Abstract
Purpose
To identify a LNCaP-specific peptide using a phage display library and evaluate its potential applications in targeted drug delivery.
Methods
Binding abilities of selected phages were evaluated by cell phage ELISA. The KYL peptide encoded by the most specific phage clone was synthesized, labeled with fluorescein, and assayed in various cell lines. A fusion peptide composed of the KYL peptide and a proapoptotic peptide d(KLAKLAK)2 was synthesized, and the cell death effect was evaluated on different cells. Moreover, the KYL peptide was conjugated to a cationic protein, protamine, to explore its potential application in siRNA delivery.
Results
One phage clone with a high binding affinity to LNCaP cells was identified. Cell phage ELISA and immunostaining demonstrated high specificity of this phage to LNCaP cells. The fluorescein-labeled KYL peptide exhibited higher binding to LNCaP cells in comparison to other cells. The fusion peptide composed of the KYL peptide and the proapoptotic peptide induced cell death in LNCaP cells, but not in PC-3 cells. The KYL peptide-protamine conjugate also efficiently delivered a fluorescein-labeled siRNA into LNCaP cells.
Conclusion
We identified a LNCaP-specific peptide and demonstrated its potential applications in targeted drug delivery to LNCaP cells.
Keywords: LNCaP, peptide ligand, phage display, prostate cancer, siRNA
INTRODUCTION
Prostate cancer is the most common form of malignancy and the second leading cause of cancer death in American men (1). Although localized prostate cancer can be diagnosed with the PSA (prostate-specific antigen) screening test and effectively treated by surgery and radiation therapy, the 5-year relative survival rate for prostate cancer metastasis is as low as 30.2%. Current standard therapies for prostate cancer include surgery, radiation and adjuvant hormonal therapy. Despite the fact that these therapies are relatively effective in the early stage, the majority of patients initially diagnosed with localized prostate cancer ultimately relapse. The major risk faced by prostate cancer patients is therefore the development to metastasis (2). Consequently, intensive efforts have been made to develop therapeutics for the treatment of late-stage prostate cancer, which requires systemic delivery of the drug.
However, lack of efficient delivery to cancer cells in vivo is the primary challenge in achieving a good therapeutic outcome in cancer therapy. As a result, a great deal of attention has been directed to targeted delivery strategies to overcome this challenge. Among the numerous targeting approaches, monoclonal antibody is the first type of ligand used for targeted drug delivery due to its high affinity. However, its application in drug delivery is limited so far, owing to the large molecular weight (150 kDa) and possible immunogenicity. Compared to antibody, peptide is a more appropriate targeting moiety due to its small molecular weight, ease of synthesis, high cellular permeability, non-immunogenicity, and flexibility in chemical conjugation (3). Small peptide ligands that bind to a specific receptor or enhance cell penetration have been conjugated to a wide variety of drug delivery systems (4), small molecule drugs (5), and nucleic acids (6) to improve the therapeutic efficacy. Peptides have also been linked to radio-labeled agents for diagnosis purposes (7,8).
Recently, peptide phage display library has been used to identify peptides that bind to specific molecular targets. Phage display biopanning is a selection technique using a library of various peptides or proteins expressed on the surface of filamentous phages. Through different biopanning procedures (in vivo or in vitro), this technology provides a valuable tool for identifying peptides that can specifically bind to target proteins (9,10), cells (4,11-15) or tissues (16,17). Biopanning has been successfully adopted to identify peptides against diverse targets via affinity selection (18-21), which resembles in essence the traditional chemical libraries but with a much diverse library containing billions of different peptides (19). Scientists generally conduct the biopanning procedure using recombinant target proteins that are immobilized in a solid phase. However, the complex and variant cell-surface landscapes on tumor cells require not only the affinity selection to the target protein, but also the affinity selection to the whole cell (18). Hence, we performed an extensive whole-cell biopanning on the PSMA (prostate specific membrane antigen)-positive prostate cancer LNCaP cells. The LNCaP cell line is the most commonly used in vitro carcinoma cell model for prostate cancer research (22,23). LNCaP cells express key prostatic biomarkers including PSA, PSMA and prostate acid phosphatase (PAP) (24). Moreover, LNCaP cells maintain the hormone-response and malignant properties. The aims of this study are to identify a LNCaP-specific peptide ligand and then evaluate its potential applications in targeted drug delivery. It is expected that the peptide identified in this study can bind to a significant proportion of prostate cancer cells.
MATERIALS AND METHODS
Materials
The M13 phage display peptide library (Ph.D.™-12) and ER2738 bacteria were purchased from New England Biolabs (Beverly, MA). LNCaP, PC-3, MCF-7, SK-BR-3 and DU145 cells were obtained from the American Type Culture Collection (Rockville, MD). The rat hepatic stellate cell line (HSC T6) was kindly provided by Dr. Scott L. Friedman (Mount Sinai School of Medicine, New York University). Horseradish peroxidase (HRP)-conjugated anti-M13 monoclonal antibody was purchased from GE Healthcare BioSciences (Piscataway, NJ). KYLAYPDSVHIW (KYL peptide) and KYLAYPDSVHIWGG-d(KLAKLAK)2(KYL-KLA peptide) were ordered from Genscript (Piscataway, NJ). d(KLAKLAK)2(KLA peptide) was purchased from Anaspec, Inc. (Fremont, CA). Protamine (grade X) was obtained from Sigma-Aldrich Corporation (St Louis, MO). N-Succinimidyl-3-(2-pyridyldithio) propionate (SPDP) was purchased from Thermo Fisher Scientific, Inc. (Pittsburgh, PA). The BLOCK-iT™ Fluorescent siRNA was obtained from Invitrogen (Carlsbad, CA). All cell culture reagents were obtained from Thermo Fisher Scientific, Inc. (Pittsburgh, PA) as well as Mediatech, Inc. (Manassas, VA).
Whole-Cell Biopanning on LNCaP Cells
The whole-cell biopanning was performed on prostate cancer cells according to the BRASIL method with minor modifications (25). First, the phage library was incubated with PC-3 cells to remove the non-specific bound phages. Briefly, PC-3 cells were detached with cold PBS including 5 mM EDTA and then suspended in RPMI-1640 medium containing 1% BSA to a density of 107 cells per ml. Ten microliters of the phage display library (~1011 pfu phages) were added into 100 μl of the cell suspension and incubated at 4°C for 1 h under shaking. The cell suspension was centrifuged through an organic phase composed of dibutyl phthalate and cyclohexane (9:1, v/v). The supernatant was collected as the precleaned phage display library, which was subsequently transferred to LNCaP cell suspension (106 cells per ml) in a 1.5 ml microcentrifuge tube. After incubation at 4°C for 3 h, the cell suspension was centrifuged through the same organic phase described above. After snap freezing in liquid nitrogen, the bottom of the micro centrifuge tube was sliced off. Phages in the cell/phage complex on the bottom were recovered by infecting ER2738 bacteria, followed by amplification for next round biopanning. Individual plaques were randomly selected, and the affinities to LNCaP cells were assayed by cell phage ELISA. DNA of the selected phages was sequenced to obtain the 12-mer peptide sequence.
