SUMMARY
Cell polarization is linked to fate determination during asymmetric division of plant stem cells, but the underlying molecular mechanisms remain unknown. In Arabidopsis, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) is polarized to control stomatal asymmetric division. A MITOGEN-ACTIVATED PROTEIN KINASE (MAPK) cascade determines terminal stomatal fate by promoting the degradation of the lineage determinant SPEECHLESS (SPCH). Here we demonstrate that a positive feedback loop between BASL and the MAPK pathway constitutes a polarity module at the cortex. Cortical localization of BASL requires phosphorylation mediated by MPK3/6. Phosphorylated BASL functions as a scaffold and recruits the MAPKKK YODA and MPK3/6 to spatially concentrate signaling at the cortex. Activated MPK3/6 reinforces the feedback loop by phosphorylating BASL, and inhibits stomatal fate by phosphorylating SPCH. Polarization of the BASL-MAPK signaling feedback module represents a mechanism connecting cell polarity to fate differentiation during asymmetric stem cell division in plants.
INTRODUCTION
Multicellular organisms use asymmetric cell division (ACD) to generate diverse cell types and to maintain stem cell populations for tissue homeostasis (Knoblich, 2008b). One of the most conserved fundamental mechanisms underlying ACD in animals is described by the intrinsic polarity model (Inaba and Yamashita, 2012; Yu et al., 2006). In the process of neuroblast ACD in Drosophila, the Par polarity complex is localized asymmetrically to the apical cortex of the neuroblast, and drives a set of fate determining factors to the opposite side to determine the asymmetric cell fates (Inaba and Yamashita, 2012; Knoblich, 2008a; Prehoda, 2009; Wu et al., 2008).
ACD is critical for plant growth and development since new cell types, tissues and even entire organs are continuously produced from a limited number of stem cells (de Smet and Beeckman, 2011; Petricka et al., 2009). Progress has been made towards deciphering the spatially coordinated regulation of ACD in plants, including intercellular mobile transcription factors (Guseman et al., 2010; Nakajima et al., 2001; Schlereth et al., 2010) and extrinsic cue-guided protein polarization (Cartwright et al., 2009; Humphries et al., 2011). Intrinsic mechanisms in plant ACD have been elusive, in part due to the lack of homologs of the conserved regulators in animals. The recent discovery of the plant polarity proteins BASL (Dong et al., 2009) and POLAR (Pillitteri et al., 2011) revealed that intrinsic elements, behaving similarly to the conserved Par proteins in animals, control stem cell ACD in Arabidopsis.
BASL is required for the development and patterning of stomata, breathing pores in plant epidermis. Both the differentiation and patterned distribution of stomata require asymmetric and orientated cell divisions (Lau and Bergmann, 2012; Pillitteri and Torii, 2012). Stomatal ACD precursors as a dispersed stem cell population have been used as a model system to study the molecular mechanisms for ACD in plants (Abrash and Bergmann, 2009). An Arabidopsis stomatal lineage originates with a precursor stem cell that undergoes ACD to produce a stomatal lineage ground cell (SLGC, the larger daughter cell), which may expand to become a pavement cell, and a meristemoid cell (the small daughter cell), which undergoes ACDs and eventually differentiates into a pair of guard cells (Bergmann and Sack, 2007) (Figure 1A). Loss-of-function basl mutants are defective in establishing stomatal division and fate asymmetries (Dong et al., 2009). Prior to ACD, the expression of BASL first appears in the nucleus, and then polarizes to one edge of the precursor cells distal to the newly formed division plane. After ACD, BASL polarity is only inherited in the SLGC and is associated with the non-stomatal fate of the expressing cell (Figure 1A) (Dong et al., 2009). How BASL is polarized and how its polarity specifies daughter cell fates remain unknown.
Figure 1. The MAPK docking motifs are indispensable for BASL localization and function.
(A) Schematic of stomatal asymmetric cell division and BASL localization (green).
(B) MAPK-docking domains (D and DEF) and putative MAPK phosphosites (S) in BASL protein. Asterisks indicate the identified MPK6 phosphosites from in vitro MPK6 kinase assays.
(C) Representative confocal image of a 2-dpg (day post growth) adaxial cotyledon of basl-2. Cell outlines are counterstained with Propidium Iodide (PI) (Magenta). Scale bar = 25 µm, others at same scale. White brackets indicate clustered stomatal lineage cells, a hallmark of basl. Inset shows representative protein localization (green channel only).
(D) GFP-labeled BASL (green, driven by the BASL promoter) in basl-2. Arrows mark protein polar accumulation.
(E) GFP-BASL_d1_d2_def in basl-2, all three MAPK docking motifs are mutated.
(F) Left: Diagram of a confocal image of GFP-BASL showing the quantification of BASL polarity by crescent length. Right: Histogram shows the polarity quantification of BASL vs. BASL_d1_d2_def and BASL_12356A. Data are mean ± SD. * significant differences relative to the native BASL (t-test, P<0.0001. n = 50 cells from two independent transgenic lines).
(G) Quantification of stomatal defects in 5-dpg adaxial cotyledons (see Experimental Procedure). Data are mean ± SD. *significantly different from the control value (BASL basl-2) (t-test, P<0.0001), n = 1,500 from two independent transgenic lines.
See also Figure S1.
During stomatal development, a canonical MAPK signaling cascade composed of the MAPK Kinase Kinase YODA (YDA), the MAPK Kinase 4 and 5 (MKK4/5), and the MAPK 3 and 6 (MPK3/6), functions in asymmetric cell fate determination and division patterns (Bergmann et al., 2004; Lukowitz et al., 2004; Wang et al., 2007). This pathway is activated by the Epidermal Patterning Factor (EPF) ligands (Abrash and Bergmann, 2010; Hara et al., 2007; Hunt and Gray, 2009; Kondo et al., 2010; Sugano et al., 2010), the Too Many Mouth (TMM) receptor (Nadeau and Sack, 2002) and the ERECTA and ER-like receptor-like kinases (including ER, ERL1 and ERL2)(Shpak et al., 2005). In the nucleus, activated MPK3/6 phosphorylate the transcription factor SPCH, promoting its degradation (Lampard et al., 2008). Since SPCH controls the expression of multiple key regulators of ACD (Lau et al., 2014) and promotes the production of stomata (MacAlister et al., 2007), MPK3/6 ultimately function to inhibit the terminal stomatal fate.
Pivotal roles of the YDA MAPK pathway in ACD have also been established in Arabidopsis embryo development (Lukowitz et al., 2004; Wang et al., 2007). The loss-of-function yda mutants have defects in the first asymmetric cell division and the basal lineage determination (Lukowitz et al., 2004). Although YDA have been implicated in promoting cell expansion and orienting the division plane in embryonic ACD (Smekalova et al., 2014), similar to the role of BASL in controlling stomatal ACD (Dong et al., 2009), no explicit molecular mechanisms have been uncovered yet.
Here, we describe a molecular mechanism that connects the MAPK signaling pathway to BASL polarization and asymmetric cell fate determination in stomatal ACD. We demonstrate that YDA physically interacts with BASL and localizes to the cell cortex of stomatal ACD cells. At the same time, BASL polarization from the nucleus to the cortical crescent also requires YDA MAPK activity, suggesting that a positive feedback loop between these two pathways establishes cell polarity. We also report that polarized BASL behaves as a scaffold protein to control the subcellular localization of not only YDA, but also of other components of the YDA MAPK cascade, including MPK3/6. We propose that the unequal inheritance of YDA and MPK3/6, associated with polarized BASL, could result in unequal inhibition of the fate-determining factor SPCH and thus produce two distinct daughter cell fates. In contrast to the intrinsic Par protein-coupled segregation of cell fate determinants in neuroblast ACD (Williams and Fuchs, 2013; Wirtz-Peitz et al., 2008) and the spatially controlled movement of transcription factors in specifying daughter cell fates during root stem cell ACD (Nakajima et al., 2001; Schlereth et al., 2010), our studies identify a mechanism linking BASL polarization to asymmetric cell fate determination through MAPK-mediated protein degradation.
