Abstract
Phage therapy is a promising option for fighting against staphylococcal infections. Two lytic phages, vB_SauM_phiIPLA-RODI (phiIPLA-RODI) and vB_SepM_phiIPLA-C1C (phiIPLA-C1C), belonging to the Myoviridae family and exhibiting wide host ranges, were characterized in this study. The complete genome sequences comprised 142,348 bp and 140,961 bp and contained 213 and 203 open reading frames, respectively. The gene organization was typical of Spounavirinae members, with long direct terminal repeats (LTRs), genes grouped into modules not clearly separated from each other, and several group I introns. In addition, four genes encoding tRNAs were identified in phiIPLA-RODI. Comparative DNA sequence analysis showed high similarities with two phages, GH15 and 676Z, belonging to the Twort-like virus genus (nucleotide identities of >84%); for phiIPLA-C1C, a high similarity with phage phiIBB-SEP1 was observed (identity of 80%). Challenge assays of phages phiIPLA-RODI and phiIPLA-C1C against planktonic staphylococcal cells confirmed their lytic ability, as they were able to remove 5 log units in 8 h. Exposure of biofilms to phages phiIPLA-RODI and phiIPLA-C1C reduced the amount of adhered bacteria to about 2 log units in both monospecies and dual-species biofilms, but phiIPLA-RODI turned out to be as effective as the mixture of both phages. Moreover, the frequencies of bacteriophage-insensitive mutants (BIMs) of Staphylococcus aureus and S. epidermidis with resistance to phiIPLA-RODI and phiIPLA-C1C were low, at 4.05 × 10−7 ± 2.34 × 10−9 and 1.1 × 10−7 ± 2.08 × 10−9, respectively. Overall, a generally reduced fitness in the absence of phages was observed for BIMs, which showed a restored phage-sensitive phenotype in a few generations. These results confirm that lytic bacteriophages can be efficient biofilm-disrupting agents, supporting their potential as antimicrobials against staphylococcal infections.
INTRODUCTION
Two staphylococcal species, Staphylococcus aureus and Staphylococcus epidermidis, are the main causes of nosocomial infections due to their ability to adhere to, colonize, and develop biofilms in medical devices and human organs (1). Staphylococcal biofilms are complex structures in which bacterial cells are surrounded by an extracellular material (polysaccharides, teichoic acids, proteins, and extracellular DNA) which confers protection against antibacterial drugs and the host immune system. In addition, growth of bacteria in a biofilm facilitates the development of antibiotic-resistant organisms (2). S. epidermidis is one of the most abundant species in the human skin microbiota, from which it easily reaches catheters, heart valves, and contact lenses. Despite being regarded as an innocuous bacterium, it is now accepted as an opportunistic pathogen and one of the most common causes of bacteremia in immunocompromised patients (3), preterm infants (4), and biofilm-related infections (5). In addition, resistance to methicillin due to the presence of the mecA gene is widely spread in hospital isolates (6). Similarly, virulence in S. aureus is mainly due to its ability to adhere to and proliferate on biotic and abiotic surfaces (7). In hospital settings, S. aureus infections affecting internal organs and implanted medical devices have become difficult to eradicate. Moreover, methicillin-resistant S. aureus (MRSA) strains are often prevalent in hospitals and have recently spread to nonrelated environments, affecting people without exposure to the health care environment (8). MRSA strains have also been isolated from foods of animal origin (9) and from livestock (10).
Phage therapy exploits the ability of phages to infect and kill bacteria in the treatment of infectious diseases. This represents a potential alternative to antibiotics to fight against multiresistant pathogenic bacteria or superbugs (11). Indeed, human trials with phages against a number of infections have confirmed their safety and shown that phage therapy can provide good results for untreatable chronic infections (12, 13). Specifically, recent results showed the efficacy of phages in animal models, such as S. aureus septicemia in mice (14) and in silkworms (15). The safety and efficacy of phage products for removal of this bacterium in a sinusitis sheep model have also been proven (16). Other applications of phages against S. aureus encompass improvements in wound healing developed on diabetic patients (17, 18) and the treatment of chronic wounds (19). Previous results also showed the ability of phages to remove biofilms formed by staphylococcal species (20–22).
In this study, we report a complete morphological and genetic characterization of two new phages infecting staphylococcal species, named vB_SauM_phiIPLA-RODI (phiIPLA-RODI) and vB_SepM_phiIPLA-C1C (phiIPLA-C1C) following the nomenclature proposed by Kropinski et al. (23). The lytic abilities of these phages, including host range and biofilm removal, were analyzed. Furthermore, the frequency of bacteriophage-insensitive mutants (BIMs) of planktonic cells was calculated for both phages, and a preliminary characterization of these resistant bacteria is also presented.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and growth conditions.
Forty-four different staphylococcal species and one Macrococcus caseolyticus strain were used in this study (Table 1). All the bacteria were isolated in Baird-Parker (BP) agar and were routinely cultured in tryptic soy broth (TSB; Scharlau, Barcelona, Spain) at 37°C with shaking or on TSB plates containing 2% (wt/vol) bacteriological agar (TSA).
TABLE 1.
Strains used in this study, with origins and phage sensitivities, expressed as efficiencies of plaque formation (EOP)
| Species | Strain | Origin | Reference | EOPa |
|
|---|---|---|---|---|---|
| phiIPLA-RODI | phiIPLA-C1C | ||||
| S. aureus | IPLA-1 | Dairy industry surfaces | 66 | 1 | — |
| IPLA-2 | Dairy industry surfaces | 66 | 0.98 ± 0.03 | — | |
| IPLA-3 | Dairy industry surfaces | 66 | 0.87 ± 0.06 | — | |
| IPLA-4 | Dairy industry surfaces | 66 | 1.03 ± 0.04 | — | |
| IPLA-5 | Dairy industry surfaces | 66 | 0.91 ± 0.01 | — | |
| IPLA-6 | Meat industry surfaces | 66 | 1.01 ± 0.03 | — | |
| IPLA-7 | Meat industry surfaces | 66 | 0.99 ± 0.03 | — | |
| IPLA-8 | Meat industry surfaces | 66 | 0.85 ± 0.11 | — | |
| IPLA-9 | Meat industry surfaces | 66 | 0.92 ± 0.23 | — | |
| IPLA-10 | Meat industry surfaces | 66 | 0.87 ± 0.07 | — | |
| IPLA-11 | Meat industry surfaces | 66 | 0.89 ± 0.06 | 0.09 ± 0.01 | |
| IPLA-12 | Meat industry surfaces | 66 | 0.79 ± 0.05 | — | |
| IPLA-13 | Meat industry surfaces | 66 | 1.02 ± 0.03 | — | |
| IPLA-14 | Meat industry surfaces | 66 | 0.96 ± 0.02 | — | |
| IPLA-15 | Meat industry surfaces | 66 | 1.02 ± 0.03 | — | |
| IPLA-16 | Meat industry surfaces | 66 | 1.23 ± 0.04 | 1.09 ± 0.07 | |
| IPLA-17 | Meat industry surfaces | 66 | 1.06 ± 0.01 | — | |
| IPLA-18 | Meat industry surfaces | 66 | 0.96 ± 0.02 | 0.39 ± 0.05 | |
| IPLA-19 | Milk sample | Unpublished data | 1.12 ± 0.03 | — | |
| 15981 | Clinical isolate | 67 | 0.99 ± 0.02 | — | |
| V329 | Bovine subclinical mastitis | 68 | 1.01 ± 0.01 | — | |
| S. epidermidis | F12 | Women's breast milk | 69 | — | 1 |
| B | Women's breast milk | 69 | 0.19 ± 0.01 | 0.78 ± 0.01 | |
| DH3LIK | Women's breast milk | 69 | — | 0.56 ± 0.07 | |
| YLIC13 | Women's breast milk | 69 | — | 0.77 ± 0.06 | |
| Z2LDC14 | Women's breast milk | 69 | — | 0.62 ± 0.03 | |
| DG2ñ | Women's breast milk | 69 | — | 0.88 ± 0.06 | |
| ASLD1 | Women's breast milk | 69 | — | 0.62 ± 0.04 | |
| LO5081 | Women's breast milk | 69 | 0.87 ± 0.04 | 1.23 ± 0.04 | |
| LX5RB4 | Women's breast milk | 69 | — | 0.89 ± 0.02 | |
| LO5RB1 | Women's breast milk | 69 | — | 0.97 ± 0.03 | |
| Staphylococcus haemolyticus | ZL89-3 | Women's breast milk | 70 | 0.91 ± 0.03 | — |
| ZL114-1 | Women's breast milk | 70 | 0.87 ± 0.02 | — | |
| Staphylococcus hominis | ZL31-13 | Women's breast milk | 70 | 0.77 ± 0.05 | — |
| ZL5-5 | Women's breast milk | 70 | 0.68 ± 0.01 | — | |
| Staphylococcus arlettae | ZL114-5 | Women's breast milk | 70 | 0.45 ± 0.03 | — |
| ZL98-5 | Women's breast milk | 70 | 0.55 ± 0.08 | 0.23 ± 0.04 | |
| Staphylococcus lugdunensis | ZL5-11 | Women's breast milk | 70 | 0.69 ± 0.04 | 0.56 ± 0.05 |
| Staphylococcus gallinarum | ZL90-5 | Women's breast milk | 70 | 0.71 ± 0.06 | 0.68 ± 0.01 |
| Staphylococcus kloosii | ZL74-2 | Women's breast milk | 70 | 0.46 ± 0.03 | 0.65 ± 0.01 |
| Staphylococcus pasteuri | ZL16-6 | Women's breast milk | 70 | 0.45 ± 0.06 | — |
| Staphylococcus xylosus | ZL61-2 | Women's breast milk | 70 | 0.65 ± 0.02 | 0.56 ± 0.01 |
| Staphylococcus saprophyticus | ZL112-15 | Women's breast milk | 70 | 0.89 ± 0.08 | 0.37 ± 0.04 |
| Staphylococcus sciuri | IPLA301 | Women's breast milk | 70 | 0.98 ± 0.05 | — |
| Macrococcus caseolyticus | IPLA101 | Dairy industry surface | Unpublished data | 0.65 ± 0.03 | 0.06 ± 0.03 |
Data are means ± standard deviations calculated for three independent experiments. —, resistance to the phage.