Cell Phage ELISA
Cell phage ELSIA was performed as reported (26). Briefly, LNCaP cells were cultured in 96-well plates at a density of 50,000 cells per well for 15 h, followed by fixation with cold methanol-acetone (1:1, v/v). Phages were suspended in RPMI-1640 medium containing 1% BSA and incubated with the fixed LNCaP cells for 1 h. After washing to remove the unbound phages, the cells were incubated with HRP-conjugated anti-M13 monoclonal antibodies for 1 h. TMB substrate was then added, and the absorbance at 450 nm was measured subsequently with a Beckman DTX 880 multimode Detector (Beckman coulter, Inc., Brea, CA).
Phage Recovery
The phage recovery procedure is similar to cell phage ELISA. LNCaP cells (5×104 cells per well) were cultured in 96-well plates for 15 h, followed by fixation with cold methanol-acetone (1:1, v/v). Phages were suspended in RPMI-1640 medium containing 1% BSA and then incubated with the LNCaP cells for 1 h. After removing the unbound phages by washing with PBS buffer, the bound phages were eluted using 0.2 M glycine-HCl buffer (pH2.2) and then neutralized with 1 M Tris-HCl buffer (pH9.1). The number of phages in the eluting buffer was titered by infecting ER2738 bacteria as per instructions from New England Biolab (Beverly, MA).
Sequencing and Analysis of the Phage Encoding Peptide
Phage DNA was isolated from selected phage clones and then sequenced with an ABI Genetic Analyzer 3100 (Applied biosystems, USA). The primer used for sequencing is 5′-CCCTCATAGTTAGCGTAACG-3′. The encoded peptide sequence was deduced from the DNA sequence.
Competitive Inhibition Study
Competitive binding inhibition between the positive phage clone and its encoded peptide was examined in this study. LNCaP cells (5×104 cells per well) were cultured and fixed with cold methanol-acetone (1:1, v/v). The fixed cells were incubated with different concentrations of the KYL peptide or a non-specific peptide for 30 min, followed by removing the unbound peptides by washing the cells with PBS. The phages were suspended in RPMI-1640 medium containing 1% BSA and then incubated with the cells for 1 h. The unbound phages were removed by washing, and the bound phage was recovered and titered as described above in the phage recovery procedure.
Immunostaining of the Bound Phages
LNCaP cells were cultured on a Lab-Tek® 8-well chamber slide overnight. The cells were fixed with cold methanolacetone (1:1, v/v) and then incubated with ~1010 pfu phages per well at room temperature for 1 h. After washing to remove the unbound phages, the cells were incubated with HRP-conjugated anti-M13 monoclonal antibodies at room temperature for 1 h. The slide was subsequently treated with DAB substrate (Invitrogen, CA) and examined with a Leica DMI3000 fluorescence microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Cellular Uptake of the Fluorescein-Labeled KYL Peptide
The KYL peptide was labeled with fluorescein using the NHS-Fluorescein Kit (Thermo scientific) as per the manufacturer’s protocol. The fluorescein-labeled KYL peptide was purified using a Bio-GelP6-DG column (Bio-Rad, CA). For the cellular uptake study, the fluorescein-labeled KYL peptide (10 μg/ml) was added to cells that were cultured and fixed on a chamber slide. After incubation at 4°C for 15 min, the cells were washed with PBS to remove the unbound fluorescein labeled KYL peptides. The cells were then examined with a Leica DMI3000 fluorescence microscope to determine the cellular uptake of the fluorescein-labeled KYL peptide.
Cell Viability Assay
LNCaP and PC-3 (1×104 cells per well) cells were cultured in 96-well plates for 12 h in RPMI-1640 medium containing 10% FBS. The cells were incubated with different concentrations of the KYL-KLA fusion peptide or the mixture of KYL and KLA peptides (KYL&KLA) for 48 h. For the endocytosis inhibition study, the cells were incubated with 20 μM KYL-KLA fusion peptide or KYL&KLA peptide mixture in the presence of 20 mM sodium azide in 100 μl RPMI-1640 medium containing 1% FBS for 2 h. Cell viability was measured using MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay.
Conjugation of the KYL Peptide to Protamine
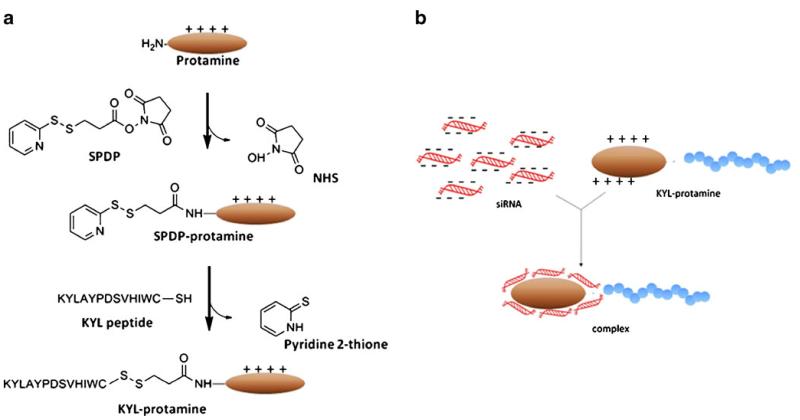
First, a N-Succinimidyl-3-(2-pyridyldithio) propionate (SPDP) group was conjugated to protamine. Two milligrams of protamine in 1 ml PBS (pH7.2) were incubated with 25 μl SPDP (100 mM in DMF) at room temperature for 1 h. The product was passed through a Sephadex G15 column twice to remove the impurities. The SPDP-protamine was freeze-dried and reconstituted in sterile water. Concentration of the SPDP-protamine was assayed using BCA protein assay. The level of SPDP-modification was determined by comparing the absorbance of pyridine 2-thione at 343 nm (E343 nm =8080M−1 cm−1) before and after the treatment with dithiothreitol (DTT).
The SPDP-protamine (150 μl, SPDP 0.2 mM) was then incubated with the KYL peptide (10 mM, 15 μl) containing an additional cysteine at the C-terminal and 16 μl glycine/NaCl buffer (100 mM, pH8.7) at 4°C for 12 h. The conjugated protamine-KYL was purified on a Sephadex G15 column. The final product was freeze-dried and dissolved in PBS buffer. Concentration of the protamine-KYL was determined using BCA protein assay.
Transfection of a Fluorescein-Labeled siRNA Using the Protamine-KYL Conjugate
LNCaP cells were seeded in 24-well plates at a density of 0.5×105 cells per well 12 h before the transfection. Two and a half micro liters of 20 μM BLOCK-iT™ Fluorescent Oligo (a fluorescein-labeled dsRNA designed for RNAi study) were mixed with 1.5 μg protamine or protamine-KYL in 100 μl serum-free RPMI-1640 medium and then incubated at room temperature for 10 min to form a complex. After washing cells with PBS, 40 μl of the transfection complexes were added to each well with 60 μl of the serum-free RPMI-1640 medium at a final concentration of 200 nM siRNA. Five hours after the transfection, the medium was removed and the cells were washed twice with PBS. Fluorescence of the FITC-fluorescein labeled siRNA was observed using a Leica DMI3000 fluorescence microscope.
