inborn SETX mutations are implicated in the development of neurodegenerative diseases ALS438 and AOA239. Our data indicate that loss of SETX results in an altered temporal activation of the innate immune response to viruses. Unlike congenital diseases that display a constitutive expression of inflammatory mediators4, 44, SETX–driven changes to the innate immune response manifest experimentally only upon infection. We postulate that the inducible phenotypes we observed in the context of SETX deficiency may be environmentally triggered with exposure to viral agents. Transient states of excessive inflammation may over time result in local dysregulation of immune homeostasis and progressive deterioration of affected tissues. We propose that infection plays an important role in the initiation or progression of AOA2 and ALS4.
ONLINE METHODS
Cells and viruses
A549, 293T, Madin-Darby canine kidney (MDCK) and SVGp12 fetal glial cells were originally obtained from the American Type Culture Collection (ATCC). Cells were maintained in Dulbecco’s minimal essential medium (DMEM) (Gibco, Life Technologies) supplemented with 2mM glutamine, 10% fetal bovine serum (FBS, Hyclone), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco, Life Technologies) at 37°C with 5% CO2. SETX-2RM (SETX-deficient) and C3ABR (wt) LCLs were maintained as described above. SETX-1RM human fibroblasts (SETX-deficient fibroblasts) and normal foreskin fibroblasts (NFF)25 were maintained in Dulbecco’s minimal essential supplemented with 2mM glutamine, 20% fetal bovine serum (FBS, Hyclone), 100 U/ml penicillin and 100 μg/ml streptomycin. A/Puerto Rico/8/1934 (PR8) was propagated in 8–10 day old embryonated chicken eggs. PR8ΔNS1 virus was propagated in NS1-expressing MDCK cells. Sendai virus (SeV) Cantell strain was propagated in 10-day old embryonated chicken eggs. KUNV was kindly provided by J. Lim. RIG-I deficient A549 cells were kindly provided by A. Garcia-Sastre.
Virus Growth Curves
A549 cells were treated with 50 nM of ON-TARGETplus SMARTpool (Thermo Scientific) SETX siRNA, Non-targeting siRNA, or were left untreated for 48 h prior to infection with either PR8 at an MOI of 0.05 pfu/cell or with PR8ΔNS1 at an MOI of 5 pfu/cell. Supernatants from PR8 and PR8ΔNS1 infections were collected at 12 and 24 hours post-infection (hpi). Where RIG-I deficient A549 or SETX-deficient primary fibroblasts were used, cells were plated the day before being infected at a low MOI (0.05 pfu/cell) with the influenza PR8 virus. Supernatants from infection were collected at 12 and 24 hours post infection. Viral titers were determined by plaque assay on MDCK cells.
For KUNV infections, wild type or SETX-deficient fibroblasts were infected with virus at an MOI of 0.05 pfu/cell. Supernatants were collected 32 hpi and virus titers were determined by plaque assay on Vero cells. Plaques were visualized by immunostaining with West Nile Virus anti-envelope clone E24 (BEI Resources), goat anti-mouse HRP conjugate and True Blue peroxidase substrate (KPL).
Immunofluorescence
A549 cells were cultured on coverslips overnight and then infected with the virus strains specified. At the indicated interval post-infection, cells were fixed in 3% paraformaldehyde (EMS) for 10 min at room temperature. Coverslips were washed in 1x PBS and blocked with blocking solution (1mg/ml BSA, 3% FBS, 0.1% Triton X100 and 1mM EDTA pH 8.0 in PBS) for 1 hour at room temperature. Cells were then probed with anti-SETX (sheep, diluted 1:400) and tubulin (mouse, diluted 1:2000) for 1 hr and detected by using the respective Alexa conjugated antibodies (Invitrogen). DNA was counterstained with DAPI.
ELISA
SETX-1RM cells and NFFs were left untreated or were treated with PR8ΔNS1 at a MOI of 100 pfu/cell and supernatants were collected at 4 and 12 hpi. Human IFN-α2, IFN-γ and CXCL10 in culture supernatants were analyzed by luminex xMAP multiplexing technology (Millipore).
