Abstract
In eukaryotes, the heat shock factors (HSFs) are recognized as the master regulator of the heat shock response. In this respect, the genes encoding the heat shock factors seem to be important for adaptation to thermal stress in organisms. Despite this, only few mammalian HSFs has been characterized. In this study, four major heat shock factor genes viz. HSF-1, 2, 4, and 5 were studied. The main objective of the present study was to characterize the cDNA encoding using conserved gene specific primers and to investigate the expression status of these buffalo HSF genes. Our RT-PCR analysis uncovered two distinct variants of buffalo HSF-1 and HSF-2 gene transcripts. In addition, we identified a variant of the HSF5 transcript in buffalo lacking a DNA-binding domain. In silico analysis of deduced amino acid sequences for buffalo HSF genes showed domain architecture similar to other mammalian species. Changes in the gene expression profile were noted by quantitative real-time PCR (qRT-PCR) analysis. We detected the transcript of buffalo HSF genes in different tissues. We also evaluated the seasonal changes in the expression of HSF genes. Interestingly, the transcript level of HSF-1 gene was found upregulated in months of high and low ambient temperatures. In contrast, the expression of the HSF-4 and 5 genes was found to be downregulated in months of high ambient temperature. This suggests that the intricate balance of different HSFs is adjusted to minimize the effect of seasonal changes in environmental conditions. These findings advance our understanding of the complex, context-dependent regulation of HSF gene expression under normal and stressful conditions.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-014-0563-y) contains supplementary material, which is available to authorized users.
Keywords: Heat shock factors, Buffalo, Variant, Expression, Seasonal
Introduction
As an integral part of agriculture, water buffaloes are recognized as the indispensable allies of small farmers in Asian countries. The domesticated water buffalo (Bubalus bubalis) is sometimes referred to as the “living tractor of the East” as it is relied upon for plowing and transportation in many parts of Asia. In India, buffaloes are primarily kept for milk and draft purposes and secondarily for meat production. The contribution of water buffalo in India’s economy is immense, which is apparent by the presence in India of about 56.7 % of the world’s buffalo population (FAOSTAT 2008). The genus Bubalis surpasses the cattle genus Bos in its ability to adapt to the hot, humid areas of muddy and swampy lands, since it thrives in the very hot areas of the tropics and subtropics such as the swamps of Southeast Asia, the marshes of southern Iraq, the valleys of flooding rivers in the Indian subcontinent, and the River Nile in Egypt (Marai and Haeeb 2010). However, little is known about the molecular basis of this tropical adaptability in buffaloes.
The heat shock factors (HSFs), the transcription factors that modulate the expression of classical heat shock genes are recognized as the central component of the eukaryotic heat shock response. The HSF family consists of at least four members (HSF-1–4) in vertebrates, whereas a single HSF is present in yeast, nematodes and the fruit fly (Akerfelt et al. 2010). HSF-1 and HSF-2 are ubiquitous among vertebrate species, whereas HSF-3 and HSF-4 have been characterized only in avian and mammalian species, respectively (Stephanou and David 2010). Although some new members have been added to the HSF gene family in verterbrates, they are still poorly understood (Akerfelt et al. 2010). HSF genes are also known to exist in different isoforms. HSF-1 and HSF-2 genes encode for at least two isoforms in mice (Sarge et al. 1991, 1994). The inclusion of exon 11 in the HSF-1 gene produces the longer HSF-1 alpha (α) isoform, while exclusion leads to the shorter beta (β) isoform (Goodson and Sarge 1995). The two isoforms of HSF2 (HSF2α and HSF2β) also contain an alternative exon 11, which is similarly flanked by two introns containing consensus-splicing sites (Goodson et al. 1995). Recently, two different HSF1 isoforms have been identified, which consist of exon9a and encode for the 28 amino acid stretch of the HSF-1 isoforms (Neueder et al. 2014).
HSFs are structurally and functionally conserved throughout the eukaryotic kingdom. The conserved domains of the distinct HSFs comprise the DNA-binding domain (DBD), the oligomerization domain (HR-A/B, HR-C), the transactivation domain, and the regulatory domain (Pirkkala et al. 2001). HSF is an essential gene in those species that have a single HSF gene (e.g., yeast and Drosophila) even under non-stress conditions (Gonsalves et al. 2011). In mice, HSF-1 is required for oogenesis, placental development, and normal growth (Zhang et al. 2002). HSF-2 is also involved in oogenesis, spermatogenesis, and brain formation (Wang et al. 2003; Kallio et al. 2002). Furthermore, mice deficient in both HSF-1 and HSF-2 are sterile with severe defects in spermatogenesis (Wang et al. 2004). HSF-1 and HSF-4 play important roles in lens and olfactory epithelium development, and mutation in HSF-4 have been found to be associated with heritable cataract formation in humans (Akerfelt et al. 2010). HSFs are widely recognized as functional participants in heat stress action. However, thus far, information on the members of the HSF gene family in Indian buffalo is not available. The present study was directed to characterize genes encoding HSFs and to investigate the expression profile of candidate HSF genes in buffalo. In this study, we identified HSF-1, 2, 4, and 5 coding sequence (CDS) and observed the distinct tissue specific expression of these genes in buffalo. We also demonstrated seasonal changes in the expression profile of these genes. Of particular note, we provide evidence that buffalo HSF-1 and HSF-2 genes exist in at least two distinct splice variants.