Treatment of DHT (5-α-dihydrotestosterone)
LNCaP cells were cultured in 5% charcoal-stripped FBS RPMI-1640 for 24 h. The cells were subsequently supplied with 5% charcoal-stripped FBS RPMI-1640 containing 2 nM DHT (or the same volume of DMSO as a control) and incubated for another 48 h. The PSMA expression on LNCaP cells was assayed by Western blot. The effects of KYL-KLA peptide on DHT-treated cells and control cells were examined by MTT assay as mentioned above.
Western Blot
The cells were lysed on ice in the RIPA lysis buffer containing protease and phosphatase inhibitor cocktails. After 5 min incubation, the cell lysate was collected and clarified by centrifugation at 4°C for 10 min at 12,000 rpm. The amount of total protein was determined using a BCA protein assay kit. Equal amount of total protein (20 μg) was loaded and separated by SDS-PAGE. The protein was transferred to a nitrocellulose membrane, blocked and probed with appropriate antibody. The protein was visualized using horseradish peroxidase-conjugated secondary antibodies and the FluorChem FC2 imaging system (Alpha Innotech, CA). Anti-PSMA antibody (Abcam, Cambridge, MA), anti-β-Actin antibody (Rockland, Gilertsville, PA), and HRP-conjugated secondary antibody (Invitrogen, Carlsbad, CA) were used in the Western blot assay.
Statistics Analysis
Data were expressed as the mean±standard deviation (SD). Difference between any two groups was determined by ANOVA. P<0.05 was considered statistically significant.
RESULTS
Identification of LNCaP-Specific Peptides Using the Ph.D.™-12 Phage Display Library
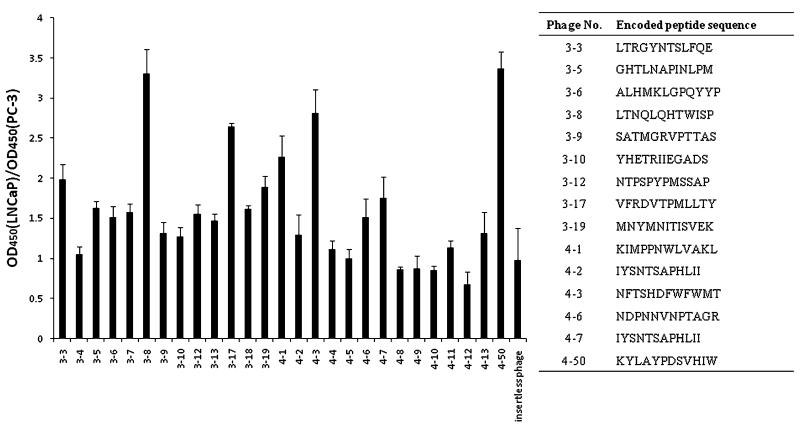
To identify peptides that specifically bind to LNCaP cells, four rounds of whole cell biopanning were performed on LNCaP cells after pre-cleaning on PC-3 cells to remove the non-specific bound phages. In each round, approximately 1011pfu of phages were incubated with LNCaP cells at 4°C for 3 h for selection. A number of phage clones after the third and fourth rounds of biopanning were randomly selected and amplified. Binding abilities of these selected phage clones were evaluated on LNCaP and PC-3 cells by cell phage ELISA (Fig. 1). The phage clone (clone 4–50) with the highest OD450 nm,LNCaP/OD450 nm,PC-3 as well as the highest binding affinity to LNCaP cells was selected for DNA sequencing, and the encoded peptide KYLAYPDSV-HIW was named the KYL peptide (the phage encoding the KYL peptide was named the KYL phage) for further studies.
Fig. 1.
Binding efficiency of randomly selected phage clones on LNCaP and PC-3 cells. Phage clones were randomly selected after the third and fourth biopanning rounds. Binding of the phages on LNCaP and PC-3 cells was examined with cell phage ELISA. Results are represented as the mean±SD (n=3).
Binding of the KYL Phage to LNCaP Cells
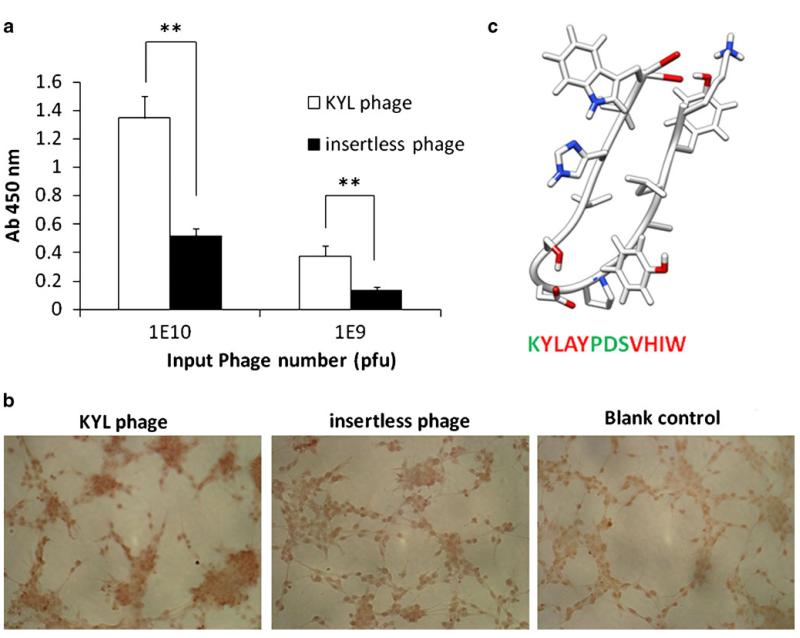
A computer-generated structure model of the KYL peptide is shown in Fig. 2c. Binding of the KYL phage to LNCaP cells is significantly higher than that of an insertless phage, which is used as a negative control because it does not express any peptide on the surface of filamentous phages (Fig. 2a). Binding of the KYL phage to LNCaP cells was also examined by immunostaining. Fixed LNCaP cells were incubated with the KYL phage and the insertless phage as the negative control. The unbound phages were removed by washing the cells with PBS buffer, and the bound phages were detected using the HRP-conjugated anti-M13 monoclonal antibody. Compared to the insertless phage, the KYL phage demonstrated a significant higher binding to LNCaP cells (Fig. 2b). This result further demonstrated the specific binding of the KYL phage to LNCaP cells. According to the computer-generated structure model (Fig. 2c), the sequence of the KYL peptide is composed of three blocks, two hydrophobic blocks (“YLAY” and “VHIW”) and one hydrophilic block (“PDS”) in the center. This model structure suggested that the KYL peptide may have a “loop-like” structure (Fig. 2c), which is a common structure in the hyper variable binding site of a Fab antibody fragment (27).
Fig. 2.
