Abstract
Lactococcal bacteriophages represent one of the leading causes of dairy fermentation failure and product inconsistencies. A new member of the lactococcal 949 phage group, named WRP3, was isolated from cheese whey from a Sicilian factory in 2011. The genome sequence of this phage was determined, and it constitutes the largest lactococcal phage genome currently known, at 130,008 bp. Detailed bioinformatic analysis of the genomic region encoding the presumed initiator complex and baseplate of WRP3 has aided in the functional assignment of several open reading frames (ORFs), particularly that for the receptor binding protein required for host recognition. Furthermore, we demonstrate that the 949 phages target cell wall phospho-polysaccharides as their receptors, accounting for the specificity of the interactions of these phages with their lactococcal hosts. Such information may ultimately aid in the identification of strains/strain blends that do not present the necessary saccharidic target for infection by these problematic phages.
INTRODUCTION
Dairy fermentations rely on the application of strains of Lactococcus lactis for the production of a wide variety of cheeses. However, these strains are under consistent pressure due to the presence of (bacterio)phages. Lactococcal phages are currently classified into 10 species or groups, based on DNA hybridization studies and morphology (1). In recent years, lactococcal phages and their hosts have become an advanced model system for studying Gram-positive phage-host interactions due to the emergence of significant data regarding key molecular players involved in phage adsorption (the phage-encoded receptor binding protein [RBP] and the host-encoded receptor) to the host and the impact of phage infection on sensitive bacterial strains (2–11). The majority of studies have focused on members of the 936 and P335 species, since these are among the most frequently isolated species in the dairy industry. Members of both of these species are believed to recognize a saccharide component of the cell wall polysaccharide (CWPS) that coats the surface of the cell (6, 10–12). Furthermore, phage 1358, which is the namesake of a rarely isolated lactococcal Siphoviridae phage species, is also predicted to recognize a CWPS component on the surface of its host, Lactococcus lactis SMQ-388 (13). The CWPS-encoding operons of currently sequenced lactococcal strains are classified into three genetic groups (types A, B, and C) (10), which have been correlated with phylogenetic subgroups of the 936 phage RBPs (10).
Confirmation that CWPS indeed acts as the receptor for phages belonging to the P335 and 936 groups was obtained by “swapping” the CWPS types produced by different lactococcal strains. In this approach, the operons encoding the two C-type CWPS were compared, and an island of three genes encoding glycosyltransferases was identified as differential between the strains L. lactis NZ9000 (a C1-type strain that is resistant to infection by the P335 phages TP901-1 and LC3 and the 936 phage Viridus JM2) and L. lactis 3107 (a C2-type strain that is sensitive to infection by the above-mentioned phages). Swapping of the CWPS types produced was achieved by introducing the variable region of the CWPS-specifying gene cluster of L. lactis 3107 into a derivative of L. lactis NZ9000, and a change in the phage sensitivity profile of the resulting strain was observed, proving that the receptor for these phages was encoded by this operon and was carbohydrate in nature (14).
In contrast to the saccharidic preference of phages of the 936, P335, and 1358 groups, the lytic c2 phages recognize a protein on the surface of the lactococcal cell, named the phage infection protein (PIP) (15). PIP is a membrane-anchored protein that is analogous to the protein receptor of the Bacillus subtilis phage SPP1, i.e., YueB (16, 17). Genome sequences for representatives of all 10 lactococcal phage species are now available, and as the number of genome sequences of rare and emerging phage species increases, the identification of the RBPs and their target material on the host cell will be possible, thus allowing an improved understanding of the phage-host interactions of a wider spectrum of lactococcal phages. One such rare species of lactococcal phages, named the 949 species, currently has two known and fully sequenced members, i.e., the prototype phage 949, which was isolated from cheese whey in 1975 (18), and L47, which was isolated from sewage in 2014 (19). In the current study, we describe the isolation and characterization of a third member of the 949 group, i.e., phage WRP3. WRP3 was isolated as part of a phage isolation study from a whey sample from a Sicilian cheese production facility in 2011. In previous analyses of the genomes of 949 and L47, two genes were proposed to encode proteins involved in docking onto the cell surface. The RBPs of phages infecting particular species of the lactic acid bacterial group typically possess a conserved amino terminus and a variable carboxyl end. In the present study, we used comparative genomics and host range analysis of the three 949 group phages as tools to identify the phage-encoded RBP. Additionally, we ascertained the nature of the receptor material for members of this group of phages. The identification of the RBP and the receptor material to which these phages attach provides a basis for the identification of a strain rotation scheme that will enhance biotechnological processes, with the potential to improve yields, product quality, and consistency for the dairy fermentation industry.
MATERIALS AND METHODS
Bacteria and phages.
L. lactis 3107 was used for the propagation of phages WRP3 and L47, while L. lactis ML8 was the propagating host of phage 949 (18). L. lactis C10 was the propagating host for P087 (20). Lactococcal host strains were grown without agitation at 30°C in M17 broth (Oxoid Ltd., Hampshire, England) supplemented with 0.5% glucose (GM17) and with 5 μg ml−1 tetracycline where relevant.
