Abstract
Small heat shock proteins (sHSPs) are a family of ATP-independent molecular chaperones which prevent cellular protein aggregation by binding to misfolded proteins. sHSPs form large oligomers that undergo drastic rearrangement/dissociation in order to execute their chaperone activity in protecting substrates from stress. Substrate-binding sites on sHSPs have been predominantly mapped on their intrinsically disordered N-terminal arms. This region is highly variable in sequence and length across species, and has been implicated in both oligomer formation and in mediating chaperone activity. Here, we present our results on the functional and structural characterization of five sHSPs in rice, each differing in their subcellular localisation, viz., cytoplasm, nucleus, chloroplast, mitochondria and peroxisome. We performed activity assays and dynamic light scattering studies to highlight differences in the chaperone activity and quaternary assembly of sHSPs targeted to various organelles. By cloning constructs that differ in the length and sequence of the tag in the N-terminal region, we have probed the sensitivity of sHSP oligomer assembly and chaperone activity to the length and amino acid composition of the N-terminus. In particular, we have shown that the incorporation of an N-terminal tag has significant consequences on sHSP quaternary structure.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-015-0570-7) contains supplementary material, which is available to authorized users.
Keywords: Small heat shock protein, Oryza sativa, Molecular chaperones, Stress response
Introduction
Small heat shock proteins (sHSPs) are a ubiquitous family of ATP-independent chaperones that constitute the first line of defence against the detrimental effects of cellular stress conditions (Hilton et al. 2013). These proteins typically have monomeric masses between 12 and 40 kDa. A characteristic feature of sHSPs is their propensity to adopt a wide range of oligomeric states, which may contain anywhere from 2 to 48 protomers (Basha et al. 2012). Under stress conditions, the oligomers dissociate and/or undergo drastic conformational changes to facilitate sHSP binding to misfolded substrate proteins in the cell. These interactions prevent the substrate proteins from aggregating irreversibly. When physiological conditions are restored, the substrates are transferred from the sHSP–substrate complexes to ATP-dependent heat shock proteins like Hsp70 for refolding (Veinger et al. 1998).
Archaeal and bacterial genomes usually encode one or two sHSPs, whereas eukaryotic genomes tend to have several sHSP genes. Plants express many more sHSPs than other eukaryotes (Kriehuber et al. 2010). For instance, the human genome contains ten sHSP genes, whereas the California poplar genome contains 36 sHSPs (Waters et al. 2008). In addition, plant sHSPs exhibit diversity in sequence and in the cellular location where they function (Waters et al. 1996). Unlike in other organisms, plant sHSPs can be organised into 12 distinct subfamilies based on their cellular localisation and chaperone function. Of these, seven subfamilies are cytoplasm/nuclear localised (CI–CVII) and five sHSP subfamilies localize to organelles like the chloroplast, endoplasmic reticulum, peroxisome and mitochondria (Siddique et al. 2008).
Among plant sHSPs, the sHSPs of rice are of particular interest, since rice is an important food crop, constituting the staple diet of more than half the world’s population. It is grown mainly in tropical and subtropical areas, and high temperature during the growing period and reproductive stage adversely affects fertility, pollen development and grain yield (Matsui et al. 2000; Peng et al. 2004). In response to different environmental conditions like heat, cold, drought, anoxia and salt stress, rice plants express 23 sHSPs (Sarkar et al. 2009). Of these, members of the CI subfamily, like Hsp16.9A (Chen et al. 2014), Hsp18.0 (Guan et al. 2004), Hsp18.0 of the CII subfamily (Chang et al. 2007) and the chloroplast-localised Hsp26.7 (Lee et al. 2000) have been investigated. Overexpression of rice Hsp16.9 (CI subfamily) in Escherichia coli conferred thermo-tolerance to the bacteria at temperatures that were otherwise considered lethal (Yeh et al. 1997). Transgenic rice lines that overexpressed Hsp17.7 (CI subfamily) displayed higher drought tolerance than the untransformed plants (Sato and Yokoya, 2008). In 2009, the expression profiles of all 23 rice sHSPs under vegetative, developmental and stress conditions were reported (Sarkar et al. 2009). Further, a genome-wide microarray-based gene expression analysis involving 25 stages of vegetative and reproductive development was performed in three rice cultivars (Ouyang et al. 2009). Data from both studies highlight the diversity in the expression of rice sHSPs and serve as a starting point for the identification of candidate genes for functional studies of the rice stress response. However, we do not yet have a complete understanding of the organelle-specific functional roles of the rice sHSPs.
Our understanding of the sHSP family is further hampered by the insufficiency in structural information. To date, there are only three crystal structures of full-length oligomers at atomic resolution (Basha et al. 2012). Of these, wheat Hsp16.9 is the only plant sHSP whose structure is known (van Montfort et al. 2001). From all available structures, it is seen that all sHSPs are defined by a core region, known as the α-crystallin domain (ACD), which is flanked on either side by the N- and C-terminal extensions. The N-terminal region displays high sequence variability and is intrinsically disordered in most structures (Kim et al. 1998; van Montfort et al. 2001; Hilario et al. 2011). The dissociation or rearrangement of the sHSP oligomer during stress exposes clusters of hydrophobic residues, located predominantly in this flexible region (Jaya et al. 2009). These residues bind to hydrophobic patches on other proteins and are central to the formation of oligomers as well as sHSP–substrate complexes. As a consequence, truncations of the N-terminal arm affect both the oligomer assembly and chaperone activity of many sHSPs (Studer et al. 2002; Sun and MacRae 2005). A study of the sHSP AgsA from Salmonella enterica demonstrated that truncations of different lengths at its termini resulted in drastic differences in the oligomeric state, which ranged from dimers to 22-mers. Furthermore, some mutants were inactive as chaperones towards some substrates, whereas others showed higher activity than the wild-type protein (Tomoyasu et al. 2010).