Global Run-On (GRO) sequencing
Transcriptionally active nuclei from infected or untreated A549 cells were prepared after swelling for 5 minutes the cells in ice-cold swelling buffer (10mM Tris (pH = 7.5), 2mM MgCl2, 3mM CaCl2). Pelleted cells were re-suspended in 1ml lysis buffer (10mM Tris (pH = 7.5), 2mM MgCl2, 3mM CaCl2, 10% glycerol, 0.5% NP40, 2U/ml SUPERaseIN (Ambion) and pipetted 20 times with a P1000 tip with the end cut off to reduce shearing. Volume was brought to 10 ml with lysis buffer and nuclei were pelleted at 600g for 5min. Nuclei were washed in 10ml lysis buffer and re-pelleted. A small aliquot was taken for Trypan blue staining to check that lysis occurred and nuclei were still intact. Nuclei were re-suspended in 1ml freezing buffer (50mM Tris-Cl (pH = 8.3), 40% glycerol, 5mM MgCl2, 0.1mM EDTA) using a P1000 tip with the end cut off and re-pelleted and re-suspended in 500μl of freezing buffer and aliquoted into 100μl aliquots and frozen in liquid nitrogen. GRO-Seq and library preparation was done as previously described26. For ChIP-seq and GRO-seq experiment validation via biologically independent experiments were done by qPCR. Samples were sequenced in accordance with manufacturer protocols on a HiSeq2000 or a HiSeq 2500 instrument.
Transcription Start Site associated RNA (tssRNA) assay
tssRNA levels were quantified using the method developed by Henriques et al33. Briefly, total RNA was extracted and large (>200 nt) and short (<200 nt) RNAs were isolated separately using miRNeasy Mini and MinElute kits (QIAGEN). Oligo-adenylation and cDNA synthesis of short RNA was performed with Superscript III and an oligo-dT primer that had an additional 5′ adaptor sequence. qPCR analysis of the short RNA was performed with forward primers specific to the 5′UTR of the gene of interest and a universal reverse primer that matched the 5′ adaptor sequence. 5s rRNA was used as a normalization standard. Statistical significance was determined with a two-tailed Student’s paired t-test
Immune stimulation of Setx−/− and wild type mice
Wild type and Setx−/− mice40 were anesthetized prior to receiving an intranasal dose of either PBS alone (n = 4 per group) or 4μg of in vitro transcribed SeV DI RNA diluted in PBS (n = 3 per group). 20μl of PBS was administered in each nostril. Six hours after administration, mice were euthanized and lungs were extracted. RNA was isolated from lungs using an RNeasy Mini kit (Qiagen) according to the manufacturer’s recommendations. cDNA was synthesized using Superscript III Reverse Transcriptase (Life Technologies) according to the manufacturer’s recommendations. All procedures were performed in compliance with procedures approved by the University of Queensland animal use committee.
Nuclear extracts and glycerol gradient sedimentation
Nuclear extracts were prepared following Dignam Roeder methods and loaded on a 5–40 % glycerol gradient for 8 h at 50000 rpm in a Beckman SW-Ti55 before being fractionated.
Lentivirus Generation and Transduction
Lentiviruses expressing wild type SETX, SETX ATPase mutant or GFP were generated by transfection of plasmids encoding the transgene in the pLX304 lentivirus backbone (Addgene) or the pLentiIII-2A-GFP backbone (ABM), VSV-G (pMD2.G, Addgene) and Gag-Pol (kind gift of M. Evans) into 293T cells at a 2.5:1.5:1 ratio. Supernatants were collected 24 and 48 hpi and viruses were concentrated using Lenti-X concentrator (Clontech) according to manufacturer’s recommendations. Optimal titers for transduction were determined by serial dilution on the appropriate cell type in the presence of 10 μg/ml Polybrene (Millipore).
Quantitative PCR (qPCR)
RNA was extracted using the RNeasy Mini Kit (QIAGEN Cat # 74106). RNA was DNase treated using the RNase free DNase kit (QIAGEN). Cells were homogenized using QIAshredder columns (QIAGEN Cat # 79656) prior to RNA extraction. Proteins were also simultaneously recovered from cell lysates by performing acetone precipitation on RNeasy spin column flow-throughs and according to manufacturer’s instructions. cDNA Synthesis was performed using SuperScript VILO Master Mix (Life Technologies Cat # 11755050) or SuperScript III First-Strand Synthesis SuperMix (Life Technologies Cat # 18080-400). qPCR was performed using SYBR green (Roche) or the LightCycler 480 Probes Master mix (Roche).