To select S. aureus IPLA16 colonies resistant to rifampin (S. aureus IPLA16-rifR), 100-μl aliquots of overnight cultures were plated onto TSA plates supplemented with 100 μg/ml of rifampin. Plates were incubated for 16 h at 37°C. Single colonies were picked up and grown in fresh TSB at 37°C with shaking for further studies.
Bacteriophages phiIPLA-RODI and phiIPLA-C1C were propagated on S. aureus IPLA1 and S. epidermidis F12, respectively, as previously described (24).
Bacteriophage isolation and propagation.
Bacteriophages were isolated from a sewage treatment plant in Colunga, Asturias, Spain. For isolation of staphylococcal phages, 1 liter of sewage was centrifuged twice at 13,600 × g for 30 min, and the supernatant was filtered using 0.45-μm and 0.22-μm cellulose acetate membrane filters sequentially (VWR, Spain). Enrichment cultures were performed by mixing 20 ml of TSB concentrated five times (5× TSB), 80 ml of filtered sewage, and 100 μl of overnight culture from one of four mixtures of S. aureus strains (mixture 1, S. aureus IPLA3, IPLA4, IPLA6, IPLA15, IPLA16, IPLA17, and IPLA18; mixture 2, S. aureus IPLA5, IPLA8, and IPLA14; mixture 3, S. aureus IPLA1, IPLA2, IPLA9, and IPLA10; and mixture 4, S. aureus IPLA7 and IPLA13). After incubation for 16 h at 37°C, the samples were centrifuged and filtered. A total of three enrichments were carried out to obtain higher phage titrations. To assess the presence of phages, 5-μl aliquots of the supernatants from the different combinations were spotted onto bacterial lawns of each of the S. aureus and S. epidermidis strains following the double-layer technique (24). The presence of an inhibition halo was considered representative of phage sensitivity. Each inhibition halo was further purified to isolate different phages. Two phages were reisolated, propagated, and purified by use of a CsCl continuous-density gradient as described previously (24). As host bacteria, S. aureus IPLA1 and S. epidermidis F12 were used for the propagation and purification of phages phiIPLA-RODI and phiIPLA-C1C, respectively.
Bacteriophage one-step growth curves, EOP, and stabilities at various pHs and temperatures.
One-step growth curves were performed with phages phiIPLA-RODI and phiIPLA-C1C, using the sensitive strains S. aureus IPLA16 and S. epidermidis LO5081, respectively, as previously described (24). Bacteriophage host ranges were determined using phiIPLA-RODI (109 PFU/ml) and phiIPLA-C1C (109 PFU/ml) in the drop test, and titrations of the phages were further carried out with all sensitive strains to differentiate between infection and lysis due to bacteriocins. The efficiency of plaque formation (EOP) was determined by dividing the phage titer on the test strain by the phage titer on the reference strain (S. aureus IPLA1 for phage phiIPLA-RODI and S. epidermidis F12 for phage phiIPLA-C1C).
The pH stability of the phage particles was tested by incubation in Britton-Robinson pH universal buffer (150 mM KCl, 10 mM KH2PO4, 10 mM sodium citrate, 10 mM H3BO3, with adjustment of the pH in the range from 3 to 11) for 3 h at room temperature. Similarly, the temperature stability was examined by incubating the phages in SM buffer (20 mg/liter Tris-HCl, 10 mg/liter MgSO4, 10 mg/liter CaCl2, 100 mg/liter NaCl, pH 7.5) at different temperatures (ranging from 40°C to 90°C) for 30 min. Phage suspensions in SM buffer at 4°C were used as controls.
Bacterium-phage challenge test with planktonic staphylococcal cultures.
Ten milliliters of TSB broth was inoculated with an overnight culture to an optical density at 600 nm (OD600) of 0.05 and then incubated at 37°C with shaking until reaching an OD600 of 0.1 (107 CFU/ml). A 100-fold dilution of the culture was infected at a multiplicity of infection (MOI) of 100 (107 PFU/ml). Infected cultures were incubated for 8 h at 37°C, and samples were taken at 2-h intervals. Phage and cell counts were performed in triplicate.
Biofilm formation and biofilm-phage challenge test.
Overnight cultures of S. aureus IPLA16-rifR and S. epidermidis LO5081 were diluted to 106 CFU/ml in fresh TSB supplemented with 0.25% glucose. Aliquots of 200 μl of each single culture or mixture of both strains (100 μl of each strain) were poured into a 96-microwell plate (Thermo Scientific, Madrid, Spain). Biofilms were grown for 24 h at 37°C. Wells were then washed twice with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4; pH 7.4). To test biofilm degradation by each phage, 100 μl of SM buffer and 100 μl of phiIPLA-RODI or phiIPLA-C1C were added to each well (109 PFU/well). To test the combined effect of both phages, 200 μl of the mixture of both phages was added to the well (109 PFU/well of each phage). SM buffer was added for control purposes. The plates were incubated for 4 h at 37°C, the supernatants were removed, and serial dilutions were plated on TSA plates. The cells that were still bound after phage treatment were collected by scratching twice with a sterile swab, suspended in 9 ml of SM buffer, and vigorously vortexed for 1 min. Serial dilutions were plated for bacterial counting. For mixed biofilms, S. epidermidis counts were calculated as the differences between total staphylococcal counts in TSA and the S. aureus IPLA16-rifR counts in TSA supplemented with 100 μg/ml of rifampin.
Alternatively, the biomass adhered to the well was observed by staining with crystal violet (0.1% [wt/vol]) as described previously (25).
Isolation and characterization of BIMs.
Bacteriophage-insensitive mutants (BIMs) resistant to phages phiIPLA-RODI and phiIPLA-C1C were obtained from strains S. aureus IPLA16 and S. epidermidis LO5081, respectively. Aliquots of 100 μl of overnight culture of each strain (108 CFU) were incubated with 100 μl of phage (109 PFU) for 10 min at 37°C. The mixture was then poured onto a 2% TSA plate and covered with 3 ml of 0.7% TSA. Plates were incubated for 16 h at 37°C. Surviving colonies were picked up and grown in fresh TSB medium for 16 h at 37°C. Bacteriophage susceptibility was tested by the drop assay (24). The BIM frequency was calculated as the ratio between the number of surviving colonies and the initial number of bacteria incubated in the presence of phage.
Surface hydrophobicity was determined according to the microbial adhesion to solvents (MATS) assay, using hexadecane (Sigma-Aldrich, Madrid, Spain) and stationary-phase cells washed with 0.15 M NaCl and adjusted to a final OD600 of 0.8 (26). Each measurement was performed in triplicate, and the assay was carried out twice with independent cultures.
The susceptibilities of S. aureus IPLA16, S. epidermidis LO5081, and their respective BIMs to several NaCl concentrations were evaluated by determining the 50% lethal dose (LD50) values, defined as the concentration of NaCl that inhibited the growth of the strain by 50% compared with the control culture of the same strain growing in standard TSB (0.5% NaCl). Overnight cultures were diluted to an OD600 of 0.05 in TSB containing different NaCl concentrations (0.5 to 20%). Aliquots of 0.2 ml were placed into 96-microwell plates (Nunclon D surface; Nunc, Roskilde, Denmark). Plates were incubated at 37°C, and growth was monitored in a Benchmark Plus microplate spectrophotometer (Bio-Rad Laboratories) until the control samples reached an OD600 of 0.9 ± 0.1. Growth rates were estimated by linear regression after plotting the ln(OD600) as a function of time during the exponential growth phase, as described previously (27). The adsorption kinetics and the adsorption rate constants (k) of the phages were calculated as previously described (28).
Electron microscopy of phage particles.
Electron microscopic examination was performed after negative staining of the phage particles with 2% uranyl acetate. Electron micrographs were taken using a JEOL 12.000 EXII transmission electron microscope (JEOL USA Inc., Peabody, MA).
DNA extraction and protein analysis.
To prepare bacterial DNA-free samples for sequence analysis, the purified phages were treated as described previously (20). Analysis of virion proteins was carried out by SDS-PAGE analysis and matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF) mass spectrometry as previously described (29).
Bacteriophage genome analysis and characterization.
The genome sequences of phages phiIPLA-RODI and phiIPLA-C1C were determined by GenProbio SRL (Parma, Italy), using an Ion Torrent personal genome machine (PGM; Life Technologies). The MIRA program (version 3.4.0) was used for assembly of genome sequences, resulting in 138-fold coverage for phiIPLA-C1C and 432-fold coverage for phiIPLA-RODI. Additional sequencing using specific primers was done to elucidate regions with ambiguities. Phage genomes were autoannotated using RAST (30) and were manually curated. BLASTX and BLASTP were used to search for similar proteins (31). Structural predictions and motif searches were performed with InterProScan (32). Putative promoters and Shine-Dalgarno sites were predicted using the software MEME (33) followed by visual inspection. ARNold (34) and TransTerm (35) were used to detect potential rho-independent terminators. Putative tRNAs were predicted using tRNAscan-SE (36) and ARAGORN (37). Genomic comparison at the nucleotide level was made with EMBOSS Stretcher (38) and with MAUVE (39), using the genome sequences available in public databases (July 2014) for phages of the Myoviridae family infecting Staphylococcus. Before the global alignments could be performed, the genomes were manually colinearized, placing the arbitrary starting point at the end of the open reading frame (ORF) preceding the large terminase subunit gene. The genome organization of the phages and comparative BLASTN figures were generated using CGView server (40). Annotation was done on the basis of homology with previously described phages.