In order to gain further insight into the plant stress response, we undertook the structural and functional characterization of rice sHSPs. To study the functional diversification of the different plant sHSP subfamilies, we chose sHSPs targeted to different organelles, viz., cytoplasm, nucleus, chloroplast, mitochondria and peroxisome. Finally, to probe the role played by the dynamic and flexible N-terminal region in mediating chaperone activity and oligomer assembly, we cloned different constructs of these sHSPs, each differing in the length of the tag in the N-terminus.
Materials and methods
Cloning of rice sHSPs
Five rice sHSPs belonging to different subfamilies were identified from the Knowledge-based Oryza Molecular biological Encyclopaedia (KOME) database. They were Hsp16 (peroxisome targeted), Hsp16.9 (cytoplasmic class I), Hsp18.6 (cytoplasmic/nuclear class III), Hsp24 (mitochondria) and Hsp26.7 (chloroplast). Full-length complementary DNAs (cDNAs) of all five rice sHSPs were purchased from the National Institute of Agrobiological Sciences, Japan. The web server SIGNALP (Petersen et al. 2011) was used to identify the signal peptide of mitochondria-targeted Hsp24. The chloroplast transit peptide of Hsp26.7 was identified by aligning its sequence with Hsp21 of Arabidopsis and pea. The primers listed in Table 1 were used to amplify the DNA sequences of the sHSP domains (without the signal peptides) on the cDNAs. Following PCR amplification, the amplified genes and their respective vectors were digested by restriction enzymes and the genes were subsequently ligated into the vectors. Constructs bearing hexa-histidine tags at their N-termini incorporated additional amino acids from the vectors at their N-termini (Table 1). All constructs expressed entire sHSP domains, including the intact N- and C-terminal regions. They differed only in the residues incorporated from the vector, which preceded the sHSP domains at their N-termini. The sequences of all the constructs were confirmed by DNA sequencing.
Table 1.
Primers used in cloning of rice sHSPs
| sHSP and KOME accession number | Vector and construct name | Additional residues from vector | Primersa |
|---|---|---|---|
| Peroxisome-Hsp16 AK105317 |
pET-28a(+) Peroxisome-Hsp16-tag |
23 | F: AGACCATGGCGGACCTCTTCTTC R: GCGAAGCTTTTATCAGAGCTTGCTGGAGAC |
| Cytoplasm-Hsp16.9 AK121025 |
pET-28a(+) Cytoplasm-Hsp16.9-tag |
23 | F: ATAGCTAGCATGTCGCTGGTGAG R: GCGGATCCTTAGCCAGAAATCTC |
| pET-21b Cytoplasm-Hsp16.9-tagless |
2 | F: GCTCATATGTCGCTGGTGAGGC R: GCGGATCCTTAGCCAGAAATCTC |
|
| Nuclear-Hsp18.6 AK119261 |
pET-28a(+) Nuclear-Hsp18.6-tagA |
23 | F: GGAGCTAGCATGACGGAGCTGTTC R: ATTGGATCCTCAGGCGATGGTGAC |
| pPROEX HTa Nuclear-Hsp18.6-tagB |
24 | F: ATCCATGGCAACGGAGCTGTTCGAC R: GCGGAATTCTCAGGCGATGGTGAC |
|
| Hsp24 AK105464 |
pPROEX HTa Mitochondria-Hsp24 |
24 | F: ATACCATGGGCGGGTCCCGTGCA R: GCCGAATTCCTACTCGACGTTGACC |
| Hsp26.7 AK063618 |
pRSET C Chloroplast-Hsp26.7-Tag |
14 | F: CGAGCTAGCCAGGAGAACAGGGACAAC R: ATTGGATCCCTACTGGACCTGCACGTC |
| pRSET A Chloroplast-Hsp26.7-tagless |
1 | F: CGCCATATGCAGGAGAACAGGGACAAC R: ATTGGATCCCTACTGGACCTGCACGTC |
F forward, R reverse primer
aRestriction sites are underlined
Cell growth and protein expression
Each clone was transformed into E. coli BL21 (DE3) cells and grown on solid Luria–Bertani (LB) agar media containing the appropriate antibiotic (34 μg/ml of kanamycin for peroxisome-Hsp16-Tag, cytoplasm-Hsp16.9-tag, nuclear-Hsp18.6-tagA; 100 μg/ml ampicillin for the other constructs). A single colony of the transformed BL21 was used to inoculate 50 ml of primary LB media, which was subsequently grown overnight. One litre of secondary LB media was inoculated with 1 % of the overnight primary culture and grown at 37 °C until optical density at 600 nm reached 0.6. In all expression systems, the genes for T7 RNA polymerase and the sHSP were under the control of the lac operator. Thus, expression of sHSPs was induced by the addition of 0.7 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and the culture was further grown at 37 °C for 5 h. The culture was centrifuged to separate the cells from the media, and the pellet obtained thus was stored at −20 °C.
Protein purification
Hexa-histidine-tagged constructs
Constructs with a hexa-histidine tag at the N-terminus (peroxisome-Hsp16-tag, cytoplasm-Hsp16.9-tag, nuclear-Hsp18.6-tagA, nuclear-Hsp18.6-tagB, mitochondria-Hsp24-tag and chloroplast-Hsp26.7-tag) were purified through immobilized metal affinity chromatography. The cell pellet was resuspended in lysis buffer containing 20 mM tris and 300 mM NaCl at pH 7.5. Cells were lysed by sonication and were centrifuged to separate the cell debris from the soluble fraction. The supernatant was loaded onto Nickel-NTA resin that had been pre-equilibrated with lysis buffer. The resin was washed with lysis buffer containing imidazole. The concentration of imidazole was increased stepwise until the required protein eluted. The presence of the protein and its purity were confirmed through sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) (Supplementary Fig. S1).
