Abstract
Escherichia coli that is unable to metabolize d-glucose (with knockouts in ptsG, manZ, and glk) accumulates a small amount of d-glucose (yield of about 0.01 g/g) during growth on the pentoses d-xylose or l-arabinose as a sole carbon source. Additional knockouts in the zwf and pfkA genes, encoding, respectively, d-glucose-6-phosphate 1-dehydrogenase and 6-phosphofructokinase I (E. coli MEC143), increased accumulation to greater than 1 g/liter d-glucose and 100 mg/liter d-mannose from 5 g/liter d-xylose or l-arabinose. Knockouts of other genes associated with interconversions of d-glucose-phosphates demonstrate that d-glucose is formed primarily by the dephosphorylation of d-glucose-6-phosphate. Under controlled batch conditions with 20 g/liter d-xylose, MEC143 generated 4.4 g/liter d-glucose and 0.6 g/liter d-mannose. The results establish a direct link between pentoses and hexoses and provide a novel strategy to increase carbon backbone length from five to six carbons by directing flux through the pentose phosphate pathway.
INTRODUCTION
The pentose phosphate (PP) pathway interconverts phospho-sugars having 3 to 7 carbon atoms principally by the action of the reversible enzymes transketolase and transaldolase. During the consumption of hexoses such as d-glucose or d-fructose, entry of carbon into this pathway provides many microorganisms, including Escherichia coli, the means to generate the reduced cofactor NADPH and to synthesize specific building-block compounds derived from intermediates of this pathway (e.g., phenylalanine, histidine, and ribose). For microorganisms having the requisite kinases and sugar transport mechanisms, the PP pathway also provides convenient entry points for the catabolism of many other sugars, including d-xylose and l-arabinose (49).
We have previously studied d-xylose and l-arabinose metabolism in E. coli that lacks the ability to metabolize d-glucose due to knockouts in the ptsG, manZ, and glk genes (1–3). Recently, small but consistent amounts (about 50 mg/liter) of d-glucose were observed as the accumulated end product when E. coli ptsG manZ glk was grown on 5 g/liter of either pentose in a defined medium (unpublished data). How might d-glucose be derived from these pentoses?
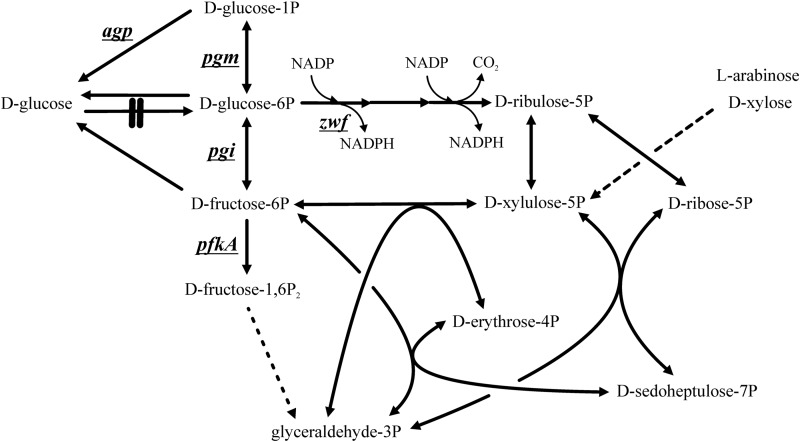
Both d-xylose and l-arabinose are converted to the common intermediate d-xylulose-5-phosphate (d-xylulose-5P) (Fig. 1), which via the PP pathway partitions to 67% d-fructose-6P and 33% d-glyceraldehyde-3P without the involvement of ATP:
| 1 |
FIG 1.
The pentose phosphate pathway and upper glycolysis of Escherichia coli, considering either l-arabinose or d-xylose as a carbon source and which form d-xylulose-5P as a common intermediate (dashed line). Genes (underlined) and the enzymes they encode are as follows: glucose-1-phosphatase (EC 3.1.3.10) (encoded by agp), phosphoglucomutase (EC 5.4.2.2) (pgm), d-glucose-6P isomerase (EC 5.3.1.9) (pgi), d-glucose-6P 1-dehydrogenase (EC 1.1.1.49) (zwf), and 6-phosphofructokinase I (EC 2.7.1.11) (pfkA). E. coli ptsG manZ glk (strain ALS1048) has gene deletions which prevent the microbe from metabolizing d-glucose (knockout indicated by double line). The conversion of d-fructose-1,6P2 to glyceraldehyde-3P is mediated by enzymes in several steps (dashed line).