Materials and methods
Sample collection
Tissue samples (brain, heart, liver, lung, heart, testis, and ovary) of water buffalo (Murrah) were collected in triplicate from the Gazipur slaughter house, New Delhi, India, with the help of an on-site veterinary officer. The fresh tissue samples were dispensed in RNA later (Sigma-Aldrich) and stored at −80 °C (ultra-low temperature freezer, Sanyo, Japan) until further use. Fresh blood samples were procured from the cattle yard of the National Dairy Research Institute, Karnal (Haryana), India. About 10 mL of peripheral whole blood sample were collected from each animal in sterile Vacutainers® (Beckton-Dickinson, Franklin Lakes, NJ, USA) containing sodium heparin as an anticoagulant. These collections were performed in accordance with the guidelines of the institute’s ethical and bio-safety committees.
cDNA preparation and DNA isolation
Total RNA was isolated from tissue samples using TRIzol reagent (Sigma-Aldrich) and treated with DNAse I (Fermentas) following the manufacturer’s instructions. RNA integrity was assessed by loading 2 ul RNA sample on 1.5 % agarose gel. Total mRNA of 2.4 ug was reverse transcribed with Superscript III cDNA synthesis kit (Invitrogen Canada Inc.) using hexamer primers according to the manufacturer’s instructions for all tissue samples and stored at −80 °C for further use. The genomic DNA was isolated from white blood cells using the standard phenol chloroform method (Sambrook and Russel 2001). The genomic DNA samples were dissolved in TE buffer (pH 8.0) and stored at −20 °C for further use. The concentrations of isolated RNA and genomic DNA samples were determined by optical density at 260 nm using NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
RT-PCR and cloning
The full-length CDS of HSF-1, 2, and 5 genes were amplified from cDNA of testicular tissue. The HSF-4 gene was amplified from genomic DNA. The primers for this study were designed manually using the conserved sequences from the Bos taurus mRNA sequence. The list of primers sets with their optimized annealing temperature is presented in Table 1. Briefly, PCR was carried out on a Veriti 96-well thermal cycler (Applied Biosystems) in a 25-μl reaction mixture containing 2.5 μl of 10× Paq5000™ reaction buffer, 1 μl of 10 mM dNTP mix (Fermentas), 0.5 of 10 μM for each primer, and 0.25 units of Stratagene Paq5000 DNA polymerase (Agilent Technologies). PCR products were electrophoresed alongside DNA molecular weight marker in 2 % agarose gel and then stained with ethidium bromide. The amplified fragments were gel purified using a PureLink Quick Gel Extraction Kit (Invitrogen Canada Inc.), subcloned into a pTZ57R/T vector (Thermo Scientific), and sequenced using the Sanger method (Scigenome). Multiple clones were sequenced to validate the obtained sequences. We deduced the HSF-1, 2, 4, and 5 gene sequences in the Murrah breed of buffalo by sequencing the cloned products that were generated throughout the study, and the resultant sequence was deposited into GenBank with the accession numbers KF658193.1, KF658194.1, KC568562, KF658195.1, KJ658339, and KJ634069.
Table 1.
Primer sequences used for end point PCR and real-time PCR experiments
| Sr. no. | Primer name | Primer sequence (5′–3′) | Size (bp) | GenBank accession number | Gene symbol | Annealing temperature |
|---|---|---|---|---|---|---|
| 1.a | HSF1F1 | CGCCTTGCCTGGTATGGATCTGC | 826 | NM_001076809.1 | HSF-1 | 55 |
| HSFIR2 | CAGTTCGGTGATGTCGGAGATGATGG | |||||
| 2.a | HSF1RTF1 | GCGCTCATCTGCTGGAGCCCG | 1482 | NM_001076809.1 | HSF-1 | 58 |
| HSFIR1 | TACGAGACAGTGGGGTCCTTGGCTT | |||||
| 3.a | HSF2F1 | TGCGCCTCGTTAACAATGAAGCAGAGTTC | 1639 | NM_001083405.1 | HSF-2 | 54 |
| HSF2R1 | GCGTCCACTTCTTGGGGATTTAGCTATCTA | |||||
| 4.a, b | HSF4F1 | CCGAGACTGCGCCATGCAGGAAG | 1651 | NM_001191202.1 | HSF-4 | 63 |
| HSF4GR1 | ATGCTGTTGACCGTGGCTCTGCCG | |||||
| 5.a, b | HSF4GF2 | CGGAAGGTGGTGAGCATCGAGCA | 1790 | NM_001191202.1 | HSF-4 | 71 |
| HSF4R2 | CGGAGATGATGGGGCCCCTGG | |||||
| 6.a, b | HSF4F2 | CTGCAGGACCCCTACTTTATCCAGTC | 2131 | NM_001191202.1 | HSF-4 | 57 |
| HSF4FLR | TGGAGGGGGTTTCAGGGTGAGGG | |||||
| 7.a | HSF5F1 | GATGGAGGAGGCGCTGCTCTCC | 286 | XM_003583530.1 | HSF-5 | 58 |
| HSF5GR2 | CTGTTGGATAGTAGGCTTGGGAGTACTG | |||||
| 8.a | HSF5RTF1 | CCAAGCCTACTATCCAACAGCTGTGCT | 891 | XM_003583530.1 | HSF-5 | 59 |
| HSF5R1 | TCCTCACTCTCTTAATTCTTCCTCCTTTGG | |||||
| 9.a | HSF1TvF1 | AGAAGTGCCTCAGCGTCGCCTGCCT | 273c | NM_001076809.1 | HSF-1 | 61 |
| HSF1TvR1 | CTGAGTCTGGGCTGCTTTTCTCTGCCTC | |||||
| 10.a | HSF2TvF1 | GATTGCAGTTTAGAGGACTTCCAAGCC | 188 c | NM_001083405.1 | HSF-2 | 59 |
| HSF2TvR1 | CCCTCTTCTGAAACTGGCTGAACTACATT | |||||
| 11.d | RPSF | TGCGAGTACTCAACACCAACATCGATGG | 184 | NM_001033614.2 | RPS18 | 60 |
| RPSR | GGATTCTGCATAATGGTGATCACACGTTCC | |||||
| 12.d | HSF1RTF1 | GCGCTCATCTGCTGGAGCCCG | 144 | NM_001076809.1 | HSF-1 | 60 |
| HSF1RTR1 | CTTCCGGAAGCCATACATGTTGAGCTG | |||||
| 13.d | HSF2RTF1 | CCTCAGAAGATCCAGTGACCATGATGGA | 168 | NM_001083405.1 | HSF-2 | 60 |
| HSF2RTR1 | CAACCAGGAGATCTGGGTCTATGCTA | |||||
| 14.d | HSF4RTF1 | ATCCGCTGGAGCCCGAGCGG | 161 | NM_001191202.1 | HSF-4 | 60 |
| HSF4RTR1 | CCACCCTGCTCGATGCTCACCAC | |||||
| 15.d | HSF5RTF1 | CCAAGCCTACTATCCAACAGCTGTGCT | 169 | XM_003583530.1 | HSF-5 | 60 |
| HSF5RTR1 | CCAATTAGAAGGCAGAAATTCAACTGGATAGG |
The table shows the primer name, primer sequences, the predicted PCR product size (base pairs, bp), GenBank accession number of the target sequence, gene symbol and unique annealing temperature (°C) that was used for the PCR amplification
aPrimer set for end point PCR
bAmplified from genomic DNA
cExpected PCR product size
dPrimer set for real-time PCR
Sequence retrieval and alignment
A comprehensive search of the sequence database on the NCBI website was carried out in order to find and compare HSF-1, 2, 4, and 5 gene orthologs among different species. Briefly, a BLASTp (Altschul et al. 1990) search was performed using protein sequences corresponding to each of the candidate buffalo HSF genes to retrieve similar sequences from other mammalian species such as Bos taurus, Capra hircus, Canis lupus, Sus scrofa, Pongo abelli, Papio anubis, Pan troglotydes, Homo sapiens, and Mus musculus (Online Resource 1). BioEdit (Hall 1999) was used to align multiple amino acid sequences by ClustalW and to identify and remove duplicate, splice variant, reading frame shift, truncated, or otherwise unusable HSF sequences from the dataset. The secondary structure of the predicted protein was ascertained using Phyre2 software (Kelly and Sternberg 2009).
