Abstract
The majority of human infections are caused by biofilms. The biofilm mode of growth enhances the pathogenicity of Staphylococcus spp. considerably, because once they adhere, staphylococci embed themselves in a protective, self-produced matrix of extracellular polymeric substances (EPSs). The aim of this study was to investigate the influence of forces of staphylococcal adhesion to different biomaterials on icaA (which regulates the production of EPS matrix components) and cidA (which is associated with cell lysis and extracellular DNA [eDNA] release) gene expression in Staphylococcus aureus biofilms. Experiments were performed with S. aureus ATCC 12600 and its isogenic mutant, S. aureus ATCC 12600 Δpbp4, deficient in peptidoglycan cross-linking. Deletion of pbp4 was associated with greater cell wall deformability, while it did not affect the planktonic growth rate, biofilm formation, cell surface hydrophobicity, or zeta potential of the strains. The adhesion forces of S. aureus ATCC 12600 were the strongest on polyethylene (4.9 ± 0.5 nN), intermediate on polymethylmethacrylate (3.1 ± 0.7 nN), and the weakest on stainless steel (1.3 ± 0.2 nN). The production of poly-N-acetylglucosamine, eDNA presence, and expression of icaA genes decreased with increasing adhesion forces. However, no relation between adhesion forces and cidA expression was observed. The adhesion forces of the isogenic mutant S. aureus ATCC 12600 Δpbp4 (deficient in peptidoglycan cross-linking) were much weaker than those of the parent strain and did not show any correlation with the production of poly-N-acetylglucosamine, eDNA presence, or expression of the icaA and cidA genes. This suggests that adhesion forces modulate the production of the matrix molecule poly-N-acetylglucosamine, eDNA presence, and icaA gene expression by inducing nanoscale cell wall deformation, with cross-linked peptidoglycan layers playing a pivotal role in this adhesion force sensing.
INTRODUCTION
Staphylococcus spp. present an important group of potentially pathogenic strains and species. According to estimates by the National Institutes of Health, about 80% of all human infections are caused by biofilms (1). The biofilm mode of growth considerably enhances the pathogenicity of Staphylococcus spp. when the biofilm is formed on the surfaces of biomaterial implants and devices, such as total knee or hip arthroplasties or pacemakers (2). Biofilm formation starts with the adhesion of individual organisms to a substratum surface. Initially, adhesion is reversible, but the bond between an adhering organism and a substratum surface rapidly matures over time to become stronger, and eventually, adhesion is irreversible (3). Adhesion is further enforced through the production of a matrix consisting of extracellular polymeric substances (EPSs) by the adhering organisms, in which they grow and find shelter against the host immune system and antibiotic treatment. The EPS composition largely depends on the bacterial strains and environmental conditions, but major components of EPSs across different species are polysaccharides, proteins, and extracellular DNA (eDNA) (4).
It is difficult to envision how adhering bacteria regulate EPS production in response to their adhesion to different surfaces. Recently, we have proposed that the bacterial response to adhesion is dictated by the magnitude of the force by which a bacterium adheres to a surface (5) and distinguished three regimes of adhesion forces (Fig. 1). In the planktonic regime, bacteria adhere weakly and, accordingly, cannot realize that they are on a surface and retain their planktonic phenotype. The opposite regime is called the lethal regime, where strong adhesion forces lead to high cell wall stresses, retarded growth, and finally, cell death. Both the planktonic regime and the lethal regime occur mostly after application of coatings, like highly hydrated and hydrophilic polymer brush coatings or positively charged quaternary ammonium coatings exerting strong, attractive electrostatic forces on adhering bacteria, which are usually negatively charged under physiological conditions (6). Most biomaterials used for implants and devices, however, exert intermediate adhesion forces on adhering bacteria, and this regime is called the interaction regime. In the interaction regime, bacteria are hypothesized to respond to the adhesion forces exerted by a surface through the production of various matrix components. Indeed, clinically, biofilms of the same strain can have different pathogenicities when formed on different biomaterials (7). For example, in abdominal wall surgery, hydrophobic surgical meshes made of polytetrafluoroethylene are more susceptible to infection than meshes made of the less hydrophobic polypropylene (8). On orthopedic biomaterials, the levels of icaA expression by Staphylococcus epidermidis and EPS production were higher on polyethylene (PE) than on polymethylmethacrylate (PMMA). Moreover, biofilms on polyethylene showed lower levels of susceptibility to gentamicin than biofilms on polymethylmethacrylate (9).
FIG 1.
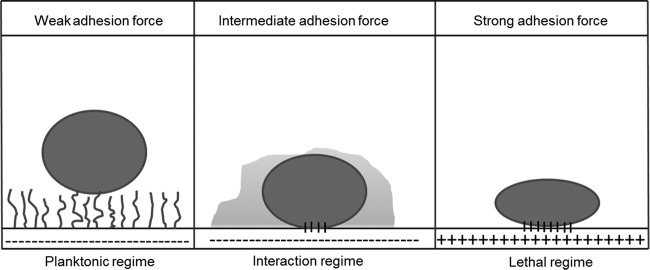
Regimented scheme for the interaction of bacteria with substratum surfaces. Weakly adhering bacteria retain the planktonic phenotype, while strongly adhering ones die upon contact. In the interaction regime, bacteria are hypothesized to respond to their adhering state with differential gene expression according to the adhesion force value that they experience (5).
Little is known, however, about the exact role of adhesion forces on the complex response of adhering bacteria in the interaction regime. A likely hypothesis is that the adhesion forces cause nanoscale cell wall deformations and membrane stresses that act as a mechanism signaling an organism to enter its adhering state. Therefore, the response of bacteria to their adhering state not only differs on different biomaterials but also depends on the rigidity of the cell wall itself, which is maintained in Gram-positive strains as a relatively thick layer of cross-linked peptidoglycan. Measuring nanoscale cell wall deformation upon bacterial adhesion to a surface is extremely difficult. Recently, a new, highly sensitive method has been proposed. That method is based on surface-enhanced fluorescence and measures cell wall deformation over a large number of adhering bacteria under the influence of the naturally occurring adhesion forces arising from a substratum surface (10). Surface-enhanced fluorescence is the phenomenon of increased fluorescence when fluorophores come closer to a reflecting metal surface. It was first described for fluorescent proteins (11) and ranges over a distance of 30 nm, beyond which it decreases exponentially with the separation distance between the fluorophore and the reflecting surface. This relationship between surface-enhanced fluorescence and separation distance was validated using fluorescent proteins attached to polymeric spacers of various lengths (12) and forms the basis for the interpretation of the surface-enhanced fluorescence of adhering fluorescent bacteria in terms of deformation of their cell wall. This method has a drawback, in that it can be applied only on reflecting metal surfaces; but it has advantages with respect to atomic force microscopy (AFM); e.g., there are no external forces applied on the adhering bacterium, and it also measures a large number of adhering bacteria simultaneously.
