Abstract
The genes and pathways that fine-tune TLR7-mediated innate inflammatory responses remain to be fully elucidated. Using an unbiased genome-scale shRNA screen, we identified the receptor TREML4 as an essential positive regulator of TLR7 signaling. Macrophages from Treml4–/– mice were hyporesponsive to TLR7 agonists and failed to produce type I interferon due to impaired phosphorylation of the transcription factor STAT1 by the MAP kinase p38 and decreased recruitment of MyD88 to TLR7. TREML4 deficiency reduced production of inflammatory cytokines and autoantibodies in SLE-prone MRL/lpr mice and inhibited the antiviral immune response to influenza. Our data identify TREML4 as a positive regulator of TLR7 signaling and provide insight into the molecular mechanisms that control antiviral immunity and the development of autoimmunity.
In humans and mice combined the Toll-like receptor (TLR) family consists of thirteen members that can be divided into two subgroups based on their cellular location1. Cell surface TLRs (TLR1, TLR2, TLR4, TLR5 and TLR6) recognize various molecules in bacteria and fungi. Intracellular TLRs (TLR3, TLR7, TLR9, TLR13) recognize nucleic acids; TLR3 recognizes double-stranded RNA, TLR7 recognizes single-stranded RNA, and TLR9 and TLR13 are receptors for double-stranded DNA and ribosomal RNA, respectively2. TLRs are an evolutionarily conserved family of type I transmembrane receptors that have an extracellular domain compromising leucine rich repeats and a cytoplasmic domain that shares significant homology with the mammalian type I IL-1 receptor3. The TLRs are germ-line encoded receptors that recognize a myriad of conserved microbial-associated molecular patterns (MAMPs) found in many different microbes, such as bacteria, fungi, viruses, and parasites. TLR recognition of these MAMPs leads to the initiation of intracellular signaling pathways that elicit the expression of inflammatory genes, such as cytokines essential for host defense. Upon ligand binding, all TLRs trigger a common signal transduction pathway that starts with the recruitment of the intracellular adaptor protein MyD88 that mediates the phosphorylation of IRAK1 by IRAK44. Phosphorylated IRAK1 associates with TRAF6, which mediates the activation of mitogen-activated protein kinases (MAPK) and subsequent activation of transcription factors that promote cytokine gene expression.
TLR activation is central to early host defense, however excessive activation of the TLR signaling pathway can contribute to chronic inflammatory diseases5. Thus, TLR signaling must be under tight positive and negative regulation to maintain immune tolerance. Recently, members of the triggering receptor expressed on myeloid cells (TREM) family have been shown to regulate innate immune responses by amplifying or dampening TLR-induced signals6-9. TREM and TREML receptors are a structurally similar family of receptors consisting of a single extracellular variable-type immunoglobulin (Ig)-like domain, a transmembrane domain and a short cytoplasmic tail lacking any known signaling motifs, which are expressed predominantly on myeloid cells10. TREM1 promotes the inflammatory response to bacteria and the TLR4 ligand LPS6. In contrast, TREM2 and TREML1 suppress TLR-induced signals and protect against autoimmunity11-13. The inhibitory TLR signals mediated by TREM2 depend upon its interaction with DAP12, a transmembrane-anchored signaling adaptor containing an ITAM within its cytoplasmic domain8,14. The ligands for TREM receptors have largely remained elusive, however several reports suggest that TREM receptors can bind to microbial and host molecules. TREML4 has been shown to bind late apoptotic and necrotic cells15 and TREML1 to fibrinogen11, whereas TREM1 and TREM2 recognize anionic ligands from bacteria16.
TLRs have also been implicated in the development and exacerbation of chronic inflammatory syndromes and autoimmunity through their inappropriate activation by endogenous self ligands, such as nucleic acids released from necrotic cells, including in the development and progression of the autoimmune disease systemic lupus erythematosus (SLE)2,5,17. Dysregulated activation of TLR7 has been implicated in the pathogenesis of SLE in humans and mice. Male BXSB mice with a Y-linked autoimmune accelerator locus (Yaa) develop spontaneous SLE-like disease due to a duplication of a 4-Mb gene segment containing TLR7 transposed to the Y chromosome18,19. This duplication is responsible for the autoimmune phenotype in Yaa male mice, because reduction of TLR7 copy number abrogated disease20. Furthermore, MRL/lpr mice that spontaneously develop SLE have significantly reduced renal disease and autoantibodies to RNA-associated autoantigens when backcrossed to TLR7-deficient mice21,22. Finally, TLR7 single nucleotide polymorphisms (SNPs) have been identified that are associated with increased transcript expression and increased risk for SLE in humans23. Together, these observations indicate that TLR7 signaling is critical for SLE-specific autoimmunity.
To identify genes required for TLR7-mediated immune cell activation, we performed a genome-scale RNA-mediated interference (RNAi)-based screen in murine macrophages and identified TREML4 is a positive regulator of TLR signaling. TREML4 was required for TLR7-mediated responsiveness to TLR7 ligands and promoted TLR7-induced activation and phosphorylation of p38 MAPK and STAT1 and the trafficking and localization of MyD88 and TLR7 to endosomes. We also show that TREML4 is critical for the antiviral immune response to the ssRNA influenza virus. Finally, TREML4 deficiency ameliorated inflammatory cytokine production and kidney disease in SLE-prone MRL/lpr mice. Thus, our findings demonstrate a critical role for TREML4 in promoting TLR7-mediated cytokine production during antiviral responses and the development of autoimmunity.
Results
TREML4 positively regulates the TLR7 pathway
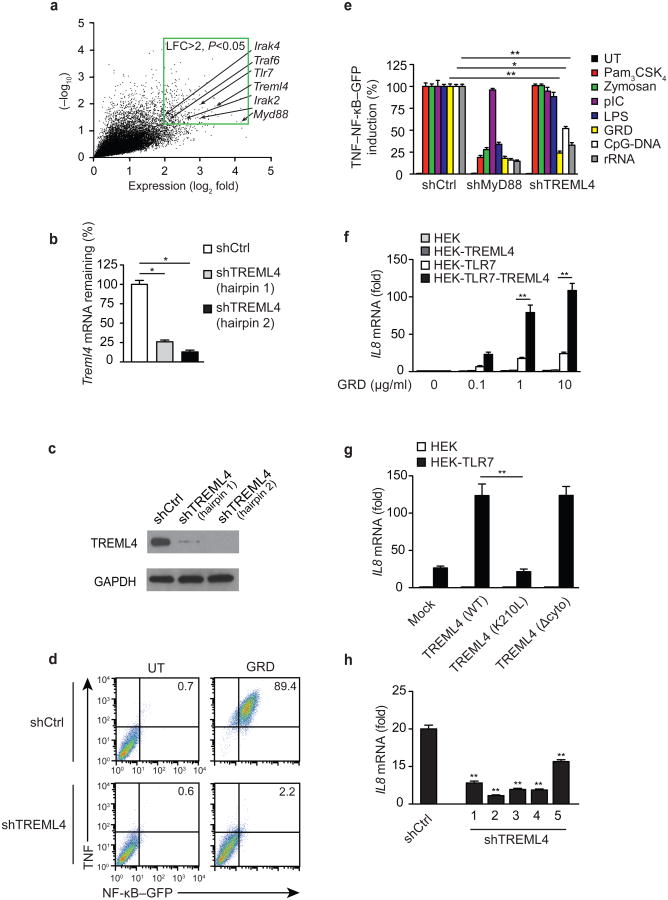
To identify genes and pathways required for TLR7-induced cell activation, we performed an unbiased genome-scale pooled lentivirus-based RNAi screen with a 40k shRNA library developed by The RNAi Consortium24,25. Mouse RAW 264.7 macrophages stably transfected with an NF-κB-responsive endothelial leukocyte adhesion molecule-1 (ELAM -1) promoter driving expression of enhanced green fluorescent protein (RAW-GFP) were transduced with a pool of ∼40,000 lentivirally encoded short hairpin RNAs (shRNAs). With this approach ∼8,000 murine genes were targeted by 5 or more independent shRNAs per gene and cells received an average of ∼0.3 shRNAs [multiplicity of infection (MOI = 0.3]. Each shRNA was introduced to ∼200 independent cells. shRNA-transduced cells were treated with the synthetic TLR7 specific agonist gardiquimod (GRD) for 6 hours and sorted by flow cytometry into two populations based on the expression of TNF and NF-κB-GFP: TNFloGFPlo and TNFhiGFPhi. The shRNA sequences were then amplified by PCR from the genomic DNA isolated from the two cell populations and hybridized to complementary microarrays. To identify the shRNAs that inhibited GRD-induced TNF and NF-κB expression (i.e. positive regulators of TLR7 signaling), we measured the overrepresentation of shRNAs that were repeatedly present in the TNFloGFPlo sorted population of RAW-GFP cells compared to the TNFhiGFPhi subset. Using this approach, we identified 257 enriched shRNAs with a log fold-difference greater than 2 and a P-value less than 0.05 (Fig. 1a and Supplementary Table 1). In the TNFloGFPlo subset we identified an enrichment of shRNAs that targeted genes with well-established roles in the TLR7 signaling pathway, such as TLR7, MyD88, IRAK2, IRAK4, TRAF6 and IRF7. In addition, we identified several genes that have not been previously described as positive regulators of TLR7 signaling. Among these, an shRNA targeting TREML4 was the 11th most enriched shRNA in our RNAi-based screen out of the ∼40,000 shRNAs tested (LFC=3.42; P=0.012; Fig. 1a and Supplementary Table 1). To validate the screen results, we independently transfected RAW-GFP reporter cells with two different shRNAs that targeted two distinct regions of TREML4 and possessed different sequences in the seed region that is known to mediate off-target effects. Quantitative PCR (qPCR) and immunoblotting in the RAW-GFP transfected cells showed that both hairpins effectively knocked down TREML4 (Fig. 1b,c), and found a significant correlation between the reduced expression of TREML4 mRNA and protein and diminished TNF and NF-κB-GFP expression in response to TLR7 stimulation (Fig. 1d). These data are the first demonstration that TREML4 is a positive regulator of TLR7 signaling. To determine whether TREML4 regulates the signaling pathways downstream of other TLRs, we transfected RAW-GFP cells with shRNA-encoding viruses to silence MyD88 or TREML4 expression and treated them with agonists for TLR2-TLR1 (Pam3CSK4), TLR2-TLR6 (zymosan), TLR3 (pIC), TLR4 (lipopolysaccharide, LPS), TLR9 (CpG-DNA) or TLR13 (rRNA). Silencing of Treml4 expression significantly reduced the ability of RAW-GFP cells to induce NF-κB-GFP and TNF expression in response to TLR7, TLR9 and TLR13 agonists, but not to other TLR ligands (Fig. 1e). These data demonstrate that TREML4 is a positive regulator for a limited sub-group of intracellular TLRs in RAW macrophages.