Statistical analysis.
Statistical analyses were performed to establish any significant differences between the control and tested strains. The differences are expressed as means ± standard deviations for three biological replicates for all assays and were determined by one-way analysis of variance (ANOVA) followed by the Bonferroni multicomparison test. P values of <0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
The sequences of phiIPLA-RODI and phiIPLA-C1C have been deposited in GenBank under accession numbers KP027446 and KP027447, respectively.
RESULTS
Bacteriophages phiIPLA-RODI and phiIPLA-C1C, two new members of the Myoviridae family that infect staphylococcal species.
Two phages were isolated from sewage after enrichment with four different mixtures of S. aureus strains. From mixtures 1 and 4, two phages were further isolated, propagated, and purified, using S. aureus IPLA1 as the host for phage phiIPLA-RODI and S. epidermidis F12 for phage phiIPLA-C1C. The host ranges of the isolated bacteriophages were determined by testing against a collection of 47 bacterial strains (Table 1). Both phages showed a wide host range, infecting 81% (phiIPLA-RODI) and 40% (phiIPLA-C1C) of the Staphylococcus strains tested. For S. aureus, all strains were infected by phiIPLA-RODI, whereas phiIPLA-C1C infected only three strains. All S. epidermidis strains were infected by phiIPLA-C1C, indicating that phage phiIPLA-C1C is more specific for S. epidermidis. In addition, 10 different species belonging to the Staphylococcus genus were also sensitive to phiIPLA-RODI, and 6 of them were sensitive to phiIPLA-C1C. Both phages were able to infect the Macrococcus caseolyticus IPLA101 strain (Table 1).
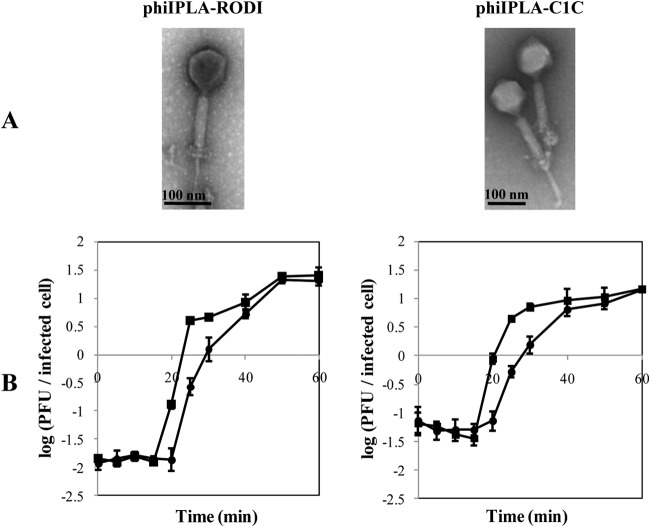
Virions of both phages were observed by transmission electron microscopy, and they showed isometric capsids and long contractile tails typical of the Myoviridae family (Fig. 1A). phiIPLA-RODI has a capsid of 73 ± 8 nm in diameter and a tail that is 95 ± 8 nm long. phiIPLA-C1C has a capsid of 88 ± 10 nm in diameter and a tail that is 110 ± 13 nm long. A double baseplate upon tail contraction, which is typical of SPO1-related phages, was clearly observed in both phages (41).
FIG 1.
(A) Transmission electron microphotographs of phages phiIPLA-RODI and phiIPLA-C1C. (B) One-step growth curves of phage phiIPLA-RODI in S. aureus IPLA16 and phage phiIPLA-C1C in S. epidermidis LO5081. Values correspond to the numbers of PFU per infected cell in chloroform-treated cultures (■) and untreated cultures (●). Each data point shows the mean ± standard deviation for three independent experiments.
One-step growth curves under standardized conditions were determined for both phages (Fig. 1B). The eclipse and latent periods of phiIPLA-RODI on S. aureus IPLA16 and of phiIPLA-C1C on S. epidermidis LO5081 were 15 and 20 min, respectively. The burst sizes were estimated to be 25 and 15 phage particles per infected cell for phiIPLA-RODI and phiIPLA-C1C, respectively (Fig. 1B).
Both phages appeared to be quite stable at temperatures below 60°C, but a total inactivation of the phages at temperatures over 70°C was observed (see Fig. S1 in the supplemental material). Concerning pH stability, a notable reduction of 3.6 log units in the phage titer was observed for phage phiIPLA-C1C at pH 11, while phiIPLA-RODI was found to be quite stable at this pH. No viable phages were recovered after incubation at pH 3 (see Fig. S1).
The genomic organization of phages phiIPLA-RODI and phiIPLA-C1C is typical of the Spounaviridae subfamily.
The genomes of phiIPLA-RODI and phiIPLA-C1C are double-stranded linear DNA molecules of 142,348 bp (carrying 213 putative ORFs) and 140,961 bp (carrying 203 putative ORFs), respectively, with ORFs preceded by potential Shine-Dalgarno sequences (see Tables S1 and S2 in the supplemental material). Up to 40 and 27 putative promoters (see Table S3), respectively, were identified, and most of them showed AT-rich sequences upstream of the −35 region and corresponded to middle or early promoters. In addition, 52 and 33 putative rho-independent terminators were predicted for phiIPLA-RODI and phiIPLA-C1C, respectively (Table S4). Two main transcriptional units were identified in both phages (Fig. 2 and 3).
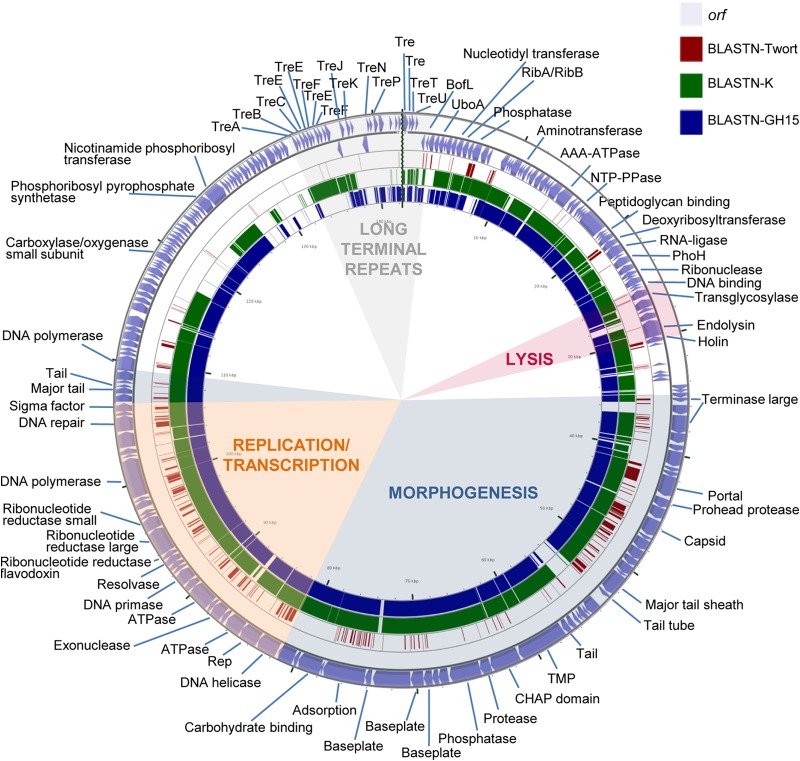
FIG 2.
Genome organization of phage phiIPLA-RODI and BLASTN comparisons. The outer ring with arrows represents the ORFs of the circularized phage. The predicted gene functions are also indicated. The different functional modules in the genome are shown by colored shading. BLASTn analysis, whose results are represented by each inner ring, was performed with the representative phages Twort (dark red), K (green), and (most similar) GH15 (blue).
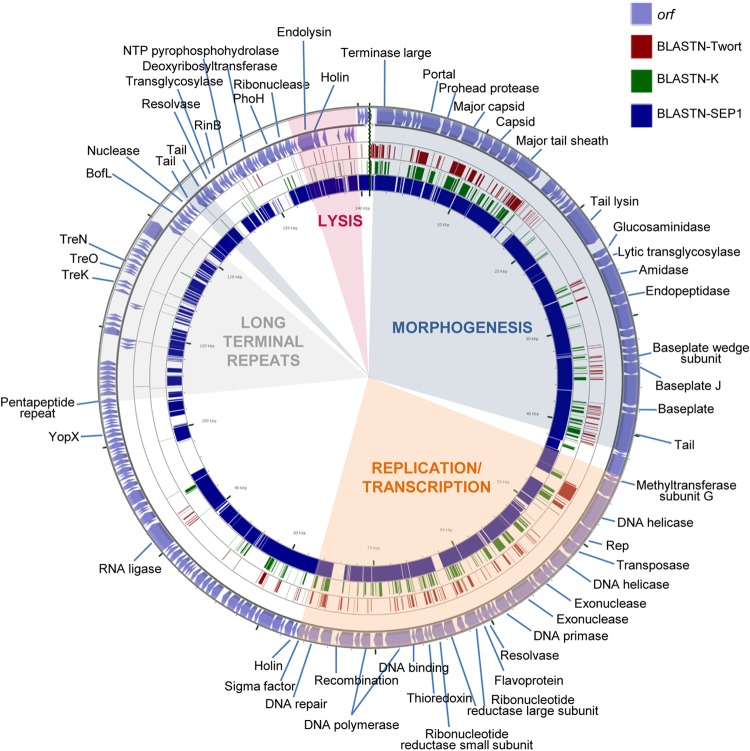
FIG 3.