Primers
Taqman Probes (Life Technologies Cat # 4331182)
SETX (Hs00209294_m1)
IFIT1 (Hs01911452_s1)
ISG15 (Hs01921425_s1)
IFNB1 (Hs01077958_s1)
EGR1 (Hs001152928_m1)
Primer sequences
Beta Actin F 5′-ACCTTCTACAATGAGCTGCG-3′
Beta Actin R 5′-CCTGGATAGCAACGTACATGG-3′
GAPDH F 5′-GCAAATTCCATGGCACCGT-3′
GAPDH R 5′-GCCCCACTTGATTTTGGAGG-3′
18S F 5′-GTAACCCGTTGAACCCCATT-3′
18S R 5′-CCATCCAATCGGTAGTAGCG-3′
PR8 HA F 5′-AAAGAAAGCTCATGGCCCAACC-3′
PR8 HA R 5′-TCCTTCTCCGTCAGCCATAGCA-3′
PR8 PB1 F 5′-TCATGAAGGGATTCAAGCCG-3′
PR8 PB1 R 5′-GGAAGCTCCATGCTGAAATTG-3′
PR8 PB2 F 5′-AGAGACGAACAGTCGATTGCCG-3′
PR8 PB2 R 5′-ATCGCTGATTCGCCCTATTGAC-3′
PR8 M1 F 5′-GCAGGGAAGAACACCGATCTTGA-3′
PR8 M1 R 5′-ACGGTGAGCGTGAACACAATCC-3′
IFIT2 F 5′-AGGCTTTGCATGTCTTGG-3′
IFIT2 R 5′-GAGTCTTCATCTGCTTGTTGC-3′
CXCL10 F 5′-CCTTATCTTTCTGACTCTAAGTGGC-3′
CXCL10 R 5′-ACGTGGACAAAATTGGCTTG-3′
CXCL9 F 5′-TGGTGTTCTTTTCCTCTTGGG-3′
CXCL9 R 5′-AACAGCGACCCTTTCTCAC-3′
TAF4 F 5′-CAGCGAATCCTCAAACAGTCA-3′
TAF4 R 5′-ACCACAGTAGTTCCTATTTGTGC-3′
ChIP and tssRNA primer sequences available upon request
siRNA transfection
Cells were transfected using Lipofectamine™ RNAiMAX Transfection Reagent (Invitrogen) according to the manufacturer’s instructions. Cells were transfected with siRNA pools targeted to either human SETX (23064, Dharmacon), TAF4 (L-006878-00-0005, Dharmacon), XRN2 (L-017622-01-0010, Dharmacon) or a control non-targeting pool (D-001810-10-05, Dharmacon) at a final siRNA concentration of 50 nM. Transfected cells were used for further assays at 48 hours post transfection and gene knockdown efficiency was determined by quantitative PCR and/or immunoblotting.
Microarray Analyses
A549 cells that were transfected with either SETX, XRN2 or control non-targeting siRNA were infected with the PR8ΔNS1 at MOI 3 in triplicate. Un-transfected cells were also infected as a control. Total RNA was isolated from infected and uninfected cells using the QIAGEN RNeasy kit. 200ng of total RNA per sample was then used to prepare labeled RNA that was hybridized to Human HT-12 v4 Expression BeadChips (Illumina). Data analysis was performed using Genespring (version 12.5) software.
Analyses for the impact of siSETX and siXRN2 treatment were conducted separately. To specifically determine the impact of SETX or XRN2 depletion on the magnitude of the cell response during infection, raw signal values of siRNA treatment conditions in uninfected and infected cells were quantile normalized before being baseline-transformed to the medians of the corresponding uninfected siRNA treated samples. To determine probesets that displayed statistically significant (p<0.01) differences in response magnitude, an analysis of variance test (ANOVA) followed by a post hoc (TUKEY HSD) test was conducted. Genes that were differentially regulated by siSETX or siXRN2 treatment were identified as genes that displayed a ≥1.5 fold change (p<0.01) in expression levels when compared to siCtrl treated cells. Where indicated, infection induced genes were identified as genes that displayed a ≥1.5 fold change (p<0.01) in virally infected siCtrl treated cells compared to uninfected siCtrl treated cells. All p-value computations were subjected to multiple testing correction using the Benjamini Hochberg method. For purposes of display in the heatmaps, genes that were represented by multiple probesets in the microarray were plotted as the averaged values of those probesets.