RESULTS
MAPK-docking motifs are critical for BASL polarity
We noticed that BASL contains three putative MAPK docking motifs, including two D-sites (Ho et al., 2003) and one DEF-site (Murphy et al., 2002) (Figure 1B and Figure S1A), which are found within other MAPK-interacting proteins (Tanoue et al., 2000). To determine the biological significance of these motifs, we introduced GFP-tagged BASL point mutants into the null mutant basl-2 (Dong et al., 2009) and assessed protein localization and function. BASL_d1_d2 (the two D-docking motifs mutated), BASL_def (the DEF site mutated) and BASL_d1_d2_def (all three sites mutated) failed to show cortical polar accumulation, in contrast to wild-type BASL (Figure 1C and 1D, Figure S1B and S1C, and Figure 1E). The fraction of cells exhibiting distinguishable polar accumulation were 3/158 for BASL_d1_d2, 5/456 for BASL_def, and 0/223 for BASL_d1_d2_def, compared to 49/72 for BASL. As BASL accumulates into distinguishable cortical crescents, we measured the length of BASL crescent relative to that of the whole cell to quantify the polarity degree of BASL variants (Figure 1F). These MAPK-docking mutants were defective in polarity formation, and also failed to complement the stomata defects of basl-2, including the key features of clustered stomatal lineage divisions and guard cells (white brackets in Figure 1C and 1E, Figure S1B and S1C, quantified in 1G). These results are consistent with our previous work showing that the polar accumulation at the cortical sites is crucial for BASL function (Dong et al., 2009). Thus, both types of MAPK-docking motif are required for BASL polarity and function.
BASL is phosphorylated by MPK3/6 in vitro
MPK3 and 6 both are highly expressed in the early stomatal lineage cells (promoter activities demonstrated in Figure S2A and S2B) and are key to stomatal development and patterning (Wang et al., 2007). We performed in vitro kinase assays using purified recombinant proteins and found that BASL was phosphorylated by constitutively active MKK5 (MKK5DD)-activated MPK3 and 6 (Figure S2C and Figure 2A, respectively). Using mass spectrometry, we identified the in vitro MPK6 phosphosites of BASL as S89, S145, S168, S235 and S246 (Figure S2D and Figure 1B, for simplicity these sites were numbered as S1, S2, S3, S5, and S6, respectively); S72 (numbered as S4) was not modified. Mutating all five modified Serine residues to Alanines (A) in BASL_12356A abrogated phosphorylation by MPK3 and 6 in vitro (Figure S2E and Figure 2B, respectively), suggesting that these sites likely represent all the MPK3/6-modified targets.
Figure 2. MAPK-mediated phosphorylation directs BASL localization and function.
(A) In vitro kinase assays using recombinant proteins, followed by western blot analysis with autoradiography ([γ-32P] ATP). Constitutively active MKK5 (MKK5DD) is included to activate MPK6 for phosphorylation of BASL.
(B) In vitro kinase assays show MKK5DD-activated MPK6 phosphorylation of BASL and BASL_12356A.
(C) Confocal image of GFP-BASL (green) in basl-2. Arrows show protein polar accumulation. Scale bar = 25 µm, others at same scale. Magenta, propidium iodide (PI)-stained cell walls.
(D) GFP-BASL_12356A in basl-2. Five MPK6 phosphosites (Ser) mutated to Ala (phospho-deficient). White brackets indicate clustered stomatal lineage cells. Inset shows representative protein localization (green channel only).
(E) GFP-BASL_123456A in basl-2. All six putative MAPK phosphosites mutated to Ala residues.
(F) GFP-BASL_123456D in basl-2. All six Ser changed to phospho-mimicking Asp (D) residues.
(G) Quantification of stomatal defects in 5-dpg adaxial cotyledons. About 1500 cells were collected from 10–12 seedlings of two independent transgenic lines. Data are mean ± SD. *significantly different from the control value (GFP-BASL basl-2) (t-test, P<0.0001).
See also Figure S2.
Phosphorylation of BASL is critical for its polarization and function in vivo
We introduced BASL_12356A into basl-2 mutant plants and found that MPK3/6-mediated phosphorylation is important for BASL partitioning out of the nucleus and polarization at the cell cortex. We observed an increased number of GFP-BASL_12356A cells that show nuclear retention (Figure 2C and 2D, 78/143 = 54.5% of GFP-BASL_12356A vs. 22/94 = 23.4% of GFP-BASL). As we cannot exclude the possibility of S72 (S4) being phosphorylated by other kinases in plants, we mutated all 6 Serines to Alanines. GFP-BASL_123456A displayed a further increase in nuclear-only phenotype (97.62%, n = 126) (Figure 2E). Careful examination of subcellular protein localization suggested that BASL phosphorylation is also important for its polarization. GFP-BASL_12356A was partially localized in the cytoplasm and along the cortex, but rarely exhibited polar distribution (Figure 2D): 9.2% of cells show visible polarity crescents, n = 65 vs. 68.1%, n = 72 cells for GFP-BASL (Figure 2C). In contrast, GFP-BASL_123456D, in which all 6 Serines are converted to phospho-mimicking Aspartic Acids (D), recovered cortical polarization and function to the wild-type level (Figure 2F). Thus, our data reveal that MPK3/6-mediated phosphorylation of BASL is required for the partitioning of the protein to the cortical pool for polarization. In addition, neither GFP-BASL_12356A nor GFP-BASL_123456A rescued the stomatal defects of basl-2 (Figure 2G), indicating that phosphorylation of these sites is required for BASL function.
To further evaluate the function and cortical polarization of BASL mutants, we added a N-terminal myristoylation modification site (Myr sequence in (Greenwood and Struhl, 1999)), which targets the fusion protein to the plasma membrane. Myr-BASL does not accumulate as crescent, but spread along the plasma membrane unevenly, so the polarity was quantified by the ratio of fluorescence intensity from the polar side relative to that of the distal side. As anticipated, Myr-BASL accumulated at the plasma membrane, was polarized, and complemented the stomatal defects of basl-2 (Figure 3A and quantified in Figure S3). However, none of the myristoylated BASL mutants (one of the MAPK-docking mutants BASL_def and the phospho-deficient mutant BASL_123456A) were polarized or rescued basl-2 stomatal defects (Figure 3B–3D and Figure S3, polarity quantified in Figure 3E). Thus, BASL’s MAPK docking sites and phosphorylation by MPK3/6 are required for its polarization and its function in stomatal development.
Figure 3. BASL phosphorylation is critical for its polarization.
(A) Confocal image of a 2-dpg cotyledons (adaxial side) expressing the Myristoylated wild-type BASL in basl-2 (Myr-GFP-BASL driven by the native promoter). Arrows indicate protein polar accumulation. Representative individual cells are shown in the inset (green only). Cells were outlined with PI staining (magenta) for quantitative analysis. Scale bar = 25 µm, others at same scale.
(B) Myr-GFP-BASL_123456A in basl-2. White brackets mark clustered stomatal lineage cells, the typical feature of BASL loss-of-function.
(C) Myr-GFP-BASL_def in basl-2.
(D) Myr-GFP-BASL_def_123456A in basl-2.
(E) Left: Diagram showing quantitative analysis of Myr-tagged protein polarity. As these proteins spread along the plasma membrane, the polarity degree was determined by the ratio of average fluorescence intensity from the segments with same length at the polarity site vs. that at the distal site. Right: Histogram showing the quantification results. Data are means ± SD (n = 50 cells). * Significant differences (t-test, P < 0.0001). n = 50 cells from two independent transgenic lines.
See also Figure S3 for complementation quantification.