Genome organization of phage phiIPLA-C1C and BLASTN comparisons. The outer ring with arrows represents the ORFs of the circularized phage. The predicted gene functions are indicated. The different functional modules in the genome are shown by colored shading. BLASTn analysis, whose results are represented by each inner ring, was performed with the representative phages Twort (dark red), K (green), and (most similar) phiIBB-SEP1 (blue).
Based on BLAST analysis and conserved domain screening, putative functions have been assigned to 93 of the predicted ORFs (44%) from phiIPLA-RODI and 80 of the predicted ORFs (39%) from phiIPLA-C1C (see Tables S1 and S2, respectively). ORFs were annotated based on the similarities of phiIPLA-RODI and phiIPLA-C1C to phage K (accession number NC_005880) and S. epidermidis phage phiIBB-SEP1 (accession number KF021268.1), respectively.
Overall, genes of both phages are organized into four functional modules, including genes for long terminal repeats (LTRs), morphogenesis, cell lysis, and replication/transcription. Furthermore, the phiIPLA-RODI genome carries a tRNA gene between orf18 and orf19, encoding tRNAMet, and three tRNA genes between orf59 and orf60, encoding tRNAAsp, tRNAPhe, and tRNATrp. The presence of a conserved sequence (TGTCAAGTTAATTT) was detected near these tRNA genes, at positions 8852 to 8865, 31999 to 32012, and 32179 to 32192, which may be binding sites for a transcriptional regulatory factor (42). No tRNA genes were identified in the phiIPLA-C1C genome.
The ends of the genomes of both phages are flanked by LTRs, which are putatively involved in the recombination of phage genomes inside the cell. These regions encode small proteins implicated in host takeover, redirecting cell metabolism to phage production (43). The exact boundaries between these LTRs and the rest of the genome have not been determined, but comparison with the terminal repeat proteins of other phages suggests that they may span from the TreA (orf192)- to the BofL (orf6)-encoding genes in phiIPLA-RODI. In this fragment, genes encoding 28 putative terminal repeat proteins were detected. In addition to the previously described TreA protein gene, the homologous TreB, TreC, TreE, TreF, TreJ, TreK, TreN, TreP, TreU, and TreT protein genes were recognized in a region of 10,330 bp. In addition, the gene for a putative group I homing HNH endonuclease (orf206), which is typical of this region, was also identified. In phiIPLA-C1C, LTRs could be expanded from the pentapeptide repeat protein gene (orf143) to the BofL gene (orf165), with a total of 11,844 bp and genes encoding 23 putative proteins. In this region, genes for three putative terminal repeat proteins, with homology to TreK (orf150), TreO (orf152), and TreN (orf155), were identified.
The morphogenesis module was split into two regions in both genomes. In phiIPLA-RODI, these regions (orf67 to orf108 and orf135 to orf139) were separated by the replication/transcription module, whereas the LTR region and the replication/transcription module were located between the two morphogenetic regions (orf1 to orf42 and orf169 to orf171) in phiIPLA-C1C. Genes encoding the large terminase subunit, portal protein, prohead protease, major capsid, major tail sheath, and tape measure protein (TMP) were identified. The large terminase subunit of phiIPLA-RODI (orf67) presented a group I intron interspaced in the gene, while this intron was not present in phiIPLA-C1C (orf2). The remainder of the proteins encoded in these regions failed to show similarity to the terminase small subunits. The TMP-encoding gene is followed by two putative genes in phiIPLA-RODI: orf95, which encodes a tail-associated protein with muralytic activity (as deduced from the presence of a CHAP domain); and orf96, which encodes a protein with a predicted endopeptidase domain. Moreover, the product of orf97 showed homology with glycerol-phosphodiester hydrolytic activities. In phiIPLA-C1C, the TMP-encoding gene is followed by four putative genes: orf28, encoding a glucosaminidase; orf29, encoding a lytic transglycosylase; orf30, encoding an amidase with a CHAP domain; and orf31, encoding a peptidase. The other genes in these modules are likely to encode baseplate, structural, and assembly proteins. Protein analysis of viral particles allowed the identification of the adsorption-associated tail protein (orf104), major tail sheath protein (orf85), capsid protein (orf78), and major tail protein (orf136) in phage phiIPLA-RODI. In phage phiIPLA-C1C, a tail protein (orf40), major tail sheath (orf18), major capsid protein (orf11), and a hypothetical protein (orf85) were also identified (Fig. 4).
FIG 4.
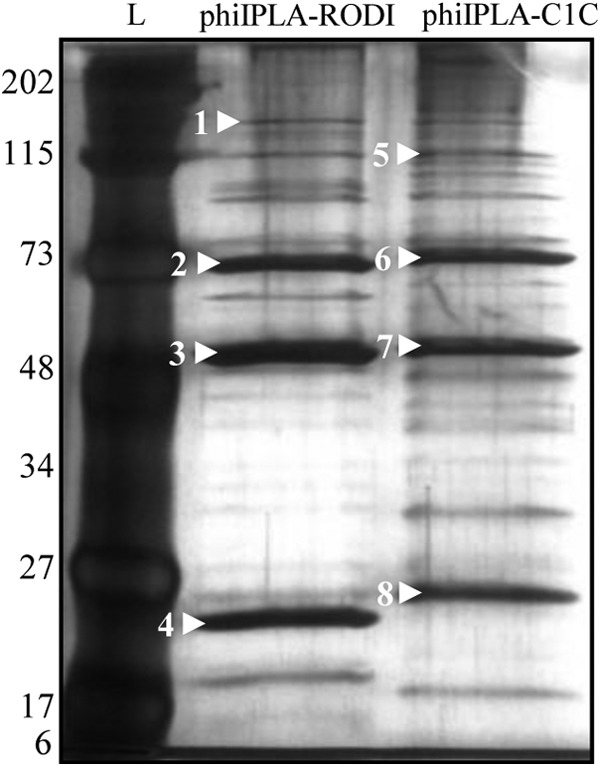
Analysis by SDS-PAGE and silver staining of the structural proteins of phages phiIPLA-RODI and phiIPLA-C1C. Protein molecular size markers (kDa) are shown on the left (lane L). Bands marked with white arrowheads were identified by mass spectrophotometry. The proteins in phage phiIPLA-RODI were the adsorption-associated tail protein (orf104) (1), the major tail sheath protein (orf85) (2), the capsid protein (orf78) (3), and the major tail protein (orf136) (4). The proteins in phage phiIPLA-C1C were the tail protein (orf40) (5), the major tail sheath (orf18) (6), the major capsid protein (orf11) (7), and a hypothetical protein (orf85) (8).
The lysis modules, containing genes involved in bacterial lysis (holin and endolysin), were located upstream of the morphogenetic module. In addition, putative transglycosylase-encoding genes (orf53 in phiIPLA-RODI and orf174 in phiIPLA-C1C), which may be involved in cell wall hydrolysis, were also identified. Moreover, in phiIPLA-C1C, a second holin gene (orf78) was located downstream of the replication/transcription module.
In the replication and transcription module, several genes related to DNA replication (DNA helicase, DNA primase, resolvase, DNA polymerase, and DNA repair protein), synthesis of DNA precursors (ribonucleotide reductase), and gene regulation (sigma factor and integration host factor) were identified. Additionally, two direct repeats of 41 nucleotides were found in the phiIPLA-RODI genome, between orf60 and orf61 (AAAAAGTACGTATTTAGAAAATAAGGAACTCTCCTATTATA). These sequences share the 27 first nucleotides with a sequence of 28 nucleotides conserved in the Myoviridae family of phages infecting Staphylococcus (except in phages Romulus, Remus, SA11, phiIBB-SEP1, and Twort). These regions are supposed to be potential binding sites for the replication initiator protein.
A group I intron associated with a VRS endonuclease was detected in the middle of the terminase large subunit gene (orf68) in phiIPLA-RODI, while in phiIPLA-C1C, two introns and one intein were identified. The group I intron GIY-YIG homing endonucleases were located interrupting the ribonucleotide reductase large subunit (orf61) and DNA polymerase (orf69) genes. The intein DOD homing endonuclease is encoded after the recombination protein (orf73). Additionally, in phiIPLA-C1C, two intronless ORFs encoding homing endonucleases (GIY-YIG and HNH) were located in intergenic regions after orf171 and orf185, respectively.
Comparative genomics.
To perform comparative genomics, genomes of Myoviridae phages infecting Staphylococcus were colinearized to start at the terminase large subunit. At the nucleotide level, phage phiIPLA-C1C shares a similarity of 80.2% with the only S. epidermidis-specific myophage, phiIBB-SEP1 (Table 2), whereas its similarity with the S. aureus phages is <55%, suggesting that phiIPLA-C1C might be specific for S. epidermidis. Phage phiIPLA-RODI is more closely related to the other phages in the database (identities of over 80%), excluding phage phiIBB-SEP1 and the representative phage Twort, in which case the similarity is <56% (Table 2).
TABLE 2.