The aim of this study was to investigate the influence of adhesion to different common biomaterials on icaA and cidA gene expression in Staphylococcus aureus biofilms. To this end, we first measured the forces of staphylococcal adhesion to different biomaterials and related these adhesion forces to the expression of the icaA and cidA genes. The ica operon is present in S. aureus and is mainly involved in the production of capsular polysaccharides upon activation (13). Recently, it has also been reported that the ica locus is also required for colonization and immunoprotection during colonization of the host (13, 14). icaA and icaD synthesize poly-N-acetylglucosamine (PNAG), which supports cell-cell and cell-surface interactions (15). cidA expression is associated with cell lysis and the release of eDNA during planktonic growth to facilitate adhesion and biofilm formation (16). Therefore, eDNA is known to act as an essential glue to maintain the integrity of both the EPS matrix and biofilms as a whole (16, 17). All experiments were performed with S. aureus ATCC 12600 and its isogenic mutant, S. aureus ATCC 12600 Δpbp4, deficient in peptidoglycan cross-linking. The higher deformability of the S. aureus ATCC 12600 Δpbp4 cell wall than the cell wall of the wild-type strain was demonstrated using surface-enhanced fluorescence.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains S. aureus ATCC 12600 and S. aureus ATCC 12600 Δpbp4 were used throughout this study. All the strains were stored at −80°C in tryptone soya broth (TSB; Oxoid, Basingstoke, United Kingdom) containing 15% glycerol. Bacteria were cultured aerobically at 37°C on blood agar or TSB agar plates with 10 μg ml−1 tetracycline. One colony was inoculated in 10 ml TSB and grown for 24 h at 37°C. The preculture was then inoculated in 10 ml fresh TSB (1:100) and cultured for 16 h. The main culture (1:100) was used for 24 h of biofilm growth, while for other experiments, staphylococci were suspended in TSB or phosphate-buffered saline (PBS; 10 mM potassium phosphate, 0.15 M NaCl, pH 7.0) to the desired density, as determined by either determination of the optical density at 578 nm (OD578; Genesys 20 visible spectrophotometer; Beun de Ronde, Abcoude, The Netherlands) or enumeration of the number of bacteria per ml using a Bürker-Türk counting chamber. A stable chromosomal mutation in S. aureus ATCC 12600 Δpbp4 was obtained by transfecting the temperature-sensitive pMAD-pbp4 plasmid, as previously described (18). The pMAD-pbp4 plasmid was obtained from M. G. Pinho, Laboratory of Bacterial Cell Biology, and S. R. Filipe, Laboratory of Bacterial Cell Surfaces and Pathogenesis, Instituto de Tecnologia Quimica e Biológica, Universidade Nova de Lisboa.
To confirm that the pbp4 deletion had an influence on cell wall deformation using surface-enhanced fluorescence, green fluorescent protein (GFP)-expressing variants (S. aureus ATCC 12600-GFP and S. aureus ATCC 12600 Δpbp4-GFP) were made by introducing the plasmid pMV158-GFP under the control of the MalP promoter into the staphylococci using electroporation and selected on TSB agar plates containing 10 μg ml−1 tetracycline.
Cell wall deformation.
The pbp4 deletion was confirmed by PCR, and its expression in both staphylococcal strains was quantified using the primer sets listed in Table 1. The main cultures were diluted 1:100 in 10 ml TSB and grown for 24 h under static conditions. Next, 1 ml of the resulting suspension was subjected to RNA isolation and cDNA synthesis procedures, as described below for icaA and cidA gene expression. To confirm that the pbp4 deletion had an influence on the cell wall deformation of the staphylococci, we applied a novel, highly sensitive method based on surface-enhanced fluorescence to demonstrate the cell wall deformation of bacteria adhering to reflecting metal surfaces (19). Briefly, staphylococci suspended in PBS (3 × 108 cells ml−1) were allowed to sediment from a 0.075-cm-high suspension volume above a stainless steel (SS) 316L substratum surface (7.6 by 1.6 cm), and the fluorescence radiance was measured as a function of time using a bio-optical imaging system (IVIS Lumina II; PerkinElmer, Inc., Hopkinton, MA, USA) at an excitation wavelength of 465 nm and an emission wavelength of between 515 and 575 nm. The IVIS system was kept at 20°C with an exposure time of 5 s, and images of the entire SS substratum surface were taken every 5 min over a period of 3 h. The average fluorescent radiance from three user-defined regions of interest (1 cm2) was determined with the Living Image software package (version 3.1; PerkinElmer Inc., USA). It was not necessary to correct the fluorescence enhancement for photobleaching because previously reported control experiments on glass showed negligible bleaching with exposure for up to 5 h (19). Staphylococcal sedimentation was monitored by direct observation, and images of adhering bacteria were taken using a metallurgical microscope equipped with a 40× objective (ULWD, CDPlan, 40PL; Olympus Co, Tokyo, Japan) connected to a charge-coupled-device camera (Basler A101F; Basler AG, Germany). The images were analyzed using software based on MATLAB developed in-house to count the number of adhering bacteria in each image. The numbers of adhering bacteria over the entire substratum surface were subsequently expressed as a percentage with respect to the total number of bacteria present in the suspension volume (0.912 ml) above the substratum.
TABLE 1.
Primer sequences for reverse transcription-qPCR used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| icaA-forward | GGAAGTTCTGATAATACTGCTG |
| icaA-reverse | GATGCTTGTTTGATTCCCTC |
| cidA-forward | AGCGTAATTTCGGAAGCAACATCCA |
| cidA-reverse | CCCTTAGCCGGCAGTATTGTTGGTC |
| gyrB-forward | GGAGGTAAATTCGGAGGT |
| gyrB-reverse | CTTGATGATAAATCGTGCCA |
The increase of the fluorescence radiance due to the sedimentation and adhesion of fluorescent staphylococci relative to the fluorescence radiance of suspended staphylococci was measured and expressed as the total fluorescence enhancement, TFE(t), according to the equation
| (1) |
in which R(t) denotes the fluorescence radiance at time t, while R0 and R(0) indicate the fluorescence radiance of a suspension in the absence of staphylococci and immediately prior to the sedimentation of staphylococci from the suspension, respectively. Whereas total fluorescence enhancement is due to a combination of increasing numbers of sedimented staphylococci and their cell wall deformation, increases in total fluorescence enhancement extending beyond the time at which sedimentation is complete are due to cell wall deformation (19). Cell wall deformation brings a larger volume of the bacterial cytoplasm closer to the surface, and therewith, more fluorophores inside the bacterium become subject to fluorescence enhancement, yielding a higher fluorescence signal. Fluorescence enhancement occurs only on reflecting substrata, and accordingly, the effects of pbp4 deletion on cell wall deformation were examined only on SS.
Staphylococcal characteristics not related to cell wall deformation.
In order to verify that other characteristics relevant for the current study were not affected by the pbp4 deletion, planktonic growth curves, biofilm formation, cell surface hydrophobicity, and the zeta potential of the bacterial cell surfaces were determined.
Planktonic growth curves.