Determination of the binding of KYL phage on LNCaP cells by cell phage ELISA (a) and immunostaining (b). (a) LNCaP cells (50,000 cells per well) were cultured in 96-well plates were fixed with cold methanol-acetone (1:1, v/v). The KYL phage and insertless phage (109 and 1010 pfu) were incubated with the cells for 1 h, and the bound phages were determined by cell phage ELISA. Results are represented as the mean±SD (n=3).*p<0.05, **p<0.01. (b) LNCaP cells cultured on Lab-TEK® 8-well chamber slides were fixed and incubated with ~1010 pfu phages at room temperature for 1 h. The cells were incubated with HRP-conjugated anti-M13 monoclonal antibody at room temperature for 1 h and then visualized using the DAB substrate. (c) Computer-generated model structure of the KYL peptide. The KYL peptide sequence was analyzed using mobile (http://mobyle.rpbs.univ-paris-diderot.fr/cgi-bin/portal.py?form=PEP-FOLD#). The structure was obtained by UCSF chimera molecular modeling system using the mobile-generated PDB file. Green color indicates hydrophilic residues, and red color indicates hydrophobic residues.
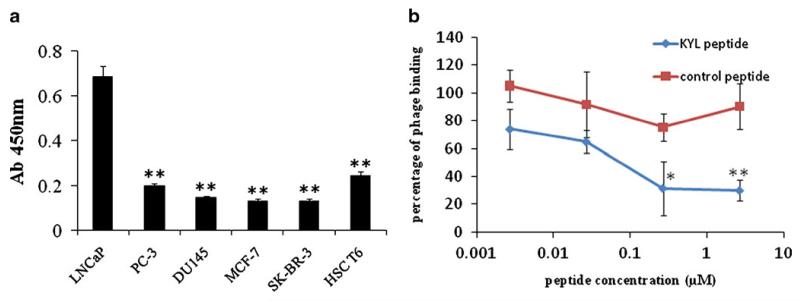
In order to evaluate the specificity of the KYL phage, we compared its binding affinity in six different cells lines, including three prostate cancer cell lines (LNCaP, PC-3, DU-145), two breast cancer cell lines (MCF-7, SK-BR-3), and a hepatic stellate cell line (HSC-T6) using cell phage ELISA. As shown in Fig. 3a, the KYL phage exhibited the highest binding to LNCaP cells in comparison to other cells. This result indicated that the KYL phage has a high binding affinity as well as a good specificity to LNCaP cells.
Fig. 3.
Binding affinity of the KYL phageon different cell lines. (a) Binding of the KYL phage on different cell lines was assayed by cell phage ELISA. The KYL phages (3×109 pfu per well) were used for this experiment. Results are represented as the mean±SD (n=3). (b) Competition of the synthetic KYL peptide with the binding of the KYL phage on LNCaP cells. Fifty thousand LNCaP cells per well were cultured and fixed with cold methanol-acetone (1:1). The cells were incubated with the synthetic KYL peptide or a scrambled peptide for 30 min, followed by washing and incubating with the KYL phages for 1 h. The bound phages were recovered and titered. Results are represented as the mean±SD (n=3). (*p<0.05, **p<0.01).
Next, we conducted a competitive inhibition study to determine whether the KYL phage binds to LNCaP cells via the specific interaction of the KYL peptide, which is expressed on the surface of filamentous phage, and LNCaP cells. LNCaP cells were pre-incubated with the synthetic KYL peptide (KYLAYPDSVHIW) or a scrambled peptide. After washing to remove the unbound peptides, the LNCaP cells were incubated with the KYL phage, and the bound phages were eluted and titered. As evident from Fig. 3b, the synthetic KYL peptide competed efficiently with the KYL phage for binding to LNCaP cells, and the competition is concentration dependant. Pre-incubation with the KYL peptide at 2.68 μM inhibited the binding affinity of the KYL phage to LNCaP cells by 70%. On the contrary, preincubation with the control peptide did not significantly inhibit the binding affinity of the KYL phage. This observation clearly indicated that the binding of the KYL phage to LNCaP cells is mediated by the KYL peptide expressed on the phage surface.
Binding of the KYL Peptide to LNCaP Cells
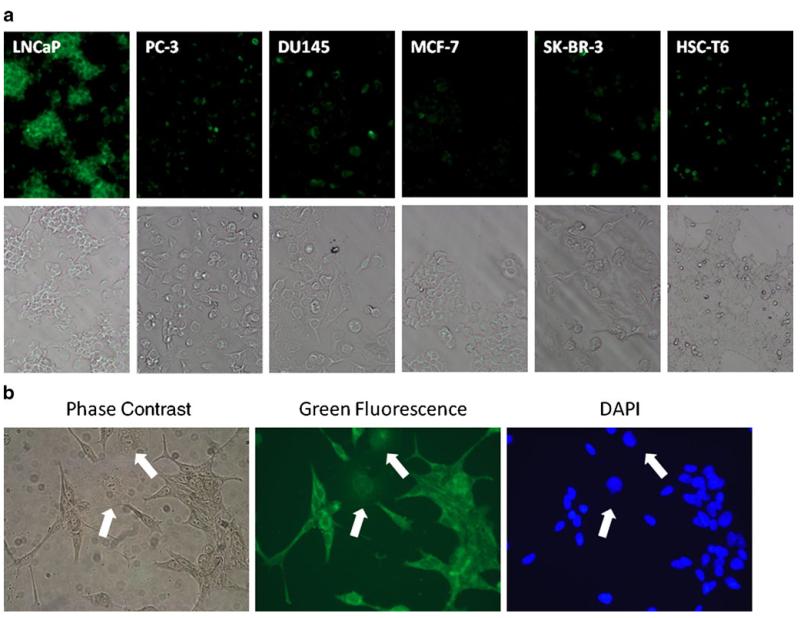
To determine whether the synthetic KYL peptide alone binds to LNCaP cells, the KYL peptide was labeled with fluorescein and then incubated with LNCaP cells and other cell lines including PC-3, DU145, MCF-7, SK-BR-3 and HSC-T6. As illustrated in Fig. 4a, a very intense fluorescence was observed on LNCaP cells, while negligible fluorescence was detected on other five cell lines. This result revealed that the synthetic KYL peptide alone has a significant higher binding on LNCaP cells in comparison to other cells tested in this study. We also co-cultured LNCaP and DU145 cells together and incubated them with the fluorescein-labeled KYL peptide. The KYL peptide only exhibits substantial staining on LNCaP cells rather than DU145 cells, suggesting the high specificity of the KYL peptide (Fig. 4b).
Fig. 4.
Binding of the KYL peptide on different cells lines. (a) Binding of the fluorescein- labeled KYL peptide on different cell lines. The fluorescein-labeled KYL peptide (10 μg/ml) was incubated with fixed cells at 4°C for 15 min. After washing, the fluorescence was examined using a Leica DMI3000 fluorescence microscope. (b) LNCaP cells and DU145 cells co-cultured in a Lab-TEK® 8-well chamber slide were fixed and stained with the fluorescein-labeled KYL peptide. The fluorescence was examined using a Leica DMI300 fluorescence microscope (white arrows indicate DU145 cells).