Comparative genomic analysis of Myoviridae phages infecting Staphylococcus, using Emboss Stretcher
| Phage | % similarity |
|
|---|---|---|
| phiIPLA-C1C | phiIPLA-RODI | |
| phiIPLA-C1C | 100 | 54.8 |
| phiIPLA-RODI | 54.8 | 100 |
| G1 | 54.6 | 83.5 |
| GH15 | 54.2 | 84.3 |
| JD007 | 54.1 | 83.5 |
| K | 53.6 | 81.1 |
| Twort | 53.4 | 54.5 |
| vB_SauM_Remus | 53.6 | 54.6 |
| vB_SauM_Romulus | 52.9 | 54.2 |
| SA11 | 54.7 | 56.2 |
| SA1 | 44.1 | 44.7 |
| ISP | 54.7 | 83.8 |
| A5W | 54.7 | 83.2 |
| Sb-1 | 54.1 | 79 |
| SA5 | 54.3 | 83.1 |
| S25-3 | 55.1 | 81.2 |
| S25-4 | 55.3 | 80.1 |
| SA012 | 54.8 | 81.6 |
| phiIBB-SEP1 | 80.2 | 55.8 |
| P4W | 55.1 | 83.6 |
| MSA6 | 54.9 | 81.7 |
| Fi200w | 55 | 84.2 |
| 676Z | 55 | 84.3 |
| A3R | 55.1 | 82.6 |
| Staph1N | 54.7 | 83.1 |
A MAUVE comparison allowed us to determine that all of these phages possess the same general modular structure, including the morphogenesis, replication/transcription, long terminal repeat, and lysis modules. The morphogenesis and replication/transcription modules are conserved among the Myoviridae phages infecting Staphylococcus (Fig. 2 and 3; see Fig. S2 in the supplemental material). The internal organization in these regions is highly dependent on the presence of homing endonucleases and transposases. Regarding the LTR region, phage phiIPLA-RODI shared homology with all the phages except Twort, phiIPLA-C1C, and phiIBB-SEP1. The phages phiIPLA-C1C and phiIBB-SEP1 possess a specific organization in the LTR region that is not shared with the other phages. The most variable regions at the nucleotide level are located upstream of the LTR region, in which differences regarding the length and gene arrangement are found (see Fig. S2).
Killing of planktonic staphylococcal cultures by phiIPLA-RODI and phiIPLA-C1C.
Phages phiIPLA-RODI and phiIPLA-C1C had the same lytic activity on S. aureus IPLA16, since no viable bacteria were detected after 8 h of incubation and a considerable decrease of the bacterial population was already achieved after 6 h of treatment (7.9 log units compared to the control). Note that both phages halted growth during the first 4 h, keeping bacterial counts at 105 CFU/ml (Fig. 5A). When S. epidermidis LO5081 was infected by either phage, no viable counts were detected after 8 h of incubation (Fig. 5B). However, phiIPLA-C1C killed host cells more quickly than phiIPLA-RODI did, and after 4 h of incubation, only 10 CFU/ml viable cells remained. As expected, the number of phages increased in all infected cultures, to about 109 PFU/ml (data not shown).
FIG 5.
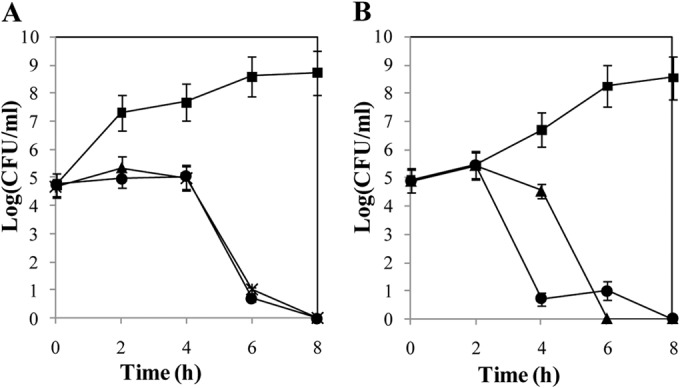
Susceptibilities of S. aureus IPLA16 (A) and S. epidermidis LO5081 (B) to phages phiIPLA-RODI and phiIPLA-C1C. Cell counts of control cultures (■) and cultures treated with phiIPLA-RODI (▲) or phiIPLA-C1C (●) are represented as log (CFU/ml). Each value corresponds to the mean ± standard deviation for three independent experiments.
phiIPLA-RODI proved to be more effective than phiIPLA-C1C for removal of mono- and dual-species staphylococcal biofilms.
To perform challenge assays against mono- and dual-species biofilms, an S. aureus IPLA16-derived strain that is resistant to rifampin (S. aureus IPLA16-rifR) was isolated, with the same phage sensitivity and biofilm formation as its parent (data not shown). S. aureus IPLA16-rifR and S. epidermidis LO5081 were grown in both mono- and dual-species biofilms and treated with the phages individually and as a mixture. Numbers of surface-adhered bacteria were successfully reduced after phage treatment (Fig. 6A). In the presence of phiIPLA-RODI, a reduction of 2.43 log units was achieved for S. aureus IPLA16-rifR, with a reduction of 1.89 log units for S. epidermidis LO5081. Phage phiIPLA-C1C showed a reduced lytic ability against both staphylococcal biofilms, with reductions in viable counts of 1.84 and 1.16 log units for S. aureus IPLA16-rifR and S. epidermidis LO5081, respectively. No significant reduction beyond that recorded for individual phages was observed on biofilms treated with a mixture of phages (Fig. 6A).
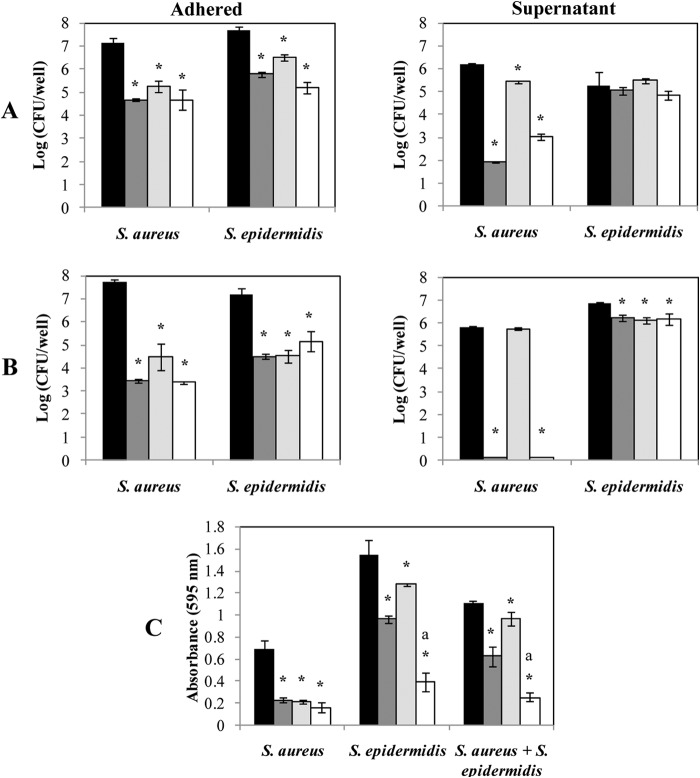
FIG 6.
Bacteriophage-mediated removal of 24-h-old S. aureus and S. epidermidis biofilms. Mono (A)- or dual (B)-species biofilms of S. aureus IPLA16-rifR and S. epidermidis LO5081 were treated with phage phiIPLA-RODI (dark gray), phage phiIPLA-C1C (light gray), or a mixture of both phages (white) for 4 h. Data on control biofilms are presented in black. Adhered cell counts and supernatant cell counts are expressed as log (CFU/well). The bacterial detection threshold was 10 log (CFU/ml). (C) Alternatively, the biomass was calculated by crystal violet staining of adhered cells after phage treatment. The absorbance was measured at a wavelength of 595 nm. Means and standard deviations were calculated for three biological replicates. Bars marked with an asterisk are significantly different from the control (ANOVA; P < 0.05), and bars marked with “a” indicate significantly different decreases in biomass between the treatment with the mixture of phages and the individual treatment with either phiIPLA-RODI or phiIPLA-C1C (ANOVA; P < 0.05).
Planktonic cells of S. aureus IPLA16-rifR were more sensitive to lysis by phage phiIPLA-RODI (reduction of 4.27 log units) than by phage phiIPLA-C1C (reduction of 0.76 log unit) (Fig. 6A). However, neither individual phage nor the phage mixture was able to kill planktonic S. epidermidis LO5081 (ANOVA; P > 0.05) (Fig. 6A).
In dual-species biofilms, treatment with phage phiIPLA-RODI showed a reduction in adhered cells of 4.27 log units for S. aureus IPLA16-rifR and of 2.66 log units for S. epidermidis LO5081 (Fig. 6B). Treatment of biofilms with phiIPLA-C1C was found to be more effective than that observed in individual biofilms, with a decrease in the adhered cells of 3.23 log units for S. aureus IPLA16-rifR and 2.64 log units for S. epidermidis LO5081. Similar to the monospecies biofilm challenge, the mixture of phages did not clearly improve the results obtained by individual phages.
Regarding the planktonic cells, the efficacy of phiIPLA-RODI was higher than that shown by phiIPLA-C1C against both strains forming the dual-species biofilm (Fig. 6B). A reduction of 5.69 log units in S. aureus IPLA16-rifR and of 0.64 log unit in S. epidermidis LO5081 was obtained. For phiIPLA-C1C, only a weak reduction in viable counts was detected for S. aureus IPLA16-rifR (Fig. 6B). Treatment with a mixture of phages gave similar results to those obtained using phiIPLA-RODI.
Crystal violet staining was used to confirm the reduction in the total biomass of phage-treated biofilms (Fig. 6C). Overall, removal of biomass by phage treatment in mono- and dual-species biofilms was in accordance with the viable count results (see above). However, in terms of total biomass, the phage treatment turned out to be more effective when a mixture of both phages was applied to both an S. epidermidis LO5081 single-species biofilm and the dual-species biofilm (Fig. 6C). Similarly, the treatment with phiIPLA-RODI was more effective than that with phiIPLA-C1C. However, in biofilms formed only by S. aureus IPLA16-rifR, all the treatments were found to have similar detachment abilities.
A phage resistance phenotype has an important fitness cost and is highly unstable.