During growth of cells having a complete glycolytic pathway, the 2 mol of d-fructose-6P formed via equation 1 readily generates 4 mol of d-glyceraldehyde-3P. For d-glucose to accumulate from pentoses in cells prevented from metabolizing d-glucose, we reasoned that some d-fructose-6P generated from these pentoses (i.e., by equation 1) is converted “back” to d-glucose and that once formed, the d-glucose is unable to reenter metabolism in the triple knockout strain. We furthermore hypothesized that even more d-glucose would accumulate from pentoses in cells that were further constrained from metabolizing d-fructose-6P or d-glucose-6P.
Because d-fructose-6P conversion to d-glyceraldehyde-3P is ubiquitous in wild-type organisms, d-glucose is not typically considered a product of d-xylose or l-arabinose metabolism, and the conversion of these pentoses to readily available d-glucose would in itself not seem to be an economically viable process. However, if the yields and rates were sufficiently large, the accumulation of hexoses directly from pentoses might advance the use of lignocellulosic hydrolysates with organisms, such as Saccharomyces cerevisiae, which metabolize d-glucose readily but are natively unable to consume pentoses. Moreover, conversion of 5-carbon saccharides into 6-carbon saccharides derived from d-fructose-6P offers a unique platform both to build carbon length and potentially to generate compounds in industrially relevant organisms such as E. coli that might not be possible under typical conditions in which products of d-fructose-6P do not accumulate.
The objectives of this study were to examine d-glucose formation from the pentoses d-xylose and l-arabinose. Specifically, we sought to identify the pathway involved in the formation of d-glucose from pentoses and to increase further the formation of d-glucose by preventing d-fructose-6P and d-glucose-6P metabolism. Finally, under the controlled conditions of a bioreactor, we examined whether elevated concentrations of d-glucose could be synthesized from either d-xylose or l-arabinose as a sole carbon source.
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli ALS1048 [MG1655 ΔptsG763::(FRT) ΔmanZ743::(FRT) Δglk-726::(FRT)] was used to construct additional strains as listed in Table 1 (2). These strains were constructed by transducing ALS1048 with the corresponding Keio (FRT)Kan deletions (4) and, if necessary, curing the Kanr using the pCP20 plasmid, which contains a temperature-inducible FLP recombinase as well as a temperature-sensitive replicon (5). All strains were verified by PCR.
TABLE 1.
E. coli strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| ALS1048 | MG1655 ΔptsG763::(FRT) ΔmanZ743::(FRT) Δglk-726::(FRT) | 2 |
| MEC132 | ALS1048 ΔpfkA775::Kan | This study |
| MEC143 | ALS1048 ΔpfkA775::(FRT) Δzwf-777::Kan | This study |
| MEC144 | ALS1048 Δzwf-777::Kan | This study |
| MEC151 | ALS1048 ΔpfkA775::(FRT) Δzwf-777::(FRT) Δmak-759::Kan | This study |
| MEC152 | ALS1048 ΔpfkA775::(FRT) Δzwf-777::(FRT) Δagp-746::Kan | This study |
| MEC178 | ALS1048 ΔpfkA775::(FRT) Δzwf-777::(FRT) Δgcd-742::Kan | This study |
| MEC180 | ALS1048 ΔpfkA775::(FRT) Δzwf-777::(FRT) ΔxylA748::Kan | This study |
| MEC319 | ALS1048 ΔpfkA775::(FRT) Δzwf-777::(FRT) Δpgm-736::Kan | This study |
| MEC320 | ALS1048 ΔpfkA775::(FRT) Δpgi-721::Kan | This study |
| MEC321 | ALS1048 ΔpfkA775::(FRT) Δzwf-777::(FRT) Δpgi-721::Kan | This study |
In one experiment, the pgi gene, encoding E. coli d-glucose-6P isomerase, was overexpressed. To construct the pTrc99A-pgi plasmid, the pgi gene was PCR amplified with primers 5′-GGGAAAGAATTCAAAAACATCAATCCAACGCAGACCGC-3′ (forward) and 5′-GGGAAAGGATCCTTAACCGCGCCACGCTTTATAGCG-3′ (reverse) using E. coli BW25113 genomic DNA as the template. The 1,671-bp PCR product was purified, restricted with EcoRI and BamHI, and ligated into the regulable expression vector pTrc99A that had also been restricted with EcoRI and BamHI to yield the plasmid pTrc99A-pgi, which was subsequently transformed into MEC320 and MEC321.