The construct mitochondria-Hsp24-tag did not bind to the Ni-NTA resin. BL21 cells overexpressing mitochondria-Hsp24-tag were lysed in buffer containing 50 mM tris, 300 mM NaCl and 0.1 % Triton X-100, at pH 7.5, and the lysate was centrifuged. Four Molar urea was added to the supernatant and the mixture was stirred for 30 min. This mixture was applied onto a pre-equilibrated Ni-NTA column and purification proceeded as described previously. We confirmed through circular dichroism (CD) spectroscopy that the protein was folded following elution.
Tagless constructs
Constructs lacking an affinity tag were purified through anion exchange chromatography. The lysis buffer contained 20 mM bis-tris, 20 mM NaCl, 1 mM DTT and 1 mM EDTA at pH 6.5. The soluble cell fraction obtained after lysis was loaded onto a Q-Sepharose anion exchange column that had been pre-equilibrated with lysis buffer. The resin was then subjected to an NaCl concentration gradient from 0 to 400 mM. The purity of the eluted proteins was confirmed through SDS-PAGE (Supplementary Fig. S1).
Size exclusion chromatography
The elution fractions containing the sHSP of interest were pooled together and concentrated by centrifugation using an Amicon Ultra-15 Centrifugal Filter Unit (Merck Millipore) having a molecular weight cut-off of 10 kDa. The concentrated proteins were subjected to size exclusion chromatography through a Sephacryl S-500 column (GE Healthcare Life Sciences) with buffer containing 20 mM tris and 300 mM NaCl, pH 7.5. The protein fractions were assessed for purity through SDS-PAGE, and the mass of the purified protein was measured by matrix-assisted laser desorption/ionization time-of-flight (MALDI/TOF) spectrometry.
Cleavage by Tobacco Etch Virus protease
The construct mitochondria-Hsp24-tag was cloned in the vector pPROEX HTa and contained a Tobacco Etch Virus (TEV) protease site two residues upstream of the sHSP domain. Following elution from the Ni-NTA column, the full-length protein was desalted by a Hitrap desalting column (GE Healthcare Life Sciences) and eluted in buffer containing 50 mM tris, 50 mM NaCl, 0.5 mM DTT and 0.5 mM EDTA, pH 8.0. The mitochondria-Hsp24-tag was incubated with TEV protease (1:10, mass ratio TEV:sHSP) at 20 °C for 24 h. Cleavage of the tag was confirmed by SDS-PAGE (Supplementary Fig. S2). The reaction mixture was subjected to size exclusion chromatography through a Superose 6 10/300 column (GE Healthcare Life Sciences) to separate the different protein components.
Biophysical characterization of sHSPs
The masses of the purified proteins were measured by MALDI/TOF spectrometry recorded in the positive ion mode after the evaporation of the solvent, and analysed by Bruker Daltonics Flex Control software. The far-UV CD spectra of the sHSPs were recorded on a Jasco J-715C spectropolarimeter at a scan rate of 50 nm/min, response time of 4 s and bandwidth of 2 nm. Dynamic light scattering (DLS) studies performed on a DynaPro dynamic light scattering systems machine using the Dynamics v6 software were used to obtain hydrodynamic radii and polydispersity of the sHSPs from the autocorrelation curves.
Chaperone activity assay
sHSPs bind to misfolded proteins and have been shown to prevent aggregation of thermally/chemically denatured substrates. The chaperone activity of the rice sHSPs was assayed using the restriction enzyme NdeI as a substrate (Lini et al. 2008). Reaction mixtures consisting of 2 units of NdeI (New England Biolabs), its commercially available buffer and 1 μg sHSP were incubated for 90 min at temperatures ranging from 37 to 55 °C. As a negative control, reaction mixtures lacking the sHSP were also incubated at the same temperatures. Following incubation, 200 ng of undigested plasmid DNA, pET-28a(+), was added to each reaction mixture and incubated at 37 °C for 5 h. All samples were then run on a 0.8 % agarose gel to determine if the restriction enzyme had digested the plasmid. The pET-28a(+) plasmid harbours an NdeI restriction site, and incubation with NdeI at 37 °C results in the digestion of the vector. An uncut, circular plasmid migrates faster (and hence further down the gel) than a cut linear plasmid on an agarose gel under the influence of an electric field. If NdeI is fully functional, restriction digestion yields a linear plasmid. However, if NdeI was thermally inactivated during the initial incubation step, it would be unable to digest the plasmid and the uncut vector would be visible in the agarose gel.
Results and discussions
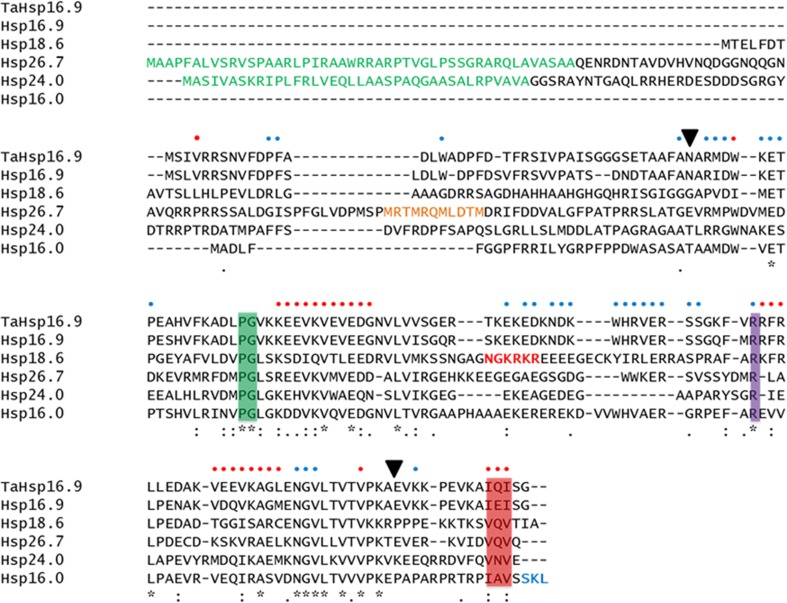
All five rice sHSPs (without the N-terminal signal peptides) were cloned, expressed in E. coli, purified and characterized. A multiple sequence alignment of all five sHSPs with wheat Hsp16.9 displayed the sequence conservation of the ACDs and variations in the lengths and sequences of the N-termini of sHSPs belonging to different subfamilies (Fig. 1). All five sHSPs contained the proline-glycine doublet which is highly conserved in all non-animal sHSPs and plays an important direct role in the subunit–subunit interactions (Fu and Chang, 2006). An arginine residue in the ACD which is conserved in sHSPs across all phyla was present in all rice sHSPs studied (van den et al. 1999). We also identified the C-terminal IXI motifs which mediate higher-order oligomerization and access to substrate-binding regions, through conserved interactions with the ACD (Hilton et al. 2013).