To genetically assess the role of MAPK signaling on BASL localization, we overexpressed either a dominant negative version of MPK6 (Bush and Krysan, 2007) (DNmpk6) or a MPK3/6 phosphatase, AP2C3 (Umbrasaite et al., 2010), which should interfere with endogenous MPK3/6 activities. As expected, we observed reduced phosphorylation of the MPK3/6 target p44/42 in AP2C3 overexpressing plants (Figure S4A). Transgenic plants overexpressing DNmpk6 or AP2C3 displayed severely clustered stomata (Figure S4B and S4C), which phenocopied the inducible loss-of-function mpk3 mpk6 mutant plants (Wang et al., 2007). GFP-BASL displayed nuclear localization and cortical polarization defects in plants overexpressing DNmpk6 and AP2C3 backgrounds (Figure 4A and Figure S4D). An elevated number of cells showed nuclear-only GFP-BASL (50/139 = 35.97% in DNmpk6, 46/110 = 41.8% in AP2C3, vs. 22/94 = 23.4% in the wild-type). In addition, the width of the BASL crescent expanded, and the occupancy along the cell periphery increased from 24.3% in the wild-type to 56.9% in DNmpk6 and 66% in AP2C3 (quantification in Figure 4B), partially phenocopying that of GFP-BASL12356A at approximately 80% and GFP-BASL_d1_d2_def at 100% (quantifications in Figure 1F. n = 50 cells from each line). We also evaluated the role of MAPK upstream activators, the MAPKKK YDA (Bergmann et al., 2004) and the ERECTA receptor-like kinases (Shpak et al., 2005), on BASL polarity. Consistently, the BASL crescent spread wider in the loss-of-function mutants yda (Figure 4C) and er erl1 erl2 (Figure S4E and S4F). In combination, these data strongly suggest that MPK3/6 phosphorylation of BASL is required for its partition outside of the nucleus and polarization at the cell cortex.
Figure 4. BASL polarity is defective when the YDA-MAPK pathway is compromised.
(A) Confocal images of GFP-BASL (green) localization in DNmpk6-mRFP (red) overexpression plants, both are driven by the BASL promoter. Left, red channel; middle: green channel showing defective polarization of GFP-BASL, and right, overly. Scale bars = 25 µm.
(B) Quantification of BASL polarity in DNmpk6 overexpression and loss-of-function yda plants (below). Representative localizations in individual cells are shown (right). Values are mean ± SD (n = 50 GFP-BASL positive cells), *significant differences (t-test, P<0.0001).
(C) Left: GFP-BASL (green) localization in yda. Cells are outlined with PI (magenta). Right: green channel only to show defective polarization of GFP-BASL. Scale bars: 25 µm.
See also Figure S4.
The MAPKKK YDA physically interacts with BASL
To investigate potential physical interactions between BASL and components of the YDA MAPK pathway, we employed in vitro pull-down assays with recombinant proteins. Although MPK3/6 phosphorylates BASL in vitro, we could not detect a stable interaction by pull-down, suggesting that they interact only transiently. However, we observed a direct interaction between recombinant 6×His-tagged BASL and Maltose Binding Protein (MBP)-tagged YDA in vitro (Figure 5A). We further confirmed their physical association using the yeast two-hybrid (Y2H) system and showed that BASL interacts with the N-terminal regulatory domain of YDA (Lukowitz et al., 2004; Wu et al., 2006) (Figure 5B and 5C). Since polarization of BASL requires phosphorylation, we assessed whether this modification affects its interaction with YDA. By Y2H, YDA displayed a stronger interaction with BASL_123456D than with BASL_123456A, suggesting that phosphorylation stabilizes YDA’s association with BASL (Figure 5C). We also assessed BASL mutants defective in MAPK-docking, since these motifs are critical for BASL cortical polarity (Figure 1E, Figure S1B and S1C). These mutants displayed a weaker interaction with YDA in both in vitro pull-down and Y2H assays (Figure 5A and Figure S5A). Thus, our results strongly suggest that the MAPK-docking motifs mediate the BASL-YDA and BASL-MPK3/6 interactions, and that BASL phosphorylation enhances its interaction with YDA.
Figure 5. BASL interacts with YDA at the cell cortex.
(A) In vitro pull-down assays using recombinant proteins show interactions between MBP-tagged YDA and BASL variants (HIS-tagged). CBB, Coomassie Brilliant Blue stain. Immunoblots were visualized with anti-6 X HIS antibody.
(B) Schematics show protein structure of YDA and variants. Mutating Lys429 to Arg (K429R) renders YDA kinase-inactive (Dominant Negative, DNyda). YDA_N, the N-terminal domain of YDA, without the kinases domain.
(C) Y2H assays of YDA_N with BASL and phospho-variants (123456A and 123456D). Top row, growth control (-Leu-Trp). Bottom row, interaction test (-Leu-Trp-His with 3-AT). Interaction strength low to high: − to +++.
(D) Confocal image showing YDA-YFP (green) localization in the stomatal lineage cells (driven by the SPCH promoter). Arrows indicate protein polar accumulation. Individual cells on right demonstrate closer view of YDA polarization. Cell outlines are marked with PI (magenta). Scale bar = 25 µm, (D–F) at same scale.
(E) DNyda-YFP (green) polarization (arrows) in the stomatal lineage cells. Individual cells with green channel only (right) show detailed localization of DNyda.
(F) Localization of DNyda-YFP in basl-2. White brackets mark clustered stomatal lineage cells. Inset shows a closer view of DNyda-YFP (green channel only) in basl-2.
(G) Top panel: quantification of YDA and DNyda polarity. Protein polarity was determined by the ratio of average fluorescence intensity from the polarity site vs. that of the distal site (same length collected). Bottom panel: histogram shows the polarity quantification data. n = 50 YFP positive cells. Values are mean ± SD, * significantly different relative to DNyda-YFP in the wild-type (t-test, P<0.0001).
(H) Co-localization of DNyda-mRFP (red) and GFP-BASL (green), both driven by the BASL promoter, in a 2-dpg cotyledon. White arrows mark protein polar accumulation. The grey panel displays 3D surface plot of GFP and mRFP generated by Image J. Scale bar = 10 µm.
(I) Co-localization of DNyda-mRFP (red) and GFP-BASL_123456A (green), both driven by the BASL promoter, in a 2-dpg basl-2 adaxial cotyledon. The grey panel (right) displays the 3D surface plot of GFP and mRFP. Scale bar = 10 µm.
See also Figure S5.
BASL recruits YDA to the cell cortex
YDA interacts strongly with the phosphorylated BASL, which polarizes to the cell cortex. To investigate the subcellular localization of YDA, we generated transgenic plants expressing YFP-tagged YDA (driven by its endogenous promoter). YDA-YFP rescued loss-of-function yda mutants and showed localization at the plasma membrane, broadly in the leaf epidermal cells (Figure S5B). To better visualize the localization of YDA-YFP in the stomatal ACD cells, we used the SPCH promoter for cell type-specific expression. Interestingly, YDA-YFP was found polarized at the cell cortex in stomatal lineage cells (Figure 5D, 5/11 independent T2 plants showed visible polarity). We quantified YDA polarity in these plants and found that 11.1% cells showed polar accumulation (n = 99 YFP positive cells).
We also created a kinase inactive version of YDA (also known as Dominant Negative, DNyda, K429R (Lampard et al., 2009)) fused to YFP, which interestingly displayed obvious polar accumulation (26.8% cells show polarity, n = 302 YFP positive) (Figure 5E). Importantly, when DNyda-YFP was introduced into basl-2 mutant plants, it was no longer polarized (Figure 5F, polarity quantified in Figure 5G). We suspect that the enhanced polarity degree of DNyda was likely due to its disrupted enzymatic activity and/or dynamics of localization. The orientation of the DNyda-YFP crescent was similar to that of YDA-YFP, both of which were reminiscent of BASL in the stomatal ACD cells (Dong et al., 2009). The co-polarization of DNyda and BASL was confirmed in plants harboring DNyda-mRFP and GFP-BASL (Figure 5H). Additionally, DNyda-mRFP lost its polarity when combined with the expression of nuclear-only BASL_123456A (Figure 5I), myristylated BASL_123456A or BASL-def, in the absence of wild-type BASL (Figure S5C and S5D). Thus, the MAPKKK YDA is polarized to the BASL crescent and its polarity requires BASL.