Phages were propagated on the appropriate L. lactis indicator strains, which had been grown to an approximate optical density at 600 nm (OD600) of 0.15 in 10 ml GM17 broth. Calcium chloride was added to a final concentration of 10 mM prior to infection of each culture with approximately 107 PFU of the relevant phage and incubation at 30°C or room temperature until lysis had occurred. The lysates were filtered through a 0.45-μm filter to remove any residual bacterial debris and stored at 4°C.
Plaque assays were performed using the previously described double-agar method (21). This method was also applied for host range analysis performed against a collection of 30 L. lactis strains (sensitive strains are listed in Table 1).
TABLE 1.
Host ranges of 949 species phagesa
| L. lactis strain | Reference | CWPS typeb | Titer (PFU/ml) on sensitive host strains infected by: |
||
|---|---|---|---|---|---|
| WRP3 | L47 | 949 | |||
| 3107 | 41 | C2 | 2.9 × 108 | 1.3 × 108 | 2.3 × 107 |
| Wg2 | 42 | C2 | 1.6 × 108 | 1.3 × 108 | 2.2 × 105 |
| 901-1 | 41 | C2 | 5.0 × 105 | 1.5 × 106 | 0 |
| H2 | 43 | C2 | 2.0 × 105 | 3.2 × 105 | 0 |
| R1K10c | C3 | 6.0 × 104 | 3.7 × 105 | 8.1 × 105 | |
| W34 | 44 | C4 | 2.0 × 104 | 5.0 × 104 | 0 |
| UC109d | A | 0 | 4.0 × 104 | 0 | |
| ML8 | 1 | B | 0 | 0 | 3.3 × 107 |
The host ranges of the three phages were assessed on 30 lactococcal strains, and only those infected by the phages are presented here, along with information on the level of susceptibility to phages and the CWPS type for each host strain.
The CWPS type was characterized by multiplex PCR.
Provided by Jan Kok, University of Groningen, The Netherlands.
Obtained from the University College Cork strain collection.
Isolation of WRP3.
Phage WRP3 was isolated as part of a phage screening of whey samples obtained from Sicilian cheese production facilities which do not add starter strains for cheese making. Specifically, phage WRP3 was isolated from the whey sampled during the production of Caciocavallo Palermitano cheese; this cheese is made with raw cow's milk transformed into wooden vats which host microbial biofilms on their surfaces that act as a source of starter lactic acid bacteria (22). The phage was isolated using the indicator strain L. lactis 3107.
DNA preparation.
DNA for sequencing of the WRP3 phage genome was extracted from 50 ml of fresh phage lysate (>108 PFU ml−1) by treatment with 1 μg ml−1 DNase and RNase at 37°C for 30 min. After centrifugation at 13,200 × g for 15 min, the lysate (50 ml) was transferred to a new tube, and polyethylene glycol 8000 (PEG) (Sigma-Aldrich, St. Louis, MO) and sodium chloride were added as dry powders, to final concentrations of 10% and 0.5 M, respectively, after which the suspension was incubated at 4°C overnight. Subsequently, the suspension was centrifuged at 17,700 × g for 15 min, and the supernatant was removed. The PEG/NaCl-induced precipitate was resuspended in 5 ml of Tris-EDTA (TE) buffer (pH 9.0) and treated with 120 μl of 20 mg ml−1 proteinase K for 20 min at 56°C, followed by treatment with SDS at a final concentration of 2% at 65°C for 20 min. Potassium acetate was added to a final concentration of 1 M, followed by incubation on ice for 20 min before centrifugation at 13,200 × g for 10 min. The supernatant was then extracted with phenol-chloroform (25:24:1 phenol:chloroform:isoamyl alcohol) (Sigma-Aldrich, St. Louis, MO) at least twice, and the resulting aqueous phase was then used to precipitate DNA by the addition of 2.5 volumes of ice-cold 96% ethanol and 0.1 volume of sodium acetate (pH 4.8). Subsequent to centrifugation, the DNA-containing pellet was washed in 70% ethanol and resuspended in 100 μl of TE buffer (pH 8.0).
Genome sequencing, assembly, and annotation.
Five micrograms of DNA was extracted and verified by nanodrop quantification. Confirmatory molecular identification tests were also conducted on the DNA extract prior to shipment to the contract sequencing facility (Macrogen Inc., Seoul, South Korea). A 77-fold sequencing coverage was obtained using pyrosequencing technology on a 454 FLX instrument. The individual sequence files generated by the 454 FLX instrument were assembled with GSassembler (454 Lifesciences, Branford, CT) to generate a consensus sequence. Quality improvement of the genome sequence involved sequencing of 23 PCR products across the entire genome to ensure correct assembly, double stranding, and the resolution of any remaining base conflicts occurring within homopolymer tracts. Protein-encoding open reading frames (ORFs) were predicted using Zcurve_V, followed by manual assessment, curation, and correction of the predicted ORFs (23). ORFs were also determined/verified using the ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/) and GeneMark (http://exon.gatech.edu/) software packages, as well as manually by looking for ATG, TTG, and GTG as potential start codons downstream of a consensus ribosome binding sequence (AGAAGGAGGT) (24). A functional annotation of ORFs was performed on the basis of BLASTP (25) analysis against the nonredundant (nr) protein database provided by the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The proposed functions of many ORFs were further confirmed by querying the Pfam protein domain database (26) and the NCBI Conserved Domain Database (27) and by performing homology prediction searches using HHPred (28). The genome was scanned for the presence of potential tRNA genes by using tRNAscan-SE (29).