Prediction of domain structure
The DNA-binding domain within the Murrah buffalo of four HSF proteins was identified using the known structural domains of human and mouse HSF proteins in UniprotKB (Apweiler et al. 2004). The domain analyses programs MARCOIL (Delorenzi and Speed 2002), PredictNLS (Cokol et al. 2000), and NetNES 1.1 (Cour et al. 2004) were used for predicting coiled-coil domains, NLS, and NES, respectively. Additionally, the conserved motifs of HSF proteins were verified by submitting their full-length amino acid sequences to the MEME web tool (Bailey et al. 2009). The MEME program was used to identify motifs in the candidate HSF protein sequences. MEME was run locally with the following parameters: number of repetitions = 1, maximum number of motifs = 28, and with optimum motif widths constrained to between 6 and 20 residues.
Quantitative PCR
The expression profile of the four HSF genes was ascertained by real-time quantitative PCR using the cDNA and gene specific primer sets (Table 1). These primers were designed to span exon-exon boundaries to ensure cDNA specificity. Melting curve analysis, gel electrophoresis, and PCR product sequencing (Scigenome) were used to verify amplification of the correct target genes. Two reference genes, RPS18 and GAPDH, were tested. RPS18 showed the most stable expression and was used for normalization of expression data. Normalized ratios were determined for all runs using the delta-delta-Ct method (ΔΔCt) (Livak and Schmittgen 2001). All qRT-PCR reactions were conducted on a Light Cycler 480 Real-Time PCR machine (Roche Diagnostics, USA). In brief, the reaction mixture consisted 2 μl of diluted cDNA template (1:2), 5 μl of 2X KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems), 0.25 μl each of forward and reverse primers (10 μM), and nuclease-free water for a final volume of 10 μl. Each sample was run in duplicate. The following cycling conditions were employed for all the genes: pre-incubation at 95 °C for 3 min, followed by 40 cycles of 10 s at 95 °C, 20 s at 60 °C, and 1 s at 72 °C. Finally, the samples were cooled down to 40 °C for 10 s.
Statistical analysis
Statistical analyses were carried out in SYSTAT v12.02 software (SYSTAT Software Inc., Chicago, IL). To determine the differences between groups, we used the analysis of variance (ANOVA). Fischer’s restricted least significant differences criterion was used to maintain the a priori type I error rate of 0.05.
Results
Buffalo HSF-1, 2, 4, and 5 gene sequence
In this study, we obtained the nucleotide 1591, 1639, 4560, and 1155 bp DNA sequence having 1575, 1602, 1467, and 1152 bp of coding sequences of HSF-1, 2, 4, and 5 genes, respectively. The amino acid sequence was deduced in silico from the ExPASy translate tool/SIB from the correct open reading frame of Murrah buffalo HSF-1, 2, 4, and 5 nucleotide sequences. Comparison of sequence homologies for the complete HSF proteins as well as conserved DNA-binding and HR-A/B domain with orthologous genes in other species is presented in Table 2. As expected, complete protein sequences of buffalo HSF-1, 2, 4, and 5 genes were found to share the highest identity >98 % with cattle. Overall, the complete protein sequence of candidate HSF genes showed >90 % identity with the majority of species and within the conserved DNA-binding and HR-A/B domain the homology increased to >96 %. We used phylogenetic analysis to further verify that the obtained buffalo HSF-1, 2, 4, and 5 gene sequences belonged to the members of the HSF gene family (Online Resource 2). The phylogenetic analysis confirmed that the isolated buffalo HSF gene sequences belongs to the HSF family.
Table 2.