The KYL Peptide Enhances the Uptake and Apoptotic Effect of a Proapoptotic Peptide
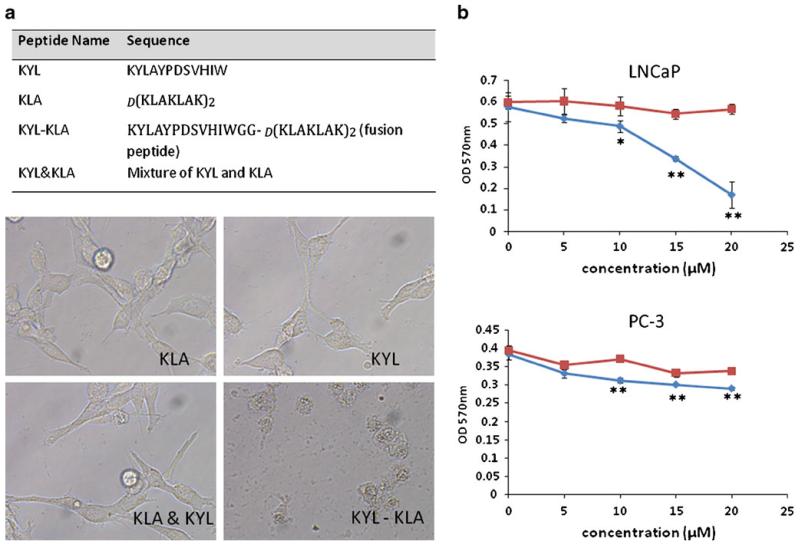
The final objective of this study is to identify a prostate cancer-specific peptide that can be applied in targeted drug delivery. Therefore, it is important to demonstrate whether the KYL peptide can be employed for cell-specific delivery of various cargos. For this purpose, a fusion peptide composed of the KYL peptide and a proapoptotic peptide d(KLAKLAK)2 were synthesized. The proapoptotic peptide is known to be capable of triggering mitochondrial disruption and inducing cell apoptosis and cell death (28). As Fig. 5a demonstrated, incubation of LNCaP cells with 20 μM KLA peptide, KYL peptide, or the mixture of KLA and KYL peptides did not exhibit any change in the cell morphology and viability. By contrast, significant cell death was observed in cells treated with the KYL-KLA fusion peptide, suggesting that the KYL peptide mediates a specific cellular uptake of the fusion protein in LNCaP cells to induce the cell death. Furthermore, we examined the effect of the KYL-KLA fusion peptide on the viability of LNCaP and PC-3 cells. The data presented in Fig. 5b indicated that the incubation of LNCaP cells with the KYL-KLA fusion peptide at 20 μM resulted in approximately 80% cell death in comparison to cells treated with the mixture of KYL and KLA peptides at the same concentration. However, the same effect of the KYL-KLA fusion peptide was not observed in PC-3 cells. These results further proved that the cell death effect of the fusion peptide in LNCaP cells is mediated by the LNCaP-specific KYL peptide, and cellular uptake of the fusion peptide in PC-3 cells is negligible.
Fig. 5.
Apoptotic effect of the KYL-KLA fusion peptide on LNCaP and PC-3 cells. (a) Sequences of the KYL peptide, KLA peptide, and KYL-KLA fusion peptide are listed. LNCaP cells were incubated with 20 μM of the KYL peptide, KLA peptide, KYL-KLA fusion peptide, or the mixture of KYL and KLA peptides for 24 h. Cell morphology was monitored with an inverted microscope. (b) LNCaP and PC-3 cells were incubated with the KYL-KLA fusion peptide and the mixture of KYL and KLA peptides at different concentrations for 24 h. Cell viability was determined by MTTassay. Results are represented as the mean±SD (n=3).
Although we did not assay the discrete binding affinity of the KYL peptide, the viability IC50 of the KYL-KLA fusion peptide on LNCaP cells is approximately 15 μM (Fig. 5b), which is similar to peptide ligands identified by other groups using the phage display technology (15,29).
Cellular Uptake of the KYL-KLA Fusion Peptide is Inhibited by Sodium Azide
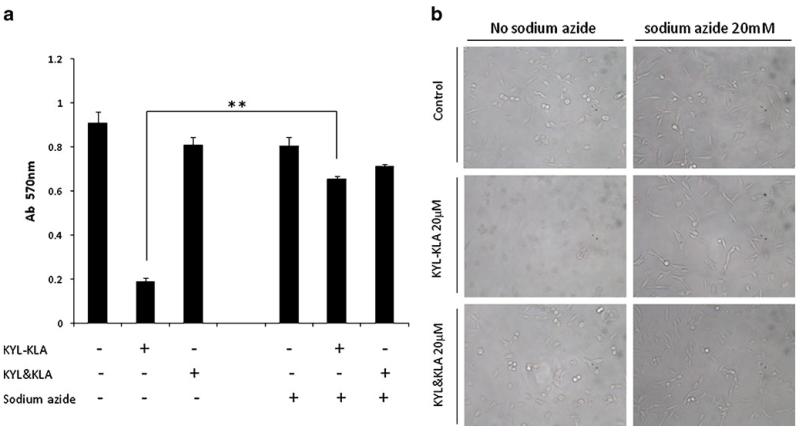
We next examined whether the cellular uptake is mediated by endocytosis. LNCaP and PC-3 cells were incubated with the KYL-KLA fusion peptide or the mixture of KYL and KLA peptides in the presence of 20 mM sodium azide, an ATPase inhibitor. As Fig. 6a showed, the KYL-KLA peptide induced cell death in the absence of sodium azide, but not in the presence of sodium azide. In comparison, sodium azide did not affect the apoptotic effect of the mixture of KYL and KLA peptides. Similar results were also observed in cell morphology (Fig. 6b). Collectively, these data suggested that the uptake of the KYL-KLA fusion peptide is mediated by endocytosis which is energy dependent.
Fig. 6.
The effect of sodium azid on the internalization of the KYL-KLA fusion peptide. (a) LNCaP cells were incubated with the KYL-KLA fusion peptide or the KYL/KLA mixture in the absence or presence of 20 mM sodium azide for 2 h. Cell viability was then determined by MTT assay. Results are represented as the mean±SD (n=3).(**p<0.01). (b) Morphology of the treated cells was monitored with an inverted microscope.