Spontaneous polarization of BASL and YDA in tobacco cells
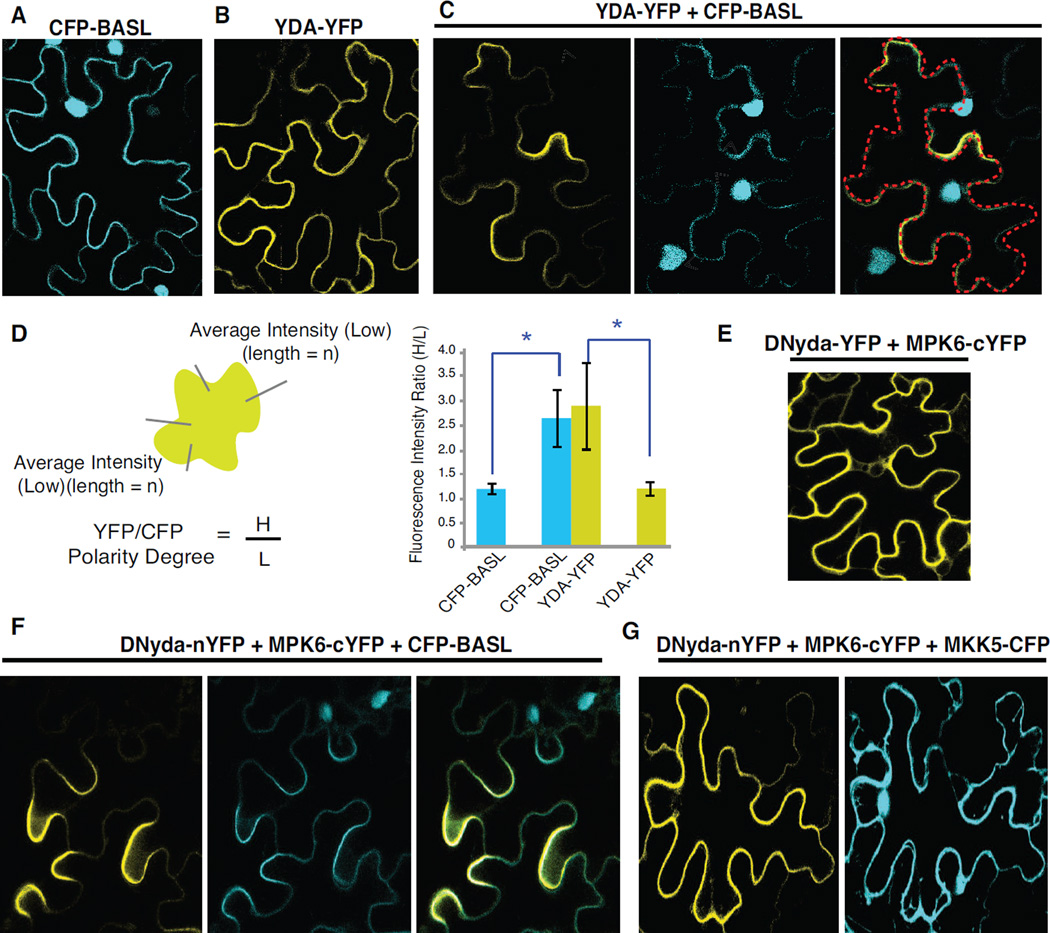
Tobacco epidermal cells are fully expanded with multiple lobes, providing a useful cellular system to evaluate the polarity construction. Although neither BASL nor YDA was polarized when individually expressed in tobacco cells (Figure 6A and 6B), co-expression of YDA-YFP and CFP-BASL was sufficient to trigger polar localization of both fusion proteins (Figure 6C, representative of one of three replicates; polarity degree quantification in Figure 6D, n = 19 cells). This suggests that the interaction between BASL and YDA triggers a spontaneous self-organizing polarization process, probably analogous to the autocatalytic clustering of the Cdc42 small GTPase with the adaptor protein Bem1 during symmetry breaking in budding yeast (Irazoqui et al., 2003; Kozubowski et al., 2008).
Figure 6. Spontaneous polarization of BASL and YDA redistributes the YDA MAPK signaling cassette.
(A) Representative confocal image of CFP-BASL overexpression (cyan) in tobacco epidermal cells. Note, expression in the nucleus and at the cell periphery (not polarized). Images were captured 2–3 days after infiltration.
(B) Overexpression of YDA-YFP (yellow) in tobacco cells.
(C) Co-expression of CFP-BASL (cyan) and YDA-YFP (yellow). Note the protein distribution change. White arrowheads indicate protein polar accumulation. Red marks cell outline.
(D) Left: a diagram showing the quantification of protein polar distribution in tobacco cells. Average intensities were taken from the fluorescence signals (YFP or CFP) along the tobacco cell periphery. The polarity factor was determined by the intensity values obtained from the polar site (H for high intensity) relative to those from the distal side with low expression levels (L). Intensity measurements were collected from a same length of H and L within a single cell. Right: quantification of induced polarity in tobacco cells (n = 18–20 cells from 3 replicated experiments). Values are mean ± SD, * significant differences (t-test, P<0.0001)
(E) Recovered split YFP signal showing DNyda-MPK6 interaction.
(F) Co-expression of CFP-BASL (cyan) with split YFP of DNyda and MPK6 (yellow).
(G) Co-expression of CFP-MKK5 (cyan) with the split YFP pair of DNyda and MPK6 (yellow).
See also Figure S6.
The co-polarization of YDA and BASL was further confirmed by Bimolecular Fluorescent Complementation (BiFC) assays using the tobacco transient expression system. When either YDA-cYFP or DNyda-cYFP was co-expressed with nYFP-BASL, we found that the split YFP fluorescence was recovered at the cell periphery, confirming their physical association in plant cells (Figure S6A), In addition, we consistently observed an uneven distribution of YFP signal along the cell cortex (Figure S6A), supporting of the asymmetric distribution of the YDA-BASL complex.
BASL spatially reorganizes the MAPK signaling pathway in plant cells
That BASL associates with both MPK3/6 and YDA suggests it may function as a scaffold/adaptor protein. Indeed, in the tobacco BiFC assays, we were able to detect BASL physical interaction with MPK6 (Figure S6B) and MKK5 (Figure S6C). Scaffold proteins often control intracellular information flow (Good et al., 2011); therefore we tested whether BASL polarizes multiple components of the MAPK cascade. We continued using tobacco epidermal cells to monitor protein localization (CFP or YFP) and interaction distribution (recovered split YFP signal). Individually tagged BASL-, YDA- (or DNyda-), and MPK6-YFP were evenly distributed (Figure 6A and Figure S6D–S6F). Co-expressing DNyda-nYFP with MPK6-cYFP generated a YFP signal, indicating a physical interaction, but did not display a biased distribution (Figure 6E). However, when CFP-BASL was also co-expressed, the localization of CFP-BASL and the interaction of YDA (or DNyda)-nYFP/MPK6-cYFP redistributed and accumulated to highly polarized cortical sites in tobacco cells (Figure 6F and Figure S6G). MPK3, which functions redundantly with MPK6 in stomata development (Wang et al., 2007), displayed similar effects (Figure S6H). This polarization did not occur when the second-tiered MAPK Kinase 5, MKK5, replaced BASL for co-expression (Figure 6G), suggesting BASL’s specific function in inducing the YDA-MPK3/6 polarity.