Comparison of the percent homologies (%) of the HSF-type DNA-binding (A) and Hydrophobic repeat (HR)-A/B (B) domain and complete HSF protein sequences (C) with that of water buffalo (Bubalus bubalis) for all species analyzed
| Species | HSF-1 | HSF-2 | HSF-4 | HSF-5 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | A | B | C | A | B | C | A | B | C | |
| Bubalus bubalis | – | – | – | – | – | – | – | – | – | – | – | – |
| Bos taurus | 99 | 100 | 98 | 99 | 100 | 99 | 99 | 100 | 98 | 100 | NA | 98 |
| Capra hircus | 99 | 99 | 97 | 99 | 100 | 98 | 99 | 100 | 98 | 100 | NA | 98 |
| Canis lupus | 97 | 100 | 92 | 99 | 100 | 98 | 99 | 93 | 92 | 100 | NA | 93 |
| Sus scrofa | 98 | 99 | 93 | 99 | 100 | 98 | 99 | 100 | 93 | 100 | NA | 92 |
| Pongo abelli | 97 | 99 | 90 | 99 | 100 | 97 | 99 | 95 | 90 | 100 | NA | 90 |
| Papio anubis | 97 | 99 | 90 | 99 | 100 | 98 | 99 | 95 | 91 | |||
| Pan troglotydes | 97 | 99 | 90 | 99 | 65 | 93 | 99 | 95 | 91 | 100 | NA | 89 |
| Homo sapiens | 97 | 99 | 90 | 99 | 100 | 97 | 99 | 95 | 91 | 100 | NA | 89 |
| Mus musculus | 97 | 97 | 88 | 97 | 100 | 94 | 99 | 88 | 86 | 100 | NA | 90 |
Bold letters represent the percent amino acid homology shared with most of the species
NA not applicable
Identification of buffalo HSF-1 and 2 splice variant
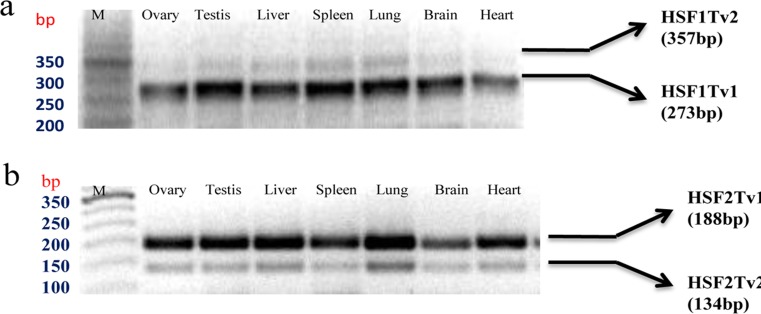
We used three strategies to explore splice variants in water buffalo HSF genes. First, we reviewed all the related articles on HSF genes available in PubMed; second, we screened all the expressed sequence tags resulting from alternative splicing of the candidate genes from the UCSC genome browser (Karolchik et al. 2014); and third, we amplified the regions of interest using cDNA prepared from various tissues (ovary, testis, liver, spleen, lung, brain, and heart). Our results of RT-PCR analysis of targeted regions shown in Fig. 1 revealed two distinct cDNA variants of HSF-1 and HSF-2 genes. The identities of these variants were verified by cloning and sequencing of both the RT-PCR products obtained in this study. The analysis of cloned sequences revealed the existence of an extra 84 bp nucleotide in the longer product of HSF-1 mRNA and an omission of 54 bp in the shorter product of HSF-2 gene compared to a previously cloned transcript (Online Resource 3). In this study, the original cloned products of these two genes were called HSF-1Tv1 and HSF-2Tv1, whereas the identified variants were named HSF1Tv2 and HSF2Tv2. In order to verify our results, we performed Blat against cow B_tau_4.6.1_genome assembly using the UCSC genome browser. The Blat result of the HSF-1 transcript variant revealed the existence of an 84 bp exon in the bovine genome assembly that corresponds to the extra nucleotide obtained in our study (Online Resource 4). This exon is bound by intronic sequences of 355 and 536 bp, both of which exhibit a GT-AG consensus sequence pattern for 5΄ splice donor and 3΄ splice acceptor sites. Similarly, the Blat result of the HSF2 sequence revealed that the gap of 54 bp corresponds to the exclusion of the exon obtained in our study (Online Resource 5). This exon is bound by intronic sequence of 8000 (approx.) and 3369 bp, both of which exhibit GT-AG consensus sequence pattern for 5΄ splice donor and 3΄ splice acceptor sites. These results show that the splice variants obtained in our study are generated by the mechanism of alternative splicing. Interestingly, we observed a very specific pattern in the expression of each variant. The HSF-1Tv1 and HSF-2Tv1 mRNA variants were found to be expressed abundantly in all the examined tissues and represent the major form. Furthermore, the expression of major variants of the HSF-2 gene was observed to be strongest for heart and brain tissues.
Fig. 1.
Detection of the buffalo HSF gene splice variant in tissues. a RT-PCR was performed for HSF-1 gene between exon 9 and 13, resulting in the amplification of a 273 bp fragment, corresponding to the original cloned product called “HSF-1Tv1” and a 357 bp fragment called “HSF-1Tv2”, containing extra 84 nucleotide. b RT-PCR was performed for HSF-2 gene between exon 10 and 12, resulting in the amplification of a 188 bp fragment, corresponding to the original cloned product called “HSF-2Tv1” and a 134 bp fragment called “HSF-2Tv2”, containing omission of 54 nucleotides. PCR products were electrophoresed on 2 % agarose gel. M indicates TrackIt ™ 50 bp DNA Ladder. The migration of size marker (bp) is shown to the left of gel
Domain organization of buffalo HSF genes
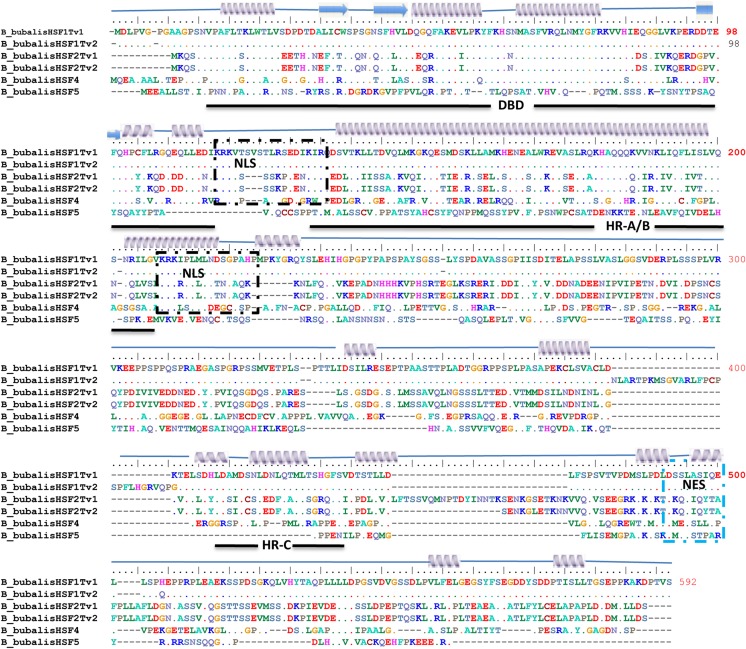
The prediction of the typical signature motifs present in buffalo HSF protein sequence was carried out by comparing the buffalo HSF proteins to those of homologous well-characterized parts of model human and mouse proteins in UniprotKB. This enabled us to analyze similar domains present in the deduced protein sequences of buffalo HSF-1, 2, 4, and 5 genes (Fig. 2). We obtained the highly conserved DNA-binding domain (DBD) of approximately 110 amino acids located in the amino terminal of HSF proteins. The length of DBD was somewhat constant with the exception of the HSF5 protein sequence. The coiled-coil structures that is a characteristic of the leucine zipper interaction domain, and is a feature of the hydrophobic repeat-A/B (HR-A/B) and hydrophobic repeat-C (HR-C) region was confirmed using the prediction tool Marcoil. The putative nuclear localization signal (NLS) and nuclear export signal (NES) motifs, required for shuttling these proteins were obtained by predict NLS and NetNES tools. We observed two clusters of basic amino acid residues in the HSF protein sequence, which might serve as a potential NLS motif. However, mutagenic studies have revealed that the one adjacent C-terminal of HR-A/B domain is functional (Vujanac et al. 2005). Similarly, NES was found close to the C-terminus in the HSF proteins. We were not able to identify the transactivation domain by any means. The results of domain prediction were further verified by employing the MEME web server (Online Resource 6 and 7). Overall, despite variability in size and sequence, we observed DBD and HR-A/B regions in HSF-1, 2, and 4 proteins.