To identify the impact of SETX or XRN2 depletion under basal conditions, raw signal values of siRNA treatment conditions in uninfected cells were quantile normalized before being baseline-transformed to the median of all samples. Analyses for SETX and XRN2 were done separately. An ANOVA followed by a post-hoc (TUKEY HSD) test was conducted to determine siRNA-regulated genes. siRNA (SETX or XRN2) regulated genes were defined as genes that displayed a ≥ 1.5 fold change (p<0.01) in XRN2- or SETX- specific siRNA treated cells compared to siCtrl treated cells. Normalized signal intensity values of a list of canonical housekeeping genes were also used to determine the overall impact of XRN2 or SETX depletion in cells. Full lists of SETX and XRN2 affected genes can be found in Supplementary Tables 1 and 2 respectively.
Functional analyses of differentially regulated genes were conducted through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com) IPA was used to identify canonical pathways that were most significant to gene lists generated from the microarray. Right-tailed Fisher’s exact test was used to calculate a p-value determining the probability that each pathway assigned to that data set is due to chance alone. In addition, we also used DAVID gene ontology analysis45 to identify genes associated with cytokine activity.
Antibodies
RNAPII (Covance, MMS-126R); RNAPII CTD Repeat (Abcam, AB12795); RNAPII (Bethyl Laboratories, A310-190A); Phospho IRF3 (Abcam, AB76493); IRF3 Total (Santa Cruz Biotech, SC9082); HA (Bethyl Laboratories, S190-108); LARP7 (Bethyl Laboratories, A303-724A); HEXIM1 (Bethyl Laboratories, A303-113A); DIS3 (Bethyl Laboratories A303-764A); TAF4 (Abcam ab69949) and a custom-made Ab kindly provided by R. Roeder; Tubulin (Cell Signaling Technology, 3873); Anti-PB2 is a custom-made antibody kindly provided by P. Palese; SETX a custom-made antibody kindly provided by MF Lavin.
Immunoblotting
Gradient gel were used based on the MW of the proteins to be evaluated followed by wet-transfer on PVDF membranes. For interaction studies we show representatives experiments, and for evaluating SiRNA downregulation of protein expression we poolled the protein extracts from 3 biological replicate for each conditions.
Chromatin immunoprecipitation (ChIP)
ChIP was conducted as previously described18. For ChIP-qPCR experiments (see below) a 10 min crosslinking time was used for both the FLAG M1 antibody (overexpression experiments) and SETX antibody. Sonication was performed in a refrigerated Bioruptor (Diagenode) and conditions were optimized to generate DNA fragments of approximately 200–1000 bp. Lysates were pre-cleared with the appropriate isotype control for 3 hours. Specific antibodies (Ab) were coupled with anti-mouse- (Dynabeads® M-280 Sheep Anti-Mouse IgG, Invitrogen 112-02), or anti-rabbit IgG bound magnetic beads (Dynabeads® M-280 Sheep Anti-Mouse IgG, Invitrogen, 112-04) for 6 hours. Antibody bound beads and chromatin were then immunoprecipitated at 4°C rotating overnight. After the wash steps, reverse crosslinking was carried out at 65°C overnight. ChIP DNA was then isolated after RNase digestion and proteinase K digestion, using the QIAGEN MinElute kit (QIAGEN, 28004) and used for downstream applications. Statistical significance of ChIP qPCR analysis was determined using two-tailed Student’s paired t-test.
Next-generation sequencing data analysis
Following adapter trimming and quality filtering, reads were mapped to the human reference genome (GRCh37, hg19) using bowtie v0.12.946 (for GRO-Seq and short RNA-Seq data) or Tophat v2.0.1047 (for directional RNA-Seq data). Unique best matches were kept and normalized to a total level of 5 million (GRO-Seq) or 30 million (directional RNA-Seq) tags per sample.
To identify genes with differences in the level of engaged RNAPII, we first counted the number of GRO-Seq tags mapping to the gene body, excluding the first 1 kb region following the transcription start site (TSS). Resulting counts were then normalized using the ‘trimmed mean of M’ method48 and statistically significant differentially expressed genes (q<0.05) between infected and uninfected cells were identified in a gene-wise log-likelihood ratio test with correction for multiple testing by Benjamini-Hochberg FDR, using the Bioconductor EdgeR package49. A list of differentially expressed genes can be found in Supplementary Table 3.