Fig. 1.
Multiple sequence alignment of wheat Hsp16.9 (TaHsp16.9), rice Hsp16.9, Hsp18.6, Hsp26.7, Hsp24 and peroxisome-Hsp16. Chloroplast transit peptide of Hsp26.7 and mitochondria signal peptide of Hsp24 are shown in green, nuclear localisation signal of Hsp18.6 in red, peroxisome-targeting signal of peroxisome-Hsp16 in blue and methionine-rich motif of Hsp26.7 in orange. Conserved residues/motifs are shown in coloured boxes: green—PG doublet, purple—conserved arginine, pink—IXI motif. Black arrowheads delineate the ACD of TaHsp16.9. Residues of wheat Hsp16.9 which mediate intra-dimer (blue dot) and inter-dimer (red dot) interactions are indicated
Hsp16, peroxisome
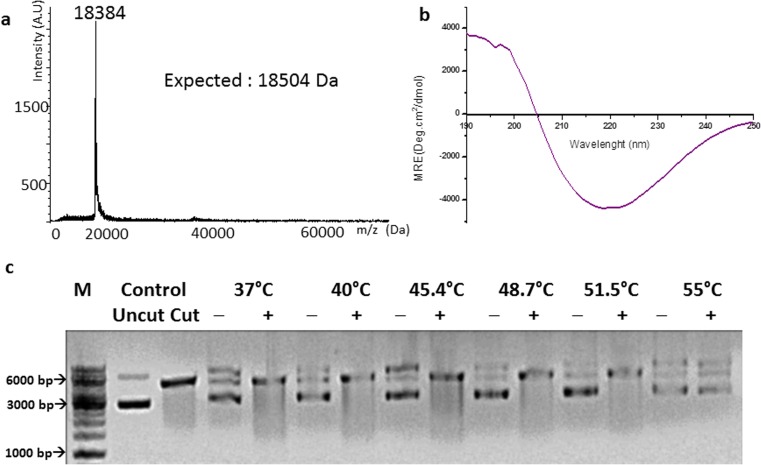
A C-terminal peroxisome-targeting amino acid triad (SKL) is present in rice Hsp16. The triad was retained when the gene was cloned into pET-28a(+). The expressed protein incorporating 23 additional residues at its N-terminus was purified. A MALDI/TOF spectrum confirmed the protein’s mass (Fig. 2a) and its CD spectrum exhibited the characteristic β-sheet curve (Fig. 2b). Peroxisome-Hsp16-tag was active as a chaperone, protecting NdeI up to a temperature of 51.5 °C (Fig. 2c). DLS studies showed that it had a hydrodynamic radius of 12.4 nm.
Fig. 2.
a MALDI/TOF spectrum of peroxisome-Hsp16-tag; b Far-UV CD spectrum of peroxisome-Hsp16-tag; c thermo-protection of NdeI by peroxisome-Hsp16-tag: NdeI incubated at each temperature with (+) and without (−) sHSP. Control migration of closed circular pET 28a(+) (uncut) and vector digested by NdeI (cut), M marker, bp base pair
Hsp15.7 in Arabidopsis is the homologue of rice Hsp16. The former was shown to be targeted to the peroxisome and strongly induced by heat and oxidative stress (Ma et al. 2006). We demonstrated the in vitro chaperone activity of rice Hsp16.
Hsp16.9, cytoplasmic class I
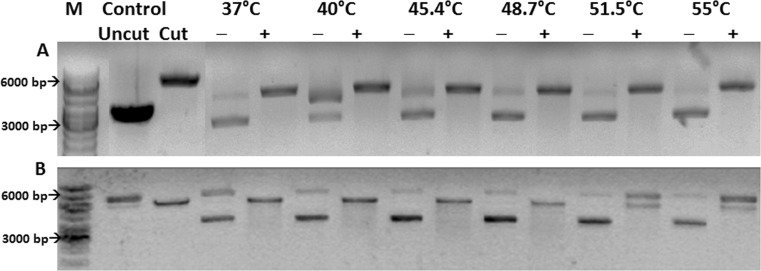
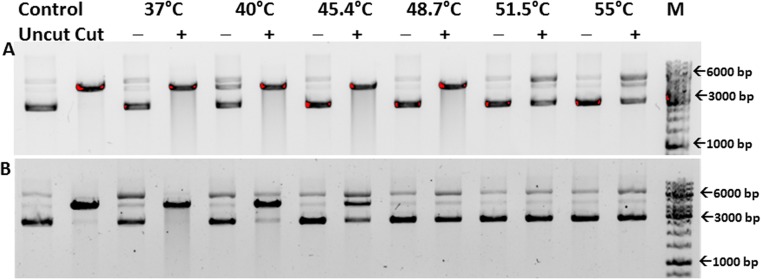
The hexa-histidine-tagged construct cytoplasm-Hsp16.9-tag incorporates 23 additional amino acids from the vector at its N-terminus. Cytoplasm-Hsp16.9-tag was shown to be active as a chaperone and could protect restriction enzyme NdeI from thermal stress up to a temperature of 55 °C (Fig. 3(A)). The untagged construct, cytoplasm-Hsp16.9-tagless containing only two additional amino acids at its N-terminus, was purified, and DLS studies indicated a hydrodynamic radius of 5.3 nm. This construct could protect NdeI from heat-induced denaturation completely up to a temperature of 48.7 °C and partially up to 55 °C (Fig. 3(B)).
