Abstract
Lethal-7 (let-7) microRNAs are the most abundant in the genome but their role in developing thymocytes is unclear. We now report that let-7 miRNAs target Zbtb16 mRNA, which encodes the lineage-specific transcription factor PLZF, to post-transcriptionally regulate PLZF expression and NKT cell effector function. Dynamic up-regulation of let-7 miRNAs during NKT thymocyte development down-regulates PLZF expression and directs terminal differentiation into interferon-γ-producing NKT1 cells. Without let-7 up-regulation, NKT thymocytes maintain high PLZF expression and terminally differentiate into IL-4-producing NKT2 and IL-17-producing NKT17 cells. Let-7 up-regulation in developing NKT thymocytes can be signaled by IL-15, vitamin D and retinoic acid. Such miRNA targeting of a lineage-specific transcription factor constitutes a new level of developmental regulation in the thymus.
Keywords: Lin28, thymus, Vitamin D Receptor, Retinoic Acid Receptor, post-transcriptional regulation
Introduction
T cell development in the thymus is regulated by both transcriptional and post-transcriptional control mechanisms. Transcriptional mechanisms of developmental regulation in the thymus have been intensively studied and have resulted in identification of lineage specific transcription factors such as ThPOK, Runx3, Foxp3, and PLZF 1–6. In contrast post-transcriptional regulatory mechanisms in the thymus have been less well studied 7. However, the discovery of microRNAs (miRNAs) as post-transcriptional regulators of gene expression has significantly advanced understanding of post-transcriptional regulation and its molecular basis 8.
The potential importance of miRNAs for T cell development in the thymus was initially revealed by Dicer-deficient mice that were devoid of all miRNAs 9. Dicer-deficiency abrogated the generation of innate-like NKT cells, suggesting that miRNAs were necessary for NKT cell development 10–12. Innate-like NKT cells play an important role in early host defenses against invading pathogens by rapidly secreting effector cytokines such as interleukin 4 (IL-4) and interferon-γ (IFN-γ) that provide early protection before a full blown adaptive immune response can be mounted 13. Innate-like NKT cells are unique in their expression of invariant αβ T cell antigen receptors (TCR) with specificity for glycolipids presented by CD1d, a non-classical MHC class Ib molecule. They develop from immature CD4+CD8+ (double positive, DP) thymocytes that are signaled by TCR engagement of CD1d-glycolipid ligands expressed on other DP thymocytes13 and their development is dependent on the zinc finger transcription factor PLZF 5,6.
While Dicer-deficiency illustrated the importance of miRNAs in the thymus, the specific miRNAs that were important for thymic development and the mechanisms by which miRNAs affected T cell differentiation remain unclear. The most abundant family of miRNAs in the genome is the let-7 family of miRNAs 14. In mammals, the let-7 miRNA family consists of 12–14 individual members encoded on different chromosomes. Initially identified as a gene in C. elegans important for development 15, let-7 has subsequently been found to encode miRNAs16 that, in mammals, potentially target genes involved in various cellular functions, including cell cycle progression, oncogenesis, and metabolism 17–21. Interestingly, hematopoietic cells in fetal mice express two proteins, LIN28A and LIN28B, which prevent the biogenesis of functional let-7 miRNAs 22–26 and are responsible for the fetal pattern of lymphopoiesis 27. LIN28 proteins bind specifically to let-7 precursor molecules, preventing their processing into functional let-7 miRNAs and promoting their degradation 22–26. In addition to binding to immature let-7 molecules and inhibiting biogenesis of functional let-7 miRNAs, LIN28 proteins bind to certain mRNAs encoding proteins in the secretory pathway 28–30. Nevertheless, the fact that LIN28 inhibition of let-7 miRNAs is limited to only fetal hematopoietic cells suggests that uninhibited expression of let-7 miRNAs might be important for hematopoietic cell development in post-natal mice.
The present study was undertaken to specifically assess post-transcriptional regulation of T cell differentiation by let-7 miRNAs in the post-natal thymus. We report that let-7 miRNAs target Zbtb16 mRNA (which encodes the transcription factor PLZF) to post-transcriptionally inhibit PLZF protein expression. During NKT cell development, let-7 expression is dynamically up-regulated, causing PLZF protein expression to be down-regulated to levels that direct NKT cells to terminally differentiate into IFN-γ-NKT1 cells rather than IL-4-producing NKT2 or IL-17-producing NKT17 cells. Moreover, we demonstrate that let-7 up-regulation during NKT cell differentiation can be signaled in the thymic medulla by exogenous stimuli such as IL-15, vitamin D, and retinoic acid. Thus, post-translational regulation of PLZF by let-7 miRNAs determines NKT effector function, providing a new perspective on NKT cell differentiation in the thymus. This study identifies miRNA targeting of a lineage specific transcription factor as a previously unrecognized level of developmental regulation in the thymus.
Results
let-7 miRNAs are expressed in thymocytes and T cells
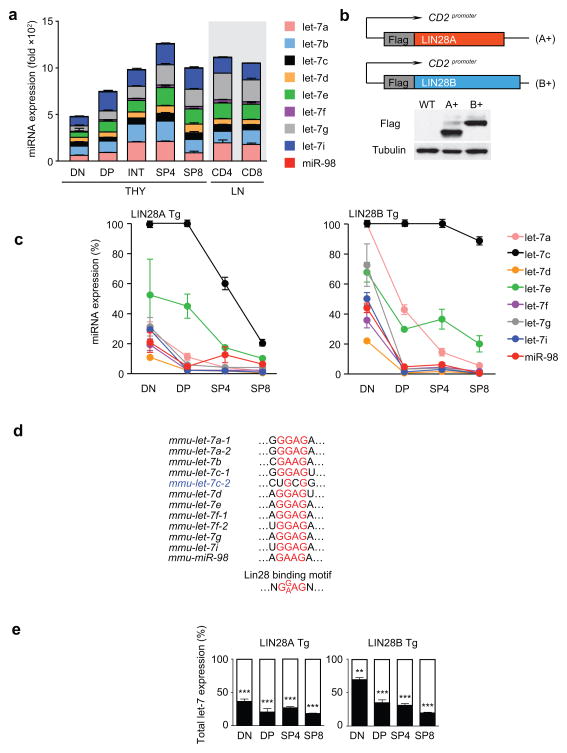
To investigate a possible regulatory role for let-7 miRNAs during T cell differentiation, we first assessed expression of individual let-7 miRNA family members in developing T cells and found that let-7 miRNAs were expressed in all thymocyte subsets and LN T cells (Fig. 1a). Total abundance of let-7 miRNAs increased during differentiation of CD4−CD8− double negative (DN) thymocytes into pre-selection double positive (DP) thymocytes, and further increased during differentiation into post-selection intermediate and single positive (SP) thymocytes (Fig. 1a).
Figure 1. let-7 miRNA expression in wild-type and LIN28 Tg thymocytes.
(a) Expression of individual let-7 miRNAs in purified thymocyte and T cell populations as determined by quantitative RT-PCR. miRNA expression is shown relative to U6 snoRNA control. Data are displayed as mean ± SE of quadruplicate samples from a representative of 3 independent experiments. (b) Schematic representation of LIN28 transgenic constructs used to generate LIN28 Tg mice (top). LIN28A (209aa) and LIN28B (247aa) protein content of total thymocytes from wild-type and LIN28 Tg mice as determined by Western blotting (bottom) with similar results in 3 independent experiments. (c) Expression of individual let-7 family members in thymocytes from LIN28A Tg and LIN28B Tg mice. Expression of let-7 family members in each LIN28 Tg thymocyte subset is expressed relative to that in the corresponding wild-type thymocyte subset. Data are displayed as mean ± SE of triplicate samples from a representative of 3 independent experiments. (d) Alignment of the distal LIN28 binding motif in let-7 pre-element sequences in let-7 family members. (e) Overall let-7 expression in thymocyte subsets from LIN28 Tg mice relative to wild-type mice (which was set equal to 100%), as determined by quantitative RT-PCR. Data are displayed as mean ± SE of quadruplicate samples and are representative of at least 3 independent experiments. Values from LIN28 Tg mice relative to wild-type mice were assessed for statistical significance.