Growth medium and conditions.
The defined medium used for the shake flask experiments contained (per liter) 1.70 g citric acid, 13.30 g KH2PO4, 4.50 g (NH4)2HPO4, 1.2 g MgSO4 · 7H2O, 13 mg Zn(CH3COO)2 · 2H2O, 1.5 mg CuCl2 · 2H2O, 15 mg MnCl2 · 4H2O, 2.5 mg CoCl2 · 6H2O, 3.0 mg H3BO3, 2.5 mg Na2MoO4 · 2H2O, 100 mg Fe(III) citrate, 4.5 mg thiamine · HCl, 8.4 mg Na2(EDTA) · 2H2O, and 5.0 g d-xylose, l-arabinose, glycerol, or d-fructose. The pH was adjusted to 7.0 with 30% (wt/vol) NaOH. Cells were routinely stored on lysogeny broth (LB) agar plates and transferred to 20 ml defined medium in a 125-ml shake flask, from which 2 ml was transferred to the 50 ml defined medium in a 250-ml shake flask used for these studies. Shake flask studies were replicated 3 to 6 times for each strain and pentose when d-glucose was detected. Statistical analyses were completed using Student's t test (two tailed, equal variance), and a P value of <0.10 was considered the criterion for significance. For larger-scale studies in a controlled bioreactor, the sequence of transfers was identical, and the 50 ml from the final shake flask was used to inoculate the larger vessel. The flasks were incubated at 37°C with an agitation of 250 rpm. Samples were stored at −20°C for subsequent analysis.
A single controlled batch process at a 1.0-liter volume was carried out using d-xylose in a 2.5-liter bioreactor (Bioflo 2000; New Brunswick Scientific Co. Edison, NJ, USA). The same defined medium was used except that the concentration of d-xylose was 20 g/liter. Air or oxygen as necessary was sparged into the fermentor with the agitation set at 500 rpm to maintain the dissolved oxygen above 40% saturation. The pH was controlled at 7.0 using 20% NaOH, and the temperature was controlled at 37°C. Antifoam C (Sigma) was used as necessary to control foaming.
Continuous processes using MEC143 operated as nitrogen (N)-limited chemostats at a 1.0-liter volume were conducted in the same 2.5-liter fermentor. To ensure N limitation and prevent contamination, the medium contained (per liter) 1.0 g (NH4)2HPO4 (15 mM N), 8.0 g d-xylose, and 40 mg kanamycin, but otherwise it remained unchanged. Four dilution rates (growth rates) were examined in the range 0.08 to 0.15 h−1, and a steady-state condition was assumed after four residence times, at which time the oxygen and CO2 concentrations in the effluent gas appeared constant. These processes were conducted at 37°C with an air flow rate of 0.5 liter/min, an agitation of 400 rpm, and a pH of 7.0. The dissolved oxygen remained above 40% saturation. A carbon balance was completed using a unit carbon formula weight for E. coli cell mass of 24.6 g/mol (6).
Analytical methods.
The optical density (OD) at 600 nm (UV-650 spectrophotometer; Beckman Instruments, San Jose, CA) was used to monitor cell growth. Liquid chromatography with a refractive index detector and a Coregel 64-H ion-exclusion column (Transgenomic Ltd., Glasgow, United Kingdom) using a mobile phase of 4 mN H2SO4 was used for analysis of sugars and acetic acid as described previously (7). For dry cell weight measurement, three 25.0-ml samples were centrifuged (8,400 × g, 10 min), and the pellets were washed by vortex mixing with 30 ml 0.9% saline solution and then centrifuged again. After repeating the washing step twice using deionized (DI) water, the cell pellets were dried at 60°C for 24 h and weighed. The concentrations of oxygen and CO2 in the off-gas were measured using a gas analyzer (Innova 1313 gas monitor; Lumasense Technologies, Ballerup, Denmark).