Fig. 3.
Thermo-protection of NdeI by cytoplasm-Hsp16.9-tag (panel A) and cytoplasm-Hsp16.9-tagless (panel B): NdeI incubated at each temperature with (+) and without (−) sHSP. Control: migration of closed circular pET 28a(+) (uncut) and vector digested by NdeI (cut), M: marker, bp: base pair
The cytoplasm-localised rice Hsp16.9 shares 81.7 % sequence identity with wheat Hsp16.9. The latter forms a double-disc dodecamer, with an outer radius of 4.7 nm (van Montfort et al. 2001). The sequence alignment of both sHSPs (Fig. 1) indicates that residues participating in intra- and inter-dimer interactions are highly conserved, and hence, it is most likely that both sHSPs adopt similar oligomeric structures. Six of the 12 N-termini of wheat Hsp16.9 are involved in forming oligomeric interfaces in the interior of the dodecamer. The dimensions of the assembly indicate that the other six unstructured N-terminal arms are tightly packed within the dodecamer. Substrate-binding regions in the buried N-termini become exposed during the temperature-dependent dissociation of wheat Hsp16.9 into dimers. In a recent computational study, the differences in the dynamics of the flexible N-terminal arms of dimers of wheat Hsp16.9 and pea Hsp18.1 have been correlated with the differences in their chaperoning efficiency (Patel et al. 2014). Thus, the N-terminal region is directly implicated in forming oligomers and sHSP–substrate complexes. Among the two rice Hsp16.9 constructs, cytoplasm-Hsp16.9-tag has a radius almost double that of its close homologue in wheat, whereas the radius of cytoplasm-Hsp16.9-tagless is comparable to wheat Hsp16.9. It is thus likely that the 23 additional residues in the N-terminus of cytoplasm-Hsp16.9-tag interfere with the formation of a tight oligomer akin to the dodecamer of wheat Hsp16.9. Both constructs show differences in their thermo-protection activity towards NdeI. Previous studies on two mutants of αA-crystallin have shown that mutations that increase the hydrodynamic radius of the assembly tend to increase chaperone activity as well (Nagaraj et al. 2012). This effect was attributed to increased accessibility of chaperone sites in the mutants (Pasta et al. 2004).
Hsp18.6, cytoplasmic/nuclear class III
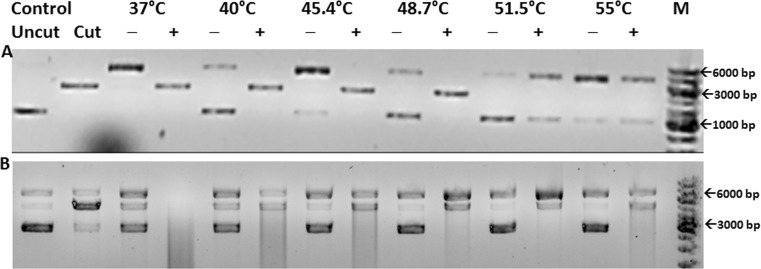
Rice Hsp18.6 was cloned in pET-28a(+) vector (construct nuclear-Hsp18.6-tagA) and, when purified, yielded well-folded protein with a hydrodynamic radius of 14.1 nm. Nuclear-Hsp18.6-tagA was able to protect NdeI from thermal inactivation up to a temperature of 48.7 °C (Fig. 4(A)). Hsp18.6 was also cloned in pPROEX HTa vector (construct Nuclear-Hsp18.6-tagB). Nuclear-Hsp18.6-tagB was active as a chaperone, protecting NdeI from thermal inactivation partially up to a temperature of 55 °C (Fig. 4(B)). Its hydrodynamic radius was determined to be 7.5 nm by DLS.
Fig. 4.
Thermo-protection of NdeI by nuclear-Hsp18.6-tagA (panel A) and nuclear-Hsp18.6-tagB (panel B): NdeI incubated at each temperature with (+) and without (−) sHSP. Control: migration of closed circular pET 28a(+) (uncut) and vector digested by NdeI (cut), M: marker, bp: base pair
Homologues of rice Hsp18.6 in tomato and tobacco harbour nuclear localisation signals and are targeted to the nucleus. The nuclear-localised Hsp16.1 in tomato forms complexes that are around 1 MDa in size, much larger than the dodecamers of CI sHSPs (Siddique et al. 2003). Our results also indicate that rice Hsp18.6 forms larger oligomers than Hsp16.9. The two constructs of rice Hsp18.6 have 23 and 24 additional residues at their N-termini, but the sequences of these residues are different. The drastic differences in the hydrodynamic radii of two constructs of the same protein confirm that the identity of the additional residues in the N-terminus plays a crucial role in the assembly of the multimer.