*, p<.05; **, p<.01; ***, p<.001; NS, not significant p>.05.
To determine if let-7 miRNAs influenced thymocyte development, we examined the developmental consequences of quantitatively reducing let-7 expression. Because individual let-7 miRNA family members are dispersed throughout the genome, our strategy for reducing let-7 expression in thymocytes took advantage of the ability of LIN28A and LIN28B fetal proteins to bind to the extended loop region of let-7 precursor molecules and prevent their processing into functionally mature miRNAs. We constructed transgenes consisting of cDNA encoding Flag-tagged LIN28A or LIN28B proteins under the control of T cell specific hCD2 promoter/enhancer elements to drive expression of LIN28 transgenic proteins specifically in thymocytes and T cells (Fig. 1b). Reflecting the initial onset of hCD2-driven transgene expression in thymocytes at the DN stage of differentiation 31, expression of individual let-7 miRNA family members was reduced in DN thymocytes from LIN28 transgenic mice (hereafter called LIN28 Tg) and was even more reduced in LIN28 Tg thymocytes at later stages (i.e. DP and SP stages) of differentiation (Fig. 1c). The major exception was an individual let-7 family member, let-7c, whose expression was relatively resistant to LIN28 Tg expression (Fig. 1c). When we examined the LIN28 binding sequences in the extended loop region of immature let-7c precursor molecules, we found that let-7c-2 precursor molecules were the only members of the family with a downstream LIN28 binding sequence (U-G-C-G) that deviated from the optimal downstream binding motif (G-G/A-A-G) (Fig. 1d). Even with this exception, overall let-7 miRNAs were significantly reduced in both LIN28A and LIN28B Tg thymocytes, with the LIN28A Tg being marginally more effective in reducing let-7 expression than the LIN28B Tg (Fig. 1e). Thus, compared to wild-type thymocytes, let-7 expression was substantially reduced in LIN28A Tg and LIN28B Tg thymocytes.
Reduced let-7 expression increases the number of IL-4+PLZFhi thymocytes
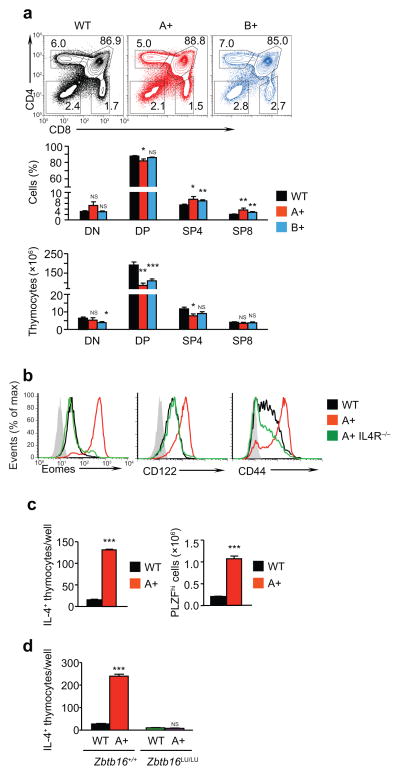
Thymocyte subpopulations defined by CD4 and CD8 expression (i.e. DN, DP and SP) were minimally altered in LIN28 Tg mice (Fig. 2a), with ~30% reduction in absolute numbers of DP thymocytes suggestive of intra-thymic stress (Fig. 2a). Because we observed similar phenotypes in LIN28A and LIN28B Tg mice, we mainly used LIN28A Tg mice for subsequent experiments. More detailed examination revealed that LIN28A Tg SP8 thymocytes were CD44hiCD122hiEomeshi memory-like cells, whereas wild-type SP8 thymocytes were CD44loCD122loEomeslo naïve cells (Fig. 2b). To determine if the memory-like phenotype of LIN28A Tg SP8 thymocytes resulted from intra-thymic IL-4 signals 32, we generated LIN28A.IL-4R−/− Tg mice in which LIN28A Tg thymocytes would be unresponsive to intra-thymic IL-4. SP8 thymocytes in LIN28A.IL4R−/− Tg mice, unlike SP8 thymocytes in Lin28A Tg mice, displayed a naive CD44loCD122loEomeslo phenotype, indicating that the appearance of memory-like SP8 thymocytes in LIN28A Tg mice was a secondary consequence of increased IL-4 signaling in the LIN28 Tg thymus (Fig. 2b).
Figure 2. Analysis of thymocytes in LIN28 Tg mice.
(a) CD4 versus CD8 expression on thymocytes from wild-type (n=12), LIN28A Tg (A+, n=7), and LIN28B Tg (B+, n=10) mice (top). Frequency (middle) and absolute number (bottom) of thymocyte sub-populations in indicated mice. Data are displayed as mean ± SE of each thymocyte subset from individual mice in 12 independent experiments. In each group, values from LIN28 Tg mice were compared to wild-type mice. (b) Phenotypic analysis of SP8 thymocytes for naïve or memory markers by intracellular staining for Eomes and surface staining for CD122 and CD44. Negative control staining is shown in gray. (c) Quantification of IL-4-producing thymocytes by ELISpot (left) and quantification of PLZFhi thymocytes by intracellular staining and flow cytometry (right). (d) Quantification of IL-4-producing thymocytes from wild-type and LIN28 Tg mice containing homozygous amounts of either wild-type or mutant PLZF. Data are displayed as mean ± SE of triplicate samples (c) or quadruplicate samples (d) from a representative of 3 independent experiments. Values from LIN28 Tg mice are compared to those from wild-type mice in each group.
*, p<.05; **, p<.01; ***, p<.001; NS, not significant p>.05.
To understand why IL-4 signaling was increased in Lin28A Tg thymi, we quantified the number of IL-4-producing thymocytes in wild-type and LIN28A Tg mice. As determined by ELISpot assay, the number of IL-4-producing thymocytes was 5–10 fold greater in LIN28A Tg thymi compared to wild-type (Fig. 2c). Because IL-4-producing thymocytes express high amounts of the transcription factor PLZF 32, we quantified the number of PLZFhi thymocytes and found that their number was also increased 5–10 fold in LIN28A Tg thymi compared to wild-type (Fig. 2c). Importantly, the generation of IL-4-producing LIN28A Tg thymocytes required PLZF expression, as IL-4-producing LIN28A Tg thymocytes did not arise in PLZF-deficient LIN28A.Zbtb16lu/lu Tg mice (Fig. 2d). Thus transgenic expression of LIN28 markedly increases the number of PLZFhi thymocytes that produce IL-4, and it is the excessive production of IL-4 that stimulates the appearance of memory-like SP8 thymocytes in LIN28 Tg mice.