Figure 1.
TREML4 positively regulates TLR7 signaling.
(a) Expression analysis of shRNAs present in the TNFloGFPlo sorted population of RAW-GFP macrophages treated with GRD for 6 h compared to the TNFhiGFPhi subset. Data within the green border represent 257 enriched shRNAs with a log fold change (LFC) >2; P<0.05. (b) Quantitative PCR analysis of Treml4 mRNA in RAW-GFP cells transduced with viruses containing shRNAs targeting Treml4 (shTREML4) or scrambled control (shCtrl). (c) Immunoblot analysis of TREML4 and GAPDH (loading control) in RAW-GFP transduced with shRNA lentiviruses targeting TREML4. (d) Flow cytometry analysis of TNF and GFP in RAW-GFP transduced with shTREML4 treated with 10 μg/mL of GRD for 6 h. (e) Flow cytometry analysis of TNF and GFP in RAW-GFP transduced with shRNA lentiviruses targeting MyD88 or TREML4 treated with specified TLR agonists for 4 h. Data presented as the percentage induction relative to shCtrl. (f) Quantitative PCR analysis of Il8 mRNA in HEK expressing TREML4 and/or TLR7 treated with GRD for 3 h. (g) Quantitative PCR analysis of Il8 mRNA in HEK transfected with TLR7 and/or TREML4(WT), TREML4(K210L), or TREML4(Δcyto) treated with 10 μg/mL GRD for 3 h. (h) Quantitative PCR analysis of Il8 mRNA in THP-1 transduced with lentivirus encoding shRNAs targeting unique sequences in the human Treml4 gene treated with 10 μg/mL GRD for 3 h. *P<0.05 and **P<0.01 (Mann-Whitney test). Data are representative of an experiment done three times (b,d-g; median and s.d. of at least triplicates).
We next tested whether TREML4 overexpression could amplify TLR signaling. Human embryonic kidney (HEK) cells stably expressing TREML4 were transfected with plasmids containing TLR2, TLR3, TLR4, TLR7, TLR9 or TLR13 cDNA and stimulated with their cognate agonists. TREML4 overexpression significantly amplified TLR7-, TLR9- and TLR13-mediated induction of Il8 mRNA in HEK cells transfected with TLR7, TLR9, or TLR13, respectively (Fig. 1f and Supplementary Fig. 1a). HEK cells overexpressing TREML4 in the absence of TLR7 overexpression did not produce Il8 mRNA in response to TLR7 agonists (Fig. 1f) indicating that TREML4 is not activated directly by TLR7 ligands.
To test if TREML4 regulates TLR7 signaling through its association with intracellular adaptors, we transfected TLR7-expressing HEK cells with plasmids encoding wild-type TREML4(WT), TREML4 lacking the cytoplasmic domain TREML4(Δcyto) or a TREML4 containing a point mutation in the transmembrane domain TREML4(K210L) and measured Il8 mRNA levels following stimulation with the TLR7 ligand GRD. We found that the cytosolic domain of TREML4 was only modestly required for TLR7 signaling (Fig. 1g). In contrast, HEK cells expressing TREML4(K210L) had a significant impairment in TLR7-induced Il8 mRNA compared to TREML4(WT) expressing HEK cells, indicating that the charged lysine residue in the TM domain is required for TREML4-mediated amplification of TLR7 signaling (Fig. 1g).
Human and mouse TREML4 share about 40% amino acid identity, however human TREML4 is truncated compared to its mouse counterpart due to insertion of a stop codon just downstream of the charged lysine residue in the TM domain. To determine whether human TREML4 regulates TLR7 signaling, we silenced TREML4 expression in the human THP-1 monocyte cell line by lentiviral transduction of Treml4-specific shRNAs. Treml4-silenced THP-1 cells had a reduced expression of Il8 mRNA following stimulation with the TLR7 ligand GRD compared to THP-1 cells transduced with control shRNAs (Fig. 1h), indicating an evolutionarily conserved function for TREML4 in the regulation of TLR7 signaling.
Because TREML4 binds to late apoptotic and necrotic cells15 and because nucleic acids can accumulate in apoptotic blebs or leak from necrotic cells, we tested whether nucleic acid ligands for TLR7, TLR9 and TLR13 could bind and trigger TREML4 signaling. Because little is known about the signaling pathways and targets downstream of TREM receptors, we constructed a chimeric protein containing the extracellular domain of TREML4 fused to the intracellular domain of the tumor-necrosis factor (TNF) receptor TNF-R1 and overexpressed it in HEK cells. The TREML4-TNF-R1 chimeric receptor allowed the quantitation of ligand binding to TREML4 through induction of the TNF-R1-dependent signaling pathway that leads to the activation of the transcription factor NF-κB and expression of Il8 mRNA in HEK cells26. Treatment with late apoptotic cells induced robust induction of Il8 mRNA in cells expressing TREML4-TNF-R1 but not those expressing DECTIN1-TNF-R1 (Supplementary Fig. 1b), which indicated a direct interaction between TREML4 and apoptotic cells. In contrast, we observed only weak induction of Il8 mRNA (<2 fold) following stimulation with GRD, CpG-DNA or rRNA compared to mock transfected cells (Supplementary Fig. 1b), suggesting that nucleic acids are not cognate ligands for TREML4. Together these results identified mouse and human TREML4 as a positive regulator of TLR7, TLR9 and TLR13 signaling.
TREML4 positively regulates TLR7, TLR9, and TLR13 signaling
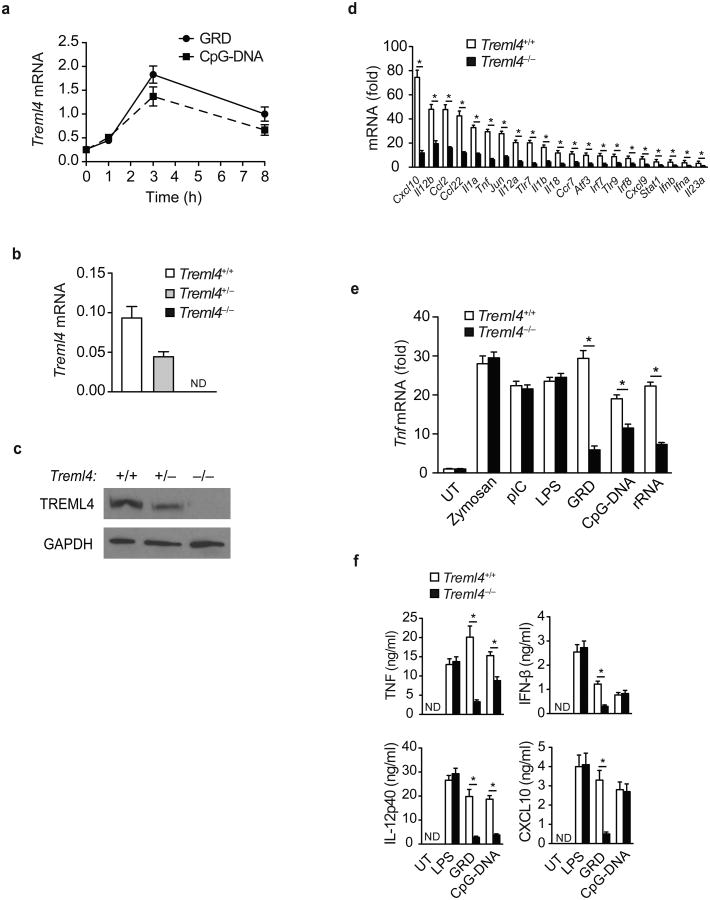
We performed qPCR analysis to examine the expression of Treml4 mRNA in various organs in wild-type B6 mice. Treml4 was mainly expressed in the spleen (Supplementary Fig. 2a). Treml4 expression was low or absent on endothelial cells (EC), B cells, T cells and NK cells, but detectable on macrophages, DCs, neutrophils, bone marrow-derived DC (BMDC) and bone marrow-derived macrophages (BMDM; Supplementary Fig. 2b), consistent with published reports suggesting restricted TREML4 expression on splenic macrophages and DCs 15,27. In addition, in vitro stimulation with the TLR7 ligand GRD induced a rapid increase of Treml4 mRNA expression in cultured splenic macrophages (Fig. 2a).
Figure 2.
Regulation of TLR7 signaling by TREML4.
(a) Quantitative PCR analysis of Treml4 mRNA in F4/80+ splenic macrophages isolated from Treml4+/+ mice treated with 10 μg/mL GRD or CpG-DNA for 1, 3, or 8 h. (b) Quantitative PCR analysis of Treml4 mRNA in the blood of Treml4+/+, Treml4+/– and Treml4–/– mice. (a,b) Data presented as copies of Treml4 mRNA normalized to copies of Gapdh mRNA. (c) Immunoblot analysis of TREML4 and GAPDH (loading control) in spleen extracts isolated from Treml4+/+, Treml4+/– and Treml4–/– mice. (d) Quantitative PCR analysis of the indicated inflammatory genes in F4/80+ splenic macrophages isolated from Treml4–/– and wild-type mice treated with 10 μg/mL GRD for 4 h. (e) Quantitative PCR analysis of Tnf mRNA in F4/80+ splenic macrophages isolated from Treml4+/+ and Treml4–/– mice treated with the specified TLR ligand for 4 h. (d,e) Data presented as fold mRNA over untreated. (f) ELISA of TNF, IL12p40, IFN-β and CXCL10 in supernatants of F4/80+ splenic macrophages isolated from Treml4+/+ and Treml4–/– mice treated with CpG-DNA (10 μg/mL), GRD (10 μg/mL) or LPS (100 ng/mL) for 18 h. (ND, none detected). *P<0.01 (Mann-Whitney test). Data are from one experiment representative of at least three independent experiments performed (b,d-f; error bars, median and s.d. of triplicates).