Curves of the planktonic growth of S. aureus ATCC 12600 and S. aureus ATCC 12600 Δpbp4 were compared. Staphylococci were suspended in 10 ml TSB to an OD578 of 0.05 and grown at 37°C under static conditions. Optical densities were subsequently measured as a function of time.
Biofilm formation and quantitation.
Biofilms were grown on SS, PMMA, and PE coupons in triplicate in a 12-well plate. After incubation for 6, 12, and 24 h at 37°C, the coupons with biofilms were carefully removed, placed into a new 12-well plate, and gently washed. The biofilms from three coupons of the same material were then suspended by repeated pipetting and pooled in 1 ml PBS. To measure the biofilm biomass, 1:10 dilutions of the pooled bacterial suspensions were prepared and the OD578s were measured.
MATH.
Microbial adhesion to hydrocarbons (MATH) was carried out while the staphylococci were in their kinetic mode (20) to reveal possible differences in adhesive cell surface properties between S. aureus ATCC 12600 and its isogenic Δpbp4 mutant. To this end, staphylococci were suspended in phosphate buffer (10 mM potassium phosphate buffer, pH 7.0) to an OD578 of 0.45 to 0.50 (initial absorbance [A0]), 150 μl hexadecane was added to 3 ml bacterial suspension, and the two-phase system was vortexed for 10 s (0.17 min) and allowed to settle for 10 min. The optical density (absorbance at time t [At]) was measured, this procedure was repeated 5 more times (with increasing vortexing times), and the results were plotted as log[(At/A0) × 100] against the vortexing time (t) to determine the rate of initial bacterial removal, Re (minutes−1), from the aqueous phase, i.e., bacterial hydrophobicity, by the kinetic MATH assay, according to the equation
| (2) |
Zeta potential.
Suspensions of the wild-type and mutant bacterial strains were prepared as mentioned above. The main cultures were centrifuged at 4,000 × g for 10 min and washed 2 times in 10 ml PBS, pH 7.0. The washed pellets were resuspended in 10 ml PBS, pH 7.0, and zeta potentials were determined by particulate microelectrophoresis (Zetasizer Nano ZS; Malvern Instruments, Worcestershire, United Kingdom) at 25°C. The experiments were repeated three times, and the data are presented as averages ± standard errors of the means.
Preparation of bacterial AFM probes and adhesion force measurements.
In order to measure adhesion forces between the S. aureus strains and different biomaterials, staphylococci were immobilized on a cantilever for atomic force microscopy (AFM), as described before (21). Bacteria were cultured as described above, with the difference that they were washed and suspended in demineralized water. Adhesion force measurements were performed at room temperature in PBS using a Dimension 3100 system (Nanoscope V; Digital Instruments, Woodbury, NY, USA). For each bacterial probe, measurements for force-distance curves were obtained with no surface delay at a 2-nN trigger threshold. Using the same bacterial probe, 15 force measurements were recorded, and three different probes were used on three random locations on each material surface. Adhesion forces were determined from the cantilever deflection data, which were converted to force values (in nanonewtons) by multiplication by the cantilever spring constant according to Hooke's law:
| (3) |
where F is the adhesion force previously measured on clean glass, Ksp is the spring constant of the cantilever, and D is the deflection of the cantilever. The spring constant of each cantilever was determined using the thermal method (22). The integrity of a bacterial probe was monitored before and after the onset of each adhesion cycle by comparing the adhesion forces measured on a clean glass surface. Whenever these adhesion forces differed by more than 0.5 nN, the last set of data obtained with that probe was discarded and a new bacterial probe was made.
icaA and cidA gene expression.
Gene expression analysis was performed on 1-, 3-, and 24-h-old biofilms. Biofilms were grown by adding to each sample 2 ml of the main culture diluted 1:100 with growth medium. Total RNA from the biofilms was isolated using a RiboPure Bacteria kit (Ambion; Invitrogen) according to the manufacturer's instructions. Traces of genomic DNA was removed using a DNA-free kit (Ambion; Applied Biosystems, Foster City, CA), and the absence of genomic DNA contamination was verified by real-time PCR prior to cDNA synthesis. cDNA synthesis was carried out using 200 ng of RNA, 4 μl 5× iScript reaction mix, and 1 μl iScript reverse transcriptase in a total volume of 20 μl (Iscript; Bio-Rad, Hercules, CA) according to the manufacturer's instructions. Real-time reverse transcription-quantitative PCR (qPCR) was performed in triplicate in a 96-well plate (AB0900; Thermo Scientific, United Kingdom) with the primer sets specific for gyrB, icaA, and cidA (Table 1). The following thermal conditions were used for all qPCRs: 95°C for 15 min and 40 cycles of 95°C for 15 s and 60°C for 20 s. The mRNA levels were quantified in relation to the level of the endogenous control gene gyrB. The levels of icaA and cidA expression in all biofilms were expressed relative to their levels of expression in biofilms grown on PE.
Production of matrix components in staphylococcal biofilms. (i) PNAG extraction and quantitation.
Extraction of PNAG from S. aureus was performed as previously described (13). Briefly, 24-h-old staphylococcal biofilms grown on SS, PMMA, and PE coupons as described above were suspended in 1 ml PBS for normalization and diluted to an OD578 of 0.75 for slime extraction. The bacterial suspension was pelleted at 4,000 × g for 15 min, the supernatant was aspirated, the pellet was resuspended in 50 μl 0.5 M EDTA (pH 8), and the pellet suspension was incubated for 5 min at 100°C on a hot plate. Cell debris was pelleted at 8,500 × g for 5 min, and 30 μl of the EPS-containing supernatant was pipetted into fresh tubes. The samples were treated with 10 μl proteinase K (20 μg ml−1) for 30 min at 37°C before quantitation. The concentrated EPS was diluted 1:100 with ultrapure water, and 20 μl was blotted on a nitrocellulose membrane using a Bio-Dot apparatus (Bio-Rad, Hercules, CA). The nitrocellulose membrane was then blocked using 1% bovine serum albumin–Tris-buffered saline (20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.05% Tween 20) for 1 h under conditions with mild shaking at room temperature. The membrane was subsequently incubated with a biotin-labeled lectin (wheat germ agglutinin [Sigma-Aldrich, St. Louis, MO, USA] isolated from Triticum vulgaris, which detects 1,4-β-N-acetyl-d-glucosamine), used as a primary antibody at a 1:1,000 dilution, for 1.5 h under conditions of mild shaking at room temperature. Finally, streptavidin-infrared dye (LI-COR Biosciences, Leusden, The Netherlands) was added as a secondary antibody at a 1:10,000 dilution for 30 min under conditions of mild shaking at room temperature. The membrane was washed 3 times for 10 min each time with Tween 20–Tris-buffered saline, and the amount of PNAG was measured using an Odyssey infrared imaging system (LI-COR Biosciences).
(ii) eDNA extraction and quantitation.