let-7 miRNAs target Zbtb16 mRNA to reduce PLZF expression
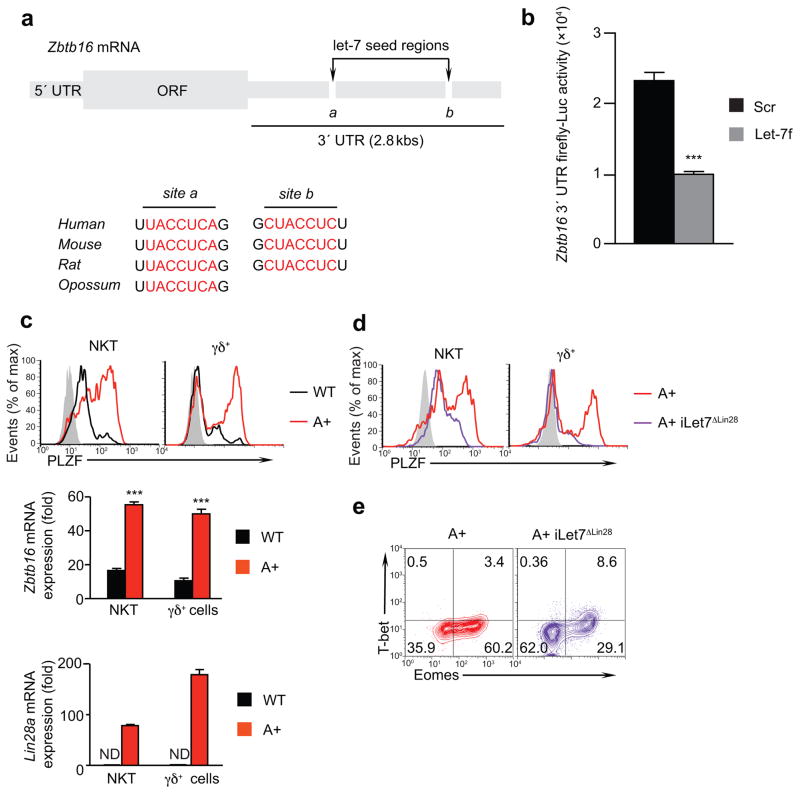
Given the increased numbers of PLZFhi thymocytes in LIN28 Tg mice, we considered that let-7 miRNAs might target Zbtb16 mRNA to inhibit expression of its protein product, the transcription factor PLZF. Consistent with such a possibility, computational analysis identified two evolutionarily conserved let-7 miRNA binding sites (seed regions) in the 3′-untranslated region (3′-UTR) of Zbtb16 mRNA (Fig. 3a). To determine if let-7 miRNAs could target the 3′-UTR of Zbtb16 mRNA and inhibit PLZF protein synthesis, we transfected 293T cells with luciferase mRNA whose 3′UTR was derived from Zbtb16 and simultaneously co-transfected either let-7 miRNA or control scrambled miRNA. Co-transfection of let-7 miRNA significantly reduced luciferase activity compared to co-transfection of control scrambled miRNA (Fig. 3b), demonstrating that let-7 miRNAs target the 3′UTR of Zbtb16 mRNA.
Figure 3. let-7 miRNAs target Zbtb16 mRNA.
(a) Schematic of Zbtb16 mRNA highlighting the two let-7 binding sites in its 3′-UTR (top). Sequence alignment of the two let-7 binding sites (site a and site b) in the 3′-UTR of Zbtb16 mRNA from several different species (bottom). (b) let-7 targeting of the Zbtb16 3′-UTR in a luciferase reporter assay. 293T cells were co-transfected with a firefly luciferase reporter along with either a let-7 miRNA or a control scrambled miRNA. Firefly luciferase activity was normalized to control renilla luciferase activity. Data are displayed as mean ± SE of quadruplicate samples from a representative of 3 independent experiments. Values from cells transfected with let-7 were compared to those transfected with scrambled miRNA. (c) PLZF protein expression in wild-type and LIN28 Tg thymocytes was determined by intracellular staining and flow cytometry (top). Control staining is shown in gray. Thymocyte content of Zbtb16 mRNA (middle) and LIN28 mRNA (bottom) was determined by quantitative RT-PCR relative to Rpl13a expression. Data are displayed as mean ± SE of quadruplicate samples from a representative of 3 independent experiments. In each group, values from LIN28 Tg mice are compared to those from wild-type mice. ND, not detected. (d) PLZF protein expression was determined by intracellular staining and flow cytometry in thymocytes from LIN28 Tg mice and LIN28.iLet7ΔLIN28 Tg mice. Control staining is shown in gray. (e) Intracellular T-bet versus Eomes expression in SP8 thymocytes from LIN28 Tg and LIN28.iLet7ΔLIN28 Tg mice.
*, p<.05; **, p<.01; ***, p<.001;
Because PLZF is the lineage determining transcription factor for NKT and innate γδ T cells 5,6,33, we measured Zbtb16 mRNA and PLZF protein expression and found that both were substantially increased in innate thymocytes from LIN28A Tg compared to wild-type mice (Fig. 3c). To investigate if PLZF expression was increased in LIN28A Tg innate thymocytes specifically because they were relatively let-7 deficient, we acutely increased the let-7 content of LIN28A Tg thymocytes with a doxycycline (dox)-inducible transgene that encoded modified pre-let-7 molecules lacking LIN28 binding sequences (iLet7ΔLin28 Tg) 21. Induction of transgenic let-7 miRNA in LIN28 mice by short term dox administration reduced PLZF expression in NKT and γδ innate thymocytes and reduced the frequency of memory-like Eomes+ SP8 thymocytes in Lin28A.iLet7ΔLin28 Tg mice (Fig. 3d, e). Thus, these findings document that reduced let-7 is responsible for increased PLZF expression in LIN28 Tg thymocytes, and identify let-7 miRNAs as specific inhibitors of PLZF expression.
Endogenous let-7 does not affect overall NKT thymocyte numbers
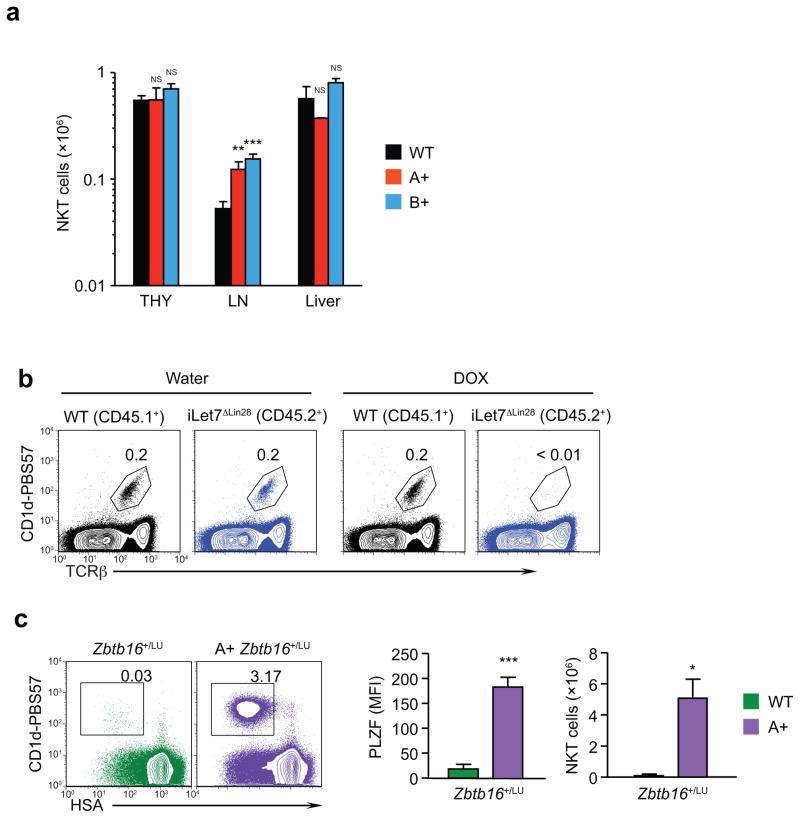
We next investigated if endogenous let-7 miRNAs reduced the overall generation of NKT cells in the thymus, in addition to preventing excessive generation of IL-4 producing NKT cells. By staining with the NKT cell specific tetramer CD1d-PBS57, we found that total NKT thymocyte numbers in LIN28 Tg and wild-type mice were very similar, although total NKT cell numbers in the periphery varied slightly (Fig. 4a). One possible reason overall NKT thymocyte numbers were not reduced by LIN28 Tg proteins might be that endogenous let-7 miRNAs were expressed in insufficient amounts, relative to PLZF, in precursor NKT thymocytes to prevent their initial commitment to differentiate into innate lineage NKT cells.