To further examine the role of TREML4 in regulating TLR7 responses, we generated TREML4-deficient (Treml4–/–) mice in which TREML4 expression was ablated by replacement of exons 2-4, which encode most of the extracellular domain, with a neomycin resistance cassette (Fig. 2b,c and Supplementary Fig. 2c). To determine whether TREML4 was required for TLR7-induced gene expression in splenic macrophages, we measured the induction of mRNA for various cytokines, chemokines and transcription factors in GRD-stimulated splenic macrophages from wild-type littermates and Treml4–/– mice. Compared to wild-type cells, Treml4–/– splenic macrophages mice had a significant impairment in the induction of 21 genes known to be activated by the TLR7 pathway (Fig. 2d), such as pro-inflammatory cytokine and chemokine expression, type I IFN and IFN-induced genes, such as Cxcl10, Cxcl9 and Stat1. We also stimulated wild-type and Treml4–/– splenic macrophages with agonists for TLR2, TLR3, TLR4, TLR7, TLR9 or TLR13. Treml4–/– splenic macrophages mice had a significant impairment in GRD, CpG-DNA and rRNA-induced Tnf mRNA expression compared to wild-type macrophages (Fig. 2e), confirming the results obtained using shRNA knockdown of TREML4 in the RAW cells (Fig. 1e). ELISA performed on the supernatants of GRD-, CpG-DNA- or LPS-stimulated splenic macrophages showed that secretion of TNF, IL-12p40, IFN-β and the IFN-inducible chemokine CXCL10 protein was significantly impaired in Treml4–/– macrophages compared to wild-type macrophages in response to GRD stimulation (Fig. 2f), while secretion of IFN-β and CXCL10 was similar, and TNF and IL-12p40 production was significantly impaired in Treml4–/– compared to wild-type macrophages in response to CpG-DNA stimulation (Fig. 2f). These results suggest that TLR7-mediated activation of the IFN pathway is TREML4-dependent, while TLR9-mediated activation of the IFN pathway is TREML4-independent. We found no differences in cytokine production between wild-type and Treml4–/– macrophages in response to LPS stimulation (Fig. 2f), indicating that TREML4 is not involved in regulating the TLR4 pathway.
We also examined whether TREML4 modified TLR-induced cytokine production in neutrophils. We found that CD11b+Ly6g+ cells (neutrophils) isolated by magnetic bead-based negative selection from the bone marrow of Treml4–/– mice had a significant impairment in GRD- and rRNA-induced Cxcl2 mRNA expression compared to wild-type neutrophils, suggesting that TLR7 and TLR13-mediated induction of Cxcl2 in neutrophils is TREML4-dependent (Supplementary Fig. 2d). In contrast, Treml4–/– neutrophils migrated normally in response to the peptide chemoattractant formyl-Methionyl-Leucyl-Phenylalanine (fMLP) in a transwell chemotaxis assay and had normal ability to phagocytose opsonized latex beads compared to wild-type neutrophils (Supplementary Fig. 2e,f), suggesting that migration and phagocytic pathways in neutrophils are TREML4-independent. Collectively, these data indicate that TREML4 is a nonredundant positive regulator of TLR7, TLR9 and TLR13 signaling in splenic macrophages and neutrophils.
TREML4 is critical for responses to ssRNA and influenza virus
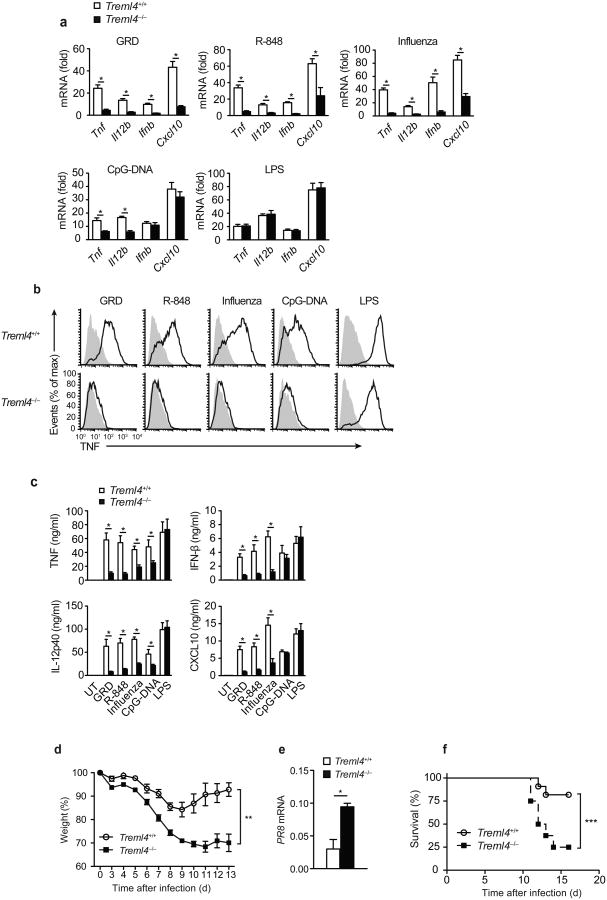
To test whether TREML4 also regulated TLR7 responses to other synthetic and natural ligands, such as the synthetic imidazoquinoline compound R-848 and the ssRNA influenza virus28, we injected GRD, R-848, CpG-DNA or LPS intravenously (i.v.) or live influenza AZ/PR8/34 (PR8) virus intranasally (i.n.) into wild-type and Treml4–/– mice and assessed the induction of TNF, IL-12p40, IFN-β and CXCL10 mRNA and protein by qPCR analysis, flow cytometry and ELISA at 8 h post-injection in the spleens and blood serum. Splenocytes from wild-type mice injected with GRD, R-848 or influenza had high expression of Tnf, Il12b, Ifnb and Cxcl10 mRNA (Fig. 3a). In contrast, splenocytes isolated from Treml4–/– mice injected with GRD, R-848 or influenza virus showed a marked impairment in the production of these inflammatory cytokines (Fig. 3a). Intracellular flow cytometry showed that F4/80hiCD11bloCD11c– splenic macrophages from GRD, R-848, influenza or CpG-DNA injected Treml4–/– mice produced significantly lower amounts of TNF protein compared to wild-type macrophages (Fig. 3b). The amounts of TNF and IL-12p40 protein in the serum were significantly reduced in Treml4–/– mice compared to wild-type mice following injection with GRD, R-848, CpG-DNA, LPS or influenza virus (Fig. 3c). Serum IFN-β and CXCL10 were significantly reduced in GRD, R-848 or influenza virus injected Treml4–/– mice as compared to wild-type mice, but were induced at similar levels as in wild-type controls by CpG-DNA or LPS (Fig. 3c), indicating that the TLR7-mediated activation of the IFN pathway in vivo is TREML4-dependent, while the TLR9- and TLR4-mediated activation of the IFN pathway in vivo is TREML4-independent.
Figure 3.
In vivo regulation of TLR7 signaling by TREML4.
(a) Quantitative PCR analysis of Tnf, Il12b, Ifnb and Cxcl10 mRNA in splenic cells from Treml4+/+ and Treml4–/– mice (n=9) injected intravenously with 100 μg/mL GRD, 100 μg/mL R-848, 100 μg/mL CpG-DNA, or 10 μg/mL LPS, or intranasally with 5 × 106 PFU live influenza virus for 8h. Data presented as fold over saline injected mice. (b) Flow cytometry analysis of intracellular TNF expression in F4/80+CD11b+CD11c– splenic macrophages isolated from Treml4+/+ and Treml4–/– mice injected and treated as in (a). (c) ELISA of TNF, IL12p40, IFN-β and CXCL10 in the sera of mice injected and treated as in (a,b). (d) Weight loss analysis of Treml4+/+ and Treml4–/– mice (n=10) injected intranasally with sublethal dose of influenza PR8 virus (34 EID/gram). (e) Quantitative PCR analysis of influenza PR8 mRNA in the lungs of mice at day 10 after influenza infection. (f) Survival of Treml4+/+ and Treml4–/– mice (n=12) following infection with LD25 influenza PR8 (50 EID/gram). *P<0.01 (Mann-Whitney test). Data are from one experiment representative of at least three independent experiments performed (a,c,e; error bars, mean and s.d. of triplicates). **P<0.0001 (two-way ANOVA). ***P<0.01 (log rank test).
To further examine the role of TREML4 in the antiviral host defense to influenza, we monitored weight loss and viral load in wild-type and Treml4–/– mice infected i.n. with a sublethal dose of influenza. Treml4–/– mice lost weight with similar kinetics compared to wild-type littermate control mice during the first week of infection (Fig. 3d). However, Treml4–/– mice continued to lose weight by day 9 post-infection, while wild-type control mice plateaued and then began to gain weight steadily (Fig. 3d). Wild-type mice cleared the influenza virus between 10-14 days after primary infection and most mice survived (Fig. 3e,f), while viral burden in the lungs of Treml4–/– mice was significantly higher than in wild-type at day 10 and a majority of the mice succumbed to infection (Fig. 3e,f). Together these data demonstrate that TREML4 is necessary to control the early antiviral response to i.n. influenza virus infection and to restrict the viral load in the lung of infected mice.
To determine whether TREML4 was required for the immune response to other pathogenic viruses, we measured the expression of Tnf mRNA by qPCR in splenic macrophages isolated from wild-type and Treml4–/– mice cultured with vesicular stomatitis virus (VSV), a ssRNA virus, or cytomegalovirus (CMV), a dsDNA virus. Treml4–/– macrophages had a significant impairment in VSV- and CMV-induced Tnf mRNA expression compared to wild-type macrophages (Supplementary Fig. 3a), suggesting that TREML4 may play an important role in the antiviral immune response to a wide range of human pathogenic viruses.
To test whether TREML4 deficiency in Treml4–/– mice had affected the cellular expression of TLR7 and/or members of the TLR7 signaling pathway, we examined the expression of TLR7 in splenic macrophages and DCs from Treml4–/– mice. As assessed by intracellular flow cytometry, TLR7 expression in Treml4–/– cells was similar to that found in wild-type cells (Supplementary Fig. 3b). In addition, we found that untreated splenic macrophages from wild-type and Treml4–/– mice expressed similar amounts of genes critical for TLR7 signaling, such as Tlr7, Myd88, Irak1, Irak3, Irak4, Traf6, Tbk1, Syk, Dap12, Irf5, Irf7, Stat1, and Stat2 (Supplementary Fig. 3c), suggesting decreased expression of TLR7 or other proteins in the TLR7 pathway does not account for the hyporesponsiveness to GRD, R-848 or influenza virus challenge observed in Treml4–/– mice. Collectively, these data demonstrate the essential role of TREML4 in the antiviral host response to influenza infection.