Electron microscopy.
Purification of the WRP3 phage by use of a CsCl gradient was performed as previously described (30). Adsorption of CsCl-purified phage or freshly produced unpurified lysate to freshly prepared carbon film floated from a freshly coated mica sheet and negative staining with 2% (wt/vol) uranyl acetate were done as described previously (31). The film was picked up with a 400-mesh copper grid (Agar Scientific, Essex, United Kingdom), and specimens were examined with a Tecnai 10 transmission electron microscope (FEI, Eindhoven, The Netherlands) operated at an acceleration voltage of 80 kV.
Lactococcal CWPS type identification.
The CWPS types of the lactococcal strains employed in the host range analysis were determined using the multiplex PCR system described previously by Mahony and colleagues (10), with classification into four categories (A, B, C, and undefined). Additionally, the dominant, C-type CWPS collection was subclassified using the multiplex PCR system devised by Ainsworth et al. to provide further information on the CWPS gene cluster-based classification of the strains and the degrees of specificity of the host preferences of the phages employed in the host range analysis (14).
CWPS receptor identification.
To investigate the possibility that WRP3 targets CWPS as its receptor material, the host ranges of 949, L47, and WRP3 were assessed on L. lactis NZ9000 (which produces the C1-type CWPS and should be insensitive to infection by these phages), L. lactis 3107 (which produces the C2-type CWPS and is a host to these phages), and an engineered yet isogenic derivative of L. lactis NZ9000 which does not produce the C1-type CWPS (as its parent NZ9000 does) but instead produces a nisin-inducible C2-type CWPS, which was previously proven to provide the saccharidic receptor for phages LC3 and TP901-1 (14).
Nucleotide sequence accession number.
The GenBank accession number for the WRP3 sequence is KM677185.
RESULTS AND DISCUSSION
Phage WRP3 is a new member of the lactococcal 949 phage species.
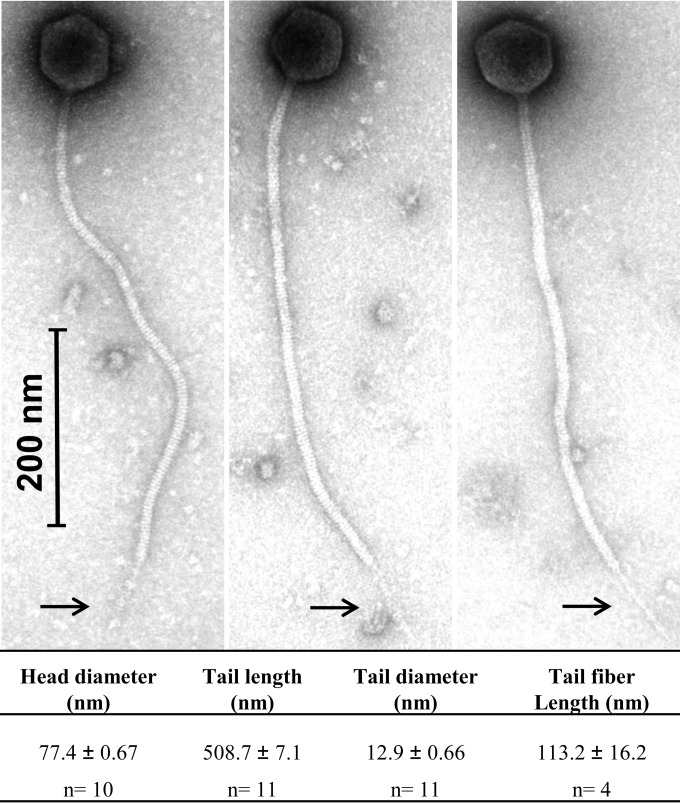
Through a combined approach of genome sequencing and electron microscopic analysis, phage WRP3 (Fig. 1) was confirmed to represent a new member of the 949 group of lactococcal phages. The tail of WRP3 measures in excess of 500 nm, which is characteristic of the 949 phage group (Fig. 1). At the distal end of the tail, a long tail fiber of approximately 110 nm is observed, a feature also observed in the 949 phage L47 (19). The tail fiber of WRP3 additionally appears to possess globular appendages along its length. Such tail fiber structures are generally believed to play a role in the recognition of the host that ultimately leads to infection of the sensitive host cell. In this study, the genome of WRP3 was sequenced in order to perform a focused comparative genomic analysis of members of the 949 group and to identify the genes that may encode the receptor binding protein and tail fiber, which are responsible for the initial interactions between the phage and its lactococcal host.
FIG 1.
Representative micrographs of WRP3. The typical long tail of approximately 510 nm and the isometric capsid of the 949-like phages can be observed. A tail fiber structure (arrows) could also be observed protruding from the tail tip in approximately 40% of the population assessed, while a globular tail fiber decoration was observed surrounding the tail fiber.