KYL Peptide Enhances the Uptake of the Protamine/siRNA Complex
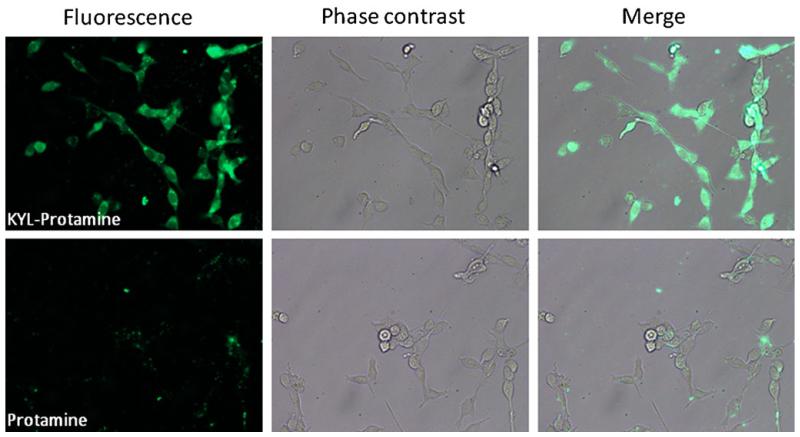
Finally, we explored the possibility of applying the KYL peptide in delivering siRNA into LNCaP cells. Protamine is a cationic peptide that has been widely used as a carrier for nucleic acid delivery (30-33). To determine whether the KYL peptide can enhance the cellular uptake of siRNA in LNCaP cells, protamine was conjugated to the KYL peptide via a disulfide bond (Fig. 7), which can be cleaved in the cytoplasm (34). A fluorescein-labeled siRNA was transfected by the protamine-KYL conjugate or protamine alone at a concentration of 200 nM. Five hours after the transfection, the cells were washed to remove the unbound siRNA. The fluorescence signal was then observed using a fluorescence microscope. As Fig. 8 illustrated, LNCaP cells incubated with the protamine-KYL/siRNA complex displayed strong and evenly distributed fluorescence in the cytoplasm, indicating a significant uptake of the fluorescein labeled siRNA. In contrast, negligible fluorescence was observed in the cells treated with the protamine/siRNA complex. This result clearly demonstrated that the KYL peptide can significantly enhance the cellular uptake of siRNA in LNCaP cells, suggesting a promising application of the KYL peptide in targeted drug delivery to prostate cancer cells.
Fig. 7.
Schematics of the KYL-protamine conjugation (a) and complexation of the KYL-protamine with siRNA (b).
Fig. 8.
KYL-conjugated protamine increases the uptake of siRNA in LNCaP cells. Two and a half microliters of the BLOCK-iT™ Fluorescent siRNA (20 μM) were mixed with 1.5 μg protamine (or the protamine-KYL conjugate) in 100 μl serum-free RPMI-1640 medium at room temperature for 10 min to form the complex. After washing cells with PBS, the siRNA complexes were added to each well at a final concentration of 200 nM. Five hours after the transfection, the fluorescence was examined using a Leica DMI3000 fluorescence microscope.
Evaluation of PSMA as the Potential Target of the KYL Peptide
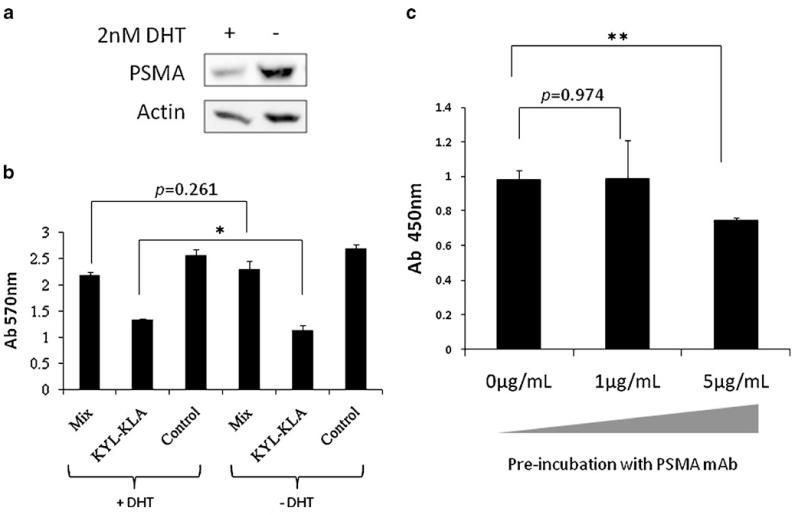
LNCaP cells are PSMA positive, and PSMA is highly expressed on most prostate cancers and the neovasculature of solid tumors in comparison to other normal tissues (24,35). Therefore, we initially speculated that PSMA could be the receptor that is responsible for the interaction with the KYL peptide. We examined this speculation by down-regulating PSMA with 5-α-dihydrotestosterone (DHT) or blocking PSMA with the anti-PSMA antibody. We incubated the apoptotic KYL-KLA fusion peptide (20 μM) with LNCaP cells that were pretreated with DHT, an androgen that decreases the PSMA expression on LNCaP cells (36). DHT treatment substantially inhibited the PSMA expression in LNCaP cells (Fig. 9a). However, there is only a minor increase in the viability of the cells treated with DHT in comparison to cells that were not treated with DHT (Fig. 9b). In another experiment, fixed LNCaP cells were pre-incubated with the anti-PSMA antibody (0, 1, 5 μg/ml) for 1 h and then incubated with 3×109 pfu KYL phages. Binding of the KYL phage was measured by cell phage ELISA. Anti-PSMA antibody showed a negligible competition effect at a low concentration (1 μg/ml) and approximately 25% competition at the concentration of 5 μg/ml (Fig. 9c). These results suggested that PSMA may not be the target for the KYL peptide.
Fig. 9.
Evaluation of PSMA as the potential target of the KYL peptide. (a) PSMA expression on DHT-treated LNCaPcells was tested by Western blot. (b) DHT-treated LNCaP cells were incubated with the KYL-KLA fusion peptide or the KYL/KLA mixture. Cell viability was then determined by MTT method. Results are represented as mean±SD (n=3). (c) The binding of KYL phage to LNCaP cells pre-incubated with 0, 1, 5 μg/mL PSMA antibody were assayed by cells phage ELISA. Results are represented as mean±SD (n=3). (*p<0.05, **p<0.01).
DISCUSSION
In this study, we identified a peptide ligand that specifically binds to prostate cancer LNCaP cells using a whole-cell biopanning procedure. The selected phage clone and its encoded peptide (KYL peptide) exhibited a high and specific binding to LNCaP cells. Moreover, the KYL peptide specifically delivered a proapoptotic peptide and a fluorescein-labeled siRNA to LNCaP cells.
Biopanning of phage display library can be performed on recombinant proteins, whole cells or living animals (also known as in vivo biopanning). Biopanning against a coated recombinant protein is easily controlled, and there is no interference from other impurities. However, the native structure and conformation of a recombinant protein may be disrupted during the coating process. As a result, the identified phage that specifically binds to the recombinant protein may not bind to the native protein on the cell surface. In the case of whole-cell biopanning, there are a great number of proteins, lipids, and carbohydrates on the cell surface that can interfere with the binding of phages to its target. Therefore, the whole-cell biopanning is more complicated and time consuming. However, the peptide identified using the whole-cell biopanning can efficiently bind to the target molecules on the cells. Thus, we used the whole-cell biopanning procedure to screen peptide ligand that can specifically bind to prostate cancer LNCaP cells.