TREML4 regulates TLR7-induced phosphorylation of MAPK and STAT1
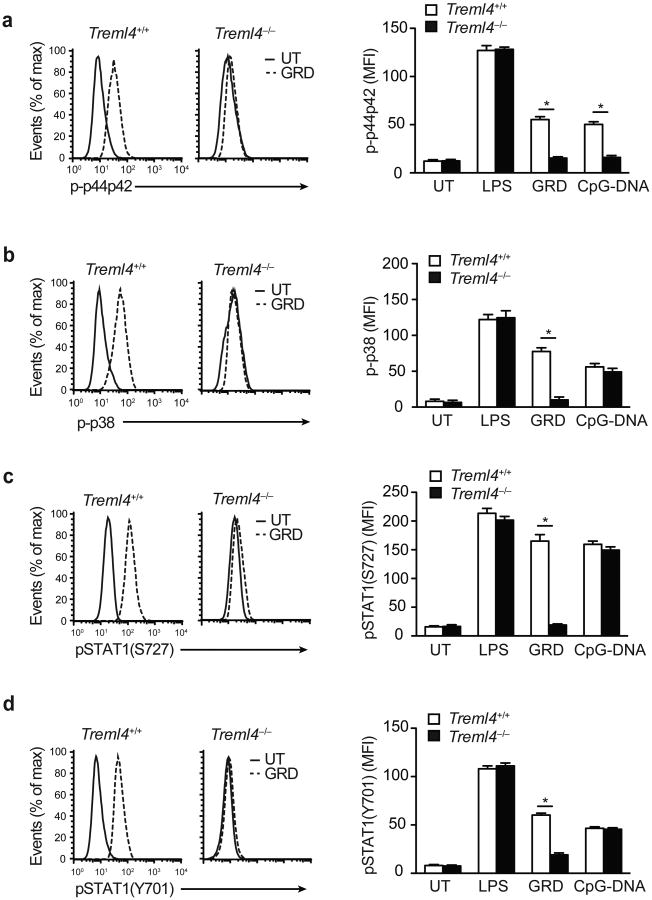
We next assessed the molecular mechanism of Treml4–/– macrophage hyporesponsiveness to TLR7 agonists. To determine whether TREML4 regulates TLR ligand-induced MAPK activation, we stimulated BMDC prepared from wild-type and Treml4–/– mice with LPS, GRD or CpG-DNA and determined phospho-p44p42 and phospho-p38 accumulation using intracellular flow cytometry. GRD stimulation caused a rapid (within 15 min) increase in phospho-p44p42 and phospho-p38 in wild-type BMDC but not in TREML4-deficient BMDC (Fig. 4a,b). Following CpG-DNA stimulation, phosphorylation of p44p42, but not p38, was reduced in Treml4–/– BMDC compared to wild-type cells (Fig. 4a,b), suggesting that TREML4 differentially regulates TLR7 and TLR9 signaling pathways. We observed no differences in the ability of LPS to activate MAPKs in wild-type and Treml4–/– BMDC (Fig. 4a,b).
Figure 4.
TLR7-induced phosphorylation of MAPKs and STAT1 is regulated by TREML4.
(a-d) Phosphoflow cytometry analysis of p44p42 (a), p38 (b), pSTAT1(S727) (c) and pSTAT1(Y701) (d) in CD11c+ BMDCs isolated from Treml4+/+ and Treml4–/– mice treated with 100 ng/mL LPS, 10 μg/mL GRD, or 10 μg/mL CpG-DNA for 30 min. Histograms are representative of one experiment of 3 independent experiments performed. Graphs represent the mean fluorescent intensity (MFI). *P<0.01 (Student's t-test; error bars, mean and s.d.).
Activation of the transcription factor STAT1 is required for the production of IFN and IFN-regulated genes. Full activation of STAT1 occurs by phosphorylation of its Ser727 and Tyr701 residues29. Ser727 phosphorylation is mediated by TLR-induced p38 MAPK, while Y701 phosphorylation is dependent on autocrine type I IFN signaling30. Thus, Ser727 phosphorylation of STAT1 is TLR-dependent and IFN-independent, while tyrosine701 phosphorylation is TLR-independent and IFN-dependent. To test whether TREML4 regulated the TLR7-induced Ser727 phosphorylation of STAT1, we stimulated wild-type and Treml4–/– BMDC with LPS, GRD or CpG-DNA and determined induction of STAT1 Ser727 phosphorylation by intracellular flow cytometry. STAT1 Ser727 phosphorylation was observed in LPS, GRD and CpG-DNA-stimulated wild-type BMDC and LPS or CpG-DNA-stimulated Treml4–/– BMDC, but not in GRD-stimulated Treml4–/– BMDC (Fig. 4c). These data indicated that the TLR7-mediated activation of p38 and STAT1 Ser727 phosphorylation were TREML4-dependent, while the TLR4- and TLR9-mediated activation of p38 and STAT1 were TREML4-independent. In addition, we found that STAT1 Tyr701 phosphorylation was significantly impaired in Treml4–/– BMDCs treated with GRD compared to wild-type BMDCs (Fig. 4d), suggesting that TREML4 also amplifies IFNAR1 signaling. Because IFN-α, IFN-β, and CXCL10 expression is induced by STAT1, these observations are consistent with the reduced induction of these genes in response to TLR7, but not TLR4 or TLR9 stimulation, and suggests that TREML4 may mediate the cross-talk between the TLR, IFN and STAT1 signaling pathways31.
TREML4 regulates MyD88 and TLR7 trafficking and localization
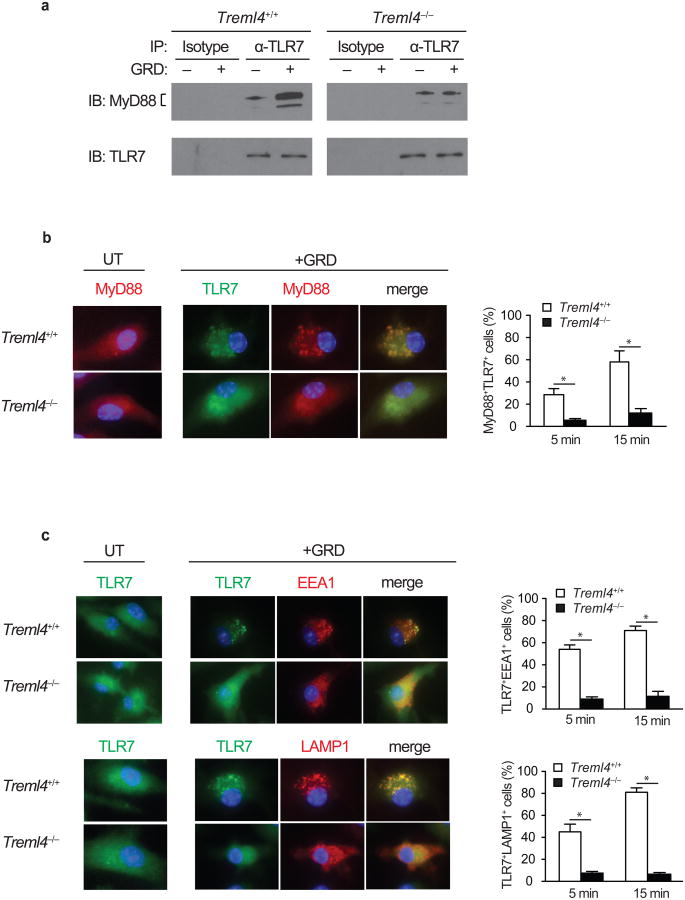
To determine whether TREML4 regulates the intracellular trafficking of MyD88 to TLR7, we examined if TLR7 and MyD88 interact following GRD stimulation. TLR7 and MyD88 co-immunoprecipitated in GRD-stimulated wild-type BMDCs, while this association was not observed in Treml4–/– BMDCs. Binding and recruitment of MyD88 to TLR7 was TREML4-dependent (Fig. 5a). We also investigated the recruitment of MyD88 to TLR7 by confocal microscopy in BMDCs stimulated with GRD for 5 or 15 minutes, and stained with fluorescently-labeled antibodies against TLR7 and Myd88. We observed a significant impairment in the trafficking and co-localization of MyD88 and TLR7 in Treml4–/– BMDC compared to wild-type BMDC (Fig. 5b), suggesting that TREML4 regulates TLR7 signaling by controlling the recruitment of MyD88 to TLR7. To investigate whether TREML4 regulates the trafficking of TLR7 from the ER to endosomal and lysosmal compartments after stimulation, we stimulated wild-type and Treml4–/– BMDCs with GRD for 5 or 15 minutes, followed by staining with fluorescently-labeled antibodies against TLR7, the endosome marker EEA1 or the lysosomal-associated membrane protein LAMP1. Confocal microscopy showed a significant impairment in the trafficking and co-localization of TLR7 to EEA1 or LAMP1 in Treml4–/– BMDC compared to wild-type BMDC (Fig. 5c). Together, these results demonstrate that TREML4 regulates TLR7 signaling by controlling the recruitment of TLR7 to endosomes and lysosomes where it interacts with MyD88 to initiate signaling.
Figure 5.
TREML4 regulates MyD88 and TLR7 trafficking and localization.
(a) Immunoblot analysis of TLR7 and MyD88 of TLR7 immunoprecipitates from BMDC generated from Treml4–/– and wild-type mice stimulated with GRD for 15 min. (b, c) Double immunofluorescence of TLR7 and MyD88, EEA1, or LAMP1 on BMDC generated from Treml4–/– and wild-type mice stimulated for 5 or 15 min. Data show a representative experiment of at least 3 independent experiments performed. *P<0.01 (b-c; Student's t-test; error bars, mean and s.d.).