The presence of sugars was confirmed by comparing samples with standards using a derivatization protocol with a gas chromatograph-mass spectrometer (GC-MS) (8). The GC-MS method was used only for identification and not quantification, in particular in those cases in which the analytes eluted closely by high-pressure liquid chromatography (HPLC) (d-mannose and d-xylose) or to confirm the absence of a sugar (e.g., d-fructose). Briefly, samples were centrifuged, the supernatant evaporated to dryness, and samples then derivatized with 700 μl hexamethyldisilazane, 200 μl anhydrous pyridine, and 10 μl trifluoroacetic acid at 60°C for 3 h. Detection of derivatized analytes was accomplished with a GC-MS (electron ionization energy of 70 eV) (HP6890/HP5973; Agilent Technologies, Inc., Santa Clara, CA, USA). One microliter was injected onto a 30 m HP-5MS column (Agilent Technologies, Inc.) in the split-flow mode at 30:1 with a 1-ml/min flow rate. The temperature profile began at 50°C for 1 min, increased at 2°C/min to 100°C, increased at 5°C/min to 250°C, and held for 5 min. The injector temperature was 250°C, MS source was 230°C, MS Quad was 150°C, and GC-MS interface was 280°C. For the N-limited chemostats, the ammonia nitrogen (NH4-N) in feed and effluent was analyzed using the colorimetric EPA method 350.1 (9).
We used nuclear magnetic resonance (NMR) to demonstrate that d-glucose was formed biologically and accumulated in the medium. Four samples were analyzed: a glucose standard, a d-glucose-6P standard, a sample from a shake flask experiment (i.e., containing d-glucose), and a d-glucose-6P standard incubated in sterile medium for 24 h at 37°C. NMR data were acquired using a Varian Inova instrument with a cryogenic probe system at 14.1 T (600 MHz 1H). The sample temperature was maintained at 25°C. Standard, natural-abundance, two-dimensional 1H,13C heteronuclear single quantum coherence (1H,13C-HSQC) spectra were acquired in the constant-time (13C-decoupled) mode. Chemical shift assignments were made by reference to database entries and published works (10–15). The 1H chemical shifts were referenced with respect to external Na+DSS− (sodium 4,4-dimethyl-4-silapentane-1-sulfonate) in D2O at 25°C (0.0 ppm). The 13C chemical shifts were referenced indirectly assuming the absolute frequency ratio 13C/1H = 0.251449530 (16). D2O was added to samples to a final concentration of approximately 7% for instrumental lock. NMR data were processed and signal intensities measured using Felix (Accelrys, San Diego, CA).
RESULTS
Formation of glucose from xylose or arabinose.
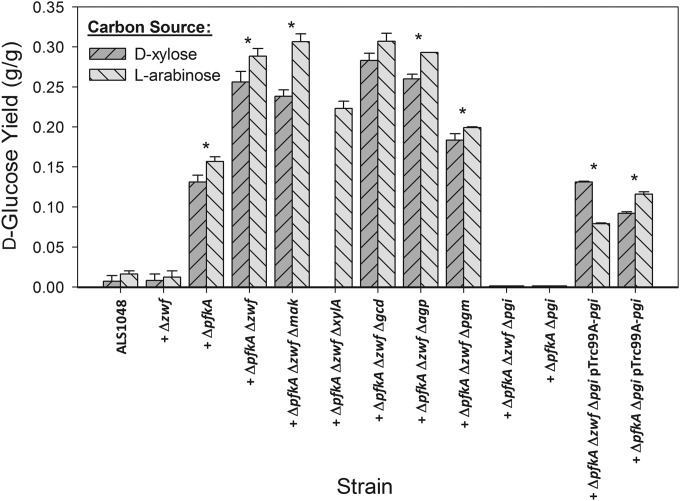
E. coli ALS1048 contains knockouts in the ptsG, manZ, and glk genes and is unable to metabolize d-glucose (2). Using this strain as a baseline for comparison, we first sought to determine whether additional d-glucose would accumulate from either d-xylose or l-arabinose if d-fructose-6P and d-glucose-6P were prevented from entering glycolysis and the PP pathway. Specifically, we first constructed strains with additional knockouts in pfkA, encoding 6P-fructokinase I (EC 2.7.1.11), and/or zwf, encoding d-glucose-6P 1-dehydrogenase (EC 1.1.1.49). During growth in shake flasks using either 5 g/liter d-xylose or 5 g/liter l-arabinose, ALS1048 and ALS1048 zwf (MEC144) accumulated d-glucose at a yield of about 0.01 g/g from either pentose (Fig. 2), while ALS1048 pfkA (MEC132) generated d-glucose at yields of 0.13 g/g from d-xylose and 0.17 g/g from l-arabinose. Eliminating both pathways in ALS1048 pfkA zwf (MEC143) resulted in the accumulation of d-glucose at yields of 0.26 to 0.29 g/g. For both pfkA knockout strains MEC132 and MEC143, we also observed the formation of 60 to 180 mg/liter d-mannose using HPLC, which was confirmed by GC-MS (see Materials and Methods). No other product such as d-fructose was identified by GC-MS. Moreover, the d-glucose and d-mannose were not metabolized within several hours after the pentose was exhausted. These results clearly show that E. coli can generate d-glucose from pentoses through d-fructose-6P, suggesting a route for the formation of 6-carbon products from 5-carbon substrates by preventing the intermediate d-fructose-6P from entering glycolysis and the PP pathway (Fig. 1). We also repeated the identical shake flask experiments using MEC143 with 5 g/liter glycerol or 5 g/liter d-fructose and observed d-glucose as a final product at a concentration of 60 mg/liter or 75 mg/liter, respectively (yield of about 0.01 g/g).