Next, we assessed the effect of BASL variants in polarizing the YDA-MPK3/6 interaction. For robust protein expression, DNyda was used as an alternative of YDA for quantitative analysis (YDA overexpression frequently triggers cell death). BASL_d1_d2, BASL_d1_d2_def, and BASL_123456A, which displayed reduced association with YDA (Figure 5A and Figure S5A), failed to induce polarization (Figure 7A, 7B and S7A). However, BASL_123456D, which interacts strongly with YDA (Figure 5C), itself did not form polarity but promoted YDA polarization (Figure S7B and 7C). Importantly, the phospho-micmicking mutant (123456D) did not recover the defective MAPK-docking-induced loss-of-polarity (d1_d2_def) (see BASL_123456D_d1_d2_def, Figure S7C). The polarity quantification data are presented in Figure 7D and Figure S7D (n = 18–20 representative cells from three replicates for each combination). Thus, BASL phosphorylation and its MAPK docking motifs both are important for the BASL-YDA interaction, and for the polarization of the BASL-YDA-MPK3/6 module.
Figure 7. Models for the feedback loop between BASL and the YDA MAPK pathway in stomatal asymmetric fate specification.
(A) Co-expression of CFP-BASL_d1_d2_def (cyan) with the split YFP pair (yellow), DNyda and MPK6.
(B) Co-expression of CFP-BASL_123456A (cyan) with the split YFP of DNyda and MPK6 (yellow).
(C) Co-expression of CFP-BASL_123456D (cyan) with the split YFP of DNyda and MPK6 (yellow).
(D) Quantification of induced polarity in tobacco cells (n = 10–12 cells, 3 replicates). Values are mean ± SD, * significant differences (t-test, P<0.0001), ** significant difference (K–S test, α=0).
(E) Confocal images of split-YFP of DNyda and MPK6 driven by the BASL promoter in a 2-dpg adaxial cotyledon. White arrows point to protein polar accumulation. Scale bar = 25 µm
(F) BASL and the YDA MAPK cascade form a feedback loop, which reinforces cell polarity in stomatal ACD precursor cells. This model does not exclude the possibility of MAPKs or other kinases phosphorylating BASL in the nucleus, cytoplasm and/or at the plasma membrane.
(G) Asymmetric MAPK activity couples cell polarity to SPCH levels and differential cell fates in stomatal ACD. In the SLGC, polarized BASL enhances YDA-MPK3/6 signaling, which increases phosphorylation and, in turn, degradation of SPCH, thus altering the SLGC fate from stomatal to a pavement cell. In the Meristemoid, BASL polarity is low or absent and the YDA-MPK3/6 inhibition on SPCH is not enforced. Stable expression of SPCH leads Meristemoid differentiation into the guard cell terminal fate.
To further demonstrate the asymmetrically distributed MAPK signaling interaction in vivo, we developed an in situ BiFC system, in which the split YFP system was introduced into Arabidopsis plants. Because co-expression of YDA and MPK6 severely suppressed the stomatal lineage initiation, similar to constitutively active YDA(Bergmann et al., 2004), we co-transformed DNyda-nYFP with MPK6-cYFP, both driven by the BASL promoter, for their expression in the stomatal ACD cells. The recovered YFP signal suggests that DNyda interacts with MPK6 in vivo, and importantly, in a similarly polarized pattern of BASL (Figure 7E). Thus, BASL association with YDA MAPK components in stomatal ACD cells induces spatially reorganization and cortical accumulation of this signaling pathway in the larger daughter cell.
DISCUSSION
The findings presented here have several important implications. First, our studies provide unique insights into a universal signaling MAPK pathway being integrated into a specific cellular event, the BASL-centered polarization process during stomatal ACD. Second, we provided strong evidence to demonstrate a forward feedback loop between the polarity protein BASL and the MAPK signaling pathway as a molecular basis for cell polarity and ACD (Figure 7F). Third, we demonstrate that the physical interaction of two proteins (BASL and YDA) is sufficient to trigger a spontaneous polarization process, a mode of action critical for cell polarity that is scarcely documented in plants. Fourth, BASL physically associates with and redistributes multiple components of the MAPK cascade, implying its scaffolding function in the spatial control of signaling flow during stomatal ACD. Finally, our data suggest that the asymmetric distribution and activation of MAPK signaling arising from the BASL-YDA polarization determines differential daughter cell fate after the asymmetric cell division.
The Working Model for MAPK-centered Cell Fate Differentiation in Plant ACD
Although the two daughter cells, Meristemoid and SLGC, both have the potential to further divide, the SLGC has already withdrawn the terminal stomatal fate, whilst the Meristemoid continues to divide and terminally differentiate into stomatal guard cells (Bergmann and Sack, 2007). The stomatal ACD cells have employed multiple regulators for daughter cell fate differentiation. Besides the SLGC-expressing BASL crescent, the other candidate is the bHLH SPCH transcription factor, the stomatal lineage determinant (MacAlister et al., 2007). High SPCH expression levels maintain stem cell behavior of the Meristemoid, and lowered SPCH expression in the nucleus is associated with SLGC and pavement cell differentiation (Robinson et al., 2011).
Our findings here coupled the enriched YDA-MPK3/6 signaling with BASL polarity, and functions of MPK3/6 have been previously linked to SPCH phosphorylation and degradation (Lampard et al., 2008). Thus, we propose a working model for how BASL polarity differentiates two daughter cell fates. The SLGC inherits BASL polarity, as well as the associated high level of YDA MAPK activity, which confers lowered expression level of SPCH and terminated stomatal formation. On the other hand, the Meristemoid, without BASL polarity, has the basal level of YDA MAPK activity, allowing SPCH to sustain at higher expression levels to drive the cell fate commitment to stomatal differentiation (Figure 7G).
In animals, the unequal segregation of cell fate determinants is driven by polarity proteins at the opposite pole, but occurs through several mechanisms, e.g., asymmetric mRNA localization (Johnstone, 2001), differential protein stability (DeRenzo C, 2003), and polarized protein localization (Hirata J, 1995; Knoblich, 1995; Lu B, 1998). Our findings reveal a molecular mechanism whereby instead of segregating the cell fate determinants, the polarity protein BASL seems to segregate a negative regulation potential (the MAPK activity) that acts upstream of the fate determinants to distinguish two daughter cells.
As the stomatal ACD defects in basl mutations are not fully penetrant (Dong et al., 2009) and BASL polarity is not entirely lost in the absence of YDA (Figure 4C), we hypothesize that other mechanisms in parallel to the BASL pathway and more players beyond the YDA MAPK cassette would contribute to the regulation of cell polarity and stomatal ACD in Arabidopsis.
Phosphorylation Turns on BASL Polarization and Function
Protein phosphorylation is one of the signature features commonly found in the process of asymmetrically distributing proteins, e.g. yeast Cdc24, (Wai et al., 2009), animal Par proteins (Goldstein and Macara, 2007) and plant PIN proteins (Friml et al., 2004; Michniewicz et al., 2007; Zourelidou et al., 2009). We have shown that without phosphorylation, BASL is sequestered in the nucleus, suggesting that MPK3/6-mediated phosphorylation to activate BASL occurs in the nucleus. Does BASL have to transit through the nucleus for phosphorylation, like the yeast proteins Ste5 (MAPK scaffold) and Cdc24 (activator of small GTPase Cdc42) do (Mahanty et al., 1999; Shimada et al., 2000)? Myr-BASL was localized at the plasma membrane, but still polarized and efficiently complemented basl mutants (Figure 3A). Considering the overall subcellular localization of MPK6-YFP in the stomatal lineage cells (both nucleus and cytoplasm, data not shown), it is expected that MPK3/6 can phosphorylate Myr-BASL at the plasma membrane. In addition, we do not exclude the possibility that other kinases may phosphorylate BASL in the nucleus or at the cortex, since mutating 5 MPK6-mediated sites in BASL (12356A) still allowed protein expression in the cytoplasm and partially polarized (Figure 2D). Future work is required to discern whether S72 (S4) is phosphorylated in vivo and what kinases play this role.