Figure 4. Effect of let-7 and PLZF on NKT thymocytes.
(a) Absolute numbers of NKT cells in the thymus and periphery of wild-type and LIN28 Tg mice. Data are displayed as mean ± SE from thymi (wild-type, n=9; LIN28A Tg, n=6; and LIN28B Tg, n=7), LNs (wild-type, n=7; LIN28A Tg, n=5; and LIN28B Tg, n=7), and livers (wild-type, n=3; LIN28A Tg, n=2; and LIN28B Tg, n=3) from mice of each strain from 9 independent experiments. In each group, values from LIN28 Tg mice are compared to those from wild-type mice. (b) Mixed bone marrow chimeras were constructed by injecting equal mixtures of wild-type and iLet7ΔLIN28 Tg bone marrow cells into lethally irradiated CD45.1 B6 host mice. Beginning on day 2 after chimera construction and continuing for 8 weeks when the mice were analyzed, mice were given either plain or doxycycline-supplemented drinking water. Protocol for chimera construction is visually displayed in Supplementary Fig. 1. Flow cytometric analyses of chimeric thymi 8 weeks later is shown. The box in each histogram identifies NKT thymocytes and numbers indicate their frequency. (c) Comparison of NKT thymocytes in wild-type and PLZFwt/lu heterozygous mice (left). Box in each histogram identifies NKT thymocytes and numbers indicate their frequency among total thymocytes. PLZF expression in NKT thymocytes was quantified as mean fluorescent intensity (MFI) (middle). Absolute NKT thymocyte numbers are shown (right). Data are displayed as mean + SE of 3–6 individual mice in 3 independent experiments. In each group, values from LIN28 Tg mice are compared to those from wild-type mice.
*, p<.05; **, p<.01; ***, p<.001; NS, not significant p>.05.
To determine if greater let-7 expression would reduce overall NKT thymocyte numbers, we increased the let-7 content of thymocytes in wild-type mice with the dox-inducible iLet7ΔLin28 Tg. In these experiments, we reconstituted lethally irradiated host mice with a mixture of bone marrow cells from donor B6 and donor iLet7ΔLin28 Tg mice, and provided doxycycline continuously in the drinking water from day 2 after bone marrow reconstitution until the mice were analyzed 8 weeks later (Fig. 4b and Supplementary Fig. 1). As determined by CD1d tetramer staining, dox treatment abrogated the generation of NKT thymocytes from iLet7ΔLin28 Tg donor cells but had no effect on the generation of NKT thymocytes from wild-type B6 donor cells within the same thymus (Fig. 4b). Thus greater expression of let-7 quantitatively inhibited NKT thymocyte generation and did so in a cell intrinsic manner.
Next, we asked if endogenous amounts of let-7 would be sufficient to impair NKT thymocyte generation in Zbtb16 heterozygous mice with reduced PLZF expression. We used Zbtb16wt/lu heterozygous mice, which only produce PLZF from one wild-type allele and have very few CD1d tetramer+ NKT thymocytes compared to wild-type mice (Fig. 4c) 6. Introduction of the LIN28A Tg into Zbtb16wt/lu heterozygous mice generated LIN28A. Zbtb16wt/lu Tg mice in which the frequency and number of NKT thymocytes were increased more than 50-fold, and their PLZF content as determined by intracellular staining was increased 10-fold, relative to PLZFwt/lu heterozygous mice (Fig. 4c). These results indicate that endogenous let-7 miRNAs are sufficient to inhibit NKT thymocyte generation in PLZF heterozygous mice, but are not sufficient to inhibit NKT thymocyte generation in PLZF homozygous mice. Thus, it is the relative amounts of let-7 versus PLZF that control the overall number of NKT thymocytes generated in the thymus.
let-7 modulates terminal NKT cell differentiation
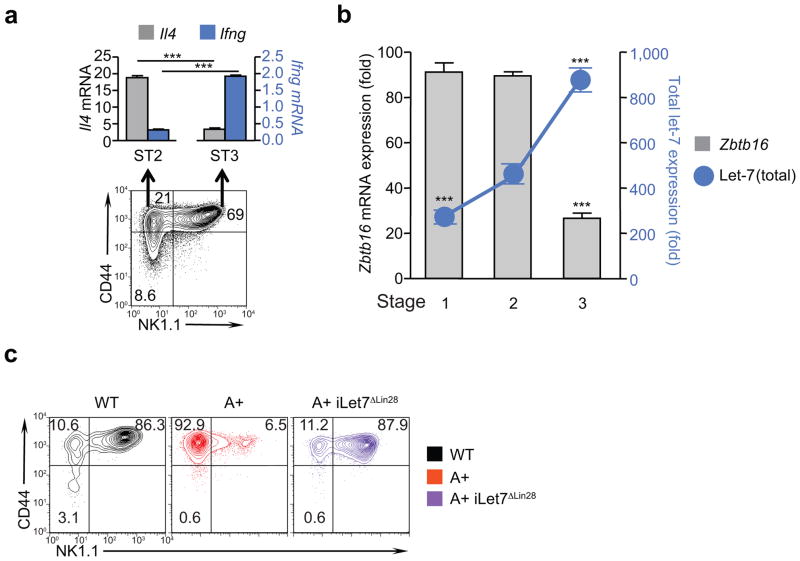
To gain greater insight into the physiological role of let-7 miRNAs during NKT thymocyte differentiation, we examined the kinetics of PLZF and let-7 expression in developing NKT thymocytes 34. Among CD24low thymocytes, the differentiation of NKT cells proceeds through three distinct stages phenotypically distinguished by CD44 and NK1.1 expression: stage 1 cells are CD44−NK1.1− precursor NKT cells; stage 2 cells are CD44+NK1.1− that predominantly produce IL-4; and stage 3 cells are CD44+NK1.1+ that primarily produce IFN-γ (Fig. 5a).
Figure 5. Reduced let-7 expression prevents generation of IFN-γ-producing NKT cells.
(a) IL-4 and IFN-γ mRNA content in purified CD24lo NKT thymocytes from B6 mice at different stages of differentiation, referred to as stages 1–3 (upper panel). Data are displayed as mean ± SE of quadruplicate samples and are representative of 3 independent experiments. Numbers in histogram indicate cell frequency within that quadrant. (b) Zbtb16 mRNA (left axis) and let-7 miRNA content (right axis) in NKT thymocytes at various stages of differentiation as determined by quantitative RT-PCR. Data are displayed as mean ± SE of quadruplicate samples from a representative of 3 independent experiments. Values of Zbtb16 mRNA and let-7 miRNA in stage 1 and stage 3 cells are each compared to their respective values in stage 2 cells. (c) CD44 and NK1.1 expression on NKT thymocytes from dox treated wild-type, LIN28A Tg, and LIN28A.iLet7ΔLIN28 Tg mice. mRNA and miRNA expression were determined relative to Rpl13a or U6 controls, respectively.
