Abstract
Shewanella loihica strain PV-4 harbors both a functional denitrification (NO3−→N2) and a respiratory ammonification (NO3−→NH4+) pathway. Batch and chemostat experiments revealed that NO2− affects pathway selection and the formation of reduced products. Strain PV-4 cells grown with NO2− as the sole electron acceptor produced exclusively NH4+. With NO3− as the electron acceptor, denitrification predominated and N2O accounted for ∼90% of reduced products in the presence of acetylene. Chemostat experiments demonstrated that the NO2−:NO3− ratio affected the distribution of reduced products, and respiratory ammonification dominated at high NO2−:NO3− ratios, whereas low NO2−:NO3− ratios favored denitrification. The NO2−:NO3− ratios affected nirK transcript abundance, a measure of denitrification activity, in the chemostat experiments, and cells grown at a NO2−:NO3− ratio of 3 had ∼37-fold fewer nirK transcripts per cell than cells grown with NO3− as the sole electron acceptor. In contrast, the transcription of nrfA, implicated in NO2−-to-NH4+ reduction, remained statistically unchanged under continuous cultivation conditions at NO2−:NO3− ratios below 3. At NO2−:NO3− ratios above 3, both nirK and nrfA transcript numbers decreased and the chemostat culture washed out, presumably due to NO2− toxicity. These findings implicate NO2− as a relevant modulator of NO3− fate in S. loihica strain PV-4, and, by extension, suggest that NO2− is a relevant determinant for N retention (i.e., ammonification) versus N loss and greenhouse gas emission (i.e., denitrification).
INTRODUCTION
Two major dissimilatory pathways determine the fate of nitrate (NO3−) in anoxic environments: denitrification and respiratory ammonification (1, 2). In denitrification, NO3− is stepwise reduced via nitrite (NO2−), nitric oxide (NO), and nitrous oxide (N2O) to dinitrogen (N2). In respiratory ammonification, NO3− is reduced via NO2− to ammonium (NH4+). Nitrite is the common intermediate of the two pathways and the branching point for both dissimilatory pathways. In the presence of NH4+, NO2− also can serve as the electron acceptor for anaerobic ammonium oxidation (anammox) (3); however, in soil environments this process appears to be less relevant than the other two processes (4). The fate of NO2− via denitrification or respiratory ammonification has great environmental impact. Denitrification forms gaseous products (N2O, N2), which are emitted from the soil, resulting in N loss, whereas respiratory ammonification generates NH4+ leading to N retention (5, 6). Atmospheric N2O is a potent greenhouse gas and an ozone-depleting agent (7, 8). Therefore, knowledge of the environmental factors that control these NO2− reduction pathways is needed to estimate, predict, and possibly manipulate N loss versus N retention.
The major sources of NO2− in the environment include NH4+ oxidation performed by nitrosifiers and NO3− reduction. In these conversions, NO2− accumulates when its production is kinetically faster than NO2− consumption (9, 10). Nitrite accumulation had been thought to occur rarely in the environment (11); however, recent observations suggested that NO2− formation can occur as a result of NO3− reduction and NH4+ oxidation under conditions that favor NO2− production over consumption (12–16). For example, high pH and abundance of NH4+ and hydroxylamine (NH2OH) affect NO2− oxidizers, causing NO2− accumulation from nitrosification (15), while oxygen intrusion and/or electron donor limitations may cause NO2− accumulation from denitrification (12). In pure-culture studies, several denitrifiers and respiratory ammonifiers were found to reduce NO3− at a higher rate than NO2−, causing dynamic changes of the NO2−:NO3− ratios in the medium (10). Additional NO2− may be generated by NO3−-to-NO2− reducers sensu stricto, which generate NO2− as an end product (12). For example, in activated sludge, the activity of NO3−-to-NO2− reducers leads to NO2− formation; however, the contribution of NO3−-to-NO2− reducers to NO2− accumulation in natural environments is uncertain (17, 18).
Even though NO2− formation occurs in diverse environments (12, 19), information regarding the impact of NO2− on dissimilatory NO3−/NO2− reduction pathways is scarce. Increased NO2− concentrations and respiratory ammonification activity have been observed in river sediments (20); however, causality was not established. A recent study reported that denitrification was the dominant pathway in a chemostat mixed culture derived from a tidal flat sediment when NO2− instead of NO3− was provided as the electron acceptor (21).
Based on genome analyses, at least three different bacterial species harbor both the denitrification and respiratory ammonification pathways: Shewanella loihica strain PV-4, Opitutus terrae strain PB90-1, and Marivirga tractuosa strain DSM 4126 (22). In Shewanella loihica strain PV-4, the functionality of both the denitrification and the respiratory ammonification pathways was confirmed (23). Recent efforts demonstrated the role of C:N ratios on pathway selection in S. loihica strain PV-4; however, the effects of NO2−:NO3− ratios were not explored. Since fluctuating NO2− concentrations are expected in many environmental systems, this work explored the effects of NO2−:NO3− ratios on pathway selection in a defined, tractable experimental system.
MATERIALS AND METHODS
Culture conditions.
Phosphate-buffered basal salts medium for the batch and chemostat experiments was prepared as described previously (24). For batch experiments, 100 ml of anoxic medium was dispensed into 160-ml serum bottles. Throughout the medium preparation procedure, anoxia was maintained by using the Hungate technique. The serum bottles were sealed with black butyl-rubber stoppers (Geo-Microbial Technologies, Inc., Ochelata, OK). The autoclaved medium was amended with vitamins (25) prepared as an anoxic, filter-sterilized, 200-fold concentrated stock solution. Lactate (60% lactate syrup; Sigma-Aldrich, St. Louis, MO) was added as the electron donor from an anoxic, autoclaved 0.5 M stock solution. A maximum of 2.0 mM lactate was required to reduce the total amount of N oxyanions added to the batch culture systems, and excess lactate (i.e., 5 mM) was added to avoid electron donor limitations (24). Various amounts of NO3− and NO2− (see below) were added to achieve a total final N oxyanion concentration of 1.0 mM. NH4+ was added to a final concentration of 0.1 mM as a source of biomass N. Sodium nitrate (≥99%; Fisher Scientific, Pittsburg, PA), sodium nitrite (≥99%; JT Baker, Phillipsburg, NJ), and ammonium chloride (≥99%; Fisher Scientific, Pittsburg, PA) were dissolved in distilled water to prepare 0.1 M NO3−, NO2−, and NH4+ stock solutions, which were degassed and autoclaved. After preparation of the culture vessels, 10% of the headspace (6 ml) was withdrawn and 6 ml of acetylene gas (99.6%; Airgas, Knoxville, TN) was added to block N2O turnover to N2 and measure N2O production as a proxy for denitrification activity (26). For the chemostat experiments, higher phosphate (25 mM) and ammonium chloride (0.5 mM) concentrations were used to increase the buffering capacity and provide reactive N for assimilation, respectively. Trace metals were added from 200-fold stock solutions to the chemostat reactor after autoclaving to avoid precipitate formation (23). All experiments were performed at room temperature (21°C).
Analytical procedures.