TREML4-deficiency reduces autoimmunity in mice
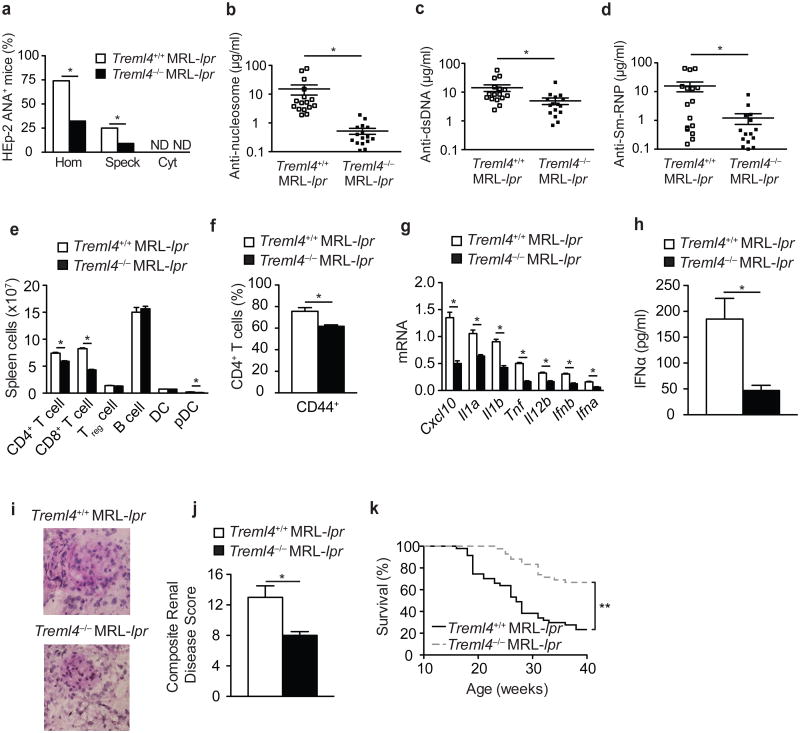
To assess the impact of TREML4 deficiency on autoimmunity, we backcrossed Treml4–/– mice for eight generations with MRL/lpr mice, which spontaneously develop an autoimmune disease with clinical features similar to human SLE, including autoantibody generation, immune cell activation and lethal glomerulonephritis. We used a HEp-2 antinuclear antibody (ANA) indirect fluorescent antibody assay for detecting antibodies to both RNA- and DNA-containing autoantigens in the sera of Treml4–/– MRL/lpr and wild-type MRL/lpr mice. At 16 weeks of age, sera from most MRL/lpr mice (∼96%) produced a homogenous (anti-DNA) or speckled (anti-RNA) pattern of nuclear ANA, while only 40% of Treml4–/– MRL/lpr serum samples showed proof of nuclear ANA production (Fig. 6a). ELISA for autoantibodies to nucleosomes, which detects anti-histones and anti-dsDNA autoantibodies, showed significantly lower concentration of autoantibodies in Treml4–/– MRL/lpr mice than in MRL/lpr mice (Fig. 6b). ELISA to quantify autoantibodies to the dsDNA and RNA-containing Sm-RNP autoantigen showed that the generation of DNA and Sm-RNP autoantibodies was significantly reduced in Treml4–/– MRL/lpr mice compare to wild-type MRL/lpr mice (Fig. 6c,d). In addition, the highly specific Crithidia luciliae immunofluorescence assay showed a substantial decrease in autoantibodies binding to the dsDNA of the C. luciliae kinetoplast in sera from Treml4–/– MRL/lpr sera (data not shown). These results are consistent with previous reports demonstrating an impairment in the generation of autoantibodies to RNA- and DNA-containing autoantigens in MRL/lpr mice deficient in TLR7 and TLR9 signaling21.
Figure 6.
Ameliorated autoimmunity in TREML4-deficient MRL/lpr mice.
(a) ANA immunofluorescence staining pattern of HEp-2 cells with sera from 16-week old MRL/lpr Treml4–/– and MRL/lpr mice. Shown is the percentage of mice positive for nuclear homogenous (Hom), nuclear speckled (Speck), or cytoplasmic (Cyt) staining pattern. (b-d) ELISA of anti-nucleosome (dsDNA/histone) (b), anti-dsDNA (c) and anti-Sm-RNP antibodies (d) in the serum of 16 week old MRL/lpr Treml4–/– and MRL/lpr mice (n=16 mice per group). (e,f) Flow cytometry analysis of immune cell distribution (e) and percentage of CD44+CD4+ activated T cells (f) in the spleen of MRL/lpr Treml4–/– and MRL/lpr mice at 16 weeks of age (n=8 mice per group). (g) Quantitative PCR analysis of cytokine mRNA in spleen of MRL/lpr Treml4–/– and MRL/lpr mice at 16 weeks of age (n=8 mice per group). (h) ELISA of IFN-α in the sera of 16 week old MRL/lpr Treml4–/– and MRL/lpr mice (n=16 mice per group). (i) Histopathologic analysis of PAS stained kidney cryosections isolated from 16 week-old MRL/lpr Treml4–/– and MRL/lpr. (j) Composite renal disease score based glomerular inflammation, glomerular IgG deposition, urine proteinuria, and blood urea nitrogen concentration (n=16 mice per group). Each parameter scored 1 to 4 for each animal, higher number indicates increased disease severity. (k) Survival of MRL/lpr (n=47) and MRL/lpr Treml4–/– (n=42) mice. *P<0.01 (e, f, j, Student's t-test; a-d, g-h, Mann-Whitney test) and **P<0.0001 (k, log rank test).
We observed a significant decrease in the number of CD4+ T cells, CD8+ T cells, and pDCs in the spleen and lymph nodes of Treml4–/– MRL/lpr mice compared to wild-type MRL/lpr mice (Fig. 6e) as well as in the number of activated CD44+CD4+ T cells in the spleen of Treml4–/– MRL/lpr mice (Fig. 6f). Splenocytes from Treml4–/– MRL/lpr mice expressed significantly lower mRNA for Cxcl10, Il1a, Il1b, Tnf, Il12b, Ifnb, and Ifna compared to wild-type MRL/lpr mice (Fig. 6g) and IFN-α concentration in the serum was significantly lower in Treml4–/– MRL/lpr mice compared to MRL/lpr mice (Fig. 6h). Glomerular size, cellularity and deposition of protein in periodic acid Schiff-stained kidney sections was reduced in Treml4–/– MRL/lpr mice compared to MRL/lpr mice (Fig. 6i). Furthermore, the composite renal disease score, which includes several parameters of nephritis such as glomerular IgG deposition, urine proteinuria and blood urea nitrogen concentration was significantly lower in Treml4–/– MRL/lpr mice compared to their MRL/lpr counterparts (Fig. 6j). Consistent with decreased kidney disease, Treml4–/– MRL/lpr mice showed significantly better survival (∼88% survival at 26 weeks) than Treml4+/+ MRL/lpr mice (∼50% survival at 26 weeks) (Fig. 6k). Finally, both male and female Treml4–/– MRL/lpr mice were equally protected against SLE, including autoantibody generation, immune cell activation, inflammatory cytokine production and nephritis (data not shown). Collectively these data demonstrate an essential role for TREML4 in the regulation of autoimmunity.
Discussion
The data presented here provide new insight into understanding signaling through the endosomal TLRs TLR7 and TLR9. We show that TREML4 positively regulates the activation of macrophages, DCs and neutrophils by amplifying TLR7-mediated production of inflammatory cytokines, such as type I IFNs during antiviral responses and autoimmune diseases. TREML4 amplifies TLR7 signaling by promoting phosphorylation and activation of the MAP kinase p38 and of the transcription factor STAT1. Furthermore, TREML4 expression was essential for the recruitment of MyD88 to TLR7 at endosomes. Together these data demonstrated an important role for TREML4 in the regulation of TLR7 signaling and that its expression in vivo is required for antiviral host defense to influenza and for promotion of TLR7-mediated autoimmune disease.
Our findings demonstrate a unique specificity for TREML4 in promoting DC, macrophage and neutrophil activation by ligands for TLR7, TLR9 and TLR13, but not other TLRs. TREML4 promoted the induction of STAT1-dependent genes, such as the type I IFN, by TLR7-, but not TLR9-stimulated DC. Full activation of STAT1 occurs by phosphorylation of the Ser727 residue by the TLR7-induced p38 MAPK and of the Tyr701 residues by autocrine IFNAR1 signaling. We found that TREML4 promoted both the TLR7-p38 MAPK-dependent phosphorylation of Ser727 and the IFNAR1-dependent phosphorylation of Tyr701, indicating that in addition to promoting TLR7 signaling, TREML4 also amplifies the IFNAR1 pathway, and suggesting that TREML4 may regulate the cross-talk between TLR, IFNAR1 and STAT1 signaling pathways31.
The ligands for TREM receptors have largely remained elusive. Our data using a chimeric TREML4-TNF-R1 reporter transfected in HEK cells suggests that none of the TLR ligands tested bind or activate TREML4 directly. While the TREML4 ligand remains unknown, we did demonstrate that amplification of TLR7 signaling by TREML4 was dependent on the charged lysine residue present in the transmembrane domain of TREML4. This cationic lysine residue is present in the transmembrane domain of several members of the TREM family, including TREM1, TREM2, TREM3 and pDC-TREM, and has been shown to be required for association to the ITAM-bearing adaptor molecule, DNAX activation protein 12 kDa (DAP12)32. TREML4 also associates with DAP12, but not DAP10 or Fc receptors15. DAP12-deficient macrophages were shown to secrete increased amounts of cytokines after TLR stimulation, indicating that DAP12-pairing receptors negatively regulate signaling through TLRs14. These data are in agreement with studies showing the DAP12-paired receptor TREM2 negatively regulates TLR signaling33. Furthermore, patients with Nasu-Hakola disease that develop bone cysts and presenile dementia were found to have loss of function mutations in either DAP12 or TREM2, indicating that this receptor-signaling adaptor pair is crucial for bone remodeling and brain function34,35. In addition, variants in TREM2 have been associated with the risk of Alzheimer's disease12,13. Clearly, DAP12 and TREM2 negatively regulate inflammatory pathways, however, what remains unclear is how DAP12 signaling downstream of TREM1 and TREML4 is involved in the positive regulation of TLR signaling pathways. One possibility is that TREM receptors may regulate TLR signals in a DAP12-independent manner. Indeed, soluble forms of TREM1 and TREM2 are released from cells activated by inflammatory stimulation, and a soluble form of TREML4 was shown to bind to dead cells36,37. These soluble TREMs may interact with unidentified ligands on cells to amplify or suppress inflammatory pathways. More studies are needed to address whether different signaling pathways are activated via soluble versus membrane bound forms of TREM receptors.
While the overexpression and genetic loss of function experiments indicated that TREML4 is essential in promoting TLR7 signaling, because the ligand for TREML4 is unknown, we were unable to induce TREML4 signaling directly and uncover its functions in the absence of TLR7 signaling. The development of specific TREML4 agonists will be very useful as a tool to decipher the key downstream signaling pathways responsible for TREML4-mediated amplification of TLR and IFN signaling, while TREML4 antagonists may have therapeutic applications in dampening inflammatory autoimmune diseases. It is also possible that no endogenous ligands exist for TREML4, and perhaps similarly to other receptors such as TREM2, its expression and signaling through adaptor molecules controls cellular functions such as cell surface transport, ectodomain shedding of soluble receptors, phagocytosis and co-stimulatory molecule expression38. Similar to mouse TREML4, we found that human TREML4 also positively regulates TLR7 signaling. Human TREML4 is highly expressed in patients with acute coronary syndrome and coronary artery calcification39,40. These results are consistent with the role of TREML4 as an activating receptor and indicate it may have a role in cardiovascular disease. Together these data suggest that increased TREML4 expression could be a useful biomarker for the early detection of chronic disease in patients.