Fig. 2.
Multiple sequence alignment of the identified heat shock factor sequences. Buffalo HSF-1, HSF-2, HSF-4 and HSF-5 protein sequences were aligned by using cluatalW2 program in BioEdit software. The secondary structure of HSF protein sequence is shown above the sequence. Arrow underneath the alignment indicates domain boundaries. Black and blue box depicts NLS and NES, respectively. Dot represents the conserved residues in the protein sequences
Pattern of expression of HSF genes in buffalo
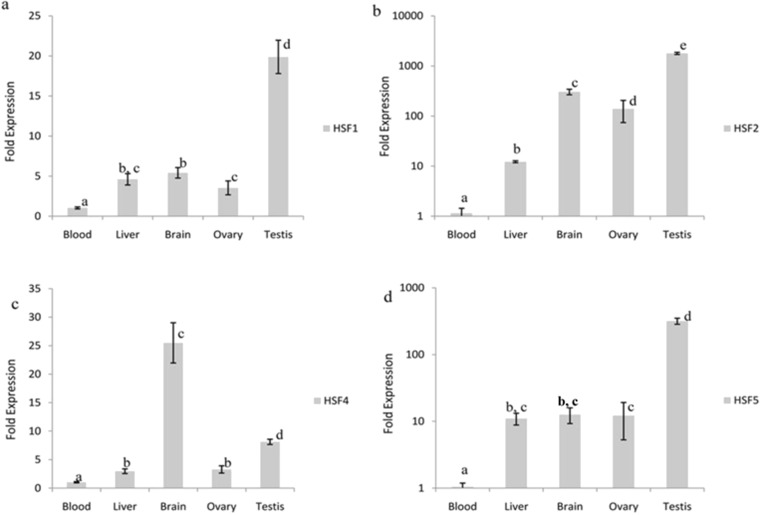
We analyzed the basal level of HSF-1, 2, 4, and 5 genes expression in a variety of tissues including blood, brain, liver, testis and ovary. We selected blood and brain, since they are crucial in maintaining homeostasis when exposed to changes in environmental temperature, the gonads because of their importance in reproduction, and the liver because of its role as the central metabolizing and detoxifying organ. The qPCR assay was performed individually for all transcripts. The overall expression profile of HSF-1, 2, 4, and 5 genes in different tissues used in this study is shown (Fig. 3). The level of expression of HSF-1, 2, 4, and 5 genes were found to vary significantly between most of the tissues examined (P < 0.001). The steady state level of HSF-1, 2, and 5 mRNA was found to be higher in testis compared to all the tissues examined (P < 0.001). Notably, the level of expression of HSF4 mRNA was found to be higher in brain compared to all other tissues (P < 0.001). Interestingly, the tissue of brain, testis, and ovary showed higher accumulations of all the HSF genes compared to blood and liver tissues. The expression of HSF2 gene was not observed in blood.
Fig. 3.
Distinct tissue specific expression of buffalo HSF-1, 2, 4, and 5 genes (a–d). RT-qPCR analysis of HSF-1, 2, 4, and 5 transcripts in the indicated tissues was performed. The level of transcripts from the gene encoding ribosomal protein RPS 18 was used as an internal reference to normalize the HSF-1, 2, 4, and 5 mRNA, respectively. The normalized expression level was then compared to that of blood sample, which was arbitrarily given the value 1. Experiments were performed from tissues collected from n = 3 animals. Bar represents ± SEM. Within the graph, values labeled with different letters differ significantly from one another
Seasonal trends in expression of buffalo HSF genes
In an attempt to further characterize the expression of buffalo HSF-1, 2, 4, and 5 genes, we examined the expression of candidate genes at three time points: autumn (October), winter (January), and summer (June), when the daytime environmental temperature and relative humidity in the north western region of the Indian subcontinent ranges from 17–30 °C (71 %), 7–19 °C (80 %), and 26–38 °C (62 %), respectively. For this analysis, we determined the expression of HSF-1, 2, 4, and 5 genes in peripheral whole blood samples of 6-month old Murrah buffalo female calves after an acclimatization period of 2–3 weeks in each season, when the level of gene expression reached a stable stage to counteract the changes in the environmental conditions prevalent in that particular season. The seasonal changes in the expression profile of HSF-1, 4, and 5 genes are shown in Fig. 4. The results show the significant effect of seasonal changes on the HSF1 mRNA concentration (P < 0.001). We observed significantly higher HSF1 mRNA levels in the summer (P < 0.05) and winter (P < 0.001) compared to the autumn. Notably, no significant difference was observed in the expression of the HSF1 gene between summer and winter seasons. Interestingly, the expression of HSF- 4 and 5 genes was higher (P < 0.001) in the autumn (P < 0.001) and winter (P < 0.001) compared to the summer. However, we did not find any significant difference in the expression of HSF-4 and 5 between the autumn and winter seasons. The expression of HSF2 gene was not observed in any season.
Fig. 4.