Analytical measurements followed established protocols (23, 24). NO3− and NO2− were measured with a Dionex ICS-2100 system (Sunnyvale, CA), and NH4+ was measured with a Dionex ICS-1100 system. For both ion chromatography systems, the limits of detection for the target analytes was ∼50 μM. N2O was quantified in headspace gas with an Agilent 3000A Micro gas chromatograph (MicroGC; Palo Alto, CA) with a limit of detection of ∼100 ppm by volume (ppmv). A dimensionless Henry's constant of 1.53 was calculated for N2O using equation 1 and parameters provided by Sander (http://www.henrys-law.org/henry-3.0.pdf).
| (1) |
where kH is the dimensionless Henry's constant, is Henry's constant under standard conditions, expressed as moles liter−1 atm−1, R is the gas constant (0.082 liters atm K−1 mol−1), T is temperature (in Kelvins), and Tθ is temperature under standard conditions (298.15 K).
This Henry's constant was adjusted for the ionic strength of the solution (27, 28). Using the corrected Henry's constant of 1.751, the aqueous-phase concentration of N2O was calculated from the headspace concentration.
Effect of NO2−:NO3− ratios on product formation.
NO2−:NO3− ratios of 0.33, 0.5, 1, and 3 were established by adding NO3− and NO2− to achieve a combined concentration of 1 mM. In addition, cultures were grown with 1 mM NO3− and 1 mM NO2− as the sole electron acceptors. The vessels were inoculated with 0.5 ml of an S. loihica strain PV-4 culture grown with 2.0 mM lactate and 1.0 mM NO3−. The initial measurements of NO2−, NO3−, NH4+, and N2O were taken immediately after inoculation, and the final measurements were taken after 5 days, when all of the initial amounts of electron acceptor(s) had been consumed. The quantitative assessment of the amounts of NH4+ and N2O determined the activity of respiratory ammonification and denitrification, respectively, under the conditions tested. Additional experiments determined that shifting NO2−:NO3− ratios, rather than changes in absolute NO2− concentration, affected denitrification and ammonification activities. S. loihica strain PV-4 batch cultures were amended with 5 mM lactate and 0.25, 0.5, 0.75, and 1.0 mM NO2−, and N2O and NH4+ amounts were determined following complete NO2− consumption.
In addition to batch cultures, chemostat experiments were performed to allow the control of the NO2−:NO3− ratios during growth. The chemostat design (460 ml total volume, 200 ml medium) and general operational parameters have been described previously (23). Anoxic medium was prepared in 2-liter glass bottles with a C:N ratio of 4.5 by adding 3.0 mM lactate and a 2.0 mM total concentration of NO3− and NO2−. This medium was supplied to the continuously stirred tank reactor at a flow rate of 20 ml h−1. NO2−:NO3− ratios of the medium were adjusted to 0 (no NO2−), 0.33, 1, and 3. To initiate the chemostat culture, lactate and NO3− were added to final concentrations of 2 mM and 1 mM, respectively, along with the vitamin and trace metal solutions. After flushing the reactor vessel with N2 gas for 1 h, S. loihica strain PV-4 cells were inoculated. The reactor was incubated for 2 days with N2 flushing before operation of the chemostat system began. The reactor headspace was flushed with a constant stream of N2 gas to prevent O2 intrusion and CO2 and N2O buildup. For all experimental conditions examined, steady-state conditions, indicated by stable dissolved carbon and nitrogen species concentrations, was reached within 72 h. In a steady-state chemostat system, the consumption of the reactants and the formation of products are quantified as rates of change. The steady-state concentration of NO2− and NO3− were maintained below the detection limit of ∼0.05 mM, and consumption rates in the reactor were calculated by multiplying the flow rate by the influent medium concentrations. The rates of NH4+ production in the reactor were calculated from the steady-state NH4+ concentrations, influent NH4+ concentrations, and the flow rate (equation 2) and served as a measure of respiratory ammonification activity.
| (2) |
where is the rate of NH4+ production (micromoles hour−1), Q is the flowrate of the chemostat (milliliters hour−1), is the steady-state concentration in the reactor (millimolar), and is the concentration of the influent medium (millimolar).
Denitrification activity was quantified by measuring N2O production rates in the headspace in the presence of acetylene. The N2 gas flow was stopped, and 26 ml (10%) of the headspace N2 was replaced with acetylene gas. N2O concentrations in the headspace then were measured four times in 40- to 50-min intervals. Since the dissolved N2O exits the system with the effluent, the loss was calculated for accurate measurement of the N2O production rates according to equation 3.
| (3) |
where LN2O is the loss of N2O with the effluent (micromoles), t1 is the time of initial N2O measurement (hours), t2 is the time of final N2O measurement (hours), and f(t) is the regression equation for the change in aqueous N2O concentration.
Using this approach, the NH4+ and N2O production rates could be directly compared. After each culture suspension sampling event, the reactor volume was adjusted to 200 ml by operating the chemostat with the outflow valve closed. Independent replicate chemostat experiments were performed for each NO2−:NO3− ratio tested. After each experiment, the reactor was disassembled, thoroughly cleaned, and sterilized.
Growth yield determination.
Dry weight measurements determined the biomass produced in defined medium amended with 5 mM lactate and 1.0 mM NO2− or 1.0 mM NO3− (22). The MicroGC was used for fast (1-min run time) headspace CO2 concentration measurements, and the cessation of CO2 formation served as a proxy for complete NO2−/NO3− consumption. Forty ml of the culture suspension was passed through a preweighed 0.22-μm membrane filter (Millipore, Billerica, MA). The filters with biomass were dried at 100°C for 24 h and cooled in a desiccator until weight consistency was achieved. For each growth condition, triplicate culture vessels were examined. The dry weight data were corrected for the weight of salts associated with the culture medium and introduced onto the membrane filter.
Nucleic acid extraction and analyses.
For cell quantification and gene expression analyses, 15-ml samples were collected from the reactor under steady-state conditions with different NO2−:NO3− ratios. The samples for expression analyses were prepared by immediately mixing 0.5-ml aliquots with 1.0 ml of RNA Protect bacterial reagent (Qiagen, Germantown, MD) and collecting the biomass by centrifugation for 10 min at 5,000 × g. The pellets were stored immediately at −80°C. The samples for cell enumeration by quantitative PCR (qPCR) were prepared by centrifuging 1.5-ml aliquots at 16,000 × g, and the pellets were immediately stored at −80°C. The extraction and purification of total RNA and reverse transcriptase quantitative PCR (RT-qPCR) followed an established protocol (23). Immediately prior to extraction, 1 μl luciferase control mRNA (Promega, Madison, WI) diluted to 1010 copies μl−1 was added to each frozen cell pellet. The recovery of the control mRNA was used to account for RNA loss during the extraction and reverse transcription procedures (29, 30). The cell pellets then were disrupted with an Omni Bead Ruptor 24 homogenizer (Omni, Kennesaw, GA) in 350 μl buffer RLT provided with the RNeasy Mini kit (Qiagen, Germantown, MD), and total RNA was extracted from the cell pellets according to the protocol provided with the kit. Extracted mRNA was purified using an RNA MinElute kit (Qiagen) after DNA was digested with the RNase-free DNase set kit (Qiagen). An 11-μl aliquot of purified RNA solution was subjected to reverse transcription using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations, and the remaining 7 μl of RNA extract was tested for DNA contamination using qPCR (23).