The rescue of the autoimmune phenotype in MRL/lpr mice by TREML4 deletion is similar to results obtained in MRL/lpr mice lacking MyD88, TLR7 and TLR9, or injected with dual TLR7-TLR9 antagonists, which have significantly lower autoantibody titers to DNA and RNA autoantigens and ameliorated disease21,41-44. We propose that in the absence of TREML4, MyD88 fails to traffic to TLR7 resulting in disruption of p38 MAPK activation and the phosphorylation of the transcription factor STAT1. Similar to STAT1-deficient mice, cells lacking TREML4 are unable to produce IFN and IFN-inducible genes, and are protected from the development of lupus disease45. Together these data demonstrate TREML4 plays a critical role in regulating TLR7 signaling and that therapeutic blocking of TREML4 holds considerable promise in preventing autoimmunity.
Although it is appealing to speculate that antagonistic anti-TREML4 therapy could be used to prevent and/or dampen lupus disease, caution may be warranted as chronic therapeutic inhibition of TLR7 and TLR9 signaling could potentially lead to increased susceptibility to infectious complications with microbial pathogens. While mice with mutations in specific TLRs or key TLR signaling molecules are more susceptible to infections, their clinical disease symptoms are often mild and occur primarily in experimental settings with high doses of pathogens. Furthermore, these mice are healthy in standard husbandry conditions, suggesting that other pathways can overcome these defects. This is true for influenza viral infection, which is controlled by at least three different classes of pattern recognition receptors, including TLR7, TLR8, the retinoic acid inducible gene-I (RIG-I) and Nod-like receptor family member, NLRP3. Influenza viral ssRNA recognition by TLR7 mediates signaling that leads to the production of antiviral cytokines, such as type I interferons, but TLR7 signaling is not required to mount an intact CD8+ T cell response. Instead NLPR3 and IL-1R signaling in DCs generates protective adaptive immunity by promoting CD8+ T cell responses to influenza virus. Lastly, increased susceptibility to viral infections has not been noted in SLE patients prescribed hydroxychloroquine, a molecule that raises lysosomal pH and inhibits TLR signaling, or in SLE patients enrolled in clinical trials receiving TLR7 and TLR9 inhibitors. Thus, while mice deficient in TREML4 are more susceptible to influenza infection, more studies are needed to fully analyze the role of TREML4 in antiviral host defense to determine whether it may be possible to therapeutically inhibit TREML4-mediated TLR7 signaling long-term for the treatment of autoimmune diseases without the potentially harmful complications that could arise from viral infections.
Methods
Reagents
Reagents were from Sigma-Aldrich unless otherwise stated. DMEM and RPMI-1640 were obtained from Invitrogen. Complete media consisted of RPMI-1640 or DMEM supplemented with 100U/ml penicillin, 100U/ml streptomycin, 2mM L-glutamine, and 10% fetal calf serum (FCS). Mouse TREM-like 4 antibody was purchased from BioLegend. All other antibodies were from eBioscience, unless otherwise specified. All TLR agonists were purchased from Invivogen. HEK293T cell line (Cat#: HCL4517) was obtained from Fisher Scientific and human THP-1 (Cat#: TIB-202) were obtained from ATCC. All cell lines were determined to be free of mycoplasma using a mycoplasma detection PCR Kit (Sigma). Influenza A/Puerto Rico/8/34 (PR8; ATCC, VR-1469) was grown in Madin Darby canine kidney cells in our laboratory.
Plasmids
Mouse Treml4 cDNAs were provided by H. Hemmi (Osaka University, Osaka, Japan) and R. Steinman (The Rockefeller University, New York, NY). Plasmids containing mouse TLR proteins were obtained from D. Golenbock (University of Massachusetts, Worcester, MA) or purchased from Invivogen.
Mice
All mice were maintained under micro-isolation in specific pathogen-free conditions at the Massachusetts General Hospital animal facility under a protocol approved by the Institutional Animal Care and Use Committee. Wild-type C57Bl/6 and SLE-prone MRL/lpr mice were purchased from The Jackson Laboratories. To generate TREML4-deficient mice, a targeting vector that contained 6239 bp of 5′ sequence upstream of the transcriptional start site, 4366 bp of 3′ sequence downstream of exon 4, and a neomycin resistance cassette replacing exons 2-4 was electroporated in C57Bl/6 ES cells. Targeted clones were selected in the presence of the antibiotic neomycin and identified by PCR and Southern blot analysis. Targeted ES clones were injected into C57BL/6 mice blastocysts by the Knockout Mouse Project (KOMP) at the University of California Davis, which resulted in several lines of chimeric mice that transmitted the disrupted allele through the germline DNA. The TREML4-deficient mice bred at normal mendelian ratios. Treml4–/– mice were backcrossed at least eight generations to Fas-deficient, SLE-prone MRL/lpr mice, at which point more than 99.8% of the genome was statistically derived from the MRL/lpr strain. These MRL/lpr Treml4–/– mice were then intercrossed to produce two cohorts of 16 MRL/lpr Treml4+/+ and 16 MRL/lpr Treml4–/– mice (32 mice total each genotype), which were analyzed at 16 weeks of age. A third cohort of 47 MRL/lpr Treml4+/+ and 42 MRL/lpr Treml4–/– mice were observed without intervention until he time of death. Moribund animals were sacrificed and were included as deaths in the analysis. Genotyping primer pairs for discriminating between wild-type Treml4 and Treml4 knockout mice: WT Fw 5′-CCCTTCCCAGTTCTGACTACC-3′; WT Rv 5′-CAAACTCAGATACTCATGCTGGCTGC-3′; KO Fw 5′-GATCAGGATGATCTGGACGAA-3′; KO Rv 5′- CATCATTGATCCAGCTCCCTA-3′. Wild-type Treml4 allele PCR is 449 bp and Treml4 mutant product is 1257 bp. Genotyping primer pairs for discriminating between wild-type Fas and lpr: WT Fw 5′-AGCATAGATTCCATTTGCT-3′ and WT Rv 5′-AGTAATGGGCTCAGTGCA-3′; lpr Fw 5′-AGCATAGATTCCATTTGCT-3′ and lpr Rv 5′-CAAATTTTATTGTTGCGACA-3′. PCR amplification conditions were: 94°C for 3 min; 30 cycles of 30 s each at 94°C, 58°C, and 72°C; followed by a 7 min incubation at 72°C.
Isolation of Bone Marrow-derived Dendritic Cells
For the enrichment of BMDCs, mice where euthanized and bone-marrow cells were harvested from the femurs and tibiae. Following treatment with RBC lysis buffer, the remaining cells were suspended at a final concentration of 1×106 cells/mL in RPMI-complete media supplemented with 10% supernatant from J558L cells as a source of GM-CSF. Cells were seeded in non-tissue culture treated Petri dishes and incubated at 37C with 5% CO2, and cells were fed every 3 days. At day 8, cells were harvested and seeded according the experimental approach.
Purification of bone marrow-derived neutrophils
Marrow cavities of the femurs of 8-week old mice were flushed with DMEM. After hypotonic lysis of red blood cells, mature neutrophils were isolated by centrifugation for 30 min at 28C and 500g over discontinuous Percoll gradients consisteing of 55%, 65% and 75% (vol/vol) Percol in PBS. Mature neutrophils were recovered at the interface of the 65% and 75% fractions. Neutrophil purity was >90% as assessed by Ly6G and CD11b staining by flow cytometry.
Pooled shRNA screen
The genome-scale pooled shRNA assays were performed using 2.5 × 107 RAW-GFP reporter cells for each replicate placed in culture medium containing 4 μg/mL polybrene and the 45k pool shRNA lentivirus was added to achieve an MOI of 0.3. After 24 hours the 45k library infected cells were split into T175 flasks and selected in culture medium containing puromycin. Five days later, 1 × 108 infected cells were treated with GRD for 6 hours, harvested, stained with antibodies to detect intracellular TNF, and sorted into two populations by flow cytometry based on low and high dual TNF and NF-κB-GFP expression. The shRNA sequences were PCR amplified from the genomic DNA isolated from the sorted cells and hybridized to complementary microarrays to assess the abundance of each shRNA usinf Dchip software.