The genome of phage WRP3 was determined to be 130,008 bp long, encompassing 178 open reading frames and four tRNA genes (corresponding to Met, Arg, Asp, and Pro), thereby representing the largest genome of sequenced phages infecting L. lactis to date (18, 19). The codon usage of the phage and that of the lactococcal strain MG1363 are quite similar; therefore, it is not expected that the tRNA genes contribute to the fitness or virulence of the phage, as they do not appear to compensate for a lack of these tRNAs in the host, in agreement with the findings for 949 and its lactococcal host (18). The GC% content of WRP3 is similar to that of phages 949 and L47 (949 species members), at 32.36%, indicating the acquisition of genetic elements from a low-GC% ancestor and a common ancestor for the members of this phage species.
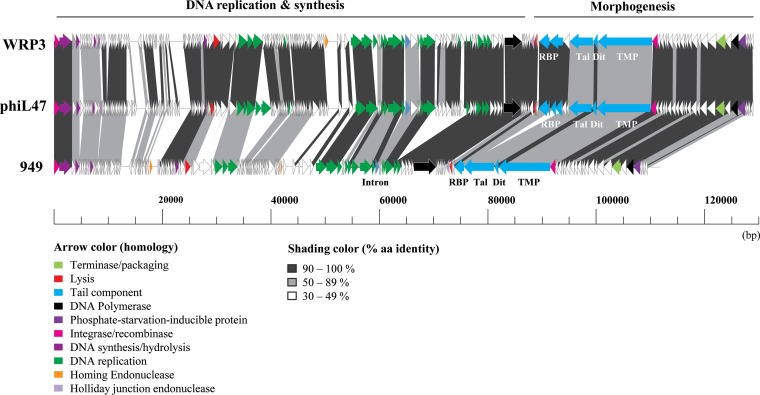
The genome is organized into four modules, organized in two head-to-head pairs of gene clusters (Fig. 2). Based on BLASTP and Pfam analyses, it was possible to assign putative functions to 51 of the 178 ORFs (29%), with many of these annotated functions specified by ORFs located within the deduced morphogenesis and DNA replication modules (Fig. 2).
FIG 2.
Genomic organization of three 949-like phages: WRP3, L47, and 949. ORFs are represented by arrows. Arrows of the same color have at least 30% identity at the amino acid level. TMP, tail tape measure protein; Dit, distal tail protein; Tal, tail-associated lysin; RBP, receptor binding protein. White arrows represent hypothetical proteins.
Molecular players in phage-host interactions of the 949 phage species.
In order to understand the phage-host interactions of the 949 phages, a focused bioinformatic analysis of the genomic regions that are proposed to encode the so-called “initiator complex” (which in the lactococcal P335 and 936 model systems is comprised of the tail tape measure protein [TMP], the distal tail [Dit] protein, and the tail-associated lysin [Tal] protein [4, 32, 33]) and the tail tip or so-called “baseplate” regions of the phages, which are proposed to interact with the cell surface receptor (2, 3, 7, 11, 34–36), was undertaken using WRP3 as the model for the species. Based on HHPred analysis, and in keeping with the conserved gene order in other lactococcal phages, e.g., TP901-1 (P335 species) and p2 (936 species), the TMP (predicted to be encoded by orf154WRP3) is recognizable by containing a C-terminal LysM motif, proposed to be involved in peptidoglycan binding and degradation, and two predicted transmembrane domains in the central region of the protein. The ensuing gene (orf153WRP3) is predicted to encode the distal tail protein of the Bacillus subtilis phage SPP1 and the lactococcal P335 phage TP901-1, based on HHPred analysis (E values of 1.2E−37 and 4.8E−30, respectively; the E value provides a measure of sequence similarity, i.e., the lower the E value, the more significant are the score and alignment). The protein encoded by orf152WRP3 has an unknown function; however, NCBI conserved domain analysis revealed similarity to a putative prophage endopeptidase (E value = 3.13E−3), which is consistent with a Tal-like (or tail lysin) function (8, 37). orf151WRP3 encodes a small protein that may perform accessory phage-host interactions, with HHPred predictions of a potential carbohydrate binding domain on this protein. Since a tail fiber structure is observed in WRP3 (Fig. 1) and L47, but not 949, it is conceivable that this gene may encode the tail fiber structure, since this gene is observed in the former two phages and does not appear to have a homologous equivalent in the 949 genome. Similarly, orf150WRP3 has an unknown function, but given its genomic position, it may encode part of the baseplate structure. Previously, the homologous equivalents of orf149/150WRP3 were both proposed as candidate genes to encode the receptor binding protein. Our analysis suggests that the phage-host adsorption and interaction functions are encoded by orf149WRP3, which appears to encode the receptor binding protein (RBP) of the baseplate, based on HHPred alignments with the baseplate protein of the coliphage T4 (E value = 2.5E−27).
To further strengthen the notion that orf149WRP3 encodes the RBP, an alignment of the sequences of the proposed RBPs of WRP3, 949, and L47 was performed. This revealed 99% amino acid (aa) sequence identity across the RBPs of WRP3 and L47, with a more distant relationship (75% aa identity) to that of 949, with the most notable divergence at the C-terminal region of the protein sequences, consistent with the identification of the C-terminally located host specificity domain of the RBPs of several phages of L. lactis and other lactic acid bacterial genera (6, 38, 39).