Seasonal differences in expression of buffalo HSF-1, 4, and 5 genes. (a–d) RT-qPCR analysis of HSF-1, 4, and 5 transcripts was performed in three different seasons as indicated. The level of transcripts from the gene encoding ribosomal protein RPS 18 was used as an internal reference to normalize the HSF-1, 4, and 5 mRNA, respectively. Summer season was chosen as calibrator arbitrarily and given the value 1. Blood samples collected from n = 3 Murrah buffalo heifers in 6 months) were used for the experiment. Bar represents ± SEM. Within the graph, values labeled with different letters differ significantly from one another
Discussion
The members of the HSF gene family have been described as transcriptional regulators involved in molecular and cellular responses to stress conditions (Morimoto 1998). Furthermore, research on humans and mouse has also shown HSFs to be an important component involved in the development and differentiation of cells under unstressed conditions (Abane and Mezger 2010). The present study investigates, for the first time the heat shock factor gene family in the economically important dairy animal. In this study, we have identified HSF-1, 2, and 4 genes in Murrah buffalo based on the draft genome sequence of Bos_tau_UMD_3.1. In addition, we found a variant of the HSF-5 gene. It was noted that it lacked a DNA-binding domain, when compared with “heat shock transcription factor-5” at the GenBank database of Loc AC_000176.1. The functionality of this gene therefore remains an enigma in buffalo. The incomplete structure of the HSF-5 gene suggests that it may be a non-functional counterpart. However, the possibility that the gene is transcribed and translated cannot be ruled out due to high conservation of its C-terminal region across the species. This aspect merits further detailed analysis.
The predicted amino acid sequence of buffalo HSFs showed strong identity to other HSFs, which is in agreement with the high levels of amino acid conservation that is characteristic of this protein family (Pirkkala et al. 2001). However, a number of specific alterations at nucleotide and amino acid levels were found to be unique to buffalo, establishing their species specific organization. Owing to >90 % sequence homology with cattle, human, and other species, buffalo HSF protein sequences showed a similar arrangement of various domains. Structural analysis of HSF genes in buffalo revealed intactness of each domain border, maintaining its reading frame even when an exon was inserted or omitted due to alternative splicing. The members of buffalo HSF genes were found to share a domain structure similar to their counterpart in other mammalian species. For example, the members of HSF-1 and HSF-2 gene orthologs contained all three major domains i.e., DBD, HR-A/B, and HR-C, whereas HSF4 gene orthologs contained only DBD and HR-A/B domains. The lack of an HR-C domain in the buffalo HSF-4 gene suggests that, similarly to other mammalian HSF-4, it is also constitutively trimerized in the cell (Akerfelt et al. 2010).
Our studies on bovine HSF-1 gene structure revealed that it comprises 14 exons including the alternatively spliced exon-10 and is located on chromosome-14. We also noted that this exon-10 is not conserved across the species and was not observed in the draft genomic sequence of humans and mice (Online Resource 4). Some authors have proposed that species specific alternative splicing events might be due to relatively recently gained “genome specific exons”, which tend to be included at low levels in spliced mRNA (Modrek and lee 2003). Our results demonstrate the low abundance of the long form of bovine HSF-1 gene splice variant. We suspect that the minor form of HSF-1 splice variant may not have a major impact on physiology, whereas the more abundant major form that is conserved across species is more likely to influence critical gene activities. As in the case of HSF-1, we have shown that the bovine HSF-2 gene contains an alternative exon-11, which is similarly flanked by an intron containing consensus-splicing sites. We noted that the HSF-2 gene splicing event is conserved across the species (Online Resource 5), which suggests that it might play an important functional role (Fiorenza et al. 1995). When HSF-2 isoforms were first discovered, the authors explained that inclusion of 54 bp exon-encoding 18 amino acid (a leucine zipper) sequences can improve the transactivation potential of the HSF-2 gene by a more favorable interaction with other components of transcriptional machinery (Goodson et al. 1995). It has been proposed recently that the leucine zipper conserved in the long isoform of the HSF-2 gene may stabilize the activation domain of the HSF-2 gene in an optimal conformation, while in the short isoforms the activation domain could be in a more relaxed conformation resulting in less efficient binding of co-factors, especially in the context of an HSF1β-HSF2β heterotrimer (Lecomte et al. 2013). We also observed that the ratios of the mRNA of bovine HSF-1 and HSF-2 splice variants are variable from one tissue to another, which suggests that each tissue may contain a specific balance of homodimer or heterotrimer of these proteins. In vitro studies have shown that overexpression of HSF-2α counteracts the inhibitory effect of HSF-2β isoform and potentiates the HSF-2 transcriptional activity for differentiation (Leppa et al. 1997). Our results revealed that the HSF-2Tv1 splice variant is predominately expressed in brain and heart, which indicates its potential role in differentiation. In addition to this tissue specific modulation of the HSF splice variant ratio, studies have shown that differential splicing of HSF-1 and HSF-2 messenger is also dependent on the type of stress (Lecomte et al. 2013). Taken together, our results demonstrate that the buffalo HSF splice variants might contribute to a new level of complexity in the regulation of target genes.
In this study, we have provided the expression profiles of water buffalo HSF genes in five important organs. We observed the expression of HSF genes in all the tissue examined except blood cells. A similar situation was found for HSF-1 and HSF-2 gene expression in mice (Fiorenza et al. 1995). However, in contrast to the observed ubiquitous expression of the HSF-4 gene in our studies, some authors have detected a selective tissue specific signal of the HSF-4 gene using northern blot in humans (Nakai et al. 1997). We owe this observation to the sensitivity of real-time PCR technique used in this study (Valasek and Repa 2005). Furthermore, expression data show distinct expression patterns of HSF genes in different tissues. This distinct tissue specific expression of HSF genes might arise from tissue specific factors of the heat shock regulatory network that regulates the heat shock response (Guisbert et al. 2013). In our analysis, we did not note any bovine HSF gene that was expressed only in a particular tissue type, but selected members of HSF genes showed higher levels of expression in specified tissue types. The expression of HSF-1, 2, and 5 in testis and HSF-4 gene in brain is noteworthy in this regard. This is similar to the expression pattern observed in mice for HSF-1, 2, and 4 genes (Fiorenza et al. 1995; Nakai et al. 1997). One possible explanation for the observed over expression of HSF-1 and 2 genes in testis may be that these genes assume a significant role in spermatogenesis. Such a role for the HSF-1 and HSF-2 gene has been elucidated by several authors (Sarge et al. 1994; Wang et al. 2004; Akerfelt et al. 2010). In contrast, the higher expression of HSF-4 gene in brain correlates with its prominent role in the development and differentiation of sensory organs (Akerfelt et al. 2007). In general, the expression of all the HSF genes was found to be higher in testis, brain, and ovary compared to blood and liver. This observation highlights the importance of HSF genes in development and differentiation.