BIMs of S. aureus IPLA16 after treatment with phiIPLA-RODI (MOI = 100) and of S. epidermidis LO5081 treated with phiIPLA-C1C (MOI = 1,000) emerged at frequencies of 4.05 × 10−7 ± 2.34 × 10−9 and 1.1 × 10−7 ± 2.08 × 10−9, respectively. To test whether resistance implies fitness costs, three phage-resistant colonies of S. aureus IPLA16 (S. aureus IPLA16-R40, S. aureus IPLA16-R53, and S. aureus IPLA16-R71) and two phage-resistant colonies of S. epidermidis LO5081 (S. epidermidis LO5081-R49 and S. epidermidis LO5081-R32) were randomly selected for further microbiological characterization (Table 3).
TABLE 3.
Fitness of S. aureus IPLA16 and S. epidermidis LO5081, and their BIMs, against phages phiIPLA-RODI and phiIPLA-C1C, respectivelya
| Strain | Growth rate (μ) (h−1) | LD50 of NaCl (%) | Biofilm formation (OD595) | % phage adsorption in 15 min | Adsorption rate constant (k) (ml/min) | Hydrophobicity (% adhesion to hexadecane) | Generation after which reversion occurred |
|---|---|---|---|---|---|---|---|
| S. aureus IPLA16 | 0.76 ± 0.02 | 8.14 ± 0.42 | 1.13 ± 0.06 | 87.67 ± 3.74 | 8.9 × 10−11 ± 7.5 × 10−12 | 85.62 ± 5.47 | |
| S. aureus IPLA16-R40 | 0.56 ± 0.02* | 6.12 ± 0.33* | 0.31 ± 0.01* | 62.66 ± 2.09* | 3.1 × 10−10 ± 7.1 × 10−11* | 44.47 ± 7.67* | 27 |
| S. aureus IPLA16-R53 | 0.59 ± 0.04* | 4.80 ± 0.23* | 0.28 ± 0.02* | 38.33 ± 6.21* | 6.5 × 10−10 ± 1.7 × 10−11* | 32.13 ± 8.37* | 27 |
| S. aureus IPLA16-R71 | 0.49 ± 0.03* | 2.82 ± 0.34* | 0.41 ± 0.02* | 39.66 ± 3.25* | 6.2 × 10−10 ± 1.2 × 10−11* | 34.77 ± 8.17* | 27 |
| S. epidermidis LO5081 | 0.74 ± 0.03 | 8.23 ± 0.74 | 9.81 ± 0.38 | 90.60 ± 8.19 | 6.5 × 10−11 ± 2.3 × 10−12 | 95.43 ± 2.06 | |
| S. epidermidis LO5081-R32 | 0.58 ± 0.02* | 8.15 ± 0.25 | 0.39 ± 0.06* | 17.60 ± 7.23* | 6.9 × 10−10 ± 1.3 × 10−10* | 33.78 ± 6.65* | 57 |
| S. epidermidis LO5081-R49 | 0.52 ± 0.06* | 8.13 ± 0.08 | 8.99 ± 0.47 | 23.00 ± 5.69* | 9.9 × 10−10 ± 1.7 × 10−10* | 77.05 ± 11.31* | 17 |
Values represent the means ± standard deviations for three biological replicates. The presence of an asterisk indicates those values that are significantly different from those of the wild-type strain (ANOVA; P < 0.05).
In phage-free liquid cultures, all the BIMs formed aggregates as observed by optical microscopy (see Fig. S3 in the supplemental material). Moreover, the growth rate of BIMs was clearly reduced compared to that of wild-type strains (Table 3). Other parameters indicative of cellular fitness, such as the ability of BIMs to grow in high NaCl concentrations and to form biofilms on polystyrene surfaces, were also determined. S. aureus IPLA16-derived BIMs were sensitive to 8% NaCl (Table 3), whereas S. epidermidis LO5081-derived BIMs showed resistance to NaCl similar to that of control cultures. Phage-resistant bacteria of both species had a reduced capacity to form biofilms on polystyrene surfaces (up to a 4-fold reduction for S. aureus IPLA16 BIMs and up to a 25-fold reduction for S. epidermidis LO5081-R33), except for S. epidermidis LO5081-R49, which showed values similar to those of the wild-type strain (Table 3).
Regardless of the phage against which BIMs had arisen, they were also resistant to either phiIPLA-RODI or phiIPLA-CIC as well as to phages vB_SauS_phiIPLA88 (44) and vB_SepS_phiIPLA5 (24), two Siphoviridae staphylococcal phages (data not shown). Cross-resistance suggested that impaired phage infection may have been due to a lower or nonexistent adsorption of phages. The kinetics of phage binding indicated that adsorption of phiIPLA-RODI and phiIPLA-C1C proceeded to 87 to 90% in 15 min for wild-type strains, while the adsorption rates of BIMs were reduced to 38 to 62% in S. aureus and 23% in S. epidermidis BIMs (Table 3), indicating that phage infection was prevented by low adsorption of phages to the bacterial surface. Changes in cell surface properties were further confirmed by the significantly less hydrophobic character of the BIMs than of the wild-type strains (Table 3).
To check whether the phage resistance phenotype of BIMs is stable without the selective pressure of phages, BIMs were subcultured for 57 generations, and their sensitivities to phages phiIPLA-RODI and phiIPLA-C1C were tested. S. aureus IPLA16-R40, IPLA16-R53, and IPLA16-R71 showed a highly unstable phenotype, and sensitive cultures (a defined transparent halo was observed) were obtained after 27 generations. More variability was observed for S. epidermidis LO5081 BIMs, as S. epidermidis LO5081-R49 reverted to the sensitive phenotype after 17 generations, while S. epidermidis LO5081-R33 lost phage resistance only after 57 generations (Table 3). Recovery of phage sensitivity was linked to the reestablishment of sensitivity to either phiIPLA-RODI or phiIPLA-C1C and the two Siphoviridae phages. Moreover, the original growth rate was also restored (data not shown).
DISCUSSION
Within the bacteriophage therapy context, we have characterized two new lytic phages, phiIPLA-RODI and phiIPLA-C1C, which show the typical wide host range of polyvalent phages, such as phage K and phi812 (45, 46), in contrast to other monovalent myophages, such as Stau2, Romulus, and Remus (28, 47), whose host range is limited to S. aureus strains, and phage phiIBB-SEP1, infecting only S. epidermidis strains (48).
From the perspective of feasibility for therapeutic application, we determined that the stabilities of phages phiIPLA-RODI and phiIPLA-C1C at various pHs and temperatures were very similar to those described for related phages, such as MSA6 (49) and Romulus and Remus (28), and therefore that these phages are suitable for design of different pharmaceutical formulations by lyophilization (50), spray drying (51), and aerosolization (52). Bioinformatic analysis of the phiIPLA-RODI and phiIPLA-C1C genomes showed the typical characteristics of the “Twort-like viruses” (Spounavirinae subfamily): strictly virulent, with large genomes (127 to 140 kb) containing LTRs, genes grouped into modules that are not clearly separated, a few genes encoding tRNAs, and the presence of group I introns (42). The genome of phage phiIPLA-C1C differs from those of the above-mentioned phages in the lack of genes encoding tRNAs, a peculiarity already observed in the S. epidermidis phage phiIBB-SEP1 (48). In addition, the nucleotide genome sequence of phiIPLA-RODI showed a lack of restriction sites for the endonucleases Sau3AI, BamHI, and BglII, but these are present in the phiIPLA-C1C genome. This appears to be a general strategy among S. aureus bacteriophages, such as Twort, K, G1, Sb-1, and MSA6, to avoid restriction by host bacteria (49, 53).
Both phages encode homing endonucleases, found within group I introns, intergenic regions, or inteins, in accordance with previous results reported for other myophages, such as T4, where 11% of the ORFs correspond to homing endonucleases (54). In T4-related phages, most of the homing endonucleases belong to the GIY-YIG and HNH families, with multiple functions, such as recombination and binding to and repair of DNA (54). Homing endonucleases encoded by phiIPLA-C1C were not identified in other phage genomes, consistent with the idea that these proteins are a recent evolutionary acquisition that may have an influence on gene arrangement and function and that, in addition, they may promote their own spread between phages (54).
Once the phages were morphologically and genetically characterized, we proceeded to study their lytic activity against staphylococcal bacteria in both planktonic cultures and preformed biofilms. The results of biofilm removal assays confirmed that phage infection in planktonic cell cultures is more efficient than that in biofilms. Overall, S. aureus IPLA16 biofilms were well infected by both phages, while S. epidermidis LO5081 biofilms were found to be more resistant to phage predation. A likely explanation is the higher content of extracellular matrix formed by S. epidermidis LO5081 than by S. aureus IPLA16, which may hinder the access of phages to bacteria. Some phages encode polysaccharide depolymerase proteins which degrade the extracellular matrix of biofilms, facilitating the access of phages to target bacteria (20). However, no genes encoding proteins with these catalytic domains were detected in either the phiIPLA-RODI or phiIPLA-C1C genome. After biofilm treatment, it was also quite surprising that detached cells were not killed by phages, except those released from S. aureus IPLA16 biofilms treated with phiIPLA-RODI. These data suggest that cells released from inside a biofilm are not susceptible to phage infection due to their unique physiological state (22). The biomass reductions of S. aureus IPLA16 biofilms by these phages were similar (67% for phage phiIPLA-RODI and 69% for phage phiIPLA-C1C) to those obtained with phages ISP, Romulus, and Remus (28). The complete eradication of biofilms by phages has not been described in the literature to date. However, prevention of biofilm formation has been achieved for S. aureus Xen29 biofilms, using a combination of phage K and modified derivatives (21). In addition, S. aureus biofilms could be eradicated efficiently with a combination of phage SAP-26 and rifampin (55).