Host range analysis of 949 group phages.
The similarity between the proposed RBPs of WRP3 and L47, partnered with the C-terminal dissimilarity of the RBP encoded by 949, is congruent with the host range profiles of 949, WRP3, and L47 (Table 1). In this study, 30 lactococcal strains were assessed for sensitivity to these three phages, and it was demonstrated that WRP3 and L47 have identical host ranges, with the exception of one strain (UC109). This may be representative of the presence of antiphage systems in L. lactis UC109 that target WRP3 more efficiently than L47, although L47 has a reduced efficiency of plaque formation (approximately 10−4) on this strain compared to that on L. lactis 3107, its propagating host. In contrast to the host ranges of WRP3 and L47, phage 949 is capable of infecting just three of the seven strains that L47 infects (and six strains that WRP3 infects), which may be a reflection of the divergent C terminus of 949's RBP relative to those of L47 and WRP3. To understand the significance of the host ranges of the phages and to determine if the phages have a preference for lactococcal strains of a particular CWPS type (A, B, or C), a multiplex PCR system previously designed for CWPS designation of lactococcal strains was employed to classify strains that were sensitive to the phages assessed in this study. Through this analysis (and using information from the study of Mahony et al. [10]), we determined that the majority of the strains possessed the C-type CWPS and that the C2 subtype dominated among the sensitive C-type strains. Therefore, it appears that the 949 group phages possess a preference for C-type CWPS-expressing lactococcal strains. The consistency with which C-type strains are preferentially recognized by the 949 phages provides evidence of a carbohydrate receptor rather than a surface protein as for the c2 group phages. To further elucidate if these phages truly recognize a carbohydrate on the surface of the lactococcal host, “CWPS swapping” was performed to assess if the phage sensitivity profile of the altered strain would be changed as expected if elements of the CWPS are the receptors for these phages.
949-like phages recognize a saccharidic receptor on the lactococcal host.
Members of the 936, P335, and 1358 species of phages are known to recognize saccharidic receptors on the host cell surface (6, 13, 14). Host range analysis revealed a strong preference of WRP3 and L47 for lactococcal strains possessing the CWPS C2 subtype, but these phages were also capable of infecting C3 and C4 subtype strains and had a limited ability to infect strains outside the CWPS C type (Table 1). L. lactis 3107 is a lactococcal C2-type CWPS-encoding strain that is host to 949 phages, while L. lactis NZ9000 is a C1-type CWPS-encoding strain that is resistant to 949 phages. CWPS-swapping experiments in which a derivative of the C1-type lactococcal strain NZ9000, now expressing the C2 CWPS instead of its native C1 CWPS, became sensitive to infection by the 949 phages demonstrated that the saccharidic C2-type CWPS is the receptor for these phages (Table 2). To eliminate the possibility of false-positive reactions, several controls were employed in this experiment: first, the two wild-type strains, L. lactis 3107 and NZ9000, were assessed for their phage sensitivity profiles; second, L. lactis NZ9000 and a derivative of this strain in which the C1-encoding glycosyltransferase gene (GT1) was mutated by introduction of a stop codon harboring the empty vector (pPTPi) only were assessed; and third, a mutant derivative of L. lactis NZ9000 harboring a construct providing the 3107-derived glycosyltransferase-encoding genes (NZ9000::GT1/pPTPi-C2) and which, when induced, should permit host range swapping to occur was also tested in both the uninduced and induced states (Table 2). As expected, only NZ9000::GT1/pPTPi-C2 permitted phage host range swapping. Furthermore, the prototypic phage of the P087 species (P087) showed a similar ability to infect L. lactis 3107, though it was incapable of infecting L. lactis NZ9000 (data not shown), and by employing the same approach as that with the 949 phages, it was possible to determine that CWPS also acts as the receptor for this phage group. The data suggest that, in agreement with the 949 phage data, host range swapping is possible for P087 in a derivative of L. lactis NZ9000 expressing the C2-type CWPS instead of the C1-type CWPS (Table 2). These data provide conclusive evidence that additional members of rare and emerging lactococcal phage species recognize cell surface polysaccharides, accounting for their particular host specificity relative to the general and widespread infection ability of c2-type phages, which recognize a proteinaceous receptor. As new species of phages continue to emerge and evolve, it is essential to develop an understanding of the interactions of lactococcal phages and their target molecules in order to be able to predict the interactions of emerging phage species, thus allowing a rapid response to emerging problems in industry.
TABLE 2.