Genomic DNA for cell quantification was extracted using a DNeasy blood and tissue kit (Qiagen, Santa Clarita, CA) as described previously (24). PCR primers used in this study are summarized in Table 1. S. loihica strain PV-4 cells were enumerated with qPCR targeting 16S rRNA genes in the genomic DNA, and gene expression analyses were performed with qPCR targeting nirK, nrfA, and luc cDNA templates. In silico PCR analysis (31) suggested that the Slo16Sf and Slo16Sr primer set amplified all nine 16S rRNA copies of S. loihica strain PV-4. Strain PV-4 possesses two different nrfA gene copies, and previous findings indicated that nrfA844 responds to N oxyanions (23). Transcripts of nrfA0505 were detected, but they were found to be of at least two orders of magnitude lower abundance than those for nrfA0844 under all growth conditions tested. qPCR was performed with an ABI ViiA7 real-time PCR system (Life Technologies) equipped with ViiA7 software v1.1 using Power SYBR green detection chemistry (Life Technologies, Carlsbad, CA) (23, 24). Plasmids carrying the strain PV-4 16S rRNA gene nirK and nrfA inserts were used to construct qPCR calibration curves. These plasmids were prepared by inserting PCR amplicons of the respective target genes into the PCR2.1 vector using the TOPO TA cloning kit (Invitrogen). Tenfold dilution series with concentrations ranging from 108 to 101 copies/μl were prepared and used as qPCR templates. The amplification efficiencies ranged from 94.5% to 103.2%. The standard deviations in the quantification cycle (Cq) values increased up to 5-fold (up to 0.71) between the template DNA dilutions of 10 and 100 copies μl−1, indicating a detection limit of about 10 copies μl−1 for all three target genes. No amplification was observed in controls without target DNA/cDNA, and the amplicons of each target exhibited consistent and expected melting curve profiles, indicating specific amplification. Both cDNA and genomic DNA samples were prepared from triplicate samples, and triplicate qPCR assays were performed for each DNA and cDNA sample. The RT-qPCR data were adjusted based on the luc control mRNA recovery, which ranged from 7.0% to 23.8% and was within the expected range (30). The corrected expression data were normalized to the cell abundance.
TABLE 1.
Primers used for RT-qPCR analyses and qPCR calibration curve parameters
| Primer | Sequence | Target gene (locus tag) | Amplicon length (bp) | Slope | y intercept | Amplification efficiency (%) | R2 | Reference |
|---|---|---|---|---|---|---|---|---|
| Slo_nirK853f | AAGGTGGGTGAGTCTGTGCT | nirK (Shew_3335) | 188 | −3.393 | 35.64 | 97.1 | 0.999 | 23 |
| Slo_nirK1040r | GGCTGGCGGAAGGTGTAT | |||||||
| Slo_nrfA1083f | GGATATCCGTCACGCTCAAT | nrfA (Shew_0844) | 226 | −3.299 | 34.93 | 101.0 | 0.992 | 24 |
| Slo_nrfA1308r | GTCCATACCCAATGCAGCTT | |||||||
| Slo_16Sf | CACACTGGGACTGAGACACG | 16S rRNA genes | 191 | −3.461 | 34.27 | 94.5 | 1.000 | 23 |
| Slo_16Sr | TGCTTCTTCTGCGAGTAACG | |||||||
| luc_refA | TACAACACCCCAACATCTTCGA | Luciferase gene control mRNA | 67 | −3.247 | 35.46 | 103.2 | 0.999 | 29 |
| luc_refB | GGAAGTTCACCGGCGTCAT |
Statistical analyses.
Statistical analyses for both phenotypic and expression analysis data were performed with SPSS 22.0 software for Mac (IBM Corp., Armonk, NY, USA). Unless otherwise mentioned, triplicate data sets were collected for each of the batch and chemostat experiments, and the Student's t tests were performed to test the null hypotheses. The RT-qPCR data were transformed to logarithmic scales for statistical analyses. P values below 0.05 were regarded as significant.
RESULTS
Batch cultivation with NO3− or NO2−.
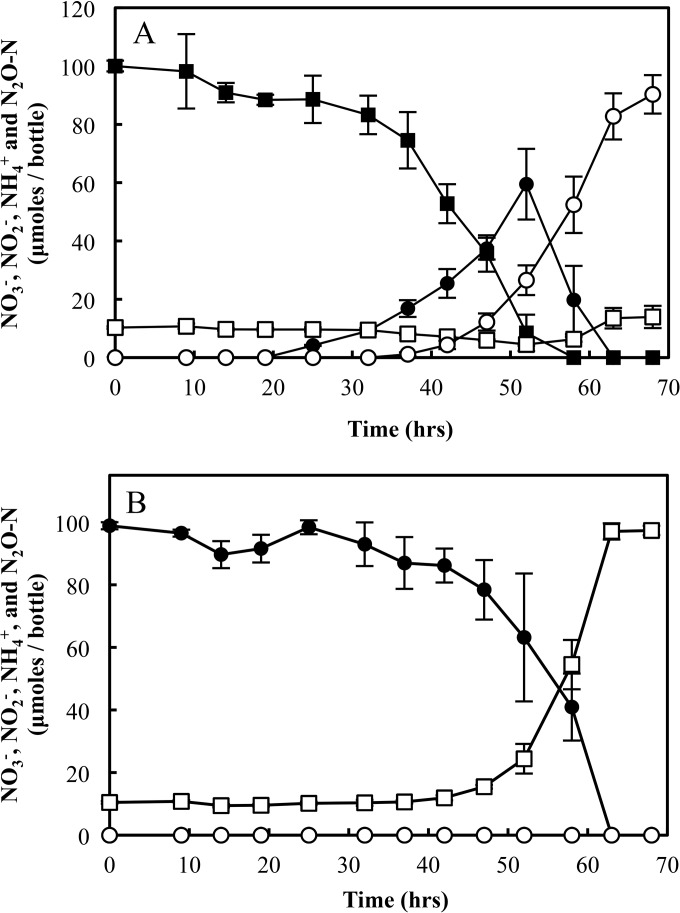
S. loihica strain PV-4 batch cultures reduced NO3− or NO2− to different products, depending on which substrate was provided (Fig. 1). An initial amount of 100.0 ± 1.9 μmol of NO3− was reduced to 90.3 ± 6.6 μmol of N2O-N after a 68-h incubation period, indicating that ∼90% of the NO3− was denitrified. Some NH4+ also was produced, and the initial amount of 10.3 ± 0.4 μmol provided in the medium increased to 14.0 ± 3.5 μmol. Cultures that received NO2− but no NO3− produced exclusively NH4+, and no N2O was observed (i.e., N2O below the detection limit of ∼100 ppmv, or 4.15 μmol liter−1 dissolved-phase concentration at equilibrium). Of the 98.9 ± 1.1 μmol of NO2− added to the cultures, 86.9 ± 1.5 μmol (87.8% ± 1.8%) was recovered as NH4+. The lack of complete recovery probably was due to NH4+-N assimilation into biomass. These batch culture studies suggested a role of NO2−:NO3− ratios for controlling NO2− fate via the respiratory ammonification or denitrification pathways.