Phage control of dual-species biofilms was investigated using S. aureus and S. epidermidis as a proof of concept to evaluate whether the mixture of both phages could be more effective than each individual phage. Our results provide evidence that phages can reduce the cell numbers of both species, but the application of a phage mixture against each of the hosts was not always more effective than the use of only one phage. Indeed, the addition of the single phage phiIPLA-RODI resulted in a reduction similar to that achieved by the mixture of phages. The low efficacy of phiIPLA-C1C may be due to the lower burst size of this phage than that of phiIPLA-RODI. Overall, these results are in agreement with those obtained by other authors (56, 57), who reported reductions of about 3 to 4 log units in viable cells from mixed biofilms treated with phages. Mixed-species biofilms are complex communities in which the physiological state of cells and the availability of phage receptors play important roles in the behavior of phages (58). These features may be altered drastically by competition with other bacterial species. In mixed cultures of Escherichia coli and Salmonella, phages against E. coli were more effective at removing this species than in monocultures of E. coli. It seems that for some bacteria, the competition with other bacterial species may enhance the effectiveness of phages, because phage bacterial resistance can decrease the competitive ability of bacteria (59). Similar data were observed in our staphylococcal biofilms, in which treatment with phages turned to be more effective in dual-species biofilms than in monospecies biofilms.
Phage resistance is often a major concern in the therapeutic application of phages, as it may compromise the efficacy of the treatment (60). In this regard, the frequencies of bacteria acquiring resistance to phages phiIPLA-RODI and phiIPLA-C1C were determined to be low, and most important, the global fitness of staphylococcal BIMs was clearly affected. This reduced fitness of phage-resistant bacteria was also observed for different species, such as Vibrio cholerae (61) and Pseudomonas aeruginosa (62). We have not determined the molecular basis of the phage resistance mechanisms in the BIMs. However, it is known that wall teichoic acid (WTA) serves as a receptor for several staphylococcal siphoviruses and myoviruses (63). Therefore, a lack of WTA in S. aureus and S. epidermidis BIMs might explain the resistance of these strains to phages belonging to the Siphoviridae and Myoviridae families. In addition, phiIPLA-RODI and phiIPLA-C1C BIMs were shown to be more sensitive to high temperatures, showed higher degrees of cell aggregation, and had a reduced capacity to form biofilms, as previously observed in S. aureus WTA-deficient mutants (64). However, we cannot disregard the presence of a capsular polysaccharide in staphylococcal BIMs, which may modify the bacterial surface properties and prevent phage adsorption to the cell (65).
The data presented in this study support the potential of the lytic bacteriophages phiIPLA-RODI and phiIPLA-C1C to be used in phage therapy. Their characterization indicates a wide host range, an adequate stability in various environmental conditions, a lack of virulence factors, the capability to remove biofilms, and a low frequency of BIMs.
Supplementary Material
ACKNOWLEDGMENTS
This research study was supported by grants AGL2012-40194-C02-01 (Ministry of Science and Innovation, Spain) and GRUPIN14-139 (Program of Science, Technology and Innovation 2013–2017, Principado de Asturias, Spain) and by the bacteriophage network FAGOMA. D.G. is a fellow of the Ministry of Science and Innovation, Spain. P.G., B.M., and A.R. are members of the FWO Vlaanderen-funded Phagebiotics research community (WO.016.14). D.V. holds a Ph.D. scholarship from the IWT Vlaanderen.
We thank R. Calvo (IPLA-CSIC) for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03560-14.
REFERENCES
- 1.Otto M. 2013. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med 64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- 2.Savage VJ, Chopra I, O'Neill AJ. 2013. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob Agents Chemother 57:1968–1970. doi: 10.1128/AAC.02008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogers KL, Fey PD, Rupp ME. 2009. Coagulase-negative staphylococcal infections. Infect Dis Clin North Am 23:73–98. doi: 10.1016/j.idc.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Hell E, Giske CG, Hultenby K, Danielsson KG, Marchini G. 2013. Attachment and biofilm forming capabilities of Staphylococcus epidermidis strains isolated from preterm infants. Curr Microbiol 67:712–717. doi: 10.1007/s00284-013-0425-3. [DOI] [PubMed] [Google Scholar]
- 5.Otto M. 2009. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat Rev Microbiol 7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hellmark B, Berglund C, Nilsdotter-Augustinsson A, Unemo M, Soderquist B. 2013. Staphylococcal cassette chromosome mec (SCCmec) and arginine catabolic mobile element (ACME) in Staphylococcus epidermidis isolated from prosthetic joint infections. Eur J Clin Microbiol Infect Dis 32:691–697. doi: 10.1007/s10096-012-1796-2. [DOI] [PubMed] [Google Scholar]
- 7.Periasamy S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, Cheung GY, Otto M. 2012. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci U S A 109:1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fessler AT, Kadlec K, Hassel M, Hauschild T, Eidam C, Ehricht R, Monecke S, Schwarz S. 2011. Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl Environ Microbiol 77:7151–7157. doi: 10.1128/AEM.00561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305-11. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kutateladze M, Adamia R. 2010. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol 28:591–595. doi: 10.1016/j.tibtech.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Miedzybrodzki R, Borysowski J, Weber-Dabrowska B, Fortuna W, Letkiewicz S, Szufnarowski K, Pawelczyk Z, Rogoz P, Klak M, Wojtasik E, Gorski A. 2012. Clinical aspects of phage therapy. Adv Virus Res 83:73–121. doi: 10.1016/B978-0-12-394438-2.00003-7. [DOI] [PubMed] [Google Scholar]
- 13.Leszczyński P, Weber-Dabrowska B, Kohutnicka M, Luczak M, Górecki A, Górski A. 2006. Successful eradication of methicillin-resistant Staphylococcus aureus (MRSA) intestinal carrier status in a healthcare worker—case report. Folia Microbiol (Praha) 51:236–238. doi: 10.1007/BF02932128. [DOI] [PubMed] [Google Scholar]
- 14.Takemura-Uchiyama I, Uchiyama J, Osanai M, Morimoto N, Asagiri T, Ujihara T, Daibata M, Sugiura T, Matsuzaki S. 2014. Experimental phage therapy against lethal lung-derived septicemia caused by Staphylococcus aureus in mice. Microbes Infect 16:512–517. doi: 10.1016/j.micinf.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Takemura-Uchiyama I, Uchiyama J, Kato S, Inoue T, Ujihara T, Ohara N, Daibata M, Matsuzaki S. 2013. Evaluating efficacy of bacteriophage therapy against Staphylococcus aureus infections using a silkworm larval infection model. FEMS Microbiol Lett 347:52–60. doi: 10.1111/1574-6968.12220. [DOI] [PubMed] [Google Scholar]
- 16.Drilling A, Morales S, Jardeleza C, Vreugde S, Speck P, Wormald PJ. 2014. Bacteriophage reduces biofilm of Staphylococcus aureus ex vivo isolates from chronic rhinosinusitis patients. Am J Rhinol Allergy 28:3–11. doi: 10.2500/ajra.2014.28.4001. [DOI] [PubMed] [Google Scholar]
- 17.Mendes JJ, Leandro C, Corte-Real S, Barbosa R, Cavaco-Silva P, Melo-Cristino J, Gorski A, Garcia M. 2013. Wound healing potential of topical bacteriophage therapy on diabetic cutaneous wounds. Wound Repair Regen 21:595–603. doi: 10.1111/wrr.12056. [DOI] [PubMed] [Google Scholar]
- 18.Chhibber S, Kaur T, Sandeep K. 2013. Co-therapy using lytic bacteriophage and linezolid: effective treatment in eliminating methicillin resistant Staphylococcus aureus (MRSA) from diabetic foot infections. PLoS One 8:e56022. doi: 10.1371/journal.pone.0056022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seth AK, Geringer MR, Nguyen KT, Agnew SP, Dumanian Z, Galiano RD, Leung KP, Mustoe TA, Hong SJ. 2013. Bacteriophage therapy for Staphylococcus aureus biofilm-infected wounds: a new approach to chronic wound care. Plast Reconstr Surg 131:225–234. doi: 10.1097/PRS.0b013e31827e47cd. [DOI] [PubMed] [Google Scholar]
- 20.Gutierrez D, Martinez B, Rodriguez A, Garcia P. 2012. Genomic characterization of two Staphylococcus epidermidis bacteriophages with anti-biofilm potential. BMC Genomics 13:228. doi: 10.1186/1471-2164-13-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly D, McAuliffe O, Ross RP, Coffey A. 2012. Prevention of Staphylococcus aureus biofilm formation and reduction in established biofilm density using a combination of phage K and modified derivatives. Lett Appl Microbiol 54:286–291. doi: 10.1111/j.1472-765X.2012.03205.x. [DOI] [PubMed] [Google Scholar]
- 22.Cerca N, Oliveira R, Azeredo J. 2007. Susceptibility of Staphylococcus epidermidis planktonic cells and biofilms to the lytic action of Staphylococcus bacteriophage K. Lett Appl Microbiol 45:313–317. doi: 10.1111/j.1472-765X.2007.02190.x. [DOI] [PubMed] [Google Scholar]
- 23.Kropinski AM, Prangishvili D, Lavigne R. 2009. Position paper: the creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ Microbiol 11:2775–2777. doi: 10.1111/j.1462-2920.2009.01970.x. [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez D, Martinez B, Rodriguez A, Garcia P. 2010. Isolation and characterization of bacteriophages infecting Staphylococcus epidermidis. Curr Microbiol 61:601–608. doi: 10.1007/s00284-010-9659-5. [DOI] [PubMed] [Google Scholar]