We further investigated the effect of seasonal acclimatization on the expression profiles of HSF-1, 2, 4 and 5 genes. Changes in gene expression in response to HS that are associated with acclimatization include elevation of the constitutive HSP system and a reduced threshold for systemic HS responses (Horowitz 2001). The regulation of HSP production is critical to cell survival. Substantial evidence suggests that expression of the heat inducible HSP70 gene is mainly under the transcriptional control of HSF-1 (Fujimoto and Nakai 2010). Our expression studies on the bovine HSF-1 gene showed a trend of increased mRNA accumulation in the summer and winter seasons, when the temperature of the region shifts to daytime averages of 26–38 and 7–19 °C, respectively. We propose that this increase in concentration of HSF-1 mRNA may alter the intricate balance between HSP and HSF-1 in favor of HSP production, resulting in a reduced threshold for systemic response to heat and cold stress encountered by animals during seasonal fluctuation in environmental temperature. In contrast, our expression data of HSF-4 and 5 genes revealed a somewhat different trend. We observed significantly decreased concentrations of HSF-4 and 5 gene transcripts in the summer. Notably, we did not observe a significant variation in the expression of these genes in the autumn and winter seasons. We hypothesize that the reduced stability of these transcripts in the summer may be due to physiologically higher body temperatures than normal. Furthermore, it has been observed that in vitro exposure of cultured cells to a temperature of few degrees above the normal results in inducible synthesis of a small set of proteins, whereas normal synthesis of a normal complement of proteins present before stress treatment is diminished (Lindquist and Craig 1988). The role of HSF-2 and HSF-4 genes in the regulation of HSP production in normal/unstressed conditions is well documented (Stephanou and David 2010). In this respect, it seems to be an alternative explanation of our observed results. Nevertheless, the control mechanisms at the level of gene transcription and translation have not been explained for these genes and ought to be the real focus of future studies.
Electronic supplementary material
Accession number of amino acid sequences of HSF genes for all species analyzed(PDF 85 kb)
Neighbor-joining phylogeny of candidate HSF genes in 10 species. The phylogenetic tree was obtained using the MEGA 5.0 software on the basis of complete amino-acid sequences HSF-1, 2, 4 and 5 genes. Rooted Neighbor–joining analysis was performed with 1000 bootstrap replication and evolutionary distances were computed using Poisson’s correction method. The tree is drawn to scale, with the branch length in the same units as those of the evolutionary distance used to infer the phylogenetic tree (PDF 2504 kb)
Sequence alignment of the cloned PCR product of identified HSF1 and HSF2 splice variants (PDF 112 kb)
Blat result of HSF1 splice variant illustrating the structure of HSF1 gene in buffalo and other species using UCSC Genome Browser as per Bos_tau_4.6.1 genome assembly (PDF 44 kb)
Blat result of HSF2 splice variant illustrating the structure of HSF2 gene in buffalo and other species using UCSC Genome Browser as per Bos_tau_4.6.1 genome assembly (PDF 31 kb)
Distribution of conserved motifs in buffalo HSF genes. All the motifs were identified by MEME using complete amino acid sequences of HSF-1, 2, 4 and 5 genes. Name of the gene and combined P-values are shown on the left side of the figure and motif sizes are indicated at the bottom of the figure. Different motifs are indicated by different colors numbered 1–28. The same color in different proteins refers to the same motif (PDF 1316 kb)
Motif sequences of identified HSF genes in buffalo by MEME tool. (PDF 2259 kb)
Acknowledgments
This work was supported by an Indian Council of Agricultural Research, New Delhi funded project on National Initiative on Climate Resilient Agriculture. The authors are grateful to the Director, National Dairy Research Institute, Karnal, India for providing all the necessary facilities during the course of this research work.
Conflict of interest
None of the authors have any conflict of interest to declare.
Contributor Information
Shardul Vikram Lal, Phone: +91-1842259505, Email: shardullal84@gmail.com.
Sachinandan De, Phone: +91-1842259505, Email: sachinandan@gmail.com.