FIG 1.
NO3−/NO2− reduction in batch cultures of S. loihica strain PV-4. All vessels received 10% acetylene (C2H2) gas in the headspace for blocking N2O reduction to N2. Cultures were amended with 5.0 mM lactate as the electron donor and 1.0 mM NO3− (A) or 1.0 mM NO2− (B) as the electron acceptor. The change in the amounts of NO3− (■), NO2− (●), NH4+ (□), and N2O-N (○) in the vessels was monitored until all NO3− and NO2− in the medium was reduced to NH4+ and N2O. The error bars represent the standard deviations from triplicate cultures.
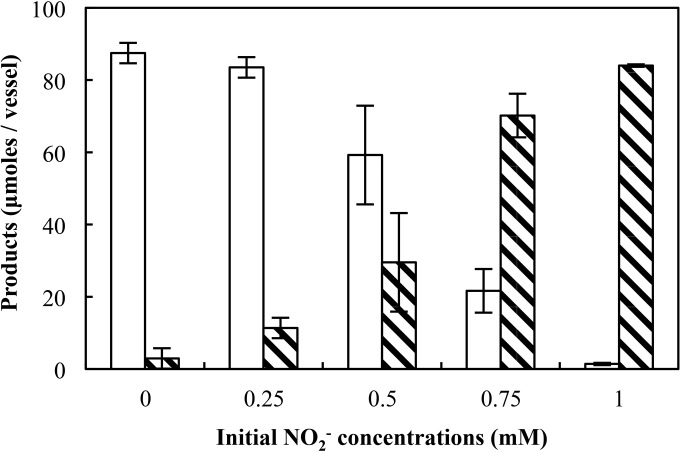
S. loihica strain PV-4 cultures amended with different NO2− concentrations (0.25, 0.5, 0.75, and 1 mM) produced predominantly NH4+, independent of the initial amount of NO2− provided (Table 2). With NO2− as the sole electron acceptor, no more than 5% of the total NO2− provided was reduced to N2O. This finding suggests that the absolute NO2− concentration (in the absence of NO3−) did not affect pathway selection in batch cultures.
TABLE 2.
Reduced product distribution in S. loihica batch cultures amended with various NO2− concentrations
| Initial NO2− concn (mM) | NH4+ produced (μmol/vessel)a | % recovered as NH4+ | N2O-N produced (μmol/vessel)a | % recovered as N2O-N |
|---|---|---|---|---|
| 0.25 | 22.9 (3.3) | 91.6 | 1.0 (0.1) | 4.0 |
| 0.5 | 41.7 (1.4) | 83.4 | 2.3 (0.1) | 4.6 |
| 0.75 | 64.8 (0.5) | 86.4 | 2.6 (0.6) | 3.5 |
| 1.0 | 86.1 (1.7) | 86.1 | 1.5 (0.1) | 1.5 |
Numbers in parentheses are standard deviations from triplicate samples.
Under batch cultivation conditions with 99.6 ± 0.8 NO3− as the electron acceptor, a biomass yield of 14.9 ± 1.5 μg (dry weight) per μmol NO3− reduced was determined (Table 3). When amended with 99.3 ± 0.3 μmol NO2−, the biomass yield was 16.2 ± 2.2 μg (dry weight) per μmol NO2− reduced. Therefore, S. loihica strain PV-4 had statistically similar (P > 0.05) growth yields from denitrification of NO3− to N2O and respiratory ammonification of NO2− to NH4+ per amount of electron acceptor consumed.
TABLE 3.
Biomass yield of S. loihica strain PV-4 upon growth on 5.0 ml lactate and 1.0 mM NO3− or 1.0 mM NO2−
| Electron acceptora |
Producta |
ΔG0′ (kJ/mol of e− accepter) | fsc | ||||
|---|---|---|---|---|---|---|---|
| Type | Amt (μmol/vessel) | N2O-N (μmol/vessel) | NH4+ (μmol/vessel) | Biomass (μmol C5H7O2N/vessel) | Biomass yieldb (mg/μmol of e− acceptor) | ||
| NO3− | 99.6 (0.8) | 87.5 (1.5) | 2.8 (1.7) | 13.1 (1.4) | 14.9 (1.5) | −342.3 | 0.458 |
| NO2− | 99.3 (0.3) | 1.4 (0.4) | 84.0 (3.8) | 14.5 (1.9) | 16.2 (2.2) | −397.3 | 0.407 |
Numbers in parentheses are standard deviations from triplicate samples.
Biomass yield was determined with the assumption that the amounts of the minor products were negligible compared to those of the major products.
fs values were calculated with the assumption that NO3− and NH4+ contributed equally to biomass (24 e− eq/mol C5H7O2N on average) (24). The following equation was used to calculate fs: fs = (e− eq to biomass)/[(e− eq to biomass) + (e− eq to e− acceptors)].
Effects of NO2−:NO3− ratios on pathway selection.
Varying the initial inputs of NO2− and NO3− in batch cultures affected the relative abundance of reduced products. The net production of NH4+ increased and the production of N2O decreased with increasing NO2−:NO3− ratios (Fig. 2). At NO2−:NO3− ratios below 1, N2O was the major product, suggesting that the denitrification pathway was favored. At a NO2−:NO3− ratio of 0.33, 83.5 ± 0.1 μmol of N2O-N and 11.4 ± 2.8 μmol of NH4+ were recovered from 94.6 ± 0.7 μmol of N oxyanions. At a NO2−:NO3− ratio of 1 (0.5 mM NO2− and 0.5 mM NO3−), 94.2 ± 0.1 μmol of N oxyanions was reduced to 59.2 ± 12.8 μmol N2O-N and 29.5 ± 13.7 μmol NH4+ (Fig. 2). At a NO2−:NO3− ratio of 3, significantly more NH4+ (70.2 ± 6.0 μmol) than N2O (21.6 ± 0.7) was produced from reduction of NO2− and NO3− (P < 0.05), indicating that higher NO2−:NO3− ratios shifted NO3−/NO2− reduction toward respiratory ammonification. These batch experiments demonstrated that the relative proportion of NO2− to NO3− affected pathway selection and reduced product formation; however, the dynamic change in the relative proportions of NO3− and NO2− (i.e., changing NO2−:NO3− ratios) (Fig. 1A) made the interpretation of the results ambiguous. For example, NO3− consumption rates exceeded NO2− utilization, causing an increase in the NO2−:NO3− ratio over time in the NO3−-fed batch cultures (Fig. 1A).
FIG 2.
Net production of N2O (white bars) and NH4+ (hatched bars) over a 5-day incubation period at 21°C in S. loihica strain PV-4 cultures amended with 5.0 mM lactate and different NO2−:NO3− ratios to achieve 1.0 mM total N-oxyanion concentration. Each bar represents the averages from triplicate samples, with the error bars representing their standard deviations.