- 25.Gutierrez D, Ruas-Madiedo P, Martinez B, Rodriguez A, Garcia P. 2014. Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS One 9:e107307. doi: 10.1371/journal.pone.0107307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellon-Fontaine MN, Rault J, Van Oss CJ. 1996. Microbial adhesion to solvents: a novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloids Surf B Biointerfaces 7:47–53. doi: 10.1016/0927-7765(96)01272-6. [DOI] [Google Scholar]
- 27.Campelo AB, Gaspar P, Roces C, Rodriguez A, Kok J, Kuipers OP, Neves AR, Martinez B. 2011. The Lcn972 bacteriocin-encoding plasmid pBL1 impairs cellobiose metabolism in Lactococcus lactis. Appl Environ Microbiol 77:7576–7585. doi: 10.1128/AEM.06107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandersteegen K, Kropinski AM, Nash JH, Noben JP, Hermans K, Lavigne R. 2013. Romulus and Remus, two phage isolates representing a distinct clade within the Twortlikevirus genus, display suitable properties for phage therapy applications. J Virol 87:3237–3247. doi: 10.1128/JVI.02763-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia P, Rodriguez I, Suarez JE. 2004. A −1 ribosomal frameshift in the transcript that encodes the major head protein of bacteriophage A2 mediates biosynthesis of a second essential component of the capsid. J Bacteriol 186:1714–1719. doi: 10.1128/JB.186.6.1714-1719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Zdobnov EM, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 33.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naville M, Ghuillot-Gaudeffroy A, Marchais A, Gautheret D. 2011. ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol 8:11–13. doi: 10.4161/rna.8.1.13346. [DOI] [PubMed] [Google Scholar]
- 35.Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. 2000. Prediction of transcription terminators in bacterial genomes. J Mol Biol 301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- 36.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers EW, Miller W. 1988. Optimal alignments in linear space. Comput Appl Biosci 4:11–17. [DOI] [PubMed] [Google Scholar]
- 39.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grant JR, Stothard P. 2008. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klumpp J, Lavigne R, Loessner MJ, Ackermann HW. 2010. The SPO1-related bacteriophages. Arch Virol 155:1547–1561. doi: 10.1007/s00705-010-0783-0. [DOI] [PubMed] [Google Scholar]
- 42.Lobocka M, Hejnowicz MS, Dabrowski K, Gozdek A, Kosakowski J, Witkowska M, Ulatowska MI, Weber-Dabrowska B, Kwiatek M, Parasion S, Gawor J, Kosowska H, Glowacka A. 2012. Genomics of staphylococcal Twort-like phages—potential therapeutics of the post-antibiotic era. Adv Virus Res 83:143–216. doi: 10.1016/B978-0-12-394438-2.00005-0. [DOI] [PubMed] [Google Scholar]
- 43.Stewart CR, Yip TK, Myles B, Laughlin L. 2009. Roles of genes 38, 39, and 40 in shutoff of host biosyntheses during infection of Bacillus subtilis by bacteriophage SPO1. Virology 392:271–274. doi: 10.1016/j.virol.2009.06.046. [DOI] [PubMed] [Google Scholar]
- 44.Garcia P, Martinez B, Obeso JM, Lavigne R, Lurz R, Rodriguez A. 2009. Functional genomic analysis of two Staphylococcus aureus phages isolated from the dairy environment. Appl Environ Microbiol 75:7663–7673. doi: 10.1128/AEM.01864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Flaherty S, Ross RP, Meaney W, Fitzgerald GF, Elbreki MF, Coffey A. 2005. Potential of the polyvalent anti-Staphylococcus bacteriophage K for control of antibiotic-resistant staphylococci from hospitals. Appl Environ Microbiol 71:1836–1842. doi: 10.1128/AEM.71.4.1836-1842.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Haddad L, Ben Abdallah N, Plante PL, Dumaresq J, Katsarava R, Labrie S, Corbeil J, St-Gelais D, Moineau S. 2014. Improving the safety of Staphylococcus aureus polyvalent phages by their production on a Staphylococcus xylosus strain. PLoS One 9:e102600. doi: 10.1371/journal.pone.0102600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsieh SE, Lo HH, Chen ST, Lee MC, Tseng YH. 2011. Wide host range and strong lytic activity of Staphylococcus aureus lytic phage Stau2. Appl Environ Microbiol 77:756–761. doi: 10.1128/AEM.01848-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melo LD, Sillankorva S, Ackermann HW, Kropinski AM, Azeredo J, Cerca N. 2014. Isolation and characterization of a new Staphylococcus epidermidis broad-spectrum bacteriophage. J Gen Virol 95:506–515. doi: 10.1099/vir.0.060590-0. [DOI] [PubMed] [Google Scholar]
- 49.Kwiatek M, Parasion S, Mizak L, Gryko R, Bartoszcze M, Kocik J. 2012. Characterization of a bacteriophage, isolated from a cow with mastitis, that is lytic against Staphylococcus aureus strains. Arch Virol 157:225–234. doi: 10.1007/s00705-011-1160-3. [DOI] [PubMed] [Google Scholar]
- 50.Merabishvili M, Vervaet C, Pirnay JP, De Vos D, Verbeken G, Mast J, Chanishvili N, Vaneechoutte M. 2013. Stability of Staphylococcus aureus phage ISP after freeze-drying (lyophilization). PLoS One 8:e68797. doi: 10.1371/journal.pone.0068797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vandenheuvel D, Singh A, Vandersteegen K, Klumpp J, Lavigne R, Van den Mooter G. 2013. Feasibility of spray drying bacteriophages into respirable powders to combat pulmonary bacterial infections. Eur J Pharm Biopharm 84:578–582. doi: 10.1016/j.ejpb.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 52.Turgeon N, Toulouse MJ, Martel B, Moineau S, Duchaine C. 2014. Comparison of five bacteriophages as models for viral aerosol studies. Appl Environ Microbiol 80:4242–4250. doi: 10.1128/AEM.00767-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andriashvili IA, Kvachadze LI, Bashakidze RP, Adamiia R, Chanishvili TG. 1986. Molecular mechanism of phage DNA protection from the restriction endonucleases of Staphylococcus aureus cells. Mol Gen Mikrobiol Virusol 1986:43–45. [PubMed] [Google Scholar]
- 54.Edgell DR, Gibb EA, Belfort M. 2010. Mobile DNA elements in T4 and related phages. Virol J 7:290. doi: 10.1186/1743-422X-7-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rahman M, Kim S, Kim SM, Seol SY, Kim J. 2011. Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin. Biofouling 27:1087–1093. doi: 10.1080/08927014.2011.631169. [DOI] [PubMed] [Google Scholar]
- 56.Carson L, Gorman SP, Gilmore BF. 2010. The use of lytic bacteriophages in the prevention and eradication of biofilms of Proteus mirabilis and Escherichia coli. FEMS Immunol Med Microbiol 59:447–455. doi: 10.1111/j.1574-695X.2010.00696.x. [DOI] [PubMed] [Google Scholar]
- 57.Sillankorva S, Neubauer P, Azeredo J. 2010. Phage control of dual species biofilms of Pseudomonas fluorescens and Staphylococcus lentus. Biofouling 26:567–575. doi: 10.1080/08927014.2010.494251. [DOI] [PubMed] [Google Scholar]
- 58.Sutherland IW, Hughes KA, Skillman LC, Tait K. 2004. The interaction of phage and biofilms. FEMS Microbiol Lett 232:1–6. doi: 10.1016/S0378-1097(04)00041-2. [DOI] [PubMed] [Google Scholar]
- 59.Harcombe WR, Bull JJ. 2005. Impact of phages on two-species bacterial communities. Appl Environ Microbiol 71:5254–5259. doi: 10.1128/AEM.71.9.5254-5259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ormala AM, Jalasvuori M. 2013. Phage therapy: should bacterial resistance to phages be a concern, even in the long run? Bacteriophage 3:e24219. doi: 10.4161/bact.24219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zahid MS, Udden SM, Faruque AS, Calderwood SB, Mekalanos JJ, Faruque SM. 2008. Effect of phage on the infectivity of Vibrio cholerae and emergence of genetic variants. Infect Immun 76:5266–5273. doi: 10.1128/IAI.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hall AR, De Vos D, Friman VP, Pirnay JP, Buckling A. 2012. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl Environ Microbiol 78:5646–5652. doi: 10.1128/AEM.00757-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia G, Corrigan RM, Winstel V, Goerke C, Grundling A, Peschel A. 2011. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J Bacteriol 193:4006–4009. doi: 10.1128/JB.01412-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vergara-Irigaray M, Maira-Litran T, Merino N, Pier GB, Penades JR, Lasa I. 2008. Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiology 154:865–877. doi: 10.1099/mic.0.2007/013292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 66.Gutierrez D, Delgado S, Vazquez-Sanchez D, Martinez B, Cabo ML, Rodriguez A, Herrera JJ, Garcia P. 2012. Incidence of Staphylococcus aureus and analysis of associated bacterial communities on food industry surfaces. Appl Environ Microbiol 78:8547–8554. doi: 10.1128/AEM.02045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penades JR, Lasa I. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol 48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 68.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol 183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delgado S, Arroyo R, Jimenez E, Marin ML, del Campo R, Fernandez L, Rodriguez JM. 2009. Staphylococcus epidermidis strains isolated from breast milk of women suffering infectious mastitis: potential virulence traits and resistance to antibiotics. BMC Microbiol 9:82. doi: 10.1186/1471-2180-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin V, Maldonado-Barragan A, Moles L, Rodriguez-Banos M, Campo RD, Fernandez L, Rodriguez JM, Jimenez E. 2012. Sharing of bacterial strains between breast milk and infant feces. J Hum Lact 28:36–44. doi: 10.1177/0890334411424729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.