LNCaP cell line is the most extensively used carcinoma cell model for prostate cancer studies (22,23). LNCaP cells were isolated from a biopsy of lymph node metastatic lesion of a human prostate adenocarcinoma. It expresses all the important prostatic markers including PSMA, PSA, AR, PAP and hK2 (22,23). Although LNCaP cells are androgen-sensitive, they gradually lose the androgen requirement for proliferation upon multiple passages, which mimics the progression of prostate cancer cells to an androgen-independent phase. Thus, LNCaP cell line is frequently used as a cell model to simulate the early stages of prostate cancer progression (37). In this study, LNCaP cells were used for the whole-cell biopanning so that we can identify a peptide that can bind to a significant proportion of prostate cancer cells.
We performed various studies, including cell ELISA, immunostaining, and competition assay, to prove that the KYL peptide specifically binds to LNCaP cells. In addition, we demonstrated that the KYL peptide can enhance the cellular uptake of therapeutic agents, including a proapoptotic peptide (Fig. 5) and a fluorescein-labeled siRNA (Fig. 8). The proapoptotic peptide is a cationic peptide that binds and disrupts mitochondrial membrane, leading to cell apoptosis and death. The proapoptotic peptide alone cannot enter eukaryotic cells except at a very high concentration (eg. 300 μM) (38). Thus, ether cell apoptosis or cell death are signs for the cellular uptake of the proapoptotic KLA peptide. As shown in Fig. 5, only the KYL-KLA fusion peptide induced cell death in LNCaP cells, but not the mixture of the KYL and KLA peptides, indicating that the KYL peptide mediates the cellular uptake of the proapototic KLA sequence in the fusion peptide. This result also excludes the possibility that the KYL peptide may just attach to the cell surface instead of internalizing into the cells. Moreover, apoptotic effect of the fusion peptide was not observed in PC-3 cells, suggesting the specificity of the KYL peptide to LNCaP cells.
RNA interference (RNAi) is a very promising approach for cancer therapy because it can potently silence a specific gene by siRNA of 21–23 nt in length (39,40). However, its therapeutic application is halted due to the lack of efficient delivery. Antibody-protamine fusion protein has been reported to deliver siRNA to cancer cells (32). In this study, we explored the possibility of using the KYL peptide as a ligand to increase the delivery of siRNA to LNCaP cells. The KYL peptide was conjugated to protamine which is used to condense siRNA. Our results demonstrated that the KYL peptide efficiently increases the uptake of a fluorescein-labeled siRNA in LNCaP cells (Fig. 8).
We initially speculated that PSMA could be the target for the KYL peptide because PSMA is highly expressed on LNCaP cells, and we used PSMA-negative PC-3 cells to preclean the non-specific-binding phages before the biopanning. However, our competition data did not support this speculation (Fig. 9). Either down-regulating (Fig. 9b) or blocking (Fig. 9c) PSMA only slightly decreased the binding affinity of the KYL peptide to LNCaP cells. Although statistical analysis revealed that this difference is significant (p<0.05), we still cautiously concluded that PSMA may not be the receptor for this KYL peptide. Nonetheless, we are planning to continue this work by using the proteomic approach to identify the receptor that accounts for the specific interaction with the KYL peptide.
In conclusion, we have successfully identified an LNCaP-specific peptide ligand (KYL peptide) using whole-cell biopanning. This peptide specifically delivers a proapoptotic peptide into LNCaP cells and triggers cell death. In addition, the KYL peptide is able to enhance the cellular uptake of siRNA in LNCaP cells. All these data suggest that the KYL peptide can be a promising ligand for targeted delivery of various therapeutic agents to prostate cancer LNCaP cells.
ACKNOWLEDGMENTS
This work was supported by awards from National Cancer Institute (NCI), NIH (1R21CA143683-01) and National Institute of Alcohol Abuse and Alcoholism (NIAAA), NIH (1R21AA017960-01A1). We also would like to express thanks for the financial support from a start-up package at the University of Missouri-Kansas City.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Mabjeesh NJ, Zhong H, Simons JW. Gene therapy of prostate cancer: current and future directions. Endocr-Relat Cancer. 2002;9:115–39. doi: 10.1677/erc.0.0090115. [DOI] [PubMed] [Google Scholar]
- 3.Tai W, Mahato R, Cheng K. The role of HER2 in cancer therapy and targeted drug delivery. J Control Release. 146:264–75. doi: 10.1016/j.jconrel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang DK, Lin CT, Wu CH, Wu HC. A novel peptide enhances therapeutic efficacy of liposomal anti-cancer drugs in mice models of human lung cancer. PLoS ONE. 2009;4:e4171. doi: 10.1371/journal.pone.0004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoneda Y, Steiniger SC, Capkova K, Mee JM, Liu Y, Kaufmann GF, et al. A cell-penetrating peptidic GRP78 ligand for tumor cell-specific prodrug therapy. Bioorg Med Chem Lett. 2008;18:1632–6. doi: 10.1016/j.bmcl.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesarone G, Edupuganti OP, Chen CP, Wickstrom E. Insulin receptor substrate 1 knockdown in human MCF7 ER+ breast cancer cells by nuclease-resistant IRS1 siRNA conjugated to a disulfide-bridged D-peptide analogue of insulin-like growth factor 1. Bioconjug Chem. 2007;18:1831–40. doi: 10.1021/bc070135v. [DOI] [PubMed] [Google Scholar]
- 7.Deutscher SL, Figueroa SD, Kumar SR. In-labeled KCCYSL peptide as an imaging probe for ErbB-2-expressing ovarian carcinomas. J Labelled Comp Radiopharm. 2009;52:583–90. doi: 10.1002/jlcr.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Xie J, Chen X. Peptide-based probes for targeted molecular imaging. Biochemistry. 49:1364–76. doi: 10.1021/bi901135x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aggarwal S, Singh P, Topaloglu O, Isaacs JT, Denmeade SR. A dimeric peptide that binds selectively to prostate-specific membrane antigen and inhibits its enzymatic activity. Cancer Res. 2006;66:9171–7. doi: 10.1158/0008-5472.CAN-06-1520. [DOI] [PubMed] [Google Scholar]
- 10.Burg MA, Pasqualini R, Arap W, Ruoslahti E, Stallcup WB. NG2 proteoglycan-binding peptides target tumor neovasculature. Cancer Res. 1999;59:2869–74. [PubMed] [Google Scholar]
- 11.Yang W, Luo D, Wang S, Wang R, Chen R, Liu Y, et al. TMTP1, a novel tumor-homing peptide specifically targeting metastasis. Clin Cancer Res. 2008;14:5494–502. doi: 10.1158/1078-0432.CCR-08-0233. [DOI] [PubMed] [Google Scholar]