Host range swapping abilities of 949-like phages and P087 between L. lactis NZ9000 and 3107a
| L. lactis strain | CWPS type | Efficiency of plaque formationb |
|||
|---|---|---|---|---|---|
| WRP3 | L47 | 949 | P087 | ||
| 3107 | C2 | 1 | 1 | 1 | 1 |
| NZ9000 | C1 | ≤2.87 × 10−8 | ≤1.83 × 10−7 | ≤4.20 × 10−6 | ≤2.51 × 10−6 |
| NZ9000/pPTPi (induced) | C1 | ≤2.87 × 10−8 | ≤1.83 × 10−7 | ≤4.20 × 10−6 | ≤2.51 × 10−6 |
| NZ9000::GT1/pPTPi (induced) | C1 | ≤2.87 × 10−8 | ≤1.83 × 10−7 | ≤4.20 × 10−6 | ≤2.51 × 10−6 |
| NZ9000::GT1/pPTPi-C2 (uninduced) | C2 | ≤2.87 × 10−8 | ≤1.83 × 10−7 | ≤4.20 × 10−6 | ≤2.51 × 10−6 |
| NZ9000::GT1/pPTPi-C2 (induced) | C2 | 7.8 × 10−2 | 1.01 × 10−1 | 3.9 × 10−2 | 6.08 × 10−2 |
Host range swapping ability is defined as the ability of phages that originally infected the lactococcal C2-type CWPS strain 3107 and were unable to infect the C1-type lactococcal strain NZ9000 to infect a derivative of NZ9000 that expresses the C2-type CWPS in place of its native C1-type CWPS.
The efficiencies of plaque formation of the phages on each strain are expressed relative to that on the original host, 3107. All results are averages for assays performed at least in triplicate.
Comparative genomic analysis of WRP3, 949, and L47.
The genomes of the three currently sequenced members of the 949 species (949, L47, and WRP3) were compared, and the WRP3 genome was shown to possess 93% nucleotide (nt) identity to the genome of phage 949 (across 80% of the former genome) and 96% nt identity to that of phage L47 (across 86% of the WRP3 genome). Comparative genomic analysis confirmed the close relationship between WRP3 and L47, despite their different origins, i.e., WRP3 is an isolate from cheese whey, while L47 is a sewage isolate. The genome of WRP3 (similar to those of 949 and L47) is organized into four gene clusters (Fig. 1). Gene modules 1 and 2, spanning orf1WRP3 to orf30WRP3 and orf31WRP3 to orf44WRP3, respectively, are arranged in a head-to-head constellation, and the putative functions of only four gene products are known. Therefore, while it is difficult to predict the overall functions of these gene modules, it is possible that the presence of genes encoding transglycosylases and the endo-DNase may indicate functions that include host cell lysis and nucleotide metabolism. Gene module 3 (orf45WRP3 to orf145WRP3) appears to represent a gene cluster encoding functions associated with DNA replication and nucleotide biosynthesis, as previously observed for the 949 and L47 genomes (18, 19). Gene module 4 of these phages (represented here by orf146WRP3 to orf178WRP3) encodes the presumed morphogenesis and packaging functions of the 949 phages. There is a high degree of similarity in this module among the three phages, with features such as the tail and presumed capsid structural protein-encoding genes (orf152WRP3, orf153WRP3, orf156WRP3, orf160WRP3, orf162WRP3, orf163WRP3, orf165WRP3, and orf167WRP3 to orf170WRP3), the presumed tail tape measure protein-encoding gene (orf154WRP3), and the receptor binding protein-encoding genes (orf149WRP3 and orf150WRP3) (see above). Additionally, a single identifiable small terminase-encoding gene (orf171WRP3) is observed adjacent to a hypothetical protein-encoding gene (orf172WRP3), which may also play a role in DNA packaging during phage assembly.
Conclusions.
It is apparent from the current work and previous studies that the majority of lactococcal phages, including members of the 949, P087, 936, P335, and 1358 groups, recognize cell wall polysaccharides on the surface of the target cell (3, 12–14). These five phage groups account for half of the 10 currently classified lactococcal phage groups (1). To date, the biochemical nature of the CWPS is known for three lactococcal strains, i.e., L. lactis MG1363, 3107, and SMQ-388, which are hosts to members of the 936, P335, and 1358 species, respectively (13, 14, 40). As increasing amounts of biochemical data emerge for the hosts of lactococcal phages, it will be possible to discern the specific saccharidic targets of these phages and more precise details regarding their interactions with the host. Given the narrow host range of the majority of the remaining lactococcal phage species whose receptors are currently unknown (P034, KSY1, 1706, and Q54 species), it seems likely that their receptor preference may also be saccharidic in nature, and with the ever-increasing number of available genome sequences, the specific phage-host interactions of a wide variety of lactococcal phages will become clear, permitting early warning systems and risk assessments to be developed for application in the dairy industry. For example, the relationships between the phylogeny of the RBPs of the 936 species phages and the CWPS types of their lactococcal hosts (A, B, or C) are currently defined and provide a rapid risk assessment tool for application in any dairy setting (10). It is envisaged that as more isolates of the rare lactococcal phage species emerge, it will be possible to derive such relationships for these as well, providing the next generation of phage prevention tools for the dairy fermentation sector.
ACKNOWLEDGMENTS
J.M. was the recipient of a Technology Innovation Development Award (TIDA) (award 14/TIDA/2287) funded by the Science Foundation Ireland (SFI). D.V.S. was supported by a Principal Investigator award (award 13/IA/1953) through the Science Foundation Ireland (SFI). W.R. was supported by a grant from the University of Palermo Perfezionamento Estero (grant 3352/2010).
We thank Jan Kok of the University of Groningen, The Netherlands, for the kind gift of lactococcal strain R1K10. Antonino Di Grigoli (University of Palermo) is also thanked for providing whey samples for phage screening.