The MAPK Docking Motifs for BASL-YDA Interaction
The major types of MAPK docking motifs, D domains and DEF domains, have been identified in many MAPK-interacting proteins for signaling efficiency and specificity (Cargnello and Roux, 2011). Diffuse cytoplasmic localization of BASL_d1_d2_def is consistent with the frequent overlap between MAPK docking motifs and the nuclear localization signal (NLS) (Cargnello and Roux, 2011), and further supports our hypothesis the MPK3/6-mediated phosphorylation of BASL occurs in the nucleus (Figure 7F). Unexpectedly, our data indicated that these docking motifs also mediate BASL’s interaction with the MAPKKK YDA. Although BASL_d1_d2_def lost its binding ability with YDA in our pull-down and Y2H assays, BASL polarity was dampened but still noticeable in yda mutants, suggesting that these three motifs might be needed for BASL’s interaction with other regulators in polarity formation.
Based on the evidence that BASL interacts with YDA, MKK5 and MPK3/6, we propose that BASL may functions as a MAPK scaffold protein in plants. Scaffold proteins are well recognized as hubs for shaping information flow and achieving signaling specificity in animal systems (Good et al., 2011). However, the putative MAPK scaffolding function in plants were revealed by two MAPKKKs (from alfalfa and Arabidopsis) for their direct binding to the downstream MAPKs (Nakagami et al., 2004; Suarez-Rodriguez et al., 2007). In addition, the polarity protein BASL’s scaffolding function would provide an additional layer of appreciation about how specific spatial-temporal control of MAPK signaling is achieved in plant stem cell asymmetric divisions.
Spontaneous Polar Accumulation of BASL-YDA
Although the molecular machinery remains elusive, our studies revealed that at least two signature features are critical for BASL-YDA polarization, as for the Par proteins, including protein phosphorylation (discussed above) and direct protein-protein interaction. The oligomerization of Par3 and dimerization of several Par proteins are required for their asymmetric distribution to cortical domains (Benton and St Johnston, 2003; McCaffrey and Macara, 2012). BASL does not bind to itself (data not shown), but BASL binds to YDA (Figure 5A and Figure S6A) and YDA interacts with itself (Figure S7E and S7F). It remains to be elucidated whether YDA oligomerization is critical for its function, but we have shown that the interaction mediated by the three MAPK-docking motifs between BASL and YDA is necessary to trigger their polarization process in plant cells (Figure 7A).
As BASL binds to YDA and MPK3/6, could MPK3/6 contribute to the polarity formation? From our tobacco assays, the polarity formation of BASL-YDA-MPK3/6 was obviously stronger than that of BASL-YDA (Figure 6F vs. Figure 6C), suggesting positive roles for MPK3/6 in the polarity module (Figure 7F). MPK3/6 may physically enhance the recruitment of YDA and BASL, and/or locally promote BASL phosphorylation for higher binding affinity with YDA.
The positive feedback loop between BASL and the YDA MAPK pathway
The MPK3/6-mediated phosphorylation of BASL and the BASL-mediated redistribution of the YDA-MPK3/6 signaling rendered a positive feedback model (Figure 7F). This positive feedback loop leads to the spatially concentrated YDA MAPK signaling in the stomatal lineage cells (Figure 7E). In addition, as BASL binds to the N-terminal auto-inhibitory domain of YDA (Figure 5C), we hypothesize that YDA activity would be elevated by the interaction with BASL, thus further promotes activated MAPKs in the nucleus to phosphorylate BASL for polarization.
Positive feedback loops amplify a small disturbance to an increased magnitude of the perturbation and have been widely accepted as a general principle to reinforce initial polarity cue and maintain cell polarity (Thompson, 2013). The well-studied symmetry breaking events mediated by small GTPases, Cdc42 in yeast and ROPs in plants, rely on cytoskeleton-dependent and -independent positive feedback loops (Johnson et al., 2011; Slaughter et al., 2009; Yang, 2008). Recent advances established that the phytohormone auxin forms a positive feedback loop with the ROP signaling during interdigitating polar growth of pavement cells in Arabidopsis (Xu et al., 2010; Yang and Lavagi, 2012). Our discovery of the BASL-YDA polarity module would encourage future investigation of whether and how MAPK upstream signaling molecules, e.g. EPF ligands, and the downstream cytoskeletal regulators (Komis et al., 2011) may function in feedback loops to establish cellular asymmetry during plant ACD.
Taken together, our studies show that the polarity protein BASL and the YDA-MPK3/6 cascade constitute a self-organizing polarity system that is essential for ACD and cell fate determination in stomata patterning. Since the YDA MAPK pathway broadly functions in plant development, e.g. embryonic asymmetric cell division (Lukowitz et al., 2004), root apical meristem integrity (Smekalova et al., 2014), and inflorescence patterning (Meng et al., 2012), it would be interesting to investigate whether the spatial organization of MAPK signaling could be commonly used to control division orientation, regional cell proliferation and fate diversification in plants.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
Arabidopsis thaliana plants were grown at 22°C on half-strength MS plates or in soil with 16-hr light/8-hr dark cycles. The ecotype Columbia (Col-0) was used as the wild-type unless otherwise noted. Mutants and alleles used in this study were: basl-2 (Dong et al., 2009), yda (Salk_105078 from the Arabidopsis Biological Resource Center, ABRC) and er erl1 erl2 (Shpak et al., 2005).
Plasmid construction
Detailed description and primers used can be found in the Supplemental Experimental Procedures.
Confocal imaging and image processing
Confocal images were captured by Leica TCS SP5 II. The Excitation/ Emission spectra for various fluorescent proteins are: CFP 458 nm/480–500nm, YFP 514 nm/ 520–540 nm, GFP 488 nm/501–528 nm, mCherry 543 nm/600–620 nm, mRFP 594 nm/ 600–620, and Propidium Iodide (PI) 543 nm/ 591–636 nm. All imaging processing was performed by Fiji software (http://fiji.sc/Fiji) and figures were assembled with Adobe Illustrator CS6.
Quantification of stomatal phenotypes in Arabidopsis
In principle, more than 12 independent transgenic T2 lines were maintained for each transgene. Two representative lines from each were established for quantitative analysis. For complementation assessment, 5-dpg (day post growth) cotyledons were stained with PI (Invitrogen) and imaged by the EC Plan-Neofluar (20×/0.5) lenses on a Carl Zeiss AXIO SCOPE A1 fluorescence microscope equipped with a ProgRes MF CCD camera (Jenoptik). Images were taken from similar central areas in the adaxial cotyledons (10–12 individual seedlings picked from two representative T2 lines). In total, 1200 to 2000 cells were collected and categorized as describe in (Dong et al., 2009). The epidermal cells were scored for 4 groups: pavement cells, guard cells, clustered guard cells and small dividing cells. The calculations represent the ratio of i cells from each category relative to the total cell number. For the limited space, we presented the two key defects of basl mutants (clustered guard cell and small dividing cells) to represent the transgene complementary effect (Figure 1G, 2G and S3). The data were subjected to normal probability test and followed by Student’s t- test.
Protein polarity quantification in Arabidopsis
For quantitative analysis of GFP-BASL polarization, confocal images were taken from 2-dpg adaxial cotyledons of 10 representative seedlings (cell outlines were imaged with PI staining). GFP-BASL forms distinctive crescent in the stomatal lineage cells. To quantify the polarity degree of BASL and variants (for Figure 1E and 1D and Figure 2D)., the length of GFP crescent (A) and the whole-cell perimeter (A+B, marked by PI) were measured Protein polarity was determined by the ratio of crescent width over the cell perimeter (A/A+B, Figure 1F). Total 50 individual cells expressing cortical BASL were selected and scored for polarity. When proteins were found spread along the plasma membrane and unevenly distributed, e.g. Myr-BASL and variants, YDA variants and those expressed in tobacco assays (Figure 6 and 7), we measured fluorescence intensity for polarity quantification. For DNyda (Figure 5G) and Myr-BASL polarity (Figure 3E), the mean values of fluorescence intensity at the plasma membrane were measured from two segments with same length; one from the DNyda or Myr-BASL enriched side (A) and the other one from the distal side (B). Polarity degree was determined by A/B. Measurements were conducted on 50 cells collected from 3-dpg adaxial cotyledons of 5 individual plants.