For GRO-Seq and directional RNA-Seq, coverage maps relative to gene features and the TSS, we calculated base coverage in 200 equally sized bins across each feature and/or 5kb flanking regions. Aggregate plots of GRO-Seq profiles across genes were prepared by calculating the geometric mean coverage for each bin. GRO-Seq travelling ratios for active genes were calculated as previously described27.
Affinity purification-Mass spectrometry (IP-MS/MS)
In brief, the 2×108 A549 cells expressing FLAG SETX or Empty Vector were infected for 4 hours with PR8ΔNS1 or left untreated. Cells were collected by scraping and washed with PBS, pelleted and re-suspended in 20 ml cold lysis buffer (0.25% NP40, 50 mM Tris–HCl pH 7.5; 200 mM NaCl, 1 mM EDTA, protease, phosphatase, inhibitors and NEM). Cells were mechanically disrupted by douncing, and sonication with a Bioruptor was performed at 4C for 5 cycles (30″ on/off). Insoluble material was then pelleted for 20 min at 15000rpm and the supernatant was pre-cleared with unspecific beads for 3 h on an overhead shaker at 4 °C. 50 ul of IP beads (anti-Flag M2 SIGMA coupled with Dynabeads) was added to the pre-cleared lysate and immunoprecipitation was performed for 3 h at 4 °C. Beads were then washed 4 times with cold 50 mM Tris–HCl pH 7.5, 200 mM NaCl, 0.25% NP40, 1 mM EDTA and 2 successive times with detergent-free buffer to avoid interference with MS analysis. Beads were eluted 3 times with 3xFLAG peptide (SIGMA) in 50 mM Tris–HCl pH 7.5, 200 mM NaCl buffer. The eluted material was subjected to acetone precipitation and resuspended proteins were analyzed via mass spectrometry after tryptic digestion.
For the analysis of Mass-spectrometry hits, initial comparisons were first made between proteins pulled down with an EV and FLAG. Proteins that demonstrated a 3-fold increase (from duplicate experiment averages) in either peptide score or area score for FLAG-pull down versus EV-pull down were then considered. Shared proteins were then selected based on a stringency setting, as having at least 3 peptides and an area score of greater than 5×106. Averages come from biological duplicates of FLAG-SETX transfections in the absence and presence of infection using PR8ΔNS1.
Helicase Assays
In brief, FLAG tagged wild type or ATPase mutant SETX was purified from 5×108 A549 cells expressing FLAG SETX or FLAG SETX Mutant. Cells were lysed in 30 ml cold lysis buffer (0.25% NP40, 50 mM Tris–HCl pH 7.5; 200 mM NaCl, 1 mM EDTA, protease, phosphatase, inhibitors and 10mM NEM). Cells were mechanically disrupted by douncing, and sonication with a Bioruptor was performed at 4C for 5 cycles (30″ on/off). The NP40 concentration was raised to 0.5% and NaCl to 300mM. The extract was rotated for 15′ in cold room. Insoluble material was then pelleted for 20 min at 15000rpm. Preclearing of the supernatant with unspecific beads for 3 h on an overhead shaker at 4 °C. 200 ul IP beads (anti-Flag M2 SIGMA coupled with Dynabeads) was added to the precleared lysate and the immunoprecipitation was performed for 3 h at 4 °C. Beads were then washed 4 times with cold 50 mM Tris–HCl pH 7.5, 300 mM NaCl, 0.25% NP40, 1 mM EDTA and 2 successive times with Helicase Buffer (20 mM HEPES-KOH (pH 7.8), 50 mM NaCl, 2 mM ATP, 1 mM MgCl2, 2 mM DTT, 100 μg/mL BSA). Aliquot of beads were quantified on coomassie gel. Immobilized-FLAG- SETX wild type, SETX-mutant and -GFP (control) were used in the unwinding assay with 200ng of synthetic DNA/RNA hybrid purchase from (IDT, Integrated DNA Technologies) and previously described in 50. After incubation at 37 °C for 1.5 h on thermomixer programmed for mixing at 700 rpm 30″on/30″ off, reactions were analyzed using the Agilent 6000 Pico Kit.