Extraction of eDNA was performed as previously described (14), but with some minor modifications. Briefly, biofilms grown for 24 h on SS, PMMA, and PE coupons as described above were suspended in 1 ml 500 mM NaCl containing 10 mM EDTA and 50 mM Tris-HCl, pH 7.5, and transferred into chilled tubes. The OD578s of the suspensions were measured for normalization, and the staphylococci were centrifuged at 4,000 × g for 15 min to separate the bacteria and eDNA. The supernatant was collected, subjected to DNA extraction twice with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1), and precipitated using 1/10 (vol/vol) 3 M sodium acetate and 2/3 (vol/vol) ice-cold isopropanol. After centrifugation (15 min, 4°C, 8,500 × g), the pellet was washed with 100% ethanol and air dried. The dried DNA pellet was dissolved in 50 μl TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5). The amount of eDNA was quantified using a CyQuant cell proliferation assay kit (Invitrogen, Molecular Probes, Eugene, OR, USA), based on a calibration curve of bacteriophage λ DNA ranging in concentration from 0 to 1,000 ng ml−1. The eDNA samples were processed according to the manufacturer's instructions, and fluorescence was measured by a fluorescence plate reader at an excitation wavelength of 485 nm and an emission wavelength of 520 nm.
Substratum surfaces, contact angle, and surface roughness measurements.
The substratum surfaces used in this study were SS, PMMA, and PE. All substratum surfaces were prepared so that they possessed a comparable surface roughness in the micrometer range (1 to 2 μm) in order to rule out possible effects of surface roughness. SS was polished using 1,200-grid SiC paper, followed by a MetaDi 3-μm diamond suspension (Buehler, Lake Bluff, IL, USA), on a polishing mat for 20 min, while the PMMA and PE surfaces were used as received. Circular coupons with a thickness of 0.5 mm and a surface area of 3.1 cm2 were made to fit into a 12-well plate, sterilized with methanol, washed with sterile PBS, and stored in sterile demineralized water until use. Water contact angles on all materials were measured at 25°C using the sessile drop technique in combination with a homemade contour monitor. The surface roughness of the biomaterials was determined by AFM (Nanoscope IV Dimension 3100 atomic force microscope) using a silicon nitride tip (probe curvature radius, 18 nm; Mikromasch, Mountain View, CA, USA).
RESULTS
Physicochemical surface properties of biomaterials.
The hydrophobicities of the biomaterials were evaluated using water contact angles. The water contact angles varied considerably among the three materials included in this study. SS was the least hydrophobic material with an average water contact angle of 33 ± 9°, followed by PMMA with an average water contact angle of 69 ± 6° and PE with an average water contact angle of 84 ± 1°. The surface roughnesses of all materials measured by AFM were in the micrometer range and amounted to 1.8 ± 0.2 μm, 2.0 ± 0.4 μm, and 1.0 ± 0.2 μm for SS, PMMA, and PE, respectively.
Effects of pbp4 deletion on cell wall deformation.
Peptidoglycan cross-links provide cell wall rigidity; therefore, effects on cell wall deformation were determined from the total fluorescence enhancement of S. aureus bacteria sedimenting and adhering to SS. The initial linear increase (1 to 2 h) in total fluorescence enhancement for S. aureus ATCC 12600-GFP and S. aureus ATCC 12600 Δpbp4-GFP was due in part to an increase in the number of sedimented bacteria (compare Fig. 2A and B), but once all staphylococci from the suspension sedimented on the surface, the slow increase in total fluorescence enhancement after 3 h was fully due to cell wall deformation. Accordingly, it can be seen that S. aureus ATCC 12600 Δpbp4-GFP deforms to a greater extent than does S. aureus ATCC 12600-GFP due to the absence of pbp4 cross-linking.
FIG 2.
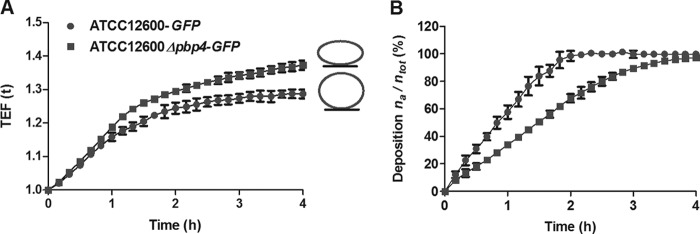
Effects of pbp4 deletion on cell wall deformation. (A) Cell wall deformation of S. aureus ATCC 12600-GFP and S. aureus ATCC 12600 Δpbp4-GFP upon adhesion to SS, as measured using surface-enhanced fluorescence. As an adhering bacterium deforms, its fluorescent intracellular content gets closer to the reflecting metal surface, yielding a surface-enhanced fluorescence that increases with increasing deformation. Each point represents the average ± standard error of the mean from three individual experiments. All differences between S. aureus ATCC 12600 and S. aureus ATCC 12600 Δpbp4 were statistically significant (P < 0.05). (B) The number of adhering S. aureus ATCC 12600-GFP and S. aureus ATCC 12600 Δpbp4-GFP bacteria on SS surfaces as a function of sedimentation time, expressed as the percentage of bacteria adhering (na) with respect to the total number of bacteria (ntot) in the suspension volume above the substratum surface. Each point represents the average ± standard error of the mean from three individual experiments. All differences between S. aureus ATCC 12600 and S. aureus ATCC 12600 Δpbp4 were statistically significant (P < 0.05).
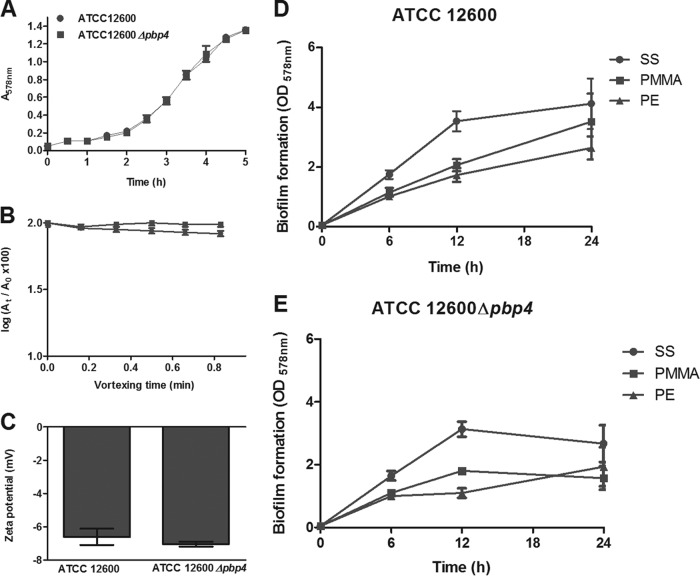
In order to establish that the pbp4 deletion affected solely the cell wall deformability of S. aureus ATCC 12600 and no other properties, planktonic growth (Fig. 3A), biofilm formation (Fig. 3D and E), cell surface hydrophobicities (determined using the MATH test in its kinetic mode [20]; Fig. 3B), and zeta potentials (Fig. 3C) for S. aureus ATCC 12600 were compared with those for S. aureus ATCC 12600 Δpbp4. The growth curves, zeta potentials, and cell surface hydrophobicities (initial removal coefficients [Res], 0.0002 min−1) of both strains were identical. Generally, S. aureus ATCC 12600 Δpbp4 formed less biofilm than S. aureus ATCC 12600. Both strains formed more biofilm on SS than on PMMA and PE at all time points at which biofilm formation was measured (Fig. 3D and E), although no statistically significant differences in the amount of biofilm on the three substratum surfaces could be established after 24 h of growth.