Transient expression in Nicotiana benthamiana
Agrobacterium tumefaciens strains GV3101 harboring binary vectors in 10 ml of LB medium with appropriate antibiotics were cultured overnight. Bacterial cells were harvested at 3500 rpm for 10 min and resuspended in 10 ml of 10mM MgCl2, followed by another step of 10 mM MgCl2 washing. Cells were then left in the medium for 3 h at room temperature prior to infiltration. Equal volumes of cell culture and p19 (to suppress gene silencing) (Voinnet et al., 2003) were mixed to reach an OD600=0.5 and infiltrated into the 4-week-old tobacco leaves. The leaf disks were excised and mounted to slides for confocal imaging 2–3 days after the infiltration. To quantify the polarity degree of CFP and YFP in tobacco cells, three replicate experiments were performed and 18–20 representative cells from each combination were scored. The absolute fluorescence intensity was measured along the cell periphery and presented in mean values. For the cells exhibiting visible polar accumulation of fluorescent proteins, the high intensity values (defined as H) and low intensity (defined as L) were collected from equal length of the cell periphery. For the cells displaying relatively uniform expression of YFP and/or CFP, the H and L values were collected by measuring 2 randomly selected peripheral segments with equal length. The polarity degree was determined by the ratios of H/L. The obtained data were tested for normal distribution, and then subject to Student’s t-test or K-S test.
Yeast two-hybrid analysis
Matchmaker II yeast two-hybrid System (Clontech) was used to detect protein-protein interactions. The manufacturer’s protocols were followed. The bait and prey plasmids were sequentially or co-transformed into yeast strain AH109. To inhibit the self-activation of YDA_N, 1mM 3-Amino-1, 2, 4-triazole was used. In addition, X-alpha-Gal (Clontech) was added into the growth medium to indicate the interaction strength.
Recombinant protein production and pull-down assay
Constructs were introduced into BL21 (DE3) dcm codon plus cells. The recombinant His-tagged BASL and variants were purified using Ni-NTA agarose (Qiagen) according to the manufacturer’s protocol. The His-tagged and maltose-binding protein (MBP)-tagged MPK3 or 6 and YDA were purified using Ni-NTA agarose (Qiagen) or Amylose Resin (New England Biolabs), respectively, according to the manufacturer’s protocol. For pull-down assays, 5 µg of purified HIS-tagged BASL (or variant) and 20 µl of Amylose Resin, which pre-absorbed 2 µg of MBP-tagged YDA proteins, were incubated in 100 µl Binding Buffer (50 mM Tris-Cl, pH 7.5, 10 mM MgCl2, 150 mM NaCl and 1mM DTT) for 30 min at 25 C°. After washing 5 times with 500 µl of Binding Buffer, the bound proteins on the Amylose Resin were separated by SDS-PAGE and visualized immunoblot with anti 6 × HIS (Sigma-Aldrich) and MBP antibody (New England Biolabs).
In vitro kinase assay and mass spectrometry analysis
The in vitro kinase assay was performed as described previously(Wang et al., 2010). Recombinant His-tagged MPK6 (10 µg) was activated by incubation with recombinant MKK5DD (1 µg) in the presence of 50 µM ATP in 50 µL of reaction buffer (25 mM Tris, pH 7.5, 10 mM MgCl2, and 1 mM DTT) at 25 C° for 30 min. Activated MPK6 was used to phosphorylate recombinant protein purified from E. coli in the same reaction buffer, with 50 mM ATP and [γ-32P] ATP (0.1 mCi per reaction). The reactions were stopped by the addition of SDS loading buffer after 30 min. Phosphorylated BASL and variants were visualized by autoradiography after separation on a 10% SDS polyacrylamide gel.
MAP kinase activity in plants
Total protein was extracted from 7-day-old seedlings by grinding in extraction buffer (100 mM HEPES, pH 7.5, 5 mM EDTA, 5 mM EGTA, 10 mM Na3VO4, 10 mM NaF, 50 mM b-glycerophosphate, 10 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 5 mg/mL leupeptin, 5 mg/mL aprotinin, and 5% glycerol). After centrifugation at 18,000 g for 40 min, supernatants were transferred into clean tubes. Protein concentrations were determined by the Bradford assay, with BSA as the standard. For immune blotting, 20 µg of total proteins were separated by electrophoresis on 10% SDS-polyacrylamide gels, and the activity of MAPK kinase activity were determined by the immune blot with phospho-P44/42 MAPK antibody (Cell Signaling Technology).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Dominique Bergman (Stanford) for the MAPK-related DNA reagents. We are grateful to Dr. Kenneth Irvine (Rutgers) and Dr. Dominique Bergmann for critical comments on the manuscript. This work is supported by grants from the U.S. National Institute of General Medical Sciences to J.D. (R01GM109080) and J.-K.Z (R01GM059138).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
This project was initiated and supervised by J.D. J.D and Y. Z. designed the experiments and Y.Z conducted the molecular and cell biological work that linked BASL polarity to the MAPK activities. P.W. identified the phosphorylation sites of BASL and performed biochemical assays related to MAPKs and YDA. W.S. genetically characterized BASL polarity in connection with YDA function and assayed the physical interaction in Y2H. J.-K.Z supervised P.W. and helped with data interpretation. J.D. wrote the manuscript. Y.Z. and J.-K.Z. assisted manuscript writing.
REFERENCES
- Abrash EB, Bergmann DC. Asymmetric cell divisions: a view from plant development. Developmental cell. 2009;16:783–796. doi: 10.1016/j.devcel.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Abrash EB, Bergmann DC. Regional specification of stomatal production by the putative ligand CHALLAH. Development (Cambridge, England) 2010;137:447–455. doi: 10.1242/dev.040931. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. A conserved oligomerization domain in drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Current biology : CB. 2003;13:1330–1334. doi: 10.1016/s0960-9822(03)00508-6. [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR. Stomatal development and pattern controlled by a MAPKK kinase. Science (New York, NY) 2004;304:1494–1497. doi: 10.1126/science.1096014. [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD. Stomatal development. Annual review of plant biology. 2007;58:163–181. doi: 10.1146/annurev.arplant.58.032806.104023. [DOI] [PubMed] [Google Scholar]
- Bush SM, Krysan PJ. Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. Journal of experimental botany. 2007;58:2181–2191. doi: 10.1093/jxb/erm092. [DOI] [PubMed] [Google Scholar]
- Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiology and molecular biology reviews : MMBR. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright HN, Humphries JA, Smith LG. PAN1: a receptor-like protein that promotes polarization of an asymmetric cell division in maize. Science (New York, NY) 2009;323:649–651. doi: 10.1126/science.1161686. [DOI] [PubMed] [Google Scholar]
- de Smet I, Beeckman T. Asymmetric cell division in land plants and algae: the driving force for differentiation. Nature Reviews Molecular Cell Biology. 2011;12:177–188. doi: 10.1038/nrm3064. [DOI] [PubMed] [Google Scholar]
- DeRenzo C, R K, Seydoux G. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature. 2003;424:685–689. doi: 10.1038/nature01887.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, MacAlister CA, Bergmann DC. BASL controls asymmetric cell division in Arabidopsis. Cell. 2009;137:1320–1330. doi: 10.1016/j.cell.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science (New York, NY) 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Developmental cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science (New York, NY) 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood S, Struhl G. Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development (Cambridge, England) 1999;126:5795–5808. doi: 10.1242/dev.126.24.5795. [DOI] [PubMed] [Google Scholar]
- Guseman JM, Lee JS, Bogenschutz NL, Peterson KM, Virata RE, Xie B, Kanaoka MM, Hong Z, Torii KU. Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8) Development (Cambridge, England) 2010;137:1731–1741. doi: 10.1242/dev.049197. [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes & development. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata JNH, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- Ho DT, Bardwell AJ, Abdollahi M, Bardwell L. A docking site in MKK4 mediates high affinity binding to JNK MAPKs and competes with similar docking sites in JNK substrates. The Journal of biological chemistry. 2003;278:32662–32672. doi: 10.1074/jbc.M304229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JA, Vejlupkova Z, Luo A, Meeley RB, Sylvester AW, Fowler JE, Smith LG. ROP GTPases Act with the Receptor-Like Protein PAN1 to Polarize Asymmetric Cell Division in Maize. The Plant Cell. 2011 doi: 10.1105/tpc.111.085597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Gray JE. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Current biology : CB. 2009;19:864–869. doi: 10.1016/j.cub.2009.03.069. [DOI] [PubMed] [Google Scholar]