Hsp24, mitochondria
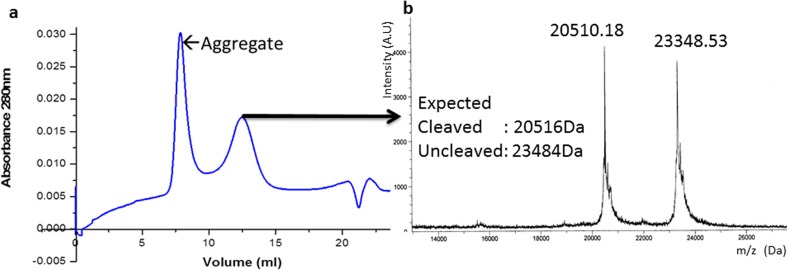
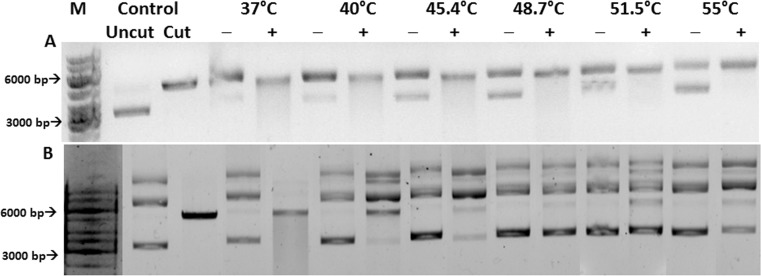
Rice Hsp24 contains an N-terminal mitochondrial targeting sequence. This signal sequence was excluded when the gene was cloned in the vector pPROEX HTa (construct mitochondria-Hsp24-tag). The purified protein had a hydrodynamic radius of 5.3 nm and could protect restriction enzyme NdeI completely up to a temperature of 48.7 °C (Fig. 6(A)). Following cleavage of the tag by TEV protease, size exclusion chromatography of the cleavage mixture yielded a single peak (Fig. 5a). However, SDS-PAGE and the MALDI/TOF spectrum of the peak revealed the presence of both the cleaved and uncleaved proteins (Fig. 5b). DLS studies of the peak indicated that it predominantly consisted of a particle with a hydrodynamic radius of 4 nm and polydispersity of 10.4 %. This particle was seen to be active as a chaperone, protecting NdeI completely at a temperature of 37 °C and partially up to 45.4 °C (Fig. 6(B)).
Fig. 6.
Thermo-protection of NdeI by mitochondria-Hsp24-tag (panel A) and mitochondria-Hsp24-after cleavage (panel B): NdeI incubated at each temperature with (+) and without (−) sHSP. Control: migration of closed circular pET 28a(+) (uncut) and vector digested by NdeI (cut), M: marker, bp: base pair
Fig. 5.
a Size exclusion chromatogram of mitochondria-Hsp24 (after cleavage). b MALDI/TOF spectrum of the eluted peak
Following cleavage of the hexa-histidine tag, mitochondrial Hsp24 appears to be composed of well-folded, mono-disperse oligomeric particles which are active as chaperones. However, this oligomer consists of both the cleaved and uncleaved proteins. It is thus likely that the two different forms of the protein cross oligomerize to form a hetero-oligomer, a phenomenon observed previously in sHSP mixtures (Studer and Narberhaus, 2000; Aquilina et al. 2013). The difference in hydrodynamic radii between the homo- and hetero-oligomeric forms (5.3 and 4 nm, respectively) suggests that the oligomeric assemblies are different. Further, both oligomers display differences in their thermo-protection activity towards the substrate NdeI.
Hsp26.7, chloroplast
Hsp21 in Arabidopsis and pea contain N-terminal chloroplast transit peptides which are cleaved off following protein import into chloroplasts. The amino terminal residues of the mature forms of both proteins following cleavage were determined (Suzuki et al. 1998). The N-terminal residue of rice Hsp26.7 following cleavage of its signal peptide was predicted to be Glu48 from a sequence alignment of HSP26.7 from rice and Hsp21 from Arabidopsis and pea (Fig. 7).
Fig. 7.

Sequence alignment of N-termini of chloroplast-localised sHSPs from Arabidopsis thaliana (At), Pisum sativum (Ps) and Oryza sativa (Os). N-termini of mature forms are shown in red. Predicted N-terminus of mature form of rice Hsp26.7 is shown in green
The sHSP domain in rice lacking the signal peptide has a molecular weight of 21.8 kDa. Two constructs of this domain were cloned: hexa-histidine-tagged chloroplast-Hsp26.7-tag with 12 amino acids at its N-terminus from the vector and chloroplast-Hsp26.7-tagless with just one additional methionine from the vector. Following purification, it was seen that chloroplast-Hsp26.7-tag and chloroplast-Hsp26.7-tagless had hydrodynamic radii of 12.4 and 8.8 nm, respectively. While chloroplast-Hsp26.7-tag protected NdeI from thermal stress up to a temperature of 51.5 °C (Fig. 8(A)), chloroplast-Hsp26.7-tagless protected NdeI completely at 37 °C and partially up to 51.5 °C (Fig. 8(B)).
Fig. 8.
Thermo-protection of NdeI by chloroplast-Hsp26.7-tag (panel A) and chloroplast-Hsp26.7-tagless (panel B): NdeI incubated at each temperature with (+) and without (−) sHSP. Control: migration of closed circular pET 28a(+) (uncut) and vector digested by NdeI (cut), M: marker, bp: base pair
Chloroplast-localised sHSPs harbour a conserved methionine-rich motif in their N-terminal arms (Harndahl et al. 1999). The sulphoxidation of the methionines in the motif causes a conformational change which is essential for the chaperone activity of this sHSP (Sundby et al. 2005). Hsp21 from Arabidopsis was seen to exist as a dodecamer. Numerous substrate-binding sites and intra-oligomer interaction sites were mapped on to its N-terminal arm (Ahrman et al. 2007). A structure model of Hsp21, from single-particle electron microscopy studies showed that it too existed as a double-disc dodecamer, albeit with a relative rotation between the two discs, when compared with the wheat Hsp16.9 dodecamer (Lambert et al. 2011). Our studies of the tagged and tagless constructs of rice Hsp26.7 confirm that the length of the N-terminal region has a direct bearing on its oligomer assembly and chaperone activity. Of the two constructs, the dimensions of chloroplast-Hsp26.7-tagless indicate it is more likely to assemble as a double-disc dodecamer, like its homologue from Arabidopsis.