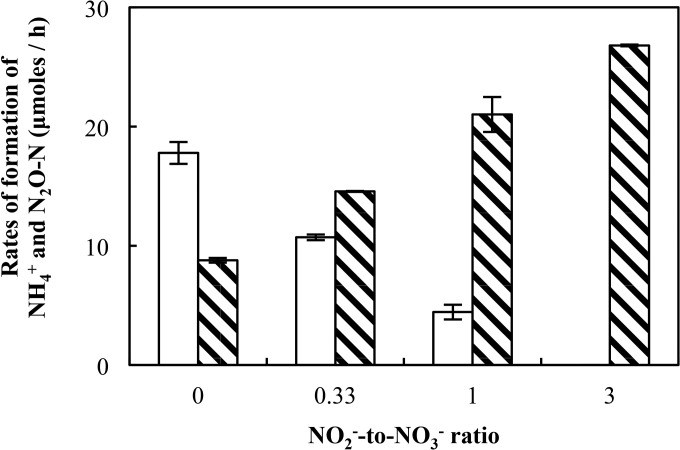
To better evaluate NO2− effects on pathway selection, chemostat experiments were performed that allowed cultivation under constant NO2−:NO3− ratios. Since the C:N ratio dominates the control of the N pathway products, chemostat experiments were conducted at a C:N ratio of 4.5 (i.e., 3 mM lactate and 2 mM NO3− plus NO2− in the feed medium), which was shown not to favor one pathway or the other (23). These C:N conditions in the reactor system revealed the effects of various NO2−:NO3− ratios on pathway selection (Fig. 3). When the electron acceptor pool consisted entirely of NO3−, N2O and NH4+ were produced at rates of 8.9 ± 0.5 μmol h−1 (17.8 ± 1.0 μmol h−1 N2O-N) and 8.8 ± 0.2 μmol h−1, respectively, indicating that approximately twice as much NO3− was reduced to N2O than to NH4+. Higher NO2−:NO3− ratios increased the net NH4+ production but decreased the N2O production, leading to the formation of significantly larger (P < 0.05) amounts of NH4+ and significantly smaller (P < 0.05) amounts of N2O. At a NO2−:NO3− ratio of 0.33 (feed rates of 29.3 ± 0.1 μmol NO3− h−1 and 9.1 ± 0.1 μmol NO2− h−1), the NH4+ production rate exceeded the N2O production rate on a per-N-atom basis by 1.36-fold ± 0.03-fold. At a NO2−:NO3− ratio of 3, NH4+ was produced at a rate of 26.8 ± 0.1 μmol h−1 and N2O formation was not observed. When 2.0 mM NO2− was provided as the sole electron acceptor in the influent medium, biomass washout occurred (i.e., the reactor failed), suggesting toxicity when NO2− was continuously supplied at a rate of 40 μM h−1. The chemostat results were consistent with the outcome of the batch culture experiments and indicated that increasing NO2−:NO3− ratios favor respiratory ammonification.
FIG 3.
Net production of N2O (white bars) and NH4+ (hatched bars) in chemostat cultures of S. loihica strain PV-4 with NO2−:NO3− ratios varying from 0 to 3 in the feed solution. The feed solution contained 5 mM lactate and combinations of NO2− and NO3− with a total N oxyanion concentration of 2.0 mM. The N oxide(s) were supplied to the reactor at ∼40 μmol h−1. Each bar represents the averages from two samples taken from separate reactors, with error bars representing the standard deviations.
nrfA and nirK expression analysis.
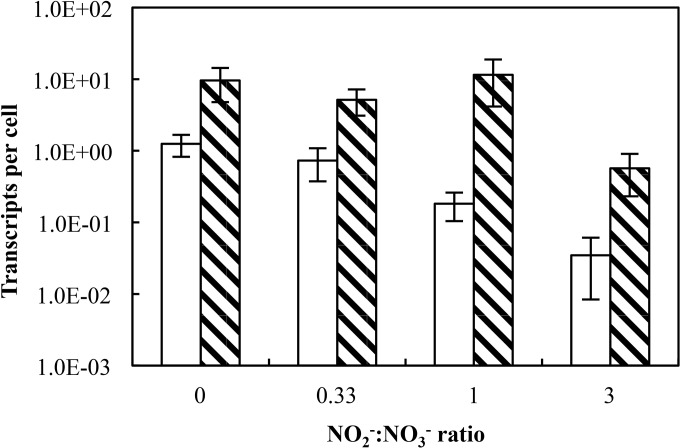
The RT-qPCR analyses of nrfA and nirK transcripts supported the phenotypic data collected from the batch and chemostat experiments (Fig. 4). The cell numbers were statistically indistinguishable at all NO2−:NO3− ratios tested (P > 0.05 by one-way analysis of variance [ANOVA]). The number of nirK transcripts was highest (1.3 ± 0.4 transcripts cell−1) when 2.0 mM NO3− was the sole electron acceptor in the feed medium and lowest (3.5 × 10−2 ± 2.6 × 10−2 transcripts cell−1) at a NO2−:NO3− ratio of 3. The numbers of nirK transcripts detected per cell were not significantly different (P > 0.05) in cells maintained in chemostats receiving medium with a NO2−:NO3− ratio of 0.33 or with NO3− as the sole electron acceptor; however, nirK expression at a NO2−:NO3− ratio of 1 (1.8 × 10−1 transcripts cell−1) was significantly lower than expression at lower NO2−:NO3− ratios. Different expression patterns were observed with nrfA, and transcription levels with NO3− as the sole electron acceptor (9.6 transcripts cell−1) were statistically indistinguishable from the transcription levels at NO2−:NO3− ratios of 0.33 and 1 (P > 0.05). The transcription of nrfA was downregulated at a NO2−:NO3− ratio of 3, with 0.6 ± 0.4 transcripts cell−1 compared to 11.5 ± 7.3 transcripts cell−1 at a NO2−:NO3− ratio of 1. This significant decrease in nrfA expression may be attributed to the repression of metabolic activity due to NO2− toxicity. As the expression of nrfA remained relatively constant, the decrease in expression of nirK at high NO2−:NO3− ratios provides an explanation for the observed decrease in denitrification activity in strain PV-4.
FIG 4.
RT-qPCR analysis of nirK (white bars) and nrfA (hatched bars) transcripts in S. loihica strain PV-4 chemostat cultures under various NO2−:NO3− ratios. The feed solution contained 5 mM lactate and combinations of NO2− and NO3− with a total N oxyanion concentration of 2.0 mM. Each bar represents the average copies of transcript measured in triplicate samples subjected to independent extraction procedures. The error bars represent the standard deviations from triplicate independent samples.