Real-Time quantitative PCR
Total RNA was extracted using the RNeasy kit and DNase treated according to the manufacturer's protocol (Qiagen), and each sample was reverse transcribed using multiscribe reverse transcriptase (Applied Biosystems). The 25-µl qPCR reaction contained 2 µl of cDNA, 12.5 µl of 2× SYBR green master mix (Applied Biosystems), and 500 nM of sense and antisense primers. Oligonucleotide primer sequences designed on the PrimerBank or Primer3 websites and obtained from Integrated DNA Technologies were as follows: human GAPDH, 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′- GAAGATGGTGATGGGATTTC-3′; human IL8, 5′-CTGGCCGTGGCTCTCTTG-3′ and 5′-CCTTGGCAAAACTGCACCTT-3′; human TREML4, 5′-GGGTGCTGTGCCTGAAGAA-3′ and 5′-AGGTACACCGACTTGGAGATG-3′; mouse TREML4, 5′-CCCTTCCCAGTTCTGACTACC-3′ and 5′-GCACACAGAAAACTGACAGCA-3′; mouse GAPDH, 5′-GGCAAATTCAACGGCACAGT-3′ and 5′-AGATGGTGATGGGCTTCCC-3′; mouse CXCL10, 5′- GCCGTCATTTTCTGCCTCA-3′ and 5′-CGTCCTTGCGAGAGGGATC-3′; mouse IL12β, 5′- TGGTTTGCCATCGTTTTGCTG-3′ and 5′-ACAGGTGAGGTTCACTGTTTCT-3′; mouse CCL2, 5′-TGGCTCAGCCAGATGCAGT-3′ and 5′-TTGGGATCATCTTGCTGGTG-3′; mouse CCL22, 5′-TACATCCGTCACCCTCTGCC-3′ and 5′-CGGTTATCAAAACAACGCCAG-3′; mouse IL1α, 5′-GCACCTTACACCTACCAGAGT-3′ and 5′-TGCAGGTCATTTAACCAAGTGG-3′; mouse TNF, 5′- CCCTCACACTCAGATCATCTTCT-3′ and 5′-GCTACGACGTGGGCTACAG-3′; mouse Jun, 5′-CCTTCTACGACGATGCCCTC-3′ and 5′-GGTTCAAGGTCATGCTCTGTTT-3′; mouse IL12α, 5′-CGCAGAGTCTCGCCATTATGAT-3′ and 5′-TGGTTTGCCATCGTTTTGCTG-3′; mouse TLR7, 5′-ATGTGGACACGGAAGAGACAA-3′ and 5′-GGTAAGGGTAAGATTGGTGGTG-3′; mouse IL1β, 5′-ACCTGTCCTGTGTAATGAAAGACG-3′ and 5′-TGGGTATTGCTTGGGATCCA-3′; mouse IL18, 5′-GTGAACCCCAGACCAGACTG-3′ and 5′-CCTGGAACACGTTTCTGAAAGA-3′; mouse CCR7, 5′-TGTACGAGTCGGTGTGCTTC-3′ and 5′-GGTAGGTATCCGTCATGGTCTTG-3′; mouse ATF3, 5′-GAGGATTTTGCTAACCTGACACC-3′ and 5′-TTGACGGTAACTGACTCCAGC-3′; mouse IRF7, 5′-GAGACTGGCTATTGGGGGAG-3′ and 5′-GACCGAAATGCTTCCAGGG-3′; mouse TLR9, 5′-ATGGTTCTCCGTCGAAGGACT-3′ and 5′-GAGGCTTCAGCTCACAGGG-3′; mouse IRF8, 5′-CGGGGCTGATCTGGGAAAAT-3′ and 5′-CACAGCGTAACCTCGTCTTC-3′; mouse CXCL9, 5′-TCCTTTTGGGCATCATCTTCC-3′ and 5′-TTTGTAGTGGATCGTGCCTCG-3′; mouse STAT1, 5′-TCACAGTGGTTCGAGCTTCAG-3′ and 5′-GCAAACGAGACATCATAGGCA-3′; mouse IFNβ, 5′-CAGCTCCAAGAAAGGACGAAC-3′ and 5′-GGCAGTGTAACTCTTCTGCAT-3′; mouse IFNα4, 5′-TGATGAGCTACTACTGGTCAGC-3′ and 5′-GATCTCTTAGCACAAGGATGGC-3′; mouse IL23α, 5′-AATAATGTGCCCCGTATCCAGT-3′ and 5′-GCTCCCCTTTGAAGATGTCAG-3′; mouse MyD88, 5′-TCATGTTCTCCATACCCTTGGT and 5′-AAACTGCGAGTGGGGTCAG-3′; mouse IRAK1, 5′-CCACCCTGGGTTATGTGCC-3′ and 5′-GAGGATGTGAACGAGGTCAGC-3′; mouse IRAK4, 5′-CATACGCAACCTTAATGTGGGG-3′ and 5′-GGAACTGATTGTATCTGTCGTCG-3′; mouse IRAK3, 5′-CTGGCTGGATGTTCGTCATATT-3′ and 5′-GGAGAACCTCTAAAAGGTCGC-3′; mouse TRAF6, 5′-AAAGCGAGAGATTCTTTCCCTG-3′ and 5′-ACTGGGGACAATTCACTAGAGC-3′; mouse TBK1, 5′-ACTGGTGATCTCTATGCTGTCA-3′ and 5′-TTCTGGAAGTCCATACGCATTG-3′; mouse SYK, 5′-CTACCTGCTACGCCAGAGC-3′ and 5′-GCCATTAAGTTCCCTCTCGATG-3′; mouse DAP12, 5′-GAGTGACACTTTCCCAAGATGC-3′ and 5′-CCTTGACCTCGGGAGACCA-3′; mouse IRF5, 5′-AGAGACAGGGAAGTACACTGAAG-3′ and 5′-TGGAAGTCACGGCTTTTGTTAAG-3′; mouse STAT2, 5′-CTGAAGGACGAACAGGATGTC-3′ and 5′-CAGGGTGGTTAATCGGCCAA-3′. Emitted fluorescence for each reaction was measured three times during the annealing-extension phase, and amplification plots were analyzed with MX4000 software, version 3.0 (Stratagene). The quantity of gene expression was generated by comparison of the fluorescence generated by each sample with standard curves of known quantities, and the calculated number of copies was divided by the number of copies of the housekeeping gene GAPDH.
Histological assessment
Histopathologic examination of kidney samples was done after routine fixation and paraffin embedding of the tissues. Tissue sections from the skin were cut and stained with Periodic acid-Shiff stain (PAS). All slides were coded and evaluated in a blinded manner with regard to identity of the sample. Pathologic changes in the kidney were graded according to the presence of glomerular, interstitial, perivascular inflammation and immune complex deposition. Scores ranging from 0 (normal) to 4 (most severely inflamed) were assigned for each of the 4 features. A minimum of 50 glomeruli were assessed to determine the glomerular index in each mouse. Sections were visualized using a Nikon Eclipse ME600 fluorescent microscope equipped with a high resolution DXM1200C Nikon digital camera. Data were analyzed using NIS-Elements software (Nikon) and Adobe Photoshop.
Autoantibody profiles
ANA and Crithidia lucilae immunofluorescence assays (Bio-Rad) were performed according to the manufacturer's instructions, with serum at 1:160-1:5120 dilution and 1:80 dilution for C. lucilae, and scored by an observer blinded to the genotype of the mice. For nucleosome and smRNP ELISAs, polystyrene plates were coated with either dsDNA and histones or smRNP complex antigens, respectively (Immunovision). After blocking with 1% BSA in PBS, serial dilutions of serum from 1:160 to 1:5120 were added. Specific antibodies were detected with HRP-conjugated goat anti-mouse IgG and absorbance at 405/630 nm was compared with monoclonal antibodies to quantitate.
Lentivirus production and infection
Plasmids encoding lentiviruses expressing shRNA molecules were obtained from The RNAi Consortium shRNA Library. The shRNA target 21-mer sequences were: shControl, CCTAAGGTTAAGTCGCCCTCG; mouse shTREML4 #1, GCAGTGCCAATACAAGCCTAA; mouse shTREML4 #2, AGAAGTAACTGTTCTCAGAAA; mouse shMyD88, GCCAGCGAGCTAATTGAGAAA; human shTREML4 #1, TGACACAGAATGACTCGGGAT; human shTREML4 #2, ACGTCTCCTATGTGGACTCTT; human shTREML4 #3, GTGCCTGAAGAACTTCACAAA; human shTREML4 #4, GAATCTACAACGCTTCCGAAA; human shTREML4 #5, CCTCCATCAATGGCTCTGAGA. Plasmids were purified with a QIAprep Miniprep kit (Qiagen). Plasmids were then transfected into HEK293T cells along with pCMV-dR8.2 dvpr and pCMV-VSVG for the production of lentivirus. RAW-GFP or THP-1 cells were placed in 24-well tissue culture dishes (2 × 105 cells per well) and were infected with 100 μl unconcentrated shRNA lentiviral supernatant and polybrene (7.5 μg/ml). Cells were spun for 30 min at 800g, media replaced, and incubated for 2 days. Infected cells were selected in complete RPMI medium containing 10% (vol/vol) FBS and puromycin (3 μg/ml) and were tested 1 week after infection. The shRNA knockdown efficiency was determined by qPCR.
Phospho-flow cytometry analysis
BMDCs were enriched as described above. BMDCs (2×106 cells/mL) were seeded in 5mL polypropelene tubes containing RPMI complete media. Cells were treated with 10 μg/mL of Gardiquimod (TLR7), 100 ng/mL LPS (TLR4) or 10U rIFNγ (Prepotech) for 25 minutes at 37C with 5% CO2. Treated cells were fixed with cold 2% paraformaldehyde for 10 minutes at 4C. Cells were washed 3 times with cold FACS buffer (PBS supplemented with 2% FCS) and permeabilized with cold 100% methanol for 20 minutes on ice. Cells were washed twice to remove methanol and stained with fluorescently labeled antibodies CD11c-APC, p42/44-Alexa488 (Y202/204; Cell Signaling), p38MAPK-PE (T180/Y182; Cell Signaling), pSTAT1 (S727; Cell Signaling clone D3B7), pSTAT1 (Y701; Cell Signaling clone 58D6). Secondary staining for pSTAT1 was performed using goat anti-Rabbit IgG fluorescently labeled with Alexa fluor 488 (Invitrogen). Samples were analyzed using Becton Dickinson FACSCalibur, data was collected using CellQuest Software and data were analyzed using FlowJo X version for Mac.
Western blots and Co-immunoprecipitation
Co-immunoprecipitation experiments were performed using BMDCs seeded in 6-well plates at a final concentration of 1×106 cells/mL in RPMI-complete media. Cells were incubated with 10 μg/mL Gardiquimod for 15 minutes at 37C with 5% CO2. Cells were harvested and lysed in 1× lysis buffer (Cell Signaling). Lysates were incubated with anti-mouse TLR7 (Fisher Scientific) or anti-mouse IgG isotype control (Santa Cruz Biotech) for 24 h at 4C. Supernatants were incubated with Protein G agarose beads overnight at 4C. Precipitated proteins are eluted, separated by SDS-PAGE and immunoblotted with anti-mouse MyD88 (Santa Cruz Biotech).
Confocal microscopy
BMDCs were seeded in confocal dishes (Mattek) at a final concentration of 1×105 cells/mL in RPMI-complete media. The following day, cells were treated with 10 μg/mL GRD for 5 minutes and 15 minutes at 37C. Cells were carefully washed with PBS and fixed with 2% PFA for 10 minutes at room temperature in the dark. Fixed cells were permeabilized with SAP buffer (2% saponin in PBS) for 1 hour at 37C. Following permeabilization, BMDCs were intracellularly stained with antibodies to mouse TLR7, MyD88, EEA1, or LAMP1 (Thermo Scientific), followed by staining with secondary antibodies fluorescently labeled with AlexaFluor488 or AlexFluor550 (Invitrogen). Cells were washed 3 times with PBS. Finally, DNA was stained with 1:1000 dilution of DAPI (Invitrogen) for 10 minutes at RT. Cells were visualized using a Nikon confocal microscope and analysis performed using ZEN software. Data analysis and processing of images was conducted using ImageJ software (NIH).
Phagocytosis assay
Latex beads (1 uM polystyrene, carboxylate modified, red fluorescent biotin labeled) were opsonized with 1 mg/mL anti-Biotin IgG and mouse serum for 1 hour at 37C. Next, they were washed twice with PBS. The opsonized latex beads were hen added to suspended neutrophils at a ratio of 25:1 and paced at 4C to determine binding or 37C to initiate phagocytosis. The samples were washed twice in PBS by centrifugation at 1500 rpm for 5 minutes. After washing, the samples were fixed with 10% formalin and viewed with a Zeiss fluorescent microscope. The data are expressed as the percentage of cells that had internalized at least one latex bead (percent phagocytic) as well as by the number of internalized latex beads per 100 phagocytees × 100 (phagocytic index).