***, p<.001.
Analysis of thymocytes at different stages of differentiation revealed that Zbtb16 mRNA expression was high in stage 1 and 2 NKT cells, but was down-regulated in stage 3 NKT cells (Fig. 5b). In contrast, endogenous let-7 expression was initially low in stage 1 and 2 NKT cells, but was up-regulated in stage 3 NKT cells (Fig. 5b). Thus, PLZF and let-7 expression both dynamically changed during NKT thymocyte development, but in reciprocal directions. To determine if Zbtb16 mRNA down-regulation in stage 3 NKT cells was caused by let-7 up-regulation, we performed a similar analysis with LIN28 Tg thymocytes and discovered that NKT thymocytes in LIN28 Tg mice differentiated into stage 2 (CD44+NK1.1−) but not stage 3 (CD44+NK1.1+) cells, indicating that Zbtb16 mRNA down-regulation did not occur in LIN28 Tg mice (Fig. 5c). The profound block in differentiation into stage 3 cells and the subsequent increase in frequency of stage 2 cells in LIN28 Tg mice were specific consequences of reduced let-7 expression because normal differentiation into stage 3 cells was restored in LIN28 Tg mice by induction of the dox-inducible iLet7ΔLin28 Tg to increase let-7 expression (Fig. 5c). Thus reduced let-7 expression results in persistently high PLZF expression which prevents differentiation into stage 3 NKT cells and results instead in terminal differentiation into functional stage 2 NKT cells. We conclude that PLZF down-regulation during NKT thymocyte development is post-transcriptionally induced by let-7 miRNAs and is required for the terminal differentiation of NKT thymocytes into stage 3 cells.
let-7 regulates the generation of NKT cell effector lineages
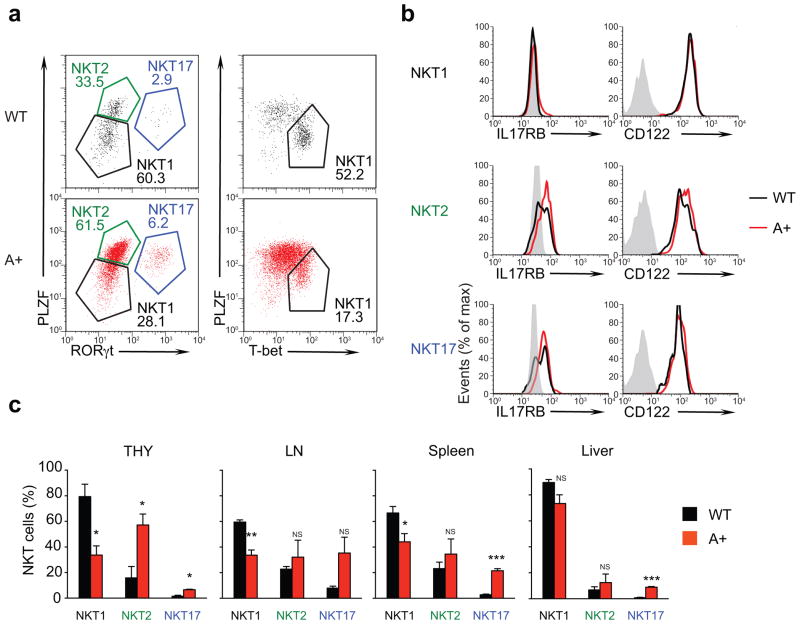
Recently, NKT thymocytes were shown to terminally differentiate into three distinct lineages of functional NKT effector cells (referred to as NKT1, NKT2, and NKT17) that differ phenotypically, express different transcription factors and produce different cytokines 35,36: NKT1 cells are phenotypically PLZFloCD44hiNK1.1+ (i.e. stage 3), express the transcription factor T-bet, and predominantly produce IFN-γ; NKT2 cells are phenotypically PLZFhiCD44hiNK1.1− (i.e. stage 2) and highly produce IL-4; and NKT17 cells resemble NKT2 cells in being PLZFhiCD44hiNK1.1− (i.e. stage 2) but express the transcription factor RORγt and produce IL-17. To assess the impact of let-7 on the generation of these three NKT effector lineages, we examined transcription factor expression by intracellular staining for PLZF, RORγt, and T-bet in NKT thymocytes from wild-type and LIN28A Tg mice (Fig. 6). In wild-type mice, NKT thymocytes consisted mostly (>60%) of NKT1 (PLZFloRORγt−Tbet+) effector cells; whereas, in LIN28 Tg mice, NKT thymocytes consisted mostly (>70%) of NKT2 (PLZFhiRORγt−Tbet−) and NKT17 (PLZFhi/intRORγt+Tbet−) effector cells (Fig. 6a). However, IL17RB and CD122 expression on different NKT effector lineages was unaffected by LIN28 Tg expression (Fig. 6b). These results indicate that let-7 miRNAs are needed for developing NKT thymocytes to terminally differentiate into NKT1 effector cells, rather than NKT2 or NKT17 effector cells.
Figure 6. Impact of let-7 on NKT effector cell lineages.
(a) NKT thymocytes were assessed for transcription factor expression by intracellular staining for PLZF vs RORγt and PLZF vs T-bet (numbers indicate frequency of gated cells), and (b) for surface expression of IL17RB and CD122. Control staining is shown in gray. (c) Frequency of NKT1, NKT2, and NKT17 effector cells among NKT cells in the thymus, LN, spleen, and liver of wild-type and LIN28 Tg mice. Data are displayed as mean ± SE of 3 individual mice from 3 independent experiments. In each group, values from LIN28 Tg mice are compared to those from wild-type mice.
*, p<.05; **, p<.01; ***, p<.001; NS, not significant, p>.05.
The thymic bias toward NKT2 and NKT17 differentiation in LIN28 Tg mice was less evident in the lymphoid periphery (LN and spleen) and was least evident in the liver (Fig. 6c and Supplementary Fig. 2), presumably reflecting differential migration and expansion of different NKT effector lineages after emigration from the thymus. Most notably, NKT cells in the liver of LIN28 Tg mice consisted predominantly of NKT1 cells, a similar observation to that in wild-type BALB/c mice (Fig. 6c and Supplementary Fig. 2) 36. Thus let-7 miRNAs are key regulators of terminal NKT cell differentiation in the thymus, but additional (probably homeostatic) mechanisms affect the frequency of NKT effector populations in the periphery.