RNA/DNA oligonucleotide sequences used: 5′-GAA UAC ACG GAA UUC GAG CUC GCC CAU CCU GGA UUG UAA AUU GUA AUG UUG UUG CGC GCG CGC UAU AGU GAG UCG UAU UAG GG -3′ Biotin - 5′-GAA TAC ACG GAA TTC GAG CTC GCC CAT CCT GGA TTG TAA ATT GTA ATG TTG TTG CGC GCG CGC TAT AGT GAG TCG TAT TAG GG -3′
Short RNA sequencing
RNA from A549 cells induced with daIRF3 and overexpressing wild type SETX (WT), SETX ATPase mutant (MUT1) or SETX RNA binding mutant (MUT2) was extracted using Trizol and ethanol precipitation. 1 ug of total RNA was then DNase treated with 1U of Baseline Zero DNase for 30mins at 37°C and cleaned up via ethanol precipitation. To facilitate ligation of adapters to capped short TSS-associated RNAs, 5′ caps were removed by treating each sample with 10U of Tobacco Acid Pyrophosphotase for 1.5hrs at 37°C, followed by reaction cleanup via ethanol precipitation. Next, small RNA libraries were prepared using the NEBNext® Small RNA Library Prep Set for Illumina kit (NEB), according to the manufacturer’s instructions and overnight (~16hrs) ligation of the 3′ SR adaptor at 16°C. Final PCR amplification and barcoding of libraries (15 cycles) was performed using the LongAmp Taq 2X MasterMix with KAPA HiFi HotStart ReadyMix (2X) and followed by purification with 1.8X of Agencourt AMPure XP beads. After analysis on an Agilent Bioanalyzer (DNA7500 kit) to verify the presence of canonical microRNA and other short RNA peaks, size-selection was performed using 3% gel cassettes on the BluePippin system (Sage Science), selecting for 135–185nt fragments. Size-selected libraries were assessed on the Agilent Bioanalyzer (High Sensitivity DNA chip), which showed a single peak for each sample corresponding to short RNA fragments ranging between 30–70nts. Finally, samples were multiplexed and sequenced on the Illumina MiSeq platform (1× 50nt reads) in accordance with manufacturer protocols.
RNA-immunoprecipitation (RIP)
Crosslinking was performed for 10 min. Sonication was performed at 4°C and conditions were optimized to generate nucleotide fragments of approximately 200–500 bp. Extracts were pre-cleared for 3 hours before FLAG and IgG control immunoprecipitation (overnight). After the wash steps, reverse crosslinking was carried out at 42°C for 1 hour and 65°C for another hour. RIP RNA was then precipitated for 2 hours at −80°C. RNA was treated with Turbo DNase (Life Technologies), followed by reverse transcription and qPCR.
Statistical methods
Statistical significance for all pairwise comparisons in qPCR assays (dCT values) was determined using a two-tailed Student’s t-test under the assumption of equal variances between groups. We did not find significant differences (FDR, q<0.05) between contrast groups in Levene’s tests of equality of variances, or departures from normality as assessed by Shapiro-Wilk tests. Unless otherwise indicated, each qPCR assay included three technical and three biological replicates.
Supplementary Material
Acknowledgments
The authors would like to thank J. Lim for the generous donation of reagents. Peter Palese, Amalio Telenti, Adolfo García-Sastre are thanked for comments and sharing material and unpublished observations. We thank Bob Roeder for kindly sharing TAF4 reagents. IM thanks LMK for years of support. We thank the Genomic facility at Icahn and the Proteomics facilities at Rockefeller University. This work was supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai. This work was contributed in part by the Canadian Institutes of Health Research Postdoctoral Fellowship (M.S.M.) and NIH U19AI106754 (I.M.).
Footnotes
AUTHOR CONTRIBUTIONS
M.S.M, A.R and J.SYH contributed equally to this study. M.S.M., L. M-G, A.M. and A.F-S. performed experiments with reporters and viruses. M.S.M, O.J.B, M.F.L, P.M. and J.M. performed experiments involving mice. A.R., N.P.M., M.T. J.N., C.M., Z.P. and I.M. performed NGS and biochemical experiments; A.R.F. and C.B. performed fractionation experiments; A.F-S, A. M., O. B.,M. L. S.H, C.B., J.Y. provided materials and insights in experimental procedures; J. SYH and H.vB performed data analysis for NGS experiments. Both H.vB and I.M supervised the project, wrote the paper with feedback from co-authors and contributed equally to this study as senior authors.