DISCUSSION
Both the phenotypic and the transcriptional analyses indicated that the concentration of NO2− relative to that of NO3− (i.e., the NO2−:NO3− ratio) is a controlling factor influencing the fate of these N oxyanions. Both the batch and chemostat experiments performed with various NO2−:NO3− ratios demonstrated that as the proportion of NO2− increases, respiratory ammonification activity supersedes denitrification activity. Several pure-culture studies report the effects of NO3− or NO2− on pathway selection and the fate of NO3−/NO2− (32–35). For example, decreased nrfA expression and subsequent accumulation of NO2− were observed in Escherichia coli chemostat cultures, with steady-state NO3− concentrations exceeding 0.2 μM (34). The experiments with S. loihica strain PV-4 demonstrated that the initial NO2− concentration did not affect pathway selection (Table 2) and suggested that the NO2−:NO3− ratio is a critical parameter determining pathway selection. Low NO3− concentrations (0.2 mM) were shown to induce denitrification pathway enzymes, including NirK, in Agrobacterium tumefaciens (35); however, S. loihica strain PV-4 batch cultures amended with 0.25 mM NO3− and 0.75 mM NO2− (i.e., a NO2−:NO3− ratio of 3) exhibited predominantly ammonification (Fig. 2). This observation also supports that the NO2−:NO3− ratio, rather than the absolute N oxyanion concentration, is the relevant control parameter. Further, the effects of NO2−:NO3− ratios were observed in strain PV-4 chemostat cultures with very low (below the limits of detection) steady-state NO3− and NO2− concentrations, emphasizing the relevance of the NO2−:NO3− ratios for pathway selection. Increased input of NO3− to chemostat cultures did not affect nrfA expression, suggesting NO3− does not inhibit nrfA gene activity, as was observed in E. coli (34). Therefore, a plausible explanation for the observations is that the NO2−:NO3− ratio, rather than the absolute concentration of NO2− or NO3−, determines pathway selection in S. loihica strain PV-4.
The reasons why S. loihica strain PV-4 prefers respiratory ammonification under high NO2−:NO3− ratios may be based on thermodynamics. S. loihica strain PV-4 produced similar amounts of biomass per micromole of NO3− denitrified to N2O and per micromole of NO2− reduced to NH4+ in batch cultures. Less biomass would be produced if S. loihica strain PV-4 denitrified NO2−, as denitrification from NO2− to N2O would involve transfer of only two electrons per N atom compared to a four-electron transfer from NO3− to N2O. NO3− reduction to NO2− is the most efficient step in terms of energy conservation in the denitrification pathway (36). Energetic considerations (37) also suggest that respiratory ammonification is more favorable under high NO2−:NO3− ratios. With lactate as the electron donor (oxidized to acetate), the ΔG0′ of NO3−-to-N2O reduction is −398.5 kJ/mol NO3−, and less free-energy change is associated with NO2−-to-N2O reduction (−231.1 kJ/mol NO2−). Respiratory ammonification of NO2− with a ΔG0′ of −450.1 kJ/mol NO2− is energetically more favorable, providing a rationale for the preference of respiratory ammonification at high NO2−:NO3− ratios.
The preference of denitrification over respiratory ammonification at low NO2−:NO3− ratios, on the other hand, cannot be explained in terms of energetics and associated growth yields, as respiratory ammonification of NO3− would yield more biomass and energy than NO3− denitrification. Possible explanations include more efficient electron transfer in denitrification than in respiratory ammonification or the kinetic properties of the enzymes involved. For example, NO2− toxicity may be a reason to favor a pathway that minimizes intermediate NO2− accumulation (10). Depending on the electron donor provided, 20 to 60% of the initial NO3− was observed as NO2− during denitrification in strain PV-4 cultures (24). Previous experiments with S. oneidensis strain MR-1 demonstrated that NO2− reduction to NH4+ proceeded only after NO3− had been stoichiometrically converted to NO2− (10). In S. loihica strain PV-4, the NO-forming NO2− reductase NirK may have faster kinetics than the NH4+-forming NO2− reductase NrfA at physiologically relevant NO2− concentrations; thus, it would prevent NO2− toxicity. When NO2− exceeds NO3− concentrations in the medium, this advantage is lost and the NO3−/NO2− reduction pathway is determined by energetics. Unfortunately, these S. loihica enzyme systems have not been characterized and kinetic data are not available to corroborate this hypothesis.
Somewhat unexpectedly, the expression levels of both nrfA and nirK decreased when the NO2−:NO3− ratio was raised to 3. Higher NO2−:NO3− ratios resulted in reactor failure, although NO2− concentrations in the reactor vessel never exceeded 2.0 mM. These observations suggest that the continuous supply of NO2− exerted stress on S. loihica strain PV-4 cells. The inhibitory effect of elevated NO2− concentrations on strain PV-4 may be the reason why the organism initially was identified as a nondenitrifier (38). NO2− toxicity was observed previously in Shewanella oneidensis strain MR-1, and no growth occurred with 4 mM NO2− provided as the electron acceptor (10). NO2− was found to be toxic to other microbial groups, including methanogens and phosphate-accumulating bacteria, at concentrations below 1 mM (39, 40), and 3 mM NO2− inhibited respiratory ammonification in Wolinella succinogenes (36). Although NO2− did not result in cell death in Agrobacterium tumefaciens, NO2− impacted O2 and NO2− metabolism (35). These findings indicate that microorganisms vary in their susceptibility to NO2− toxicity. Therefore, a continuous supply of NO2− without NO3− may select for organisms tolerant to elevated NO2− concentrations, and this selection process may bias the outcome of the competition between denitrifiers and respiratory ammonifiers. Kraft et al. (21) observed increased denitrification activity in a tidal flat mixed community when NO2− was replaced with NO3− as the electron acceptor. In the chemostat seeded with the tidal flat mixed community, phylogenetically diverse populations contributed to denitrification and respiratory ammonification (21). These populations may vary in their susceptibility to NO2− toxicity, and continuous NO2− feeding without NO3− may have selected for organisms with greater NO2− tolerance. A mixture of organic compounds and/or sulfide were provided as electron donors to the tidal flat sediment chemostat (21), suggesting that a diversity of organisms was supported. In pure-culture studies of denitrifiers and respiratory ammonifiers, growth yields varied greatly depending on the bacterial strain examined and the electron donors used (36). It is possible that the dominant denitrifying population in the mixed-culture chemostat community utilized NO2− more efficiently than NO3−, as observed in pure-culture studies with P. stutzeri (36), and has an advantage over ammonifying strains when grown with NO2−.