mTEC-derived signals up-regulate let-7 in NKT thymocytes
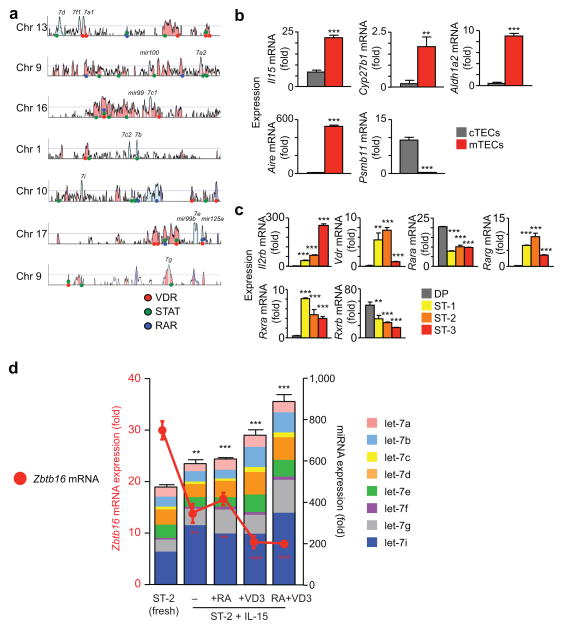
Next we investigated the signals that induce let-7 up-regulation in developing NKT thymocytes. To assess the possibility that up-regulation of multiple let-7 encoding genes on different chromosomes was coordinated by exogenous stimuli in the thymic medulla, we performed computational analyses to identify let-7 associated conserved noncoding sequences (CNS) that were conserved between humans and mice (Fig. 7a). We searched let-7 associated CNS for the presence of binding motifs for Signal Transducers and Activators of Transcription (STAT), Vitamin D Receptor (VDR), and Retinoic Acid Receptors (RAR) because: (a) let-7 gene expression in nematodes is up-regulated by the DAF-12 steroid receptor whose mammalian homologues are VDR and RAR 37,38; and (b) NKT thymocyte differentiation requires IL-15 which signals the translocation of STAT5 molecules to the nucleus where they activate gene expression. This search identified 121 different let-7 associated CNS that contain multiple binding sequences for STAT, VDR, and RAR (Fig. 7a).
Figure 7. Signals that up-regulate let-7 expression in developing NKT thymocytes.
(a) Vista conservation analysis of human and murine let-7 genomic regions. Peaks represent degree of conservation. Colored circles indicate potential binding sites for VDR, Vitamin D Receptor (red); STAT, Signal Transducer and Activator of Transcription (green); and RAR, Retinoic Acid Receptors (blue). (b) Gene expression in purified cTECs and mTECs as determined by quantitative RT-PCR. As indicators of cell purity, the Aire gene is only expressed in mTECs, whereas the Psmb11 gene encoding the β5t proteosomal protein is only expressed in cTECs. Data are displayed as mean ± SE of quadruplicate samples from a representative of 3 independent experiments. For each gene, values from mTECs are compared to cTECs. (c) Gene expression in DP thymocytes and NKT thymocytes at different stages of differentiation as determined by quantitative RT-PCR. Data are displayed as mean ± SE of quadruplicate samples from a representative of 3 independent experiments. For each gene, values in developing NKT thymocytes are compared to that of DP thymocytes. (d) Purified CD44hiNK1.1− thymocytes at NKT developmental stage 2 were analyzed fresh or after 48h culture under various conditions and assessed for expression of Zbtb16 mRNA (left axis) and let-7 miRNAs (right axis) by quantitative RT-PCR. Data are displayed as mean ± SE of quadruplicate samples from a representative of 3 independent experiments. Values for Zbtb16 mRNA and let-7 miRNAs from stage 2 NKT cells after 48h culture are compared to their values in fresh stage 2 NKT cells. mRNA and miRNA expression was determined relative to Rpl13a or U6 controls, respectively.
*, p<.05; **, p<.01; ***, p<.001.
To determine if signals in the thymic medulla could induce let-7 expression in NKT thymocytes, we examined whether medullary thymic epithelial cells (mTECs), which support NKT cell differentiation 39,40, expressed ligands or inducers of STAT, VDR, and RAR. mTECs expressed mRNAs encoding the cytokine IL-15 (which signals through STAT5), the enzyme Cyp27b1 (which produces the active form of vitamin D) 41, and the enzyme Aldh1a2 (which produces the active form of retinoic acid) 41 (Fig. 7b). In turn, CD1d tetramer+ NKT thymocytes expressed Il2rb mRNA which encodes CD122, the limiting component of surface IL-15 receptors, as well as mRNAs encoding VDR and the retinoic acid receptors RARA, RARG, RXRA and RXRB (Fig. 7c) 41.
In addition, ex vivo stimulation with IL-15, vitamin D3 and retinoic acid substantially up-regulated let-7 miRNA expression and reduced Zbtb16 mRNA expression in stage 2 CD44hiNK1.1− PLZFhi NKT cells (Fig. 7d), experimentally demonstrating that these stimuli signal developing stage 2 NKT thymocytes to up-regulate let-7 and down-regulate PLZF expression.
We conclude that phenotypically stage 2 (CD44hiNK1.1−PLZFhi) thymocytes include developmentally intermediate cells with the potential to terminally differentiate into alternative effector cell fates: if let-7 miRNAs are up-regulated, PLZF expression is down-regulated and NKT thymocytes terminally differentiate into NKT1 effector cells; if let-7 miRNAs are not up-regulated, PLZF expression is not down-regulated and NKT thymocytes terminally differentiate into NKT2 and NKT17 effector cells (schematized in Supplementary Fig. 3).
Discussion
This study uncovers a new level of developmental regulation in the thymus that is mediated by endogenous miRNA targeting of a lineage-specifying transcription factor. This study reveals that let-7 miRNAs target the 3′UTR of Zbtb16 mRNA to inhibit expression of PLZF protein, the zinc finger transcription factor that is required by innate lineage T cells for their generation in the thymus. However, endogenous let-7 miRNAs are not expressed in sufficient amounts relative to PLZF to interfere with the initial commitment of precursor thymocytes to differentiate into innate-like NKT cells. However, as NKT thymocyte differentiation progresses, endogenous let-7 miRNA expression is dynamically up-regulated to levels that down-regulate PLZF, with the result that developing NKT thymocytes terminally differentiate into NKT1 cells that predominantly produce IFN-γ. Without let-7 up-regulation, PLZF expression remains high throughout NKT thymocyte differentiation and results in terminal differentiation into IL-4-producing NKT2 and IL-17-producing NKT17 cells. Thus, dynamic up-regulation of let-7 miRNAs during NKT development is required for terminal differentiation into NKT effector cells that predominantly produce IFN-γ, instead of IL-4 or IL-17. Dynamic up-regulation of let-7 miRNA expression can be signaled by exogenous stimuli such as IL-15, Vitamin D, and Retinoic Acid in the thymic medulla. We conclude that endogenous let-7 miRNAs, by down-regulating PLZF expression, regulate terminal NKT thymocyte differentiation and NKT cytokine effector function.
The let-7 miRNA family consists of individual members encoded by multiple genes on different chromosomes 14. Each gene encoding a let-7 family member transcribes precursor miRNA molecules with seed regions that specifically bind to a spectrum of let-7 target mRNAs such as Zbtb16 mRNA 24. A recent study using retroviral transduction of LIN28b cDNA into adult bone marrow cells demonstrated that LIN28B proteins impose a fetal pattern of T and B lymphopoiesis on adult retrogenic mice, including increased numbers of NKT cells and memory phenotype SP8 thymocytes 27. Using transgenes to force LIN28A and LIN28B protein expression in post-natal thymocytes, our present study supports these observations and significantly extends them to substantially enhance understanding of NKT cell differentiation. Notably, the present study reveals that PLZF expression must be actively down-regulated in developing NKT thymocytes for differentiation into IFN-γ-producing NKT cells, and that the down-regulation of PLZF is post-transcriptionally induced by let-7 miRNAs.
The mechanism by which let-7 genes are coordinately up-regulated at stage 2 of NKT thymocyte differentiation merits consideration. We found that developing NKT thymocytes at differentiation stage 2 up-regulate let-7 miRNAs in response to stimulation by active vitamin D, retinoic acid, and IL-15. The involvement of VDR signaling in let-7 up-regulation during NKT thymocyte development explains two previous observations relating Vitamin D and NKT cell development 42,43. First, let-7 up-regulation occurs in stage 2 NKT cells with high surface CD44 levels, and surface CD44 proteins have been shown to bind serum protein gc-globulin, also known as Vitamin D binding protein, which is the major carrier of active vitamin D 42. Second, VDR-deficiency partially arrests NKT thymocyte development at stage 2 of differentiation 43, although an additional effect of VDR-deficiency was diminished IL-4 production relative to wild-type mice 44. Thus, we suggest that VDR signaling contributes to let-7 up-regulation in stage 2 NKT cells because they are CD44hi and can access Vitamin D by binding soluble Vitamin D binding protein.