FIG 3.
Effects of expression of pbp4 in S. aureus ATCC 12600 on strain characteristics not related to cell wall deformation. (A) Planktonic growth curves (fully overlapping) of S. aureus ATCC 12600 and S. aureus ATCC 12600 Δpbp4 at 37°C. (B) The optical density {log[(At/A0) × 100]} as a function of the vortexing time for the removal of S. aureus ATCC 12600 and its isogenic mutant, S. aureus ATCC 12600 Δpbp4, from the aqueous phase (10 mM potassium phosphate buffer, pH 7.0) by hexadecane. The absence of removal indicates a hydrophilic cell surface. Each point represents the average ± standard error of the mean from three individual experiments with separately grown staphylococcal cultures. None of the differences between S. aureus ATCC 12600 and S. aureus ATCC 12600 Δpbp4 were statistically significant. (C) Zeta potentials of S. aureus ATCC 12600 and S. aureus ATCC 12600 Δpbp4 in PBS, pH 7.0. Each point represents the average ± standard error of the mean from three individual experiments with separately grown staphylococcal cultures. None of the differences between S. aureus ATCC 12600 and S. aureus ATCC 12600 Δpbp4 were statistically significant. (D and E) Biofilm formation by S. aureus ATCC 12600 and S. aureus ATCC 12600 Δpbp4, expressed as the OD578, after 6, 12, and 24 h of growth on SS, PMMA, and PE.
Forces of S. aureus adhesion to different biomaterials.
The adhesion forces of S. aureus ATCC 12600 and S. aureus ATCC 12600 Δpbp4 were measured using an AFM equipped with a bacterial probe, as recently advocated by Alsteens et al. (23). For S. aureus ATCC 12600 (Fig. 4A), the strongest adhesion forces were observed on the PE surface (4.9 ± 0.5 nN), and the adhesion forces on the more hydrophilic PMMA (3.1 ± 0.7 nN) and SS (1.3 ± 0.2 nN) surfaces decreased in a statistically significant manner (P < 0.05). The adhesion forces of the Δpbp4 mutant were significantly weaker (P < 0.05) than those of S. aureus ATCC 12600 (Fig. 4B).
FIG 4.
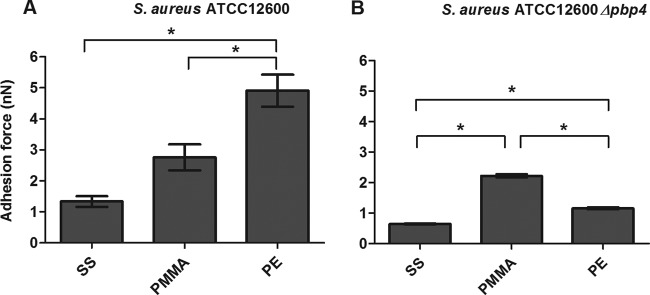
Forces of S. aureus adhesion to different biomaterials. The forces of S. aureus ATCC 12600 (A) and S. aureus ATCC 12600 Δpbp4 (B) adhesion to SS, PMMA, and PE are shown. Each bar represents the average of 135 adhesion force curves measured with 9 different bacterial probes taken from three separately grown staphylococcal cultures. Error bars represent the standard errors of the means. *, significant differences (P < 0.05) in forces of staphylococcal adhesion to different biomaterials (two-tailed, two-sample equal variance Student's t test).
Production of matrix components and gene expression in relation to staphylococcal adhesion forces in 24-h-old biofilms.
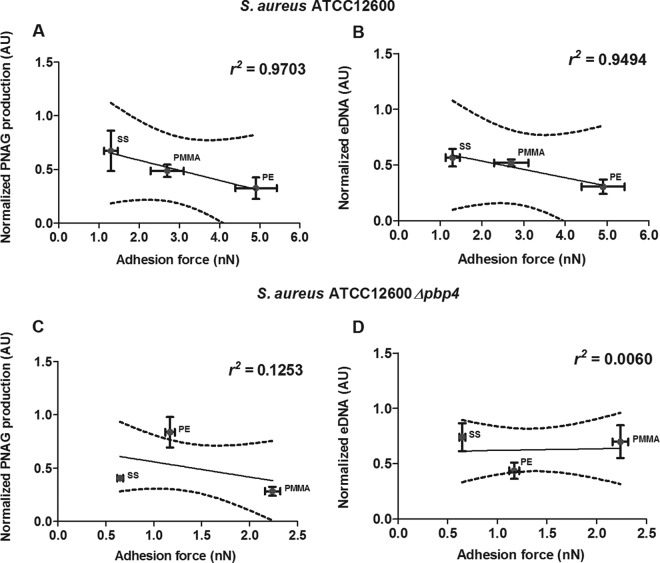
The level of PNAG production normalized with respect to the amount of biofilm formed decreased with increasing adhesion force toward the more hydrophobic PE surface in a statistically significant manner (P < 0.05) (Fig. 5A). Normalized amounts of eDNA in 24-h-old S. aureus ATCC 12600 biofilms decreased as well with increasing adhesion force (P < 0.05) (Fig. 5B). However, for 24-h-old S. aureus ATCC 12600 Δpbp4 biofilms, neither PNAG production nor eDNA presence related in a significant way to the forces of S. aureus ATCC 12600 Δpbp4 adhesion to different biomaterials (Fig. 5C and D).
FIG 5.
S. aureus PNAG production and eDNA presence versus adhesion forces. (A and C) Normalized PNAG production in 24-h-old S. aureus ATCC 12600 (A) and S. aureus ATCC 12600 Δpbp4 (C) biofilms as a function of the adhesion force. (B and D) Normalized eDNA presence in 24-h-old S. aureus ATCC 12600 (B) and S. aureus ATCC 12600 Δpbp4 (D) biofilms as a function of the adhesion force. Linear regression analysis was performed for all graphs to analyze the correlation between PNAG production, eDNA presence, and adhesion force. The solid lines represent the best fit to a linear function, while r2 values represent the correlation coefficients. The dotted lines enclose the 95% confidence intervals. The amounts of PNAG and eDNA were normalized to the amount of biofilm formed on each substratum, and each point represents the average ± standard error of the mean from three individual experiments with separately grown staphylococcal cultures. AU, arbitrary units.