Accession Numbers
Sequencing and array data have been deposited in the NCBI GEO repository, accession number GSE52937.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Pichlmair A, Sousa CRE. Innate recognition of viruses. Immunity. 2007;27(3):370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Ward PA. New approaches to the study of sepsis. Embo Mol Med. 2012;4(12):1234–1243. doi: 10.1002/emmm.201201375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandes M, Klauschen F, Kuchen S, Germain RN. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154(1):197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38(8):910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312(5775):879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 6.Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007;282(21):15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 7.Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141(4):668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11(6):426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138(1):114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon KR, Bandle R, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153(5):988–999. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14(4):211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung AC, Cramer P. A movie of RNA polymerase II transcription. Cell. 2012;149(7):1431–1437. doi: 10.1016/j.cell.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Buratowski S. Progression through the RNA Polymerase II CTD Cycle. Mol Cell. 2009;36(4):541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauberth SM, Nakayama T, Wu XL, Ferris AL, Tang ZY, Hughes SH, et al. H3K4me3 Interactions with TAF3 Regulate Preinitiation Complex Assembly and Selective Gene Activation. Cell. 2013;152(5):1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17(3):251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Bio. 2011;12(5):283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horng T, Medzhitov R. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9(10):692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 18.Marazzi I, Ho JSY, Kim J, Manicassamy B, Dewell S, Albrecht RA, et al. Suppression of the antiviral response by an influenza histone mimic. Nature. 2012;483(7390):428–433. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468(7327):1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination. Mol Cell. 2011;42(6):794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuce O, West SC. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol Cell Biol. 2013;33(2):406–417. doi: 10.1128/MCB.01195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24(5):735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 23.Kim KY, Levin DE. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell. 2011;144(5):745–756. doi: 10.1016/j.cell.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Downes CP, Wolf CR, Lane DP. Cellular Responses to Stress. Princeton University Press; 2014. [Google Scholar]
- 25.Suraweera A, Becherel OJ, Chen P, Rundle N, Woods R, Nakamura J, et al. Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage. J Cell Biol. 2007;177(6):969–979. doi: 10.1083/jcb.200701042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Core LJ, Waterfall JJ, Lis JT. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc Regulates Transcriptional Pause Release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chopra VS, Hendrix DA, Core LJ, Tsui C, Lis JT, Levine M. The polycomb group mutant esc leads to augmented levels of paused Pol II in the Drosophila embryo. Mol Cell. 2011;42(6):837–844. doi: 10.1016/j.molcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hazelbaker DZ, Marquardt S, Wlotzka W, Buratowski S. Kinetic Competition between RNA Polymerase II and Sen1-Dependent Transcription Termination. Mol Cell. 2013;49(1):55–66. doi: 10.1016/j.molcel.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porrua O, Libri D. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat Struct Mol Biol. 2013;20(7):884–891. doi: 10.1038/nsmb.2592. [DOI] [PubMed] [Google Scholar]
- 31.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27(3):339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Jin DJ, Burgess RR, Richardson JP, Gross CA. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(4):1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, et al. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell. 2013;52(4):517–528. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valen E, Preker P, Andersen PR, Zhao X, Chen Y, Ender C, et al. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat Struct Mol Biol. 2011;18(9):1075–1082. doi: 10.1038/nsmb.2091. [DOI] [PubMed] [Google Scholar]
- 35.Chen WY, Zhang J, Geng H, Du Z, Nakadai T, Roeder RG. A TAF4 coactivator function for E proteins that involves enhanced TFIID binding. Genes Dev. 2013;27 (14):1596–1609. doi: 10.1101/gad.216192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright KJ, Tjian R. Wnt signaling targets ETO coactivation domain of TAF4/TFIID in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(1):55–60. doi: 10.1073/pnas.0811914106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Y, Liu S, Lausen J, Woodrell C, Cho S, Biris N, et al. A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription. Nat Struct Mol Biol. 2007;14(7):653–661. doi: 10.1038/nsmb1258. [DOI] [PubMed] [Google Scholar]
- 38.Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74(6):1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nature Genetics. 2004;36(3):225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- 40.Becherel OJ, Yeo AJ, Stellati A, Heng EY, Luff J, Suraweera AM, et al. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet. 2013;9(4):e1003435. doi: 10.1371/journal.pgen.1003435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45(6):814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee TI, Young RA. Transcriptional Regulation and Its Misregulation in Disease. Cell. 2013;152(6):1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science. 2002;296(5576):2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 44.Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, et al. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am J Hum Genet. 2007;81 (4):713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3) doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3) doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10(3):1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.