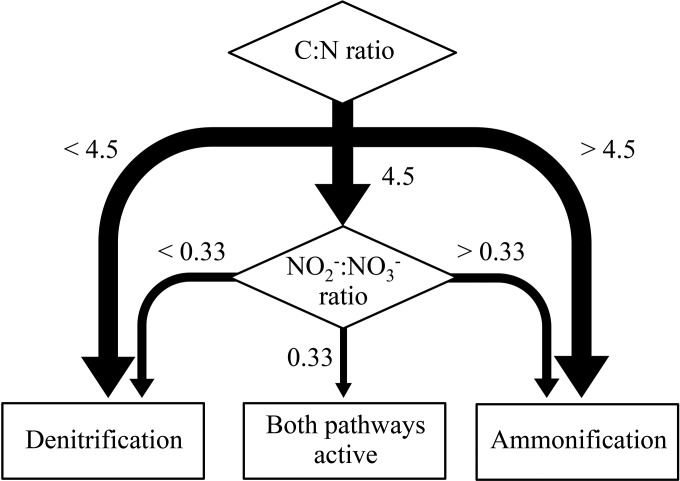
Prior chemostat experiments with strain PV-4 and NO3− as the sole electron acceptor demonstrated that the simultaneous production of NH4+ and N2O occurred only when the C:N ratio of the influent medium was 4.5 (23). The effects of various NO2−:NO3− ratios on denitrification versus respiratory ammonification pathway selection were apparent only in chemostats operated at a C:N ratio of 4.5. Apparently, the C:N ratio control dominates over the NO2−:NO3− ratio for pathway selection (Fig. 5). Respiratory ammonification dominates at C:N ratios above 4.5, and denitrification dominates at C:N ratios below 4.5 regardless of the NO2−:NO3− ratio the culture is experiencing. These observations suggest that the role of NO2−:NO3− ratios in determining the fate of NO3−/NO2− is secondary to the role of the C:N ratio. Nevertheless, NO2− may play a relevant role as a modulator of denitrification versus respiratory ammonification activities under conditions where neither electron donors nor N oxyanions are available in excess. As the results from the batch experiments suggested, NO2−:NO3− ratios also become important when organic electron donor and N oxyanions are provided in nonlimiting amounts, regardless of the C:N ratio. Such conditions may occur following N fertilizer application events in agricultural soils. NO3− reduction may exceed NO2− consumption and subsequently increase the NO2−: NO3− ratio, leading to a condition favoring respiratory ammonification.
FIG 5.
Hierarchical effects of C:N and NO2−:NO3− ratios on the fate of NO2−/NO3− via denitrification and/or respiratory ammonification in Shewanella loihica strain PV-4 chemostat cultures.
Certain environmental conditions may favor high rates of aerobic NH4+ oxidation and lower rates of NO2− oxidation to NO3−, which can lead to elevated NO2− concentrations. For example, elevated NO2− concentrations have been observed in an alkaline fluvo-aquic loam soil for >200 h after application of an ammonium sulfate solution (21). The transient formation of up to 67.2 mg NO2− kg soil−1 was observed after application of 100 mg urea kg−1 (14). In semiarid pine forest soils, first winter rains caused transient formation of NO2−, and the NO2− concentrations greatly exceeded those of NO3− (13, 14, 41). Our results suggest that the fluctuating NO2−:NO3− ratios in such environments can influence the activities of microbial community members involved in NO3− and NO2− reduction. Further investigations of NO3− and NO2− reduction in environmental systems experiencing fluctuating NO2−:NO3− ratios is warranted to more comprehensively understand the parameters controlling the fate of N oxyanions in anoxic environments.
In summary, the growth experiments with various NO2−/NO3− supplies to S. loihica strain PV-4 revealed that NO2− affects denitrification and respiratory ammonification pathway selection. In both batch and chemostat cultures, the increase in NO2− content in the electron acceptor pool favored respiratory ammonification over denitrification, and transcriptional analyses of nirK and nrfA genes supported the phenotypic observations. However, the chemostat experiments demonstrated that respiratory ammonification dominated even at low NO2−:NO3− ratios when sufficient electron donor was available, indicating that the C:N ratio has higher-level control over NO2−/NO3− fate. Pure-culture studies have limitations to predict the behavior of natural microbial assemblages. Nevertheless, the results obtained with S. loihica strain PV-4 add to our understanding of environmental parameters governing the fate of NO3− and NO2− in anoxic ecosystems. The pure-culture experiments allowed the observation of the isolated effect of various NO2−:NO3− ratios on pathway selection (i.e., denitrification and respiratory ammonification) in the absence of competition.
ACKNOWLEDGMENT
This research was supported by the U.S. Department of Energy, Office of Biological and Environmental Research, Genomic Science Program, award DE-SC0006662.
REFERENCES
- 1.Burgin AJ, Hamilton SK. 2007. Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front Ecol Environ 5:89–96. doi: 10.1890/1540-9295(2007)5[89:HWOTRO]2.0.CO;2. [DOI] [Google Scholar]
- 2.Tiedje JM, Sexstone AJ, Myrold DD, Robinson JA. 1982. Denitrification: ecological niches, competition and survival. Antonie Van Leeuwenhoek 48:569–583. [DOI] [PubMed] [Google Scholar]
- 3.Francis CA, Beman JM, Kuypers MMM. 2007. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J 1:19–27. doi: 10.1038/ismej.2007.8. [DOI] [PubMed] [Google Scholar]
- 4.Long A, Heitman J, Tobias C, Philips R, Song B. 2013. Co-occurring anammox, denitrification, and codenitrification in agricultural soils. Appl Environ Microbiol 79:168–176. doi: 10.1128/AEM.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laima MJC, Girard MF, Vouve F, Blanchard GF, Gouleau D, Galois R, Richard P. 1999. Distribution of adsorbed ammonium pools in two intertidal sedimentary structures. Mar Ecol Prog Ser 182:29–35. doi: 10.3354/meps182029. [DOI] [Google Scholar]
- 6.Fitzhugh RD, Lovett GM, Venterea RT. 2003. Biotic and abiotic immobilization of ammonium, nitrite, and nitrate in soils developed under different tree species in the Catskill Mountains, New York, USA. Glob Change Biol 9:1591–1601. doi: 10.1046/j.1365-2486.2003.00694.x. [DOI] [Google Scholar]
- 7.Ravishankara AR, Daniel JS, Portmann RW. 2009. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- 8.Lashof DA, Ahuja DR. 1990. Relative contributions of greenhouse gas emissions to global warming. Nature 344:529–531. doi: 10.1038/344529a0. [DOI] [Google Scholar]
- 9.Betlach MR, Tiedje JM. 1981. Kinetic explanation for accumulation of nitrite, nitric oxide, and nitrous oxide during bacterial denitrification. Appl Environ Microbiol 42:1074–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz-Garcia C, Murray AE, Klappenbach JA, Stewart V, Tiedje JM. 2007. Respiratory nitrate ammonification by Shewanella oneidensis MR-1. J Bacteriol 189:656–662. doi: 10.1128/JB.01194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paul EA, Clarke FE. 1996. Soil microbiology and biochemistry, 2nd ed Academic Press, San Diego, CA. [Google Scholar]
- 12.Philips S, Laanbroek H, Verstraete W. 2002. Origin, causes and effects of increased nitrite concentrations in aquatic environments. Rev Environ Sci Biotechnol 1:115–141. doi: 10.1023/A:1020892826575. [DOI] [Google Scholar]
- 13.Gelfand I, Yakir D. 2008. Influence of nitrite accumulation in association with seasonal patterns and mineralization of soil nitrogen in a semi-arid pine forest. Soil Biol Biochem 40:415–424. doi: 10.1016/j.soilbio.2007.09.005. [DOI] [Google Scholar]