PLZF is the zinc finger transcription factor responsible for differentiation of innate lineage T cells, but its pattern of expression during innate lineage differentiation is quite unlike that of other lineage specifying transcription factors in the thymus. RUNX3 and ThPOK specify CD8 and CD4 lineage differentiation, respectively, but neither’s expression declines during lineage-specific differentiation in the thymus. That is, RUNX3 expression continually increases throughout differentiation of CD8 committed RUNX3+ thymocytes into functional CD8 cytotoxic T cells 2,45, and ThPOK expression continually increases throughout differentiation of CD4 committed ThPOK+ thymocytes into mature CD4 helper T cells 3,4,45. In contrast PLZF expression declines during lineage specific differentiation as a result of post-transcriptional regulation by let-7 miRNAs. Thus, the unusual behavior of PLZF for a lineage-specific transcription factor is due to let-7. Notably, our study also explains why a PLZF transgene was previously found to skew NKT thymocyte development toward IL-4-producing NKT2 cells 46, i.e. because the PLZF transgene encoded Zbtb16 cDNA lacking the let-7 target sequence so that its expression could not be down-regulated.
The present study suggests that phenotypically CD44hiNK1.1−PLZFhi stage 2 thymocytes actually consist of two distinct subsets at different stages of development: stage 2 ‘intermediate’ cells are immature cells that terminally differentiate into various effector cell fates (NKT2, NKT1,7 or NKT1) depending on let-7 expression; whereas stage 2 ‘mature’ cells are terminally differentiated NKT2 or NKT17 effector cells (see Supplementary Fig. 3). In our perspective, intermediate stage 2 cells are the cells in which NKT effector fate is decided based on their PLZF expression levels. Persistence of PLZF expression drives stage 2 intermediate cells to terminally differentiate into IL-4-producing NKT2 cells and IL-17-producing NKT17 cells, whereas let-7-induced down-regulation of PLZF expression drives stage 2 intermediate cells to terminally differentiate into IFN-γ-producing NKT1 cells.
In conclusion, let-7 miRNA inhibition of PLZF expression is a necessary component of NKT thymocyte development, with NKT cells acquiring different effector functions depending on whether or not PLZF expression is down-regulated by let-7. As a secondary consequence of their regulation of NKT cell effector function, let-7 miRNAs also affect the differentiation of SP8 thymocytes into either naive or memory-like CD8 cytotoxic T cells. Thus, endogenous let-7 miRNA targeting of the innate lineage transcription factor PLZF is an important mechanism of developmental regulation in the thymus.
Online Methods
Animals
FLAG-tagged cDNAs encoding mouse LIN28A and LIN28B proteins were cloned into hCD2-based cassettes and used to generate LIN28A and LIN28B transgenic mice. iLet7-Tg and M2rtTA double transgenic mice were kindly provided by Dr. G.Q. Daley (Harvard Medical School) 21 and PLZFwt/lu mice by Dr. A. Bendelac (University of Chicago) 6. C57BL/6 (B6) mice were obtained from Frederick Cancer Research and Development Center. IL-4Rα−/− mice were purchased from The Jackson Laboratory. Animal experiments were approved by the National Cancer Institute Animal Care and Use Committee, and all mice were cared for in accordance with National Institutes of Health guidelines.
Bone Marrow Chimeras
Radiation bone marrow chimeras were generated by reconstituting lethally irradiated (950 Rads) host mice with a total of 10–15×106 T cell-depleted bone marrow cells 6h after irradiation. Doxycycline was added to the drinking water on the second day after bone marrow reconstitution. Chimeric mice were analyzed 8 weeks after reconstitution.
Doxycycline-mediated induction of let-7g expression
Mice were fed 2mg/ml of doxycycline in drinking water supplemented with 10mg/ml of sucrose 21.
Flow cytometry and protein immunoblotting
Monoclonal antibodies with the following specificities were used: CD4 (GK1.5), CD8α (53-6.7), CD122 (TM-β1), CD44 (IM7), TCRβ (H57-597), TCRγδ (GL3), Ly-51 (BP-1), NK1.1 (PK136), and T-bet (04-46) from BD Pharmingen; Eomes (Dan11mag), T-bet (4B10), and RORγt (B2D) from eBioscience; CD24 (M1/69), CD326 (G8.8), CD45.2 (104) from BioLegend; IL-17RB (752101) from R&D Systems; UEA from Vector Laboratories; and streptavidin from BD Pharmingen. Antibodies against PLZF (D-9) from Santa Cruz Biotechnology, Inc were directly conjugated to Alexa-647 or Alexa-594 using APEX antibody labeling kit (Invitrogen). Antibodies with FLAG specificity (Sigma-Aldrich) were used for protein immunoblotting. PBS57-loaded or unloaded CD1d tetramers were provided by the Tetramer Core Facility of the US National Institutes of Health. Doublets and dead cells were removed from data analysis. For intracellular staining, live cells were first stained for surface proteins, then fixed and permeabilized, and then stained for intracellular proteins according to manufacturer’s instructions (eBioscience). Data were analyzed with software designed by the Division of Computer Research and Technology of the US National Institutes of Health or with FlowJo software by TreeStar.
Cell sorting and in vitro culture
CD4+ and CD8+ lymph node T cells were isolated by antibody-mediated depletion of CD8+ and immunoglobulin-positive cells or CD4+ and immunoglobulin-positive cells using magnetic beads (BioMag Qiagen). Thymocytes were electronically sorted to purify DN (CD4−CD8−TCRβ−), DP (CD4+CD8+TCRβlow), INT (CD4+CD8lowCD69+CCR7+), SP4 (CD4+CD8−TCRβhi) and SP8 (CD4−CD8+TCRβhi) cells. For purification of different NKT subsets in the thymus, thymocytes were stained with PBS57-loaded CD1d-PE tetramers, incubated with anti-PE microbeads, and then purified on a MACS column (Miltenyi Biotec), followed by electronic sorting of PBS57CD1d+HSA−CD44−NK1.1− (stage1), PBS57CD1d+HSA−CD44+NK1.1− (stage 2) and PBS57CD1d+HSA−CD44+NK1.1+ (stage 3) cells. For isolation of total NKT or γδ thymocytes, cells were electronically sorted for PBS57CD1d+ or GL3+ cells. Thymic epithelial cells (TECs) were sorted from 11–13 day old B6 mice. Thymi were harvested and gently tweezed to release thymocytes. Thymic remnants were digested with Collagensas/Dispase (Roche) and DNAse I (Roche) to release TECs. Cells were then stained for CD45.2, EpCAM, Ly51 and UEA and electronically sorted by a FACSAria. cTECs were gated as CD45.2−EpCAM+Ly51+UEA− and mTECs were gated as CD45.2−EpCAM+UEA+Ly51−.
For in vitro cell cultures, sorted PBS57CD1d+HSA−CD44+NK1.1− NKT cells were cultured in medium for 48 hours. As indicated, IL-15 (50ng/ml; R&D System) alone or in combination with 1α,25-dihydroxyvitamin D3 (100nM; Sigma-Aldrich) and/or all-trans-retinoic acid (100nM; Sigma-Aldrich) were added to the culture. ELISpot assay for IL-4 was done according to manufacturer’s instructions (Mabtech) using 150×103 cells/well of total freshly isolated thymocytes stimulated in the presence of ionomycin (500nM; Sigma-Aldrich) and PMA (10ng/ml; Sigma-Aldrich).