References
- Abane R, Mezger V. Roles of heat shock factors in gametogenesis and development. FEBS J. 2010;277(20):4150–4172. doi: 10.1111/j.1742-4658.2010.07830.x. [DOI] [PubMed] [Google Scholar]
- Akerfelt M, Trouillet D, Mezger V, Sistonen L. Heat shock factors at a crossroad between stress and development. Ann N Y Acad Sci. 2007;1113:15–27. doi: 10.1196/annals.1391.005. [DOI] [PubMed] [Google Scholar]
- Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, Donovan C, Redaschi N, Yeh LS. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004;32(90001):115D–1119D. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, William S, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cour TL, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17(6):527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Delorenzi M, Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- FAO statistics . FAO statistical yearbook. FAO: Rome; 2008. [Google Scholar]
- Fiorenza MT, Farkas T, Dissing M, Kolding D, Zimarino V. Complex expression of murine heat shock transcription factors. Nucleic Acids Res. 1995;23:467–474. doi: 10.1093/nar/23.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Nakai A. The heat shock factor family and adaptation to proteotoxic stress. FEBS J. 2010;277:4112–4125. doi: 10.1111/j.1742-4658.2010.07827.x. [DOI] [PubMed] [Google Scholar]
- Gonsalves SE, Moses AM, Razak Z, Robert F, Westwood JT. Whole-genome analysis reveals that active heat shock factor binding sites are mostly associated with non-heat shock genes in Drosophila melanogaster. PLoS ONE. 2011;6(1):15934. doi: 10.1371/journal.pone.0015934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson ML, Sarge KD. Regulated expression of heat shock factor 1 isoforms with distinct leucine zipper arrays via tissue-dependent alternative splicing. Biochem Biophys Res Commun. 1995;211:943–949. doi: 10.1006/bbrc.1995.1903. [DOI] [PubMed] [Google Scholar]
- Goodson ML, Park-Sarge OK, Sarge KD. Tissue-dependent expression of heat shock factor 2 isoforms with distinct transcriptional activities. Mol Cell Biol. 1995;15:5288–5293. doi: 10.1128/mcb.15.10.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisbert G, Czyz DM, Richter K, McMullen PD, Morimo RI. Identification of a tissue-selective heat shock response regulatory network. PLoS Genet. 2013;9(4):e1003466. doi: 10.1371/journal.pgen.1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Horowitz M. Heat acclimation: phenotypic plasticity and cues to the underlying molecular mechanisms. J Therm Biol. 2001;26:357–363. doi: 10.1016/S0306-4565(01)00044-4. [DOI] [Google Scholar]
- Kallio M, Chang Y, Manuel M, Alastalo TP, Rallu M, Gitton Y, Pirkkala L, Loones MT, Paslaru L, Larney S, Hiard S, Morange M, Sistonen L, Mezger V. Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J. 2002;21(11):2591–2601. doi: 10.1093/emboj/21.11.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hinrichs AS, Learned K, Lee BT, Li CH, Raney BJ, Rhead B, Rosenbloom KR, Sloan CA, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ. The UCSC genome browser database: 2014 update. Nucleic Acids Res. 2014;42(1):D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LA, Sternberg MJE. Protein structure prediction on the web: a case study using the Phyre server. Nat Protocol. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Lecomte S, Reverdy L, Le QC, Le MF, Amon A. Unraveling complex interplay between heat shock factor 1 and 2 splicing isoforms. PLoS ONE. 2013;8(2):56–85. doi: 10.1371/journal.pone.0056085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppa S, Pirlakkala L, Saarento H, Sarge KD, Sistonen L. Overexpression of HSF2-β inhibits hemin-induced heat shock gene expression and erythroid differentiation in K562 cells. J Biol Chem. 1997;272:15293–15298. doi: 10.1074/jbc.272.24.15293. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T) Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marai IFM, Haeeb AAM. Buffalo’s biological functions as affected by heat stress—a review. Livest Sci. 2010;127:89–109. doi: 10.1016/j.livsci.2009.08.001. [DOI] [Google Scholar]
- Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Nagata K. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17(1):469–481. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neueder A, Achilli F, Moussaoui S, Bates GP (2014) Novel isoforms of heat shock transcription factor 1, HSF1γα and HSF1γβ, regulate chaperone protein gene transcription. J Biol Chem [DOI] [PMC free article] [PubMed]
- Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. pp. 8.18–8.25. [Google Scholar]
- Sarge KD, Zimarino V, Holm K, Wu C, Morimoto RI. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA binding ability. Genes Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Sarge OP, Kirby JD, Mayo KE, Morimoto RI. Expression of heat shock factor 2 in mouse testis: potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol Reprod. 1994;50:1334–1343. doi: 10.1095/biolreprod50.6.1334. [DOI] [PubMed] [Google Scholar]
- Stephanou A, David SL. Transcriptional modulation of heat shock protein gene expression. Biochem Res Int. 2010 doi: 10.1155/2011/238601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek MA, Repa JJ. The power of real- time PCR. Adv Physiol Educ. 2005;29:151–159. doi: 10.1152/advan.00019.2005. [DOI] [PubMed] [Google Scholar]
- Vujanac M, Fenaroli A, Zimarino V. Constitutive nuclear import and stress-regulated nucleocytoplasmic shuttling of mammalian heat-shock factor 1. Traffic. 2005;6(3):214–229. doi: 10.1111/j.1600-0854.2005.00266.x. [DOI] [PubMed] [Google Scholar]
- Wang GJ, Zhang DM, Mivechi NF. Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis. 2003;36:48–61. doi: 10.1002/gene.10200. [DOI] [PubMed] [Google Scholar]
- Wang G, Ying Z, Jin X, Tu N, Zhang Y, Phillips M, Moskophidis D, Mivechi NF. Essential requirement for both HSF1 and HSF2 transcriptional activity in spermatogenesis and male fertility. Genesis. 2004;38(2):66–80. doi: 10.1002/gene.20005. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Huang L, Zhang J, Moskophidis D, Mivechi NF. Targeted disruption of HSF1 leads to lack of thermotolerance and defines tissue-specific regulation for stress-inducible HSP molecular chaperones. J Cell Biochem. 2002;86:376–393. doi: 10.1002/jcb.10232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accession number of amino acid sequences of HSF genes for all species analyzed(PDF 85 kb)
Neighbor-joining phylogeny of candidate HSF genes in 10 species. The phylogenetic tree was obtained using the MEGA 5.0 software on the basis of complete amino-acid sequences HSF-1, 2, 4 and 5 genes. Rooted Neighbor–joining analysis was performed with 1000 bootstrap replication and evolutionary distances were computed using Poisson’s correction method. The tree is drawn to scale, with the branch length in the same units as those of the evolutionary distance used to infer the phylogenetic tree (PDF 2504 kb)
Sequence alignment of the cloned PCR product of identified HSF1 and HSF2 splice variants (PDF 112 kb)
Blat result of HSF1 splice variant illustrating the structure of HSF1 gene in buffalo and other species using UCSC Genome Browser as per Bos_tau_4.6.1 genome assembly (PDF 44 kb)
Blat result of HSF2 splice variant illustrating the structure of HSF2 gene in buffalo and other species using UCSC Genome Browser as per Bos_tau_4.6.1 genome assembly (PDF 31 kb)
Distribution of conserved motifs in buffalo HSF genes. All the motifs were identified by MEME using complete amino acid sequences of HSF-1, 2, 4 and 5 genes. Name of the gene and combined P-values are shown on the left side of the figure and motif sizes are indicated at the bottom of the figure. Different motifs are indicated by different colors numbered 1–28. The same color in different proteins refers to the same motif (PDF 1316 kb)
Motif sequences of identified HSF genes in buffalo by MEME tool. (PDF 2259 kb)