- 14.Shen QR, Ran W, Cao ZH. 2003. Mechanisms of nitrite accumulation occurring in soil nitrification. Chemosphere 50:747–753. doi: 10.1016/S0045-6535(02)00215-1. [DOI] [PubMed] [Google Scholar]
- 15.Smith RV, Burns LC, Doyle RM, Lennox SD, Kelso BHL, Foy RH, Stevens RJ. 1997. Free ammonia inhibition of nitrification in river sediments leading to nitrite accumulation. J Environ Qual 26:1049–1055. doi: 10.2134/jeq1997.00472425002600040016x. [DOI] [Google Scholar]
- 16.Stevens RJ, Laughlin RJ, Malone JP. 1998. Soil pH affects the processes reducing nitrate to nitrous oxide and di-nitrogen. Soil Biol Biochem 30:1119–1126. doi: 10.1016/S0038-0717(97)00227-7. [DOI] [Google Scholar]
- 17.Wilderer PA, Jones WL, Dau U. 1987. Competition in denitrification systems affecting reduction rate and accumulation of nitrite. Water Res 21:239–245. doi: 10.1016/0043-1354(87)90056-X. [DOI] [Google Scholar]
- 18.Glass C, Silverstein J. 1998. Denitrification kinetics of high nitrate concentration water: pH effect on inhibition and nitrite accumulation. Water Res 32:831–839. doi: 10.1016/S0043-1354(97)00260-1. [DOI] [Google Scholar]
- 19.Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, Daims H. 2006. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol 8:1487–1495. doi: 10.1111/j.1462-2920.2006.01033.x. [DOI] [PubMed] [Google Scholar]
- 20.Kelso B, Smith RV, Laughlin RJ, Lennox SD. 1997. Dissimilatory nitrate reduction in anaerobic sediments leading to river nitrite accumulation. Appl Environ Microbiol 63:4679–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraft B, Tegetmeyer HE, Sharma R, Klotz MG, Ferdelman TG, Hettich RL, Geelhoed JS, Strous M. 2014. The environmental controls that govern the end product of bacterial nitrate respiration. Science 345:676–679. doi: 10.1126/science.1254070. [DOI] [PubMed] [Google Scholar]
- 22.Sanford RA, Wagner DD, Wu Q, Chee-Sanford JC, Thomas SH, Cruz-Garcia C, Rodríguez G, Massol-Deyá A, Krishnani KK, Ritalahti KM, Nissen S, Konstantinidis KT, Löffler FE. 2012. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc Natl Acad Sci U S A 109:19709–19714. doi: 10.1073/pnas.1211238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon S, Cruz-Garcia C, Sanford R, Ritalahti KM, Löffler FE. 31 October 2014. Denitrification versus respiratory ammonification: environmental controls of two competing dissimilatory NO3−/NO2− reduction pathways in Shewanella loihica strain PV-4. ISME J doi: 10.1038/ismej.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon S, Sanford RA, Löffler FE. 2013. Shewanella spp. use acetate as an electron donor for denitrification but not ferric iron or fumarate reduction. Appl Environ Microbiol 79:2818–2822. doi: 10.1128/AEM.03872-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolin EA, Wolfe RS, Wolin MJ. 1964. Viologen dye inhibition of methane formation by Methanobacillus omelianskii. J Bacteriol 87:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshinari T, Hynes R, Knowles R. 1977. Acetylene inhibition of nitrous oxide reduction and measurement of denitrification and nitrogen fixation in soil. Soil Biol Biochem 9:177–183. doi: 10.1016/0038-0717(77)90072-4. [DOI] [Google Scholar]
- 27.Schumpe A. 1993. The estimation of gas solubilities in salt solutions. Chem Eng Sci 48:153–158. doi: 10.1016/0009-2509(93)80291-W. [DOI] [Google Scholar]
- 28.Schumpe A, Quicker G, Deckwer W-D. 1982. Gas solubilities in microbial culture media. Adv Biochem Eng Biotechnol 24:1–38. [Google Scholar]
- 29.Johnson DR, Lee PKH, Holmes VF, Alvarez-Cohen L. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl Environ Microbiol 71:3866–3871. doi: 10.1128/AEM.71.7.3866-3871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amos BK, Ritalahti KM, Cruz-Garcia C, Padilla-Crespo E, Löffler FE. 2008. Oxygen effect on Dehalococcoides viability and biomarker quantification. Environ Sci Technol 42:5718–5726. doi: 10.1021/es703227g. [DOI] [PubMed] [Google Scholar]
- 31.Bikandi J, Millan RS, Rementeria A, Garaizar J. 2004. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR and endonuclease restriction. Bioinformatics 20:798–799. doi: 10.1093/bioinformatics/btg491. [DOI] [PubMed] [Google Scholar]
- 32.Ferguson S. 1994. Denitrification and its control. Antonie Van Leeuwenhoek 66:89–110. doi: 10.1007/BF00871634. [DOI] [PubMed] [Google Scholar]
- 33.Overton TW, Griffiths L, Patel MD, Hobman JL, Penn CW, Cole JA, Constantinidou C. 2006. Microarray analysis of gene regulation by oxygen, nitrate, nitrite, FNR, NarL and NarP during anaerobic growth of Escherichia coli: new insights into microbial physiology. Biochem Soc Trans 34:104–107. doi: 10.1042/BST0340104. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Gunsalus RP. 2000. The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than to nitrite. J Bacteriol 182:5813–5822. doi: 10.1128/JB.182.20.5813-5822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergaust L, Shapleigh J, Frostegård Å, Bakken L. 2008. Transcription and activities of NOx reductases in Agrobacterium tumefaciens: the influence of nitrate, nitrite and oxygen availability. Environ Microbiol 10:3070–3081. doi: 10.1111/j.1462-2920.2007.01557.x. [DOI] [PubMed] [Google Scholar]
- 36.Strohm TO, Griffin B, Zumft WG, Schink B. 2007. Growth yields in bacterial denitrification and nitrate ammonification. Appl Environ Microbiol 73:1420–1424. doi: 10.1128/AEM.02508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madigan MT, Martinko JM, Stahl DA, Clark DP. 2010. Biology of microorganisms, 13th ed Pearson/Benjamin Cummings, San Francisco, CA. [Google Scholar]
- 38.Gao H, Obraztova A, Stewart N, Popa R, Fredrickson JK, Tiedje JM, Nealson KH, Zhou J. 2006. Shewanella loihica sp. nov., isolated from iron-rich microbial mats in the Pacific Ocean. Int J Syst Evol Microbiol 56:1911–1916. doi: 10.1099/ijs.0.64354-0. [DOI] [PubMed] [Google Scholar]
- 39.Meinhold J, Arnold E, Isaacs S. 1999. Effect of nitrite on anoxic phosphate uptake in biological phosphorus removal activated sludge. Water Res 33:1871–1883. doi: 10.1016/S0043-1354(98)00411-4. [DOI] [Google Scholar]
- 40.Klüber HD, Conrad R. 1998. Inhibitory effects of nitrate, nitrite, NO and N2O on methanogenesis by Methanosarcina barkeri and Methanobacterium bryantii. FEMS Microbiol Ecol 25:331–339. [Google Scholar]
- 41.Van Cleemput O, Samater A. 1995. Nitrite in soils: accumulation and role in the formation of gaseous N compounds. Fertil Res 45:81–89. doi: 10.1007/BF00749884. [DOI] [Google Scholar]