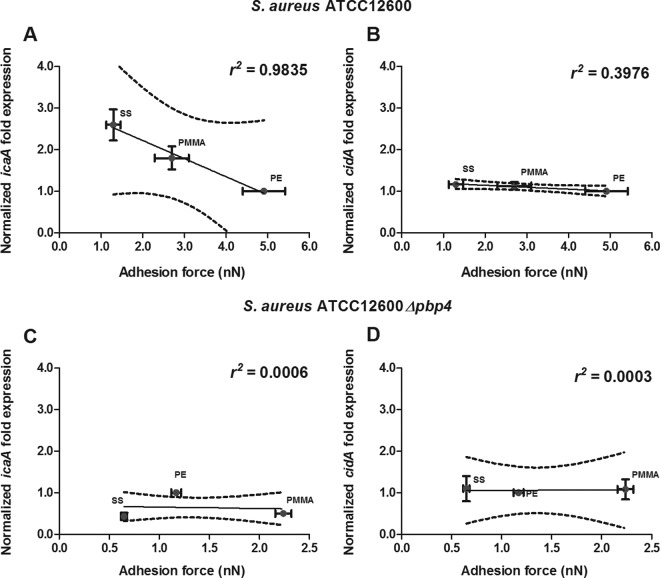
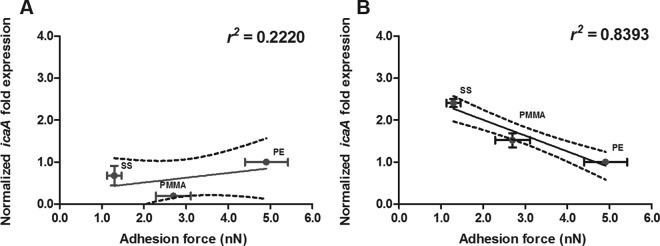
In Fig. 6 we have plotted the staphylococcal adhesion forces on the different biomaterials versus the levels of icaA and cidA gene expression in 24-h-old biofilms, as icaA and cidA are responsible for the production of PNAG and eDNA, respectively. In S. aureus ATCC 12600, icaA gene expression decreased as adhesion forces increased (Fig. 6A), in line with the level of PNAG production. cidA gene expression did not follow a trend similar to that of icaA expression in 24-h-old biofilms but was equally expressed on all the biomaterials irrespective of the forces of adhesion to the different biomaterials experienced (Fig. 6B). In S. aureus ATCC 12600 Δpbp4, lacking peptidoglycan cross-linking, expression of neither icaA nor cidA related to its force of adhesion to the different biomaterials (Fig. 6C and D).
FIG 6.
S. aureus icaA and cidA gene expressions versus adhesion forces in 24-h-old biofilms. (A and C) Normalized icaA expression in 24-h-old S. aureus ATCC 12600 (A) and S. aureus ATCC 12600 Δpbp4 (C) biofilms as a function of the adhesion force. (B and D) Normalized cidA expression in 24-h-old S. aureus ATCC 12600 (B) and S. aureus ATCC 12600 Δpbp4 (D) biofilms as a function of the adhesion force. Linear regression analysis was performed for all graphs to analyze the correlation between gene expression and adhesion force. The solid lines represent the best fit to a linear function, while r2 values represent the correlation coefficients. The dotted lines enclose the 95% confidence intervals. The levels of icaA and cidA expression were normalized to the level of gyrB expression and are presented as normalized fold expression with respect to the results for PE. Each point represents the average ± standard error of the mean from three individual experiments with separately grown staphylococcal cultures.
icaA gene expression in relation to staphylococcal adhesion forces in 1- and 3-h-old biofilms of S. aureus ATCC 12600.
In order to assess the speed at which gene expression is regulated by the adhesion forces that an adhering bacterium experiences, icaA gene expression was also assessed in 1-h and 3-h-old biofilms of S. aureus ATCC 12600 and plotted against the adhesion forces (Fig. 7). In 1-h-old biofilms, icaA gene expression did not show any relation to adhesion force (Fig. 7A), but in 3-h-old biofilms (Fig. 7B), a relation between icaA gene expression and adhesion force similar to that seen in 24-h-old biofilms was observed (compare Fig. 7B and 6A).
FIG 7.
icaA gene expression versus adhesion forces in 1-h and 3-h-old biofilms of S. aureus ATCC 12600. Normalized icaA expression in 1-h-old (A) and 3-h-old (B) S. aureus biofilms is shown as a function of the adhesion force. Linear regression analysis was performed to analyze the correlation between icaA gene expression and adhesion force. The solid lines represent the best fit to a linear function, while r2 values represent the correlation coefficients. The dotted lines enclose the 95% confidence intervals. The level of icaA expression was normalized to that of gyrB expression and is presented as the normalized fold expression with respect to the results for PE. Each point represents the average ± standard error of the mean from three individual experiments with separately grown staphylococcal cultures.
DISCUSSION
In this study, we hypothesized that the adhesion forces sensed by S. aureus upon adhesion to different biomaterials regulate the expression of two important genes, icaA and cidA, known to contribute to the formation of the self-produced EPS matrix. Over the range of adhesion forces of between 1 and 5 nN, icaA gene expression decreased with increasing adhesion force in 3-h and 24-h-old biofilms but not in 1-h-old ones, while for cidA gene expression, no influence of adhesion forces was found. Moreover, the levels of production of the EPS matrix components PNAG and eDNA decreased with increasing adhesion forces experienced by S. aureus ATCC 12600 on different biomaterials, making it unlikely that cidA expression solely regulates eDNA release. The differences in eDNA presence in biofilms grown on SS, PMMA, and PE can be caused by the autolysin atl gene. This gene produces two functional proteins responsible for regulating growth, cell lysis, and biofilm formation (24). The expression of the alt gene occurs under several external stress conditions (25), including adhesion, as a potential trigger for DNA release. Since matrix components (PNAG and eDNA) provide an important means through which bacteria can evade the host immune response and antibiotic attack, we can speculate from the results in this study that the pathogenicity of S. aureus biofilms is regulated in part by the adhesion forces arising from the substratum to which they adhere.
Bacterial behavior has been found to be extremely sensitive to minor differences in adhesion forces. In S. aureus, invasive isolates exhibited mean forces of adhesion to a fibronectin-coated substratum 0.28 nN higher than those of noninvasive control isolates (26). Moreover, strains of Listeria monocytogenes with forces of adhesion to the silicon nitride tip of an AFM cantilever stronger than 0.38 nN were found to be more pathogenic than strains with weaker adhesion forces (27), coinciding with our conclusion on the impact of adhesion forces on S. aureus gene expression and associated pathogenicity. In the current study, we measured the adhesion forces between S. aureus and different biomaterial surfaces by the use of bacterial probe AFM. This method has been applied more often, but concerns as to whether contact is established by a single organism or multiple ones have been raised. In the past (28), we have noticed that multiple contacts seldom or never happen because bacteria attached to the cantilever are unlikely to be equidistant to the substratum surface within the small distance range of interaction forces. In addition, the bacterial probe is contacting the surface at an angle of 15 degrees, which makes multiple contacts less probable. Multiple contact points, however, would become evident from double contour lines when a bacterial probe is used for imaging. Routine checks on probes have never yielded double contour lines, and hence, it is safe to assume that our bacterial probes do not yield multiple contact points.