- Inaba M, Yamashita YM. Asymmetric stem cell division: precision for robustness. Cell stem cell. 2012;11:461–469. doi: 10.1016/j.stem.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Irazoqui JE, Gladfelter AS, Lew DJ. Scaffold-mediated symmetry breaking by Cdc42p. Nature cell biology. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Jin M, Lew DJ. Symmetry breaking and the establishment of cell polarity in budding yeast. Current opinion in genetics & development. 2011;21:740–746. doi: 10.1016/j.gde.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annual Review of Genetics. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of Asymmetric Stem Cell Division. Cell. 2008a;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008b;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Komis G, Illes P, Beck M, Samaj J. Microtubules and mitogen-activated protein kinase signalling. Current opinion in plant biology. 2011;14:650–657. doi: 10.1016/j.pbi.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Kondo T, Kajita R, Miyazaki A, Hokoyama M, Nakamura-Miura T, Mizuno S, Masuda Y, Irie K, Tanaka Y, Takada S, et al. Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant & cell physiology. 2010;51:1–8. doi: 10.1093/pcp/pcp180. [DOI] [PubMed] [Google Scholar]
- Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Current biology : CB. 2008;18:1719–1726. doi: 10.1016/j.cub.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, Lukowitz W, Ellis BE, Bergmann DC. Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. The Plant cell. 2009;21:3506–3517. doi: 10.1105/tpc.109.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, Macalister CA, Bergmann DC. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science (New York, NY) 2008;322:1113–1116. doi: 10.1126/science.1162263. [DOI] [PubMed] [Google Scholar]
- Lau OS, Bergmann DC. Stomatal development: a plant's perspective on cell polarity, cell fate transitions and intercellular communication. Development (Cambridge, England) 2012;139:3683–3692. doi: 10.1242/dev.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Davies KA, Chang J, Adrian J, Rowe MH, Ballenger CE, Bergmann DC. Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science (New York, NY) 2014 doi: 10.1126/science.1256888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, R M, Jan LY, Jan YN. Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell. 1998;95:225–235. doi: 10.1016/s0092-8674(00)81753-5. [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell. 2004;116:109–119. doi: 10.1016/s0092-8674(03)01067-5. [DOI] [PubMed] [Google Scholar]
- MacAlister CA, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445:537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- Mahanty SK, Wang Y, Farley FW, Elion EA. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG. Signaling pathways in cell polarity. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Wang H, He Y, Liu Y, Walker JC, Torii KU, Zhang S. A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. The Plant cell. 2012;24:4948–4960. doi: 10.1105/tpc.112.104695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nature cell biology. 2002;4:556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD. Control of stomatal distribution on the Arabidopsis leaf surface. Science (New York, NY) 2002;296:1697–1700. doi: 10.1126/science.1069596. [DOI] [PubMed] [Google Scholar]
- Nakagami H, Kiegerl S, Hirt H. OMTK1, a novel MAPKKK, channels oxidative stress signaling through direct MAPK interaction. The Journal of biological chemistry. 2004;279:26959–26966. doi: 10.1074/jbc.M312662200. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, van Norman JM, Benfey PN. Symmetry Breaking in Plants: Molecular Mechanisms Regulating Asymmetric Cell Divisions in Arabidopsis. Cold Spring Harbor Perspectives in Biology. 2009;1:a000497–a000497. doi: 10.1101/cshperspect.a000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Peterson KM, Horst RJ, Torii KU. Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. The Plant cell. 2011;23:3260–3275. doi: 10.1105/tpc.111.088583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU. Mechanisms of stomatal development. Annual review of plant biology. 2012;63:591–614. doi: 10.1146/annurev-arplant-042811-105451. [DOI] [PubMed] [Google Scholar]
- Prehoda KE. Polarization of Drosophila Neuroblasts During Asymmetric Division. Cold Spring Harbor Perspectives in Biology. 2009;1:a001388–a001388. doi: 10.1101/cshperspect.a001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S, Barbier de Reuille P, Chan J, Bergmann D, Prusinkiewicz P, Coen E. Generation of spatial patterns through cell polarity switching. Science (New York, NY) 2011;333:1436–1440. doi: 10.1126/science.1202185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464:913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Gulli MP, Peter M. Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nature cell biology. 2000;2:117–124. doi: 10.1038/35000073. [DOI] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science (New York, NY) 2005;309:290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]
- Slaughter BD, Smith SE, Li R. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harbor perspectives in biology. 2009;1:a003384. doi: 10.1101/cshperspect.a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smekalova V, Luptovciak I, Komis G, Samajova O, Ovecka M, Doskocilova A, Takac T, Vadovic P, Novak O, Pechan T, et al. Involvement of YODA and mitogen activated protein kinase 6 in Arabidopsis post-embryogenic root development through auxin up-regulation and cell division plane orientation. The New phytologist. 2014;203:1175–1193. doi: 10.1111/nph.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant physiology. 2007;143:661–669. doi: 10.1104/pp.106.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463:241–244. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nature cell biology. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- Thompson BJ. Cell polarity: models and mechanisms from yeast, worms and flies. Development (Cambridge, England) 2013;140:13–21. doi: 10.1242/dev.083634. [DOI] [PubMed] [Google Scholar]
- Umbrasaite J, Schweighofer A, Kazanaviciute V, Magyar Z, Ayatollahi Z, Unterwurzacher V, Choopayak C, Boniecka J, Murray JA, Bogre L, et al. MAPK phosphatase AP2C3 induces ectopic proliferation of epidermal cells leading to stomata development in Arabidopsis. PloS one. 2010;5:e15357. doi: 10.1371/journal.pone.0015357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant journal : for cell and molecular biology. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- Wai SC, Gerber SA, Li R. Multisite phosphorylation of the guanine nucleotide exchange factor Cdc24 during yeast cell polarization. PloS one. 2009;4:e6563. doi: 10.1371/journal.pone.0006563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. The Plant cell. 2007;19:63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Du Y, Li Y, Ren D, Song CP. Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. The Plant cell. 2010;22:2981–2998. doi: 10.1105/tpc.109.072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SE, Fuchs E. Oriented divisions, fate decisions. Current opinion in cell biology. 2013;25:749–758. doi: 10.1016/j.ceb.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Jansen G, Zhang J, Thomas DY, Whiteway M. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes & development. 2006;20:734–746. doi: 10.1101/gad.1375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PS, Egger B, Brand AH. Asymmetric stem cell division: lessons from Drosophila. Seminars in cell & developmental biology. 2008;19:283–293. doi: 10.1016/j.semcdb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell. 2010;143:99–110. doi: 10.1016/j.cell.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Cell polarity signaling in Arabidopsis. Annual review of cell and developmental biology. 2008;24:551–575. doi: 10.1146/annurev.cellbio.23.090506.123233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lavagi I. Spatial control of plasma membrane domains: ROP GTPase-based symmetry breaking. Current opinion in plant biology. 2012;15:601–607. doi: 10.1016/j.pbi.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Kuo CT, Jan YN. Drosophila neuroblast asymmetric cell division: recent advances and implications for stem cell biology. Neuron. 2006;51:13–20. doi: 10.1016/j.neuron.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Zourelidou M, Muller I, Willige BC, Nill C, Jikumaru Y, Li H, Schwechheimer C. The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development (Cambridge, England) 2009;136:627–636. doi: 10.1242/dev.028365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.