Transwell chemotaxis assasy
Chemotaxis was assayed in 96-well chemotaxis chambers with polycarbonate membranes (pore size, 5 uM; Neuroprobe). CyQUANT dye mix was used for quantification of migrating cells. After 2 h of incubation, fluorescence was measured in a FluoroSkan Ascent fluorescent plate reader. fMLP was placed in the bottom well of a Neuroprobe chemotaxis chamber. Cells were spun (100 μl, containing on average 4 × 104 cells), then were resuspended in 50 μl DMEM and placed on top of the filter in the chemotaxis chamber. After 3 h of incubation, the number of cells migrating to the lower chamber was determined with CyQUANT. The chemotactic index was determined by division of the number of migrating cells (as determined by CyQUANT fluorescence) by the number of input cells and the results were normalized to 1 on the basis of migration of untreated cells.
Infection with influenza
Mice were infected with a live PR8 influenza virus at 30-50 EID/gram in 50 µl PBS per mouse intranasally. Some mice were euthanized on day 10 after infection, and the lungs were harvested and the viral load determined by qPCR using the following primers: forward primer, 5′-CGGTCCAAATTCCTGCTGA-3′; and reverse primer, 5′-CATTGGGTTCCTTCCATCCA-3-3′. Other mice were monitored for weight loss and survival daily for 14–18 days.
Statistical analysis
Statistical calculations were performed using a statistical software package (GraphPad Prism 5.0d). For comparisons of two groups, means ± SE were analyzed by the two-tailed unpaired Student t-test with the Bonferroni correction applied when making multiple comparisons. For comparisons of greater than two groups, significance was determining using the one- or two-way analysis of variance (ANOVA) with correction or Mann-Whitney Test. For survival studies, significance was determined using the log rank test.
Supplementary Material
Acknowledgments
We thank B. Luo, M. Okamoto, and members of the Genetic Perturbation Platform (Broad Institute) and The RNAi Consortium (TRC) for shRNA lentiviral libraries and technical assistance. The vector, ES cell, and TREML4-deficient mouse strain used for this research project was generated by the trans-NIH Knock-Out Mouse Project (KOMP) and obtained from the KOMP Repository (www.komp.org). NIH grants to Velocigene at Regeneron Inc (U01HG004085) and the CSD Consortium (U01HG004080) funded the generation of gene-targeted ES cells for 8500 genes in the KOMP Program and archived and distributed by the KOMP Repository at UC Davis and CHORI (U42RR024244). Supported by the National Institutes of Allergy and Infectious Diseases (R01-AI084884 to T.K.M.; U24 AI082660 to J.E.K.; and T32-AI007061 to Z.G.R.-O.), the National Institutes of Arthritis, Musculoskeletal and Skin Diseases (K01-AR051367 to T.K.M.; K01-AR066716 to Z.G.R.-O), the National Institute of Diabetes and Digestive and Kidney Diseases (F32-DK097891 to W.F.P.), the Lupus Research Institute (T.K.M. and N.H.), the Alliance for Lupus Research (T.K.M. and N.H.) and the American Society of Nephrology (W.F.P.).
Footnotes
Author Contributions: T.K.M., N.H., Z.G.R.-O, A.P., and W.F.P. planned the research, analyzed and interpreted data and wrote the manuscript; Z.G.R.-O., A.P., and T.K.M. did most of the experiments; A.P., J.W.G, and W.F.P. performed and analyzed ELISA, PCR and mouse pathology studies; M.T. helped with mouse breeding, genotyping, and shRNA lentiviral production; J.E.K, A.D.L., D.E.R., and G.S.C. analyzed and interpreted data; T.K.M, J.E.K. and M.T. contributed to the generation of TREML4-deficient mice; and all authors participated in editing the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.O'Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 2.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 3.Gay NJ, Keith FJ. Drosophila Toll and IL-1 receptor. Nature. 1991;351:355–356. doi: 10.1038/351355b0. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 5.Kono DH, Baccala R, Theofilopoulos AN. TLRs and interferons: a central paradigm in autoimmunity. Curr Opin Immunol. 2013;25:720–727. doi: 10.1016/j.coi.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchon A, Facchetti F, Weigand MA, Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410:1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- 7.Turnbull IR, et al. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol. 2006;177:3520–3524. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- 8.Hamerman JA, et al. Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J Immunol. 2006;177:2051–2055. doi: 10.4049/jimmunol.177.4.2051. [DOI] [PubMed] [Google Scholar]
- 9.Ornatowska M, et al. Functional genomics of silencing TREM-1 on TLR4 signaling in macrophages. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1377–1384. doi: 10.1152/ajplung.00140.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ford JW, McVicar DW. TREM and TREM-like receptors in inflammation and disease. Curr Opin Immunol. 2009;21:38–46. doi: 10.1016/j.coi.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Washington AV, et al. TREM-like transcript-1 protects against inflammation-associated hemorrhage by facilitating platelet aggregation in mice and humans. J Clin Invest. 2009;119:1489–1501. doi: 10.1172/JCI36175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerreiro R, et al. TREM2 variants in Alzheimer's disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamerman JA, Tchao NK, Lowell CA, Lanier LL. Enhanced Toll-like receptor responses in the absence of signaling adaptor DAP12. Nat Immunol. 2005;6:579–586. doi: 10.1038/ni1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemmi H, et al. A new triggering receptor expressed on myeloid cells (Trem) family member, Trem-like 4, binds to dead cells and is a DNAX activation protein 12-linked marker for subsets of mouse macrophages and dendritic cells. J Immunol. 2009;182:1278–1286. doi: 10.4049/jimmunol.182.3.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.N'Diaye EN, et al. TREM-2 (triggering receptor expressed on myeloid cells 2) is a phagocytic receptor for bacteria. J Cell Biol. 2009;184:215–223. doi: 10.1083/jcb.200808080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lovgren T, Eloranta ML, Bave U, Alm GV, Ronnblom L. Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum. 2004;50:1861–1872. doi: 10.1002/art.20254. [DOI] [PubMed] [Google Scholar]
- 18.Pisitkun P, et al. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian S, et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci U S A. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deane JA, et al. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27:801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen SR, et al. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Berland R, et al. Toll-like receptor 7-dependent loss of B cell tolerance in pathogenic autoantibody knockin mice. Immunity. 2006;25:429–440. doi: 10.1016/j.immuni.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 23.Wang CM, et al. Genetic variations in Toll-like receptors (TLRs 3/7/8) are associated with systemic lupus erythematosus in a Taiwanese population. Sci Rep. 2014;4:3792. doi: 10.1038/srep03792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez-Ortiz ZG, et al. The scavenger receptor SCARF1 mediates the clearance of apoptotic cells and prevents autoimmunity. Nat Immunol. 2013;14:917–926. doi: 10.1038/ni.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemmi H, et al. Treml4, an Ig superfamily member, mediates presentation of several antigens to T cells in vivo, including protective immunity to HER2 protein. J Immunol. 2012;188:1147–1155. doi: 10.4049/jimmunol.1102541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 29.Ramsauer K, et al. p38 MAPK enhances STAT1-dependent transcription independently of Ser-727 phosphorylation. Proc Natl Acad Sci U S A. 2002;99:12859–12864. doi: 10.1073/pnas.192264999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luu K, et al. STAT1 plays a role in TLR signal transduction and inflammatory responses. Immunol Cell Biol. 2014;92:761–769. doi: 10.1038/icb.2014.51. [DOI] [PubMed] [Google Scholar]
- 31.Ivashkiv LB. Cross-regulation of signaling by ITAM-associated receptors. Nat Immunol. 2009;10:340–347. doi: 10.1038/ni.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klesney-Tait J, Turnbull IR, Colonna M. The TREM receptor family and signal integration. Nat Immunol. 2006;7:1266–1273. doi: 10.1038/ni1411. [DOI] [PubMed] [Google Scholar]
- 33.Paradowska-Gorycka A, Jurkowska M. Structure, expression pattern and biological activity of molecular complex TREM-2/DAP12. Hum Immunol. 2013;74:730–737. doi: 10.1016/j.humimm.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Paloneva J, et al. Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat Genet. 2000;25:357–361. doi: 10.1038/77153. [DOI] [PubMed] [Google Scholar]
- 35.Paloneva J, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71:656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibot S, et al. A soluble form of the triggering receptor expressed on myeloid cells-1 modulates the inflammatory response in murine sepsis. J Exp Med. 2004;200:1419–1426. doi: 10.1084/jem.20040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piccio L, et al. Identification of soluble TREM-2 in the cerebrospinal fluid and its association with multiple sclerosis and CNS inflammation. Brain. 2008;131:3081–3091. doi: 10.1093/brain/awn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinberger G, et al. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci Transl Med. 2014;6:243ra286. doi: 10.1126/scitranslmed.3009093. [DOI] [PubMed] [Google Scholar]
- 39.Sen SK, et al. Integrative DNA, RNA, and protein evidence connects TREML4 to coronary artery calcification. Am J Hum Genet. 2014;95:66–76. doi: 10.1016/j.ajhg.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silbiger VN, et al. Novel genes detected by transcriptional profiling from whole-blood cells in patients with early onset of acute coronary syndrome. Clin Chim Acta. 2013;421:184–190. doi: 10.1016/j.cca.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Christensen SR, et al. Toll-like receptor 9 controls anti-DNA autoantibody production in murine lupus. J Exp Med. 2005;202:321–331. doi: 10.1084/jem.20050338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herlands RA, Christensen SR, Sweet RA, Hershberg U, Shlomchik MJ. T cell-independent and toll-like receptor-dependent antigen-driven activation of autoreactive B cells. Immunity. 2008;29:249–260. doi: 10.1016/j.immuni.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nickerson KM, et al. TLR9 regulates TLR7- and MyD88-dependent autoantibody production and disease in a murine model of lupus. J Immunol. 2010;184:1840–1848. doi: 10.4049/jimmunol.0902592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrat FJ, Meeker T, Chan JH, Guiducci C, Coffman RL. Treatment of lupus-prone mice with a dual inhibitor of TLR7 and TLR9 leads to reduction of autoantibody production and amelioration of disease symptoms. Eur J Immunol. 2007;37:3582–3586. doi: 10.1002/eji.200737815. [DOI] [PubMed] [Google Scholar]
- 45.Thibault DL, et al. IRF9 and STAT1 are required for IgG autoantibody production and B cell expression of TLR7 in mice. J Clin Invest. 2008;118:1417–1426. doi: 10.1172/JCI30065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.