Biofilm formation starts with the adhesion of so-called linking-film bacteria, which provides the groundwork for further biofilm growth. In essence, only these linking-film bacteria are capable of sensing a substratum surface, since all organisms appearing in a biofilm later adhere to neighboring organisms, yet we found that the relationship between icaA gene expression and adhesion force in 3-h-old biofilms of S. aureus ATCC 12600 (Fig. 7B) was similar to that in 24-h-old biofilms (compare Fig. 7B and 6A), while in 1-h-old biofilms, this relationship was still lacking (Fig. 7A), as bacteria may not have adapted within 1 h to the substratum to which they adhered. This shows that gene expression is a time-dependent process and that stable expression occurs only after 3 h and lasts minimally for 24 h of biofilm growth. This raises the important question of how organisms appearing later in a biofilm, due to either growth or progressive coadhesion, sense the adhesion forces arising from a substratum. Clearly, the range of all attractive or repulsive forces arising from a substratum surface is limited to few 10s of nanometers, making it impossible for later organisms to directly sense a surface. Much more clearly, they experience adhesion forces from neighboring organisms with which they coadhere (29). This implies that a means of communication must be available within a biofilm through which substratum information is passed to bacteria in a biofilm that are not in direct contact with the substratum.
Expression of the icaA gene but not the cidA gene decreased with an increase in the adhesion forces experienced by adhering staphylococci. Adhesion forces arising from substratum surfaces have recently been demonstrated to induce nanoscopic cell wall deformation, yielding membrane stresses (21). Deformation of lipid bilayers has been shown to result in the opening of mechanosensitive channels involved in adhesion force sensing, as they transduce a mechanical force into chemical signals (30). Note that also for Pseudomonas aeruginosa, surface-associated organisms have been found to produce more pili than their planktonic counterparts, suggesting that a localized mechanical signal, i.e., cell wall stress arising from the surface association, plays a pivotal role in regulating genes associated with surface adhesion (31). Cell wall stress and the resulting deformation are extremely difficult to measure due to the rigidity of the peptidoglycan layer, and therefore, we employed an isogenic mutant, S. aureus ATCC 12600 Δpbp4-GFP, lacking peptidoglycan cross-linking and confirmed the greater deformability of the isogenic mutant (Fig. 2) using surface-enhanced fluorescence (32). Surface-enhanced fluorescence can be measured only on reflecting surfaces and was thus performed only on SS. Importantly, due to the extreme sensitivity of surface-enhanced fluorescence measurements, other wild-type strains have also been shown to deform upon adhesion to a surface (19). As an important aspect of surface-enhanced fluorescence, the number of bacteria involved in a single analysis is much larger than the number that can be determined by any other microscopic method presently used, like AFM, while surface-enhanced fluorescence also measures deformation under the naturally occurring adhesion forces that are not present under an applied force, as in AFM (21). Therefore, it can be anticipated that differences in adhesion forces between S. aureus and various substratum surfaces may actually induce different degrees of cell wall deformation, which supports our hypothesis that adhesion forces cause nanoscale cell wall deformations and membrane stresses that act as mechanisms signaling an organism to enter its adhering state.
cidA expression did not relate to adhesion forces, possibly because cidA membrane proteins program cell death on the basis of the oxidation and reduction state of the cell membrane (33) rather than its deformation, suggesting that other environmental conditions, like pH, nutrient availability, biofilm age, or antimicrobial stress, influence DNA release (34). The peptidoglycan layer, which ensures the rigidity of the bacterial cell wall, appears to be of pivotal importance in adhesion force sensing, as its deformation is directly transmitted to the membrane. In the isogenic mutant S. aureus ATCC 12600 Δpbp4, which lacks cross-linked peptidoglycan and therefore possesses a softer cell wall, adhesion force sensing appears to be ineffective, as no relation between adhesion forces and gene expression was found.
Deletion of pbp4 from S. aureus ATCC 12600 did not have an effect on planktonic growth, cell surface hydrophobicity, or zeta potential and had only a small effect on biofilm formation (Fig. 3). However, it may be considered strange that the amount of biofilm of both strains formed on different materials bore no significant relation to the forces experienced by these linking-film organisms. This can be explained by the fact that bacteria adhere only once they experience attractive forces that exceed the prevailing detachment forces in a given environment. The current experiments were carried out under static conditions rather than under flow conditions, which implies that a detachment force of virtually zero was operating during adhesion, which made any adhesion force large enough for a bacterium to remain adhering. In this respect, it is not surprising that S. aureus ATCC 12600 Δpbp4 had an ability to form biofilm similar to that of its parent strains, as both its cell surface hydrophobicity and its zeta potential were similar to those of the parent strain (Fig. 3). Importantly, for the development of biofilms in the presence of weak adhesion forces, biofilms even form on highly hydrated, polymer brush coatings, which exert very small adhesion forces in the subnanonewton range that were found to be insufficient for adhering bacteria to even realize that they were in an adhering state (35).
In conclusion, S. aureus reacts to its adhering state on the basis of the magnitude of the adhesion forces that it experiences to be arising from the substratum surface to which it adheres. This response predominantly involves icaA gene expression and the production of EPS matrix components (PNAG and eDNA), both of which decrease with increasing adhesion forces. Increasing adhesion forces bring an adhering organism closer to the lethal regime, which might be a reason why less EPS is produced by organisms experiencing stronger adhesion forces. In addition, our data also suggest that the mechanical properties of the cell wall provided by the peptidoglycan layer surrounding the cell membrane serve as an important tool for the adhesion force-sensing capacity in S. aureus.
ACKNOWLEDGMENTS
This study was entirely funded by the University Medical Center Groningen, Groningen, The Netherlands.
We are grateful to Mariana G. Pinho, Laboratory of Bacterial Cell Biology, and Sergio R. Filipe, Laboratory of Bacterial Cell Surfaces and Pathogenesis, Instituto de Tecnologia Quimica e Biológica, Universidade Nova de Lisboa, for providing the pMAD-pbp4 plasmid.
H.J.B. is also a director of a consulting company, SASA BV. We declare no potential conflicts of interest with respect to authorship and/or publication of this article.
The opinions and assertions contained herein are those of the authors and are not construed as necessarily representing the views of the funding organization or the authors' employers.