- 12.Wu C, Lo SL, Boulaire J, Hong ML, Beh HM, Leung DS, et al. A peptide-based carrier for intracellular delivery of proteins into malignant glial cells in vitro. J Control Release. 2008;130:140–5. doi: 10.1016/j.jconrel.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y, Lillo AM, Steiniger SC, Liu Y, Ballatore C, Anichini A, et al. Targeting heat shock proteins on cancer cells: selection, characterization, and cell-penetrating properties of a peptidic GRP78 ligand. Biochemistry. 2006;45:9434–44. doi: 10.1021/bi060264j. [DOI] [PubMed] [Google Scholar]
- 14.Jager S, Jahnke A, Wilmes T, Adebahr S, Vogtle FN, Delima-Hahn E, et al. Leukemia-targeting ligands isolated from phage-display peptide libraries. Leukemia. 2007;21:411–20. doi: 10.1038/sj.leu.2404548. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura S, Takahashi S, Kamikatahira H, Kuroki Y, Jaalouk DE, O’Brien S, et al. Combinatorial targeting of the macropinocytotic pathway in leukemia and lymphoma cells. J Biol Chem. 2008;283:11752–62. doi: 10.1074/jbc.M708849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joyce JA, Laakkonen P, Bernasconi M, Bergers G, Ruoslahti E, Hanahan D. Stage-specific vascular markers revealed by phage display in a mouse model of pancreatic islet tumorigenesis. Cancer Cell. 2003;4:393–403. doi: 10.1016/s1535-6108(03)00271-x. [DOI] [PubMed] [Google Scholar]
- 17.Newton JR, Kelly KA, Mahmood U, Weissleder R, Deutscher SL. In vivo selection of phage for the optical imaging of PC-3 human prostate carcinoma in mice. Neoplasia. 2006;8:772–80. doi: 10.1593/neo.06331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shuklaand GS, Krag DN. Phage display selection for cell-specific ligands: development of a screening procedure suitable for small tumor specimens. J Drug Target. 2005;13:7–18. doi: 10.1080/10611860400020464. [DOI] [PubMed] [Google Scholar]
- 19.Smithand GP, Petrenko VA. Phage display. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 20.Pero SC, Shukla GS, Armstrong AL, Peterson D, Fuller SP, Godin K, et al. Identification of a small peptide that inhibits the phosphorylation of ErbB2 and proliferation of ErbB2 over-expressing breast cancer cells. Int J Cancer. 2004;111:951–60. doi: 10.1002/ijc.20306. [DOI] [PubMed] [Google Scholar]
- 21.Karasseva NG, Glinsky VV, Chen NX, Komatireddy R, Quinn TP. Identification and characterization of peptides that bind human ErbB-2 selected from a bacteriophage display library. J Protein Chem. 2002;21:287–96. doi: 10.1023/a:1019749504418. [DOI] [PubMed] [Google Scholar]
- 22.Sobeland RE, Sadar MD. Cell lines used in prostate cancer research: a compendium of old and new lines-part 2. J Urol. 2005;173:360–72. doi: 10.1097/01.ju.0000149989.01263.dc. [DOI] [PubMed] [Google Scholar]
- 23.Sobeland RE, Sadar MD. Cell lines used in prostate cancer research: a compendium of old and new lines-part 1. J Urol. 2005;173:342–59. doi: 10.1097/01.ju.0000141580.30910.57. [DOI] [PubMed] [Google Scholar]
- 24.Israeli RS, Powell CT, Corr JG, Fair WR, Heston WD. Expression of the prostate-specific membrane antigen. Cancer Res. 1994;54:1807–11. [PubMed] [Google Scholar]
- 25.Giordano RJ, Cardo-Vila M, Lahdenranta J, Pasqualini R, Arap W. Biopanning and rapid analysis of selective interactive ligands. Nat Med. 2001;7:1249–53. doi: 10.1038/nm1101-1249. [DOI] [PubMed] [Google Scholar]
- 26.Liang S, Lin T, Ding J, Pan Y, Dang D, Guo C, et al. Screening and identification of vascular-endothelial-cell-specific binding peptide in gastric cancer. J Mol Med. 2006;84:764–73. doi: 10.1007/s00109-006-0064-2. [DOI] [PubMed] [Google Scholar]
- 27.Bruccoleri RE, Haber E, Novotny J. Structure of antibody hypervariable loops reproduced by a conformational search algorithm. Nature. 1988;335:564–8. doi: 10.1038/335564a0. [DOI] [PubMed] [Google Scholar]
- 28.Mai JC, Mi Z, Kim SH, Ng B, Robbins PD. A proapoptotic peptide for the treatment of solid tumors. Cancer Res. 2001;61:7709–12. [PubMed] [Google Scholar]
- 29.Arap MA, Lahdenranta J, Mintz PJ, Hajitou A, Sarkis AS, Arap W, et al. Cell surface expression of the stress response chaperone GRP78 enables tumor targeting by circulating ligands. Cancer Cell. 2004;6:275–84. doi: 10.1016/j.ccr.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Bruckheimer E, Harvie P, Orthel J, Dutzar B, Furstoss K, Mebel E, et al. In vivo efficacy of folate-targeted lipid-protamine-DNA (LPD-PEG-Folate) complexes in an immunocompetent syngeneic model for breast adenocarcinoma. Cancer Gene Ther. 2004;11:128–34. doi: 10.1038/sj.cgt.7700662. [DOI] [PubMed] [Google Scholar]
- 31.Zhang JS, Li S, Huang L. Cationic liposome-protamine-DNA complexes for gene delivery. Meth Enzymol. 2003;373:332–42. doi: 10.1016/S0076-6879(03)73021-3. [DOI] [PubMed] [Google Scholar]
- 32.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–17. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 33.Dinauer N, Lochmann D, Demirhan I, Bouazzaoui A, Zimmer A, Chandra A, et al. Intracellular tracking of protamine/antisense oligonucleotide nanoparticles and their inhibitory effect on HIV-1 transactivation. J Control Release. 2004;96:497–507. doi: 10.1016/j.jconrel.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 34.Mannervik B, Axelsson K, Sundewall AC, Holmgren A. Relative contributions of thioltransferase-and thioredoxin-dependent systems in reduction of low-molecular-mass and protein disulphides. Biochem J. 1983;213:519–23. doi: 10.1042/bj2130519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silver DA, Pellicer I, Fair WR, Heston WD, Cordon-Cardo C. Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin Cancer Res. 1997;3:81–5. [PubMed] [Google Scholar]
- 36.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–15. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 37.Dozmorov MG, Hurst RE, Culkin DJ, Kropp BP, Frank MB, Osban J, et al. Unique patterns of molecular profiling between human prostate cancer LNCaP and PC-3 cells. Prostate. 2009;69:1077–90. doi: 10.1002/pros.20960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellerby HM, Arap W, Ellerby LM, Kain R, Andrusiak R, Rio GD, et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat Med. 1999;5:1032–8. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 39.Sithanandam G, Fornwald LW, Fields J, Anderson LM. Inactivation of ErbB3 by siRNA promotes apoptosis and attenuates growth and invasiveness of human lung adenocarcinoma cell line A549. Oncogene. 2005;24:1847–59. doi: 10.1038/sj.onc.1208381. [DOI] [PubMed] [Google Scholar]
- 40.Liu TG, Yin JQ, Shang BY, Min Z, He HW, Jiang JM, et al. Silencing of hdm2 oncogene by siRNA inhibits p53-dependent human breast cancer. Cancer Gene Ther. 2004;11:748–56. doi: 10.1038/sj.cgt.7700753. [DOI] [PubMed] [Google Scholar]