REFERENCES
- 1.Deveau H, Labrie SJ, Chopin MC, Moineau S. 2006. Biodiversity and classification of lactococcal phages. Appl Environ Microbiol 72:4338–4346. doi: 10.1128/AEM.02517-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sciara G, Bebeacua C, Bron P, Tremblay D, Ortiz-Lombardia M, Lichiere J, van Heel M, Campanacci V, Moineau S, Cambillau C. 2010. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc Natl Acad Sci U S A 107:6852–6857. doi: 10.1073/pnas.1000232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spinelli S, Desmyter A, Verrips CT, de Haard HJ, Moineau S, Cambillau C. 2006. Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat Struct Mol Biol 13:85–89. doi: 10.1038/nsmb1029. [DOI] [PubMed] [Google Scholar]
- 4.Bebeacua C, Tremblay D, Farenc C, Chapot-Chartier MP, Sadovskaya I, van Heel M, Veesler D, Moineau S, Cambillau C. 2013. Structure, adsorption to host, and infection mechanism of virulent lactococcal phage p2. J Virol 87:12302–12312. doi: 10.1128/JVI.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricagno S, Campanacci V, Blangy S, Spinelli S, Tremblay D, Moineau S, Tegoni M, Cambillau C. 2006. Crystal structure of the receptor-binding protein head domain from Lactococcus lactis phage bIL170. J Virol 80:9331–9335. doi: 10.1128/JVI.01160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tremblay DM, Tegoni M, Spinelli S, Campanacci V, Blangy S, Huyghe C, Desmyter A, Labrie S, Moineau S, Cambillau C. 2006. Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site. J Bacteriol 188:2400–2410. doi: 10.1128/JB.188.7.2400-2410.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins B, Bebeacua C, Mahony J, Blangy S, Douillard FP, Veesler D, Cambillau C, van Sinderen D. 2013. Structure and functional analysis of the host recognition device of lactococcal phage Tuc2009. J Virol 87:8429–8440. doi: 10.1128/JVI.00907-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stockdale SR, Mahony J, Courtin P, Chapot-Chartier MP, van Pijkeren JP, Britton RA, Neve H, Heller KJ, Aideh B, Vogensen FK, van Sinderen D. 2013. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J Biol Chem 288:5581–5590. doi: 10.1074/jbc.M112.444901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ainsworth S, Zomer A, Mahony J, van Sinderen D. 2013. Lytic infection of Lactococcus lactis by bacteriophages Tuc2009 and c2 triggers alternative transcriptional host responses. Appl Environ Microbiol 79:4786–4798. doi: 10.1128/AEM.01197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahony J, Kot W, Murphy J, Ainsworth S, Neve H, Hansen LH, Heller KJ, Sorensen SJ, Hammer K, Cambillau C, Vogensen FK, van Sinderen D. 2013. Investigation of the relationship between lactococcal host cell wall polysaccharide genotype and 936 phage receptor binding protein phylogeny. Appl Environ Microbiol 79:4385–4392. doi: 10.1128/AEM.00653-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veesler D, Spinelli S, Mahony J, Lichiere J, Blangy S, Bricogne G, Legrand P, Ortiz-Lombardia M, Campanacci V, van Sinderen D, Cambillau C. 2012. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc Natl Acad Sci U S A 109:8954–8958. doi: 10.1073/pnas.1200966109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupont K, Janzen T, Vogensen FK, Josephsen J, Stuer-Lauridsen B. 2004. Identification of Lactococcus lactis genes required for bacteriophage adsorption. Appl Environ Microbiol 70:5825–5832. doi: 10.1128/AEM.70.10.5825-5832.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farenc C, Spinelli S, Vinogradov E, Tremblay D, Blangy S, Sadovskaya I, Moineau S, Cambillau C. 2014. Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein. J Virol 88:7005–7015. doi: 10.1128/JVI.00739-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ainsworth S, Sadovskaya I, Vinogradov E, Courtin P, Guerardel Y, Mahony J, Grard T, Cambillau C, Chapot-Chartier MP, van Sinderen D. 2014. Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. mBio 5:e00880-14. doi: 10.1128/mBio.00880-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valyasevi R, Sandine WE, Geller BL. 1991. A membrane protein is required for bacteriophage c2 infection of Lactococcus lactis subsp. lactis C2. J Bacteriol 173:6095–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baptista C, Santos MA, Sao-Jose C. 2008. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. J Bacteriol 190:4989–4996. doi: 10.1128/JB.00349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sao-Jose C, Lhuillier S, Lurz R, Melki R, Lepault J, Santos MA, Tavares P. 2006. The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J Biol Chem 281:11464–11470. doi: 10.1074/jbc.M513625200. [DOI] [PubMed] [Google Scholar]