REFERENCES
- 1.National Institutes of Health. 1999. SBIR/STTR study and control of microbial biofilms. National Institutes of Health, Bethesda, MD: http://grants.nih.gov/grants/guide/pa-files/PA-99-084.html. [Google Scholar]
- 2.Darouiche RO. 2004. Treatment of infections associated with surgical implants. N Engl J Med 350:1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- 3.Monroe D. 2007. Looking for chinks in the armor of bacterial biofilms. PLoS Biol 5:e307. doi: 10.1371/journal.pbio.0050307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 5.Busscher HJ, van der Mei HC. 2012. How do bacteria know they are on a surface and regulate their response to an adhering state? PLoS Pathog 8:e1002440. doi: 10.1371/journal.ppat.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kügler R, Bouloussa O, Rondelez F. 2005. Evidence of a charge-density threshold for optimum efficiency of biocidal cationic surfaces. Microbiology 151:1341–1348. doi: 10.1099/mic.0.27526-0. [DOI] [PubMed] [Google Scholar]
- 7.Tiller JC, Liao CJ, Lewis K, Klibanov AM. 2001. Designing surfaces that kill bacteria on contact. Proc Natl Acad Sci U S A 98:5981–5985. doi: 10.1073/pnas.111143098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelsman AF, van Dam GM, van der Mei HC, Busscher HJ, Ploeg RJ. 2010. In vivo evaluation of bacterial infection involving morphologically different surgical meshes. Ann Surg 251:133–137. doi: 10.1097/SLA.0b013e3181b61d9a. [DOI] [PubMed] [Google Scholar]
- 9.Nuryastuti T, Krom BP, Aman AT, Busscher HJ, van der Mei HC. 2011. Ica-expression and gentamicin susceptibility of Staphylococcus epidermidis biofilm on orthopedic implant biomaterials. J Biomed Mater Res A 96:365–371. doi: 10.1002/jbm.a.32984. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Busscher HJ, Swartjes J, Chen Y, Harapanahalli AK, Norde W, van der Mei HC, Sjollema J. 2014. Residence-time dependent cell wall deformation of different Staphylococcus aureus strains on gold measured using surface-enhanced-fluorescence. Soft Matter 10:7638–7646. doi: 10.1039/C4SM00584H. [DOI] [PubMed] [Google Scholar]
- 11.Lakowicz JR, Geddes CD, Gryczynski I, Malicka J, Gryczynski Z, Aslan K, Lukomska J, Matveeva E, Zhang J, Badugu R, Huang J. 2004. Advances in surface-enhanced fluorescence. J Fluoresc 14:425–441. doi: 10.1023/B:JOFL.0000031824.48401.5c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Lakowicz JR. 2006. Enhanced fluorescence of Cy5-labeled oligonucleotides near silver island films: a distance effect study using single molecule spectroscopy. J Phys Chem B 110:22557–22562. doi: 10.1021/jp060402e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun 67:5427–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, Ausubel FM, Sifri CD. 2007. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog 3:e57. doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arciola CR, Baldassarri L, Montanaro L. 2001. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microbiol 39:2151–2156. doi: 10.1128/JCM.39.6.2151-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A 104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominiak DM, Nielsen JL, Nielsen PH. 2011. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ Microbiol 13:710–721. doi: 10.1111/j.1462-2920.2010.02375.x. [DOI] [PubMed] [Google Scholar]
- 18.Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Busscher HJ, van der Mei HC, Sjollema J. 2013. Surface enhanced bacterial fluorescence and enumeration of bacterial adhesion. Biofouling 29:11–19. doi: 10.1080/08927014.2012.742074. [DOI] [PubMed] [Google Scholar]
- 20.Lichtenberg D, Rosenberg M, Sharfman N, Ofek I. 1985. A kinetic approach to bacterial adherence to hydrocarbon. J Microbiol Methods 4:141–146. doi: 10.1016/0167-7012(85)90029-6. [DOI] [Google Scholar]
- 21.Chen Y, Harapanahalli AK, Busscher HJ, Norde W, van der Mei HC. 2014. Nanoscale cell wall deformation impacts long-range bacterial adhesion forces on surfaces. Appl Environ Microbiol 80:637–643. doi: 10.1128/AEM.02745-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burnham NA, Chen X, Hodges CS, Matei GA, Thoreson EJ, Roberts CJ, Davies MC, Tendler SJB. 2002. Comparison of calibration methods for atomic-force microscopy cantilevers. Nanotechnology 14:1–6. doi: 10.1088/0957-4484/14/1/301. [DOI] [Google Scholar]
- 23.Alsteens D, Beaussart A, El-Kirat-Chatel S, Sullan RMA, Dufrêne YF. 2013. Atomic force microscopy: a new look at pathogens. PLoS Pathog 9:e1003516. doi: 10.1371/journal.ppat.1003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh VK. 2014. High level expression and purification of Atl, the major autolytic protein of Staphylococcus aureus. Int J Microbiol 2014:615965. doi: 10.1155/2014/615965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vollmer W, Joris B, Charlier P, Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 26.Yongsunthon R, Fowler VG Jr, Lower BH, Vellano FP III, Alexander E, Reller LB, Corey GR, Lower SK. 2007. Correlation between fundamental binding forces and clinical prognosis of Staphylococcus aureus infections of medical implants. Langmuir 23:2289–2292. doi: 10.1021/la063117v. [DOI] [PubMed] [Google Scholar]
- 27.Park B-J, Haines T, Abu-Lail NI. 2009. A correlation between the virulence and the adhesion of Listeria monocytogenes to silicon nitride: an atomic force microscopy study. Colloids Surf B Biointerfaces 73:237–243. doi: 10.1016/j.colsurfb.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 28.Younes JA, van der Mei HC, van den Heuvel E, Busscher HJ, Reid G. 2012. Adhesion forces and coaggregation between vaginal staphylococci and lactobacilli. PLoS One 7:e36917. doi: 10.1371/journal.pone.0036917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer C, Moritz R, Kirschner C, Borchard W, Maibaum R, Wingender J, Flemming H-C. 1999. The role of intermolecular interactions: studies on model systems for bacterial biofilms. Int J Biol Macromol 26:3–16. doi: 10.1016/S0141-8130(99)00057-4. [DOI] [PubMed] [Google Scholar]
- 30.Kumamoto CA. 2008. Molecular mechanisms of mechanosensing and their roles in fungal contact sensing. Nat Rev Microbiol 6:667–673. doi: 10.1038/nrmicro1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowles KN, Gitai Z. 2010. Surface association and the MreB cytoskeleton regulate pilus production, localization and function in Pseudomonas aeruginosa. Mol Microbiol 76:1411–1426. doi: 10.1111/j.1365-2958.2010.07132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao Q, Qiu T, Chu PK. 2012. Surfaced-enhanced cellular fluorescence imaging. Prog Surf Sci 87:23–45. doi: 10.1016/j.progsurf.2012.03.001. [DOI] [Google Scholar]
- 33.Ranjit DK, Endres JL, Bayles KW. 2011. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J Bacteriol 193:2468–2476. doi: 10.1128/JB.01545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan JB, Izano EA, Gopal P, Karwacki MT, Kim S, Bose JL, Bayles KW, Horswill AR. 2012. Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio 3(4):e00198-12. doi: 10.1128/mBio.00198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong B, Jiang H, Manolache S, Wong ACL, Denes FS. 2007. Plasma-mediated grafting of poly(ethylene glycol) on polyamide and polyester surfaces and evaluation of antifouling ability of modified substrates. Langmuir 23:7306–7313. doi: 10.1021/la0633280. [DOI] [PubMed] [Google Scholar]