FIG 2.
Comparison of E. coli strains for the production of d-glucose from 5 g/liter l-arabinose or 5 g/liter d-xylose. All strains are derived from ALS1048 (MG1655 ptsG manZ glk) and have additional gene knockouts as indicated. Strains which accumulated d-glucose were studied in 3 to 6 replicate cultures grown at 50 ml in a 250-ml shake flask, and error bars show the standard error of the measurements from these replicate samples. An asterisk indicates a significant difference (P < 0.10) in yield of d-glucose from d-xylose compared to from l-arabinose.
Identification of key enzymes involved in glucose formation.
By preventing d-glucose utilization in E. coli while simultaneously blocking entry of d-fructose-6P into glycolysis and reentry into the PP pathway, significant d-glucose formed from d-xylose or l-arabinose (Fig. 2). We therefore sought next to clarify the pathway that E. coli uses to convert d-fructose-6P to d-glucose by constructing additional knockout strains.
The formation of some d-mannose during the accumulation of d-glucose suggests the involvement of d-mannose as a pathway intermediate. Also, one possible route for d-glucose formation from d-fructose-6P would be via d-fructose. The enzyme mannokinase (EC 2.7.1.4), encoded by mak, is known to phosphorylate d-mannose and d-fructose (17). We therefore hypothesized that mannokinase might be involved in the accumulation of d-glucose from pentoses via the conversion of d-fructose-6P to d-fructose. However, E. coli ALS1048 pfkA zwf mak (MEC151) did not show any difference in d-glucose formation from either pentose compared to MEC143 (Fig. 2). Also, d-mannose was formed as before (230 to 270 mg/liter), supporting the conclusion that mannokinase is not involved in the formation of either d-mannose or d-glucose from pentoses.
Another pathway that potentially could serve to form d-glucose is via the enzyme xylose isomerase (EC 5.3.1.5). In addition to interconverting d-xylose and d-xylulose, the E. coli xylose isomerase interconverts d-fructose and d-glucose (18–20), but less efficiently (21). Though the Km values for d-fructose, d-glucose, and d-xylose have not been reported for the E. coli enzyme, the kcat values for d-glucose and d-fructose are similar to those for the enzymes from other organisms (22), suggesting that d-fructose and d-glucose both readily serve as substrates for this isomerization. We therefore knocked out the xylA gene to form strain ALS1048 pfkA zwf xylA (MEC180). Of course, the xylA knockout also renders this strain unable to consume d-xylose, and therefore only the conversion of l-arabinose to d-glucose could be examined. The deletion of xylose isomerase reduced the d-glucose yield only slightly to 0.23 g/g (Fig. 2), and d-mannose was detected at 170 to 190 mg/liter, corresponding to a yield of about 0.03 g/g. These results suggest that xylose isomerase does not play a significant role in the formation of both d-glucose and d-mannose from pentoses.
We next examined one possible route through which d-glucose could be utilized. The gcd gene, encoding glucose dehydrogenase (EC 1.1.5.2), is able to convert d-glucose into d-glucono-1,5-lactone, which can then spontaneously form gluconate (23), although pyrroloquinoline quinone appears to be necessary for this conversion in E. coli (24). In order to determine whether d-glucose accumulation is influenced by glucose dehydrogenase, we constructed ALS1048 pfkA zwf gcd (MEC178). MEC178 formed d-glucose from d-xylose (yield of 0.28 g/g) or from l-arabinose (0.30 g/g), and also formed 110 to 150 mg/liter d-mannose from either pentose (0.03 g/g), indicating that this route does not significantly affect hexose formation (Fig. 2).