Conclusions
We have studied five rice sHSPs that vary in their sizes, chaperone activity and cellular localisation. The physiological expression of these sHSPs is triggered by sundry stress factors (Sarkar et al. 2009). sHSPs targeted to different organelles in rice are of different sizes, indicating differences in quaternary structures. The hydrodynamic radii obtained for various constructs range from 4 to 14.1 nm. The sHSPs also show variations in the temperature (40 to 55 °C) up to which they confer thermo-protection on the substrate NdeI. From our studies on the different constructs of Hsp16.9, Hsp26.7 and Hsp24, we observe that constructs that display higher hydrodynamic radii tend to protect NdeI at higher temperatures. The incorporation of additional residues at the N-termini of these proteins causes the formation of expanded assemblies. We postulate that such assemblies undergo temperature-dependant dissociation/rearrangement more readily and hence protect NdeI at higher temperatures. We have also seen the formation of a hetero-oligomer between two forms of the mitochondria-localised Hsp24. This cross oligomer forms mono-disperse particles and is active as a chaperone.
Our findings highlight the role played by the flexible N-terminal region of sHSPs in facilitating oligomerization and chaperone activity. We have shown that the inclusion of an N-terminal tag has drastic consequences on the quaternary structure of sHSPs. The assembly of the oligomer is sensitive to both the length and sequence of the affinity tag used. These results provide guidelines for designing constructs for functional and structural studies on sHSPs. The results of the characterization of sHSPs targeted to the cytoplasm, nucleus, chloroplast, mitochondria and peroxisome reflect the diversity in structure and function of plant sHSPs belonging to different subfamilies.
Electronic supplementary material
SDS-PAGE of purified rice sHSPs. a Peroxisome-Hsp16-tag; b Cytoplasm-Hsp16.9-tag; c Cytoplasm-Hsp16.9-tagless; d Nuclear-Hsp18.6-tagA; e Nuclear-Hsp18.6-tagB; f Chloroplast-Hsp26.7-tag; and g Chloroplast-Hsp26.7-tagless (TIFF 93 kb)
SDS-PAGE showing cleavage of mitochondria-Hsp24-tag. Lane 1: TEV protease, Lane 2: Purified mitochondria-Hsp24-tag, Lane 3: Mitochondria-Hsp24-tag after TEV cleavage (TIFF 146 kb)
Acknowledgements
This work was funded by a grant from the Department of Science and Technology, Government of India. N.M. thanks the Council of Scientific and Industrial Research for a Senior Research Fellowship. K.R. thanks the Department of Biotechnology, Government of India, for a Post-Doctoral Research Associate Fellowship. Protein masses were determined by MALDI/TOF experiments performed at the Proteomics Facility of our institute, supported by the Department of Biotechnology, Government of India.
Conflict of interest
The authors declare no conflict of interest.
References
- Ahrman E, Lambert W, Aquilina JA, Robinson CV, Emanuelsson CS. Chemical cross-linking of the chloroplast localized small heat-shock protein, Hsp21, and the model substrate citrate synthase. Protein Sci. 2007;16:1464–1478. doi: 10.1110/ps.072831607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquilina JA, Shrestha S, Morris AM, Ecroyd H. Structural and functional aspects of hetero-oligomers formed by the small heat shock proteins alphaB-crystallin and HSP27. J Biol Chem. 2013;288:13602–13609. doi: 10.1074/jbc.M112.443812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha E, O'Neill H, Vierling E. Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PFL, Jinn TL, Huang WK, Chen YS, Chang HM, Wang CW. Induction of a cDNA clone from rice encoding a class II small heat shock protein by heat stress, mechanical injury, and salicylic acid. Plant Sci. 2007;172:64–75. doi: 10.1016/j.plantsci.2006.07.017. [DOI] [Google Scholar]
- Chen X, et al. Expression and interaction of small heat shock proteins (sHSPs) in rice in response to heat stress. Biochim Biophys Acta. 2014;1844:818–828. doi: 10.1016/j.bbapap.2014.02.010. [DOI] [PubMed] [Google Scholar]
- Fu X, Chang Z. Identification of a highly conserved pro-gly doublet in non-animal small heat shock proteins and characterization of its structural and functional roles in Mycobacterium tuberculosis Hsp16.3. Biochemistry (Mosc) 2006;71(Suppl 1):S83–S90. doi: 10.1134/S0006297906130141. [DOI] [PubMed] [Google Scholar]
- Guan JC, Jinn TL, Yeh CH, Feng SP, Chen YM, Lin CY. Characterization of the genomic structures and selective expression profiles of nine class I small heat shock protein genes clustered on two chromosomes in rice (Oryza sativa L.) Plant Mol Biol. 2004;56:795–809. doi: 10.1007/s11103-004-5182-z. [DOI] [PubMed] [Google Scholar]
- Harndahl U, Hall RB, Osteryoung KW, Vierling E, Bornman JF, Sundby C. The chloroplast small heat shock protein undergoes oxidation-dependent conformational changes and may protect plants from oxidative stress. Cell Stress Chaperones. 1999;4:129–138. doi: 10.1379/1466-1268(1999)004<0129:TCSHSP>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilario E, Martin FJ, Bertolini MC, Fan L. Crystal structures of Xanthomonas small heat shock protein provide a structural basis for an active molecular chaperone oligomer. J Mol Biol. 2011;408:74–86. doi: 10.1016/j.jmb.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Hilton GR, Lioe H, Stengel F, Baldwin AJ, Benesch JL. Small heat-shock proteins: paramedics of the cell. Top Curr Chem. 2013;328:69–98. doi: 10.1007/128_2012_324. [DOI] [PubMed] [Google Scholar]
- Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci U S A. 2009;106:15604–15609. doi: 10.1073/pnas.0902177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- Kriehuber T, Rattei T, Weinmaier T, Bepperling A, Haslbeck M, Buchner J. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 2010;24:3633–3642. doi: 10.1096/fj.10-156992. [DOI] [PubMed] [Google Scholar]