Prediction of miRNA targets and alignment of let-7 miRNA genes
The following URLs were used: http://www.targetscan.org/mmu_61/for TargetScan (release 6.2) to predict let-7 targeting PLZF mRNA, http://genome.lbl.gov/vista/index.shtml for conservation analysis between human and mouse genomic regions containing let-7 genes. Genomatics software package was used to predict nuclear factor binding motifs in these conserved regions.
Luciferase assay
For measuring luciferase activity, 293T cells were co-transfected with PLZF-3′UTR hLuc-hRLuc vector (GeneCopoeia) and either scrambled control microRNA or let-7 containing vectors using Lipofectamine 2000 (Invitrogen). Luciferase activity was assessed after 48hr with the Luc-Pair miR Luciferase Assay Kit (GeneCopoeia).
Isolation of RNA and qPCR
Total RNA was isolated by using miRNeasy Mini Kit (Qiagen) and genomic DNA was digested using DNA-free kit (Ambion). cDNA was synthesized using the SuperScript III kit (Invitrogen) with oligo(dT) priming and amplification of gene-specific products was achieved using TaqMan probes (Applied Biosystems) and are listed (Supplementary Table 1). miRNAs cDNA was synthesized using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) and amplification primers are listed (Supplementary Table 1).
Statistical analysis
P values were determined by Student’s two-tailed t-test.
Supplementary Material
Acknowledgments
We thank N. Taylor and J.-H. Park for critical reading of the manuscript; G.Q. Daley (Harvard Medical School) for iLet7ΔLIN28 and M2rtTA double transgenic mice; A. Bendelac for Zbtb16wt/lu mice; the Tetramer Core Facility of the US National Institutes of Health for tetramer reagents; A. Adams and L. Granger for expert flow cytometry; and J.A. Williams for generously providing cDNA reagents. This research was supported by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Footnotes
Author contributions
L.A.P. designed the study, performed experiments, analyzed data, and contributed to the writing of the manuscript; R.E., S.J., T.K., T.M., M.Y.K., S.O.S., and T.G. performed experiments and analyzed data; A.A., L.F. generated experimental mice; A.S. designed the study, analyzed data, and wrote the manuscript.
References
- 1.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 2.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 3.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 4.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 5.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant’Angelo DB. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahama Y, Singer A. Post-transcriptional regulation of early T cell development by T cell receptor signals. Science. 1992;258:1456–1462. doi: 10.1126/science.1439838. [DOI] [PubMed] [Google Scholar]
- 8.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 9.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedeli M, Napolitano A, Wong MP, Marcais A, de Lalla C, Colucci F, Merkenschlager M, Dellabona P, Casorati G. Dicer-dependent microRNA pathway controls invariant NKT cell development. J Immunol. 2009;183:2506–2512. doi: 10.4049/jimmunol.0901361. [DOI] [PubMed] [Google Scholar]
- 11.Zhou L, Seo KH, He HZ, Pacholczyk R, Meng DM, Li CG, Xu J, She JX, Dong Z, Mi QS. Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development. Proc Natl Acad Sci U S A. 2009;106:10266–10271. doi: 10.1073/pnas.0811119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo KH, Zhou L, Meng D, Xu J, Dong Z, Mi QS. Loss of microRNAs in thymus perturbs invariant NKT cell development and function. Cell Mol Immunol. 2010;7:447–453. doi: 10.1038/cmi.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 14.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Meneely PM, Herman RK. Lethals, steriles and deficiencies in a region of the X chromosome of Caenorhabditis elegans. Genetics. 1979;92:99–115. doi: 10.1093/genetics/92.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 17.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 19.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 20.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, Shah SP, Tanwar PS, Mermel CH, Beroukhim R, Azam M, Teixeira J, Meyerson M, Hughes TP, Llovet JM, Radich J, Mullighan CG, Golub TR, Sorensen PH, Daley GQ. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Consortium D, Investigators M, Altshuler D, Daley GQ. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faehnle CR, Walleshauser J, Joshua-Tor L. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature. 2014;514:252–256. doi: 10.1038/nature13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335:1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, Kim VN. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151:765–777. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX, Yan BY, Nathanson JL, Hutt KR, Lovci MT, Kazan H, Vu AQ, Massirer KB, Morris Q, Hoon S, Yeo GW. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hafner M, Max KE, Bandaru P, Morozov P, Gerstberger S, Brown M, Molina H, Tuschl T. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA. 2013;19:613–626. doi: 10.1261/rna.036491.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greaves DR, Wilson FD, Lang G, Kioussis D. Human CD2 3′-flanking sequences confer high-level, T cell-specific, position-independent gene expression in transgenic mice. Cell. 1989;56:979–986. doi: 10.1016/0092-8674(89)90631-4. [DOI] [PubMed] [Google Scholar]
- 32.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, von Boehmer H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U S A. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 35.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol. 2013;25:161–167. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mooijaart SP, Brandt BW, Baldal EA, Pijpe J, Kuningas M, Beekman M, Zwaan BJ, Slagboom PE, Westendorp RG, van Heemst DC. elegans DAF-12, Nuclear Hormone Receptors and human longevity and disease at old age. Ageing Res Rev. 2005;4:351–371. doi: 10.1016/j.arr.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 38.Shen Y, Wollam J, Magner D, Karalay O, Antebi A. A steroid receptor-microRNA switch regulates life span in response to signals from the gonad. Science. 2012;338:1472–1476. doi: 10.1126/science.1228967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elewaut D, Shaikh RB, Hammond KJ, De Winter H, Leishman AJ, Sidobre S, Turovskaya O, Prigozy TI, Ma L, Banks TA, Lo D, Ware CF, Cheroutre H, Kronenberg M. NIK-dependent RelB activation defines a unique signaling pathway for the development of V alpha 14i NKT cells. J Exp Med. 2003;197:1623–1633. doi: 10.1084/jem.20030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sivakumar V, Hammond KJ, Howells N, Pfeffer K, Weih F. Differential requirement for Rel/nuclear factor kappa B family members in natural killer T cell development. J Exp Med. 2003;197:1613–1621. doi: 10.1084/jem.20022234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mora JR, Iwata M, von Andrian UH. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat Rev Immunol. 2008;8:685–698. doi: 10.1038/nri2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McVoy LA, Kew RR. CD44 and annexin A2 mediate the C5a chemotactic cofactor function of the vitamin D binding protein. J Immunol. 2005;175:4754–4760. doi: 10.4049/jimmunol.175.7.4754. [DOI] [PubMed] [Google Scholar]
- 43.Yu S, Cantorna MT. The vitamin D receptor is required for iNKT cell development. Proc Natl Acad Sci U S A. 2008;105:5207–5212. doi: 10.1073/pnas.0711558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu S, Zhao J, Cantorna MT. Invariant NKT cell defects in vitamin D receptor knockout mice prevents experimental lung inflammation. J Immunol. 2011;187:4907–4912. doi: 10.4049/jimmunol.1101519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raberger J, Schebesta A, Sakaguchi S, Boucheron N, Blomberg KE, Berglof A, Kolbe T, Smith CI, Rulicke T, Ellmeier W. The transcriptional regulator PLZF induces the development of CD44 high memory phenotype T cells. Proc Natl Acad Sci U S A. 2008;105:17919–17924. doi: 10.1073/pnas.0805733105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.