- 18.Samson JE, Moineau S. 2010. Characterization of Lactococcus lactis phage 949 and comparison with other lactococcal phages. Appl Environ Microbiol 76:6843–6852. doi: 10.1128/AEM.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavanagh D, Guinane CM, Neve H, Coffey A, Ross RP, Fitzgerald GF, McAuliffe O. 2014. Phages of non-dairy lactococci: isolation and characterization of ΦL47, a phage infecting the grass isolate Lactococcus lactis ssp. cremoris DPC6860. Front Microbiol 4:417. doi: 10.3389/fmicb.2013.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villion M, Chopin MC, Deveau H, Ehrlich SD, Moineau S, Chopin A. 2009. P087, a lactococcal phage with a morphogenesis module similar to an Enterococcus faecalis prophage. Virology 388:49–56. doi: 10.1016/j.virol.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 21.Lillehaug D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J Appl Microbiol 83:85–90. doi: 10.1046/j.1365-2672.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- 22.Settanni L, Di Grigoli A, Tornambe G, Bellina V, Francesca N, Moschetti G, Bonanno A. 2012. Persistence of wild Streptococcus thermophilus strains on wooden vat and during the manufacture of a traditional Caciocavallo type cheese. Int J Food Microbiol 155:73–81. doi: 10.1016/j.ijfoodmicro.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 23.Guo FB, Zhang CT. 2006. ZCURVE_V: a new self-training system for recognizing protein-coding genes in viral and phage genomes. BMC Bioinformatics 7:9. doi: 10.1186/1471-2105-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 11:731–753. doi: 10.1101/gr.GR-1697R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL. 2004. The Pfam protein families database. Nucleic Acids Res 32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 31.Deasy T, Mahony J, Neve H, Heller KJ, van Sinderen D. 2011. Isolation of a virulent Lactobacillus brevis phage and its application in the control of beer spoilage. J Food Prot 74:2157–2161. doi: 10.4315/0362-028X.JFP-11-262. [DOI] [PubMed] [Google Scholar]
- 32.Bebeacua C, Lai L, Vegge CS, Brondsted L, van Heel M, Veesler D, Cambillau C. 2013. Visualizing a complete Siphoviridae member by single-particle electron microscopy: the structure of lactococcal phage TP901-1. J Virol 87:1061–1068. doi: 10.1128/JVI.02836-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGrath S, Neve H, Seegers JF, Eijlander R, Vegge CS, Brondsted L, Heller KJ, Fitzgerald GF, Vogensen FK, van Sinderen D. 2006. Anatomy of a lactococcal phage tail. J Bacteriol 188:3972–3982. doi: 10.1128/JB.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bebeacua C, Bron P, Lai L, Vegge CS, Brondsted L, Spinelli S, Campanacci V, Veesler D, van Heel M, Cambillau C. 2010. Structure and molecular assignment of lactococcal phage TP901-1 baseplate. J Biol Chem 285:39079–39086. doi: 10.1074/jbc.M110.175646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spinelli S, Campanacci V, Blangy S, Moineau S, Tegoni M, Cambillau C. 2006. Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901-1. J Biol Chem 281:14256–14262. doi: 10.1074/jbc.M600666200. [DOI] [PubMed] [Google Scholar]
- 36.Sciara G, Blangy S, Siponen M, McGrath S, van Sinderen D, Tegoni M, Cambillau C, Campanacci V. 2008. A topological model of the baseplate of lactococcal phage Tuc2009. J Biol Chem 283:2716–2723. doi: 10.1074/jbc.M707533200. [DOI] [PubMed] [Google Scholar]
- 37.Kenny JG, McGrath S, Fitzgerald GF, van Sinderen D. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J Bacteriol 186:3480–3491. doi: 10.1128/JB.186.11.3480-3491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kot W, Hammer K, Neve H, Vogensen FK. 2013. Identification of the receptor-binding protein in lytic Leuconostoc pseudomesenteroides bacteriophages. Appl Environ Microbiol 79:3311–3314. doi: 10.1128/AEM.00012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duplessis M, Levesque CM, Moineau S. 2006. Characterization of Streptococcus thermophilus host range phage mutants. Appl Environ Microbiol 72:3036–3041. doi: 10.1128/AEM.72.4.3036-3041.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapot-Chartier MP, Vinogradov E, Sadovskaya I, Andre G, Mistou MY, Trieu-Cuot P, Furlan S, Bidnenko E, Courtin P, Pechoux C, Hols P, Dufrene YF, Kulakauskas S. 2010. Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J Biol Chem 285:10464–10471. doi: 10.1074/jbc.M109.082958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braun V, Hertwig S, Neve H, Geis A, Teuber M. 1989. Taxonomic differentiation of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization, and protein profiles. J Gen Microbiol 135:2551–2560. [Google Scholar]
- 42.Schafer A, Geis A, Neve H, Teuber M. 1991. Bacteriophage receptors of Lactococcus lactis subsp. ‘diacetylactis’ F7/2 and Lactococcus lactis subsp. cremoris Wg2-1. FEMS Microbiol Lett 62:69–73. [DOI] [PubMed] [Google Scholar]
- 43.Boyce JD, Davidson BE, Hillier AJ. 1995. Spontaneous deletion mutants of the Lactococcus lactis temperate bacteriophage BK5-T and localization of the BK5-T attP site. Appl Environ Microbiol 61:4105–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Josephsen J, Andersen N, Behrndt H, Brandsborg E, Christiansen G, Hansen MB, Hansen S, Waagner Nielsen E, Vogensen FK. 1994. An ecological study of lytic bacteriophages of Lactococcus lactis subsp. cremoris isolated in a cheese plant over a five year period. Int Dairy J 4:123–140. doi: 10.1016/0958-6946(94)90064-7. [DOI] [Google Scholar]