- Lambert W, et al. Subunit arrangement in the dodecameric chloroplast small heat shock protein Hsp21. Protein Sci. 2011;20:291–301. doi: 10.1002/pro.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Won SH, Lee HS, Miyao M, Chung WI, Kim IJ, Jo J. Expression of the chloroplast-localized small heat shock protein by oxidative stress in rice. Gene. 2000;245:283–290. doi: 10.1016/S0378-1119(00)00043-3. [DOI] [PubMed] [Google Scholar]
- Lini N, Rehna EA, Shiburaj S, Maheshwari JJ, Shankernarayan NP, Dharmalingam K. Functional characterization of a small heat shock protein from Mycobacterium leprae. BMC Microbiol. 2008;8:208. doi: 10.1186/1471-2180-8-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Haslbeck M, Babujee L, Jahn O, Reumann S. Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol. 2006;141:47–60. doi: 10.1104/pp.105.073841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Omasa K, Horie T. High temperature at flowering inhibits swelling of pollen grains, a driving force for thecae dehiscence in rice (Oryza sativa L.) Plant Prod Sci. 2000;3:430–434. doi: 10.1626/pps.3.430. [DOI] [Google Scholar]
- Nagaraj RH, Panda AK, Shanthakumar S, Santhoshkumar P, Pasupuleti N, Wang B, Biswas A. Hydroimidazolone modification of the conserved Arg12 in small heat shock proteins: studies on the structure and chaperone function using mutant mimics. PLoS One. 2012;7:e30257. doi: 10.1371/journal.pone.0030257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Chen J, Xie W, Wang L, Zhang Q. Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol Biol. 2009;70:341–357. doi: 10.1007/s11103-009-9477-y. [DOI] [PubMed] [Google Scholar]
- Pasta SY, Raman B, Ramakrishna T, Rao Ch M. The IXI/V motif in the C-terminal extension of alpha-crystallins: alternative interactions and oligomeric assemblies. Mol Vis. 2004;10:655–662. [PubMed] [Google Scholar]
- Patel S, Vierling E, Tama F. Replica exchange molecular dynamics simulations provide insight into substrate recognition by small heat shock proteins. Biophys J. 2014;106:2644–2655. doi: 10.1016/j.bpj.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, et al. Rice yields decline with higher night temperature from global warming. Proc Natl Acad Sci U S A. 2004;101:9971–9975. doi: 10.1073/pnas.0403720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Sarkar NK, Kim YK, Grover A. Rice sHSP genes: genomic organization and expression profiling under stress and development. BMC Genomics. 2009;10:393. doi: 10.1186/1471-2164-10-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yokoya S. Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat-shock protein, sHSP17.7. Plant Cell Rep. 2008;27:329–334. doi: 10.1007/s00299-007-0470-0. [DOI] [PubMed] [Google Scholar]
- Siddique M, et al. Tomato heat stress protein Hsp16.1-CIII represents a member of a new class of nucleocytoplasmic small heat stress proteins in plants. Cell Stress Chaperones. 2003;8:381–394. doi: 10.1379/1466-1268(2003)008<0381:THSPHR>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M, Gernhard S, von Koskull-Doring P, Vierling E, Scharf KD. The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones. 2008;13:183–197. doi: 10.1007/s12192-008-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer S, Narberhaus F. Chaperone activity and homo- and hetero-oligomer formation of bacterial small heat shock proteins. J Biol Chem. 2000;275:37212–37218. doi: 10.1074/jbc.M004701200. [DOI] [PubMed] [Google Scholar]
- Studer S, Obrist M, Lentze N, Narberhaus F. A critical motif for oligomerization and chaperone activity of bacterial alpha-heat shock proteins. Eur J Biochem. 2002;269:3578–3586. doi: 10.1046/j.1432-1033.2002.03049.x. [DOI] [PubMed] [Google Scholar]
- Sun Y, MacRae TH. Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundby C, Harndahl U, Gustavsson N, Ahrman E, Murphy DJ. Conserved methionines in chloroplasts. Biochim Biophys Acta. 2005;1703:191–202. doi: 10.1016/j.bbapap.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Suzuki TC, Krawitz DC, Vierling E. The chloroplast small heat-shock protein oligomer is not phosphorylated and does not dissociate during heat stress in vivo. Plant Physiol. 1998;116:1151–1161. doi: 10.1104/pp.116.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T, Tabata A, Nagamune H. Investigation of the chaperone function of the small heat shock protein-AgsA. BMC Biochem. 2010;11:27. doi: 10.1186/1471-2091-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Den IP, Norman DG, Quinlan RA. Molecular chaperones: small heat shock proteins in the limelight. Curr Biol. 1999;9:R103–R105. doi: 10.1016/S0960-9822(99)80061-X. [DOI] [PubMed] [Google Scholar]
- van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- Veinger L, Diamant S, Buchner J, Goloubinoff P. The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J Biol Chem. 1998;273:11032–11037. doi: 10.1074/jbc.273.18.11032. [DOI] [PubMed] [Google Scholar]
- Waters ER, Lee GJ, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–338. doi: 10.1093/jxb/47.3.325. [DOI] [Google Scholar]
- Waters ER, Aevermann BD, Sanders-Reed Z. Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperones. 2008;13:127–142. doi: 10.1007/s12192-008-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CH, Chang PF, Yeh KW, Lin WC, Chen YM, Lin CY. Expression of a gene encoding a 16.9-kDa heat-shock protein, Oshsp16.9, in Escherichia coli enhances thermotolerance. Proc Natl Acad Sci U S A. 1997;94:10967–10972. doi: 10.1073/pnas.94.20.10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SDS-PAGE of purified rice sHSPs. a Peroxisome-Hsp16-tag; b Cytoplasm-Hsp16.9-tag; c Cytoplasm-Hsp16.9-tagless; d Nuclear-Hsp18.6-tagA; e Nuclear-Hsp18.6-tagB; f Chloroplast-Hsp26.7-tag; and g Chloroplast-Hsp26.7-tagless (TIFF 93 kb)
SDS-PAGE showing cleavage of mitochondria-Hsp24-tag. Lane 1: TEV protease, Lane 2: Purified mitochondria-Hsp24-tag, Lane 3: Mitochondria-Hsp24-tag after TEV cleavage (TIFF 146 kb)