We next examined whether the formation of d-glucose was the result of the hydrolysis of either d-glucose-1P or d-glucose-6P. d-Fructose-6P is converted to d-glucose-6P by d-glucose-6P isomerase encoded by pgi, d-glucose-6P is converted to d-glucose-1P by phosphoglucomutase encoded by pgm, and d-glucose-1P can be dephosphorylated by d-glucose 1-phosphatase encoded by agp (Fig. 1). The d-glucose yield was unchanged as a result of the agp knockout (ALS1048 pfkA zwf agp [MEC152]) and was 0.18 to 0.20 g/g with a pgm knockout (ALS1048 pfkA zwf pgm [MEC319]). Both MEC152 and MEC319 accumulated 130 to 170 mg/liter d-mannose from 5 g/liter of either pentose. However, d-glucose and d-mannose formation was completely eliminated as a result of the pgi knockout (ALS1048 pfkA zwf pgi [MEC321]) (Fig. 2). Because MEC319 showed a slight reduction in d-glucose yield, the results do not exclude the possibility of some d-glucose-1P hydrolysis resulting in d-glucose formation, although the lower observed glucose yield in MEC319 than in MEC143 could also be simply due to the cells' reduced ability to form necessary metabolites from d-glucose-1P and UDP–d-glucose. The complete elimination of d-glucose formation as a result of a pgi deletion supports the conclusion that the hydrolysis of d-glucose-6P is the principal final step by which d-glucose is formed from pentoses.
Two additional experiments were conducted to confirm the role of pgi in d-glucose formation. First, because the pgi knockout blocks d-glucose-6P formation (Fig. 1), the zwf knockout should not affect d-glucose formation in a pgi knockout. In other words, the ptsG manZ glk pfkA pgi strain should also be unable to accumulate d-glucose. We therefore examined ALS1048 pfkA pgi (MEC320) and indeed observed no d-glucose formation from either d-xylose or l-arabinose (Fig. 2). Second, we transformed both MEC320 and MEC321 with pTrc99A-pgi expressing native d-glucose-6P isomerase, and these strains regained the ability to accumulate d-glucose from either d-xylose or l-arabinose. MEC320 pTrc99A-pgi attained a yield of 0.09 g/g, and MEC321 pTrc99A-pgi attained a yield of 0.13 g/g.
Interestingly, knockout strains which generated more than 0.05 g/g d-glucose accumulated significantly more d-glucose from l-arabinose than from d-xylose (P < 0.10) (Fig. 2), except MEC178 (ALS1048 pfkA zwf gcd), for which there was no significant difference. For example, MEC143 (ALS1048 pfkA zwf) generated 27% more d-glucose from l-arabinose than from d-xylose.
Finally, we confirmed that d-glucose was the biological product from both pentoses by comparing the NMR spectra of d-glucose and d-glucose-6P and also by demonstrating that d-glucose could not have formed from d-glucose-6P by chemical hydrolysis within the medium or during the HPLC procedure at the temperatures used (Fig. 3). The NMR results confirm that extracellular d-glucose and not d-glucose-6P was the biological product of d-xylose or l-arabinose metabolism in these knockout strains.
FIG 3.
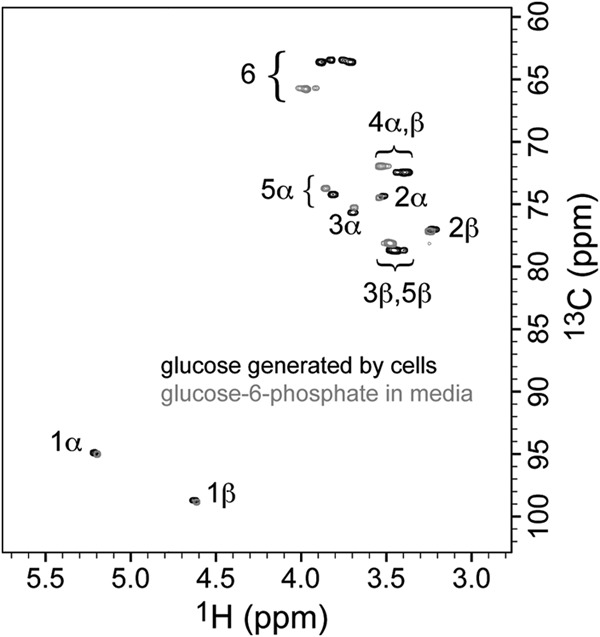
Confirmation of d-glucose production from xylose, using an overlay of the two-dimensional, 1H,13C-HSQC NMR spectra of a sample of product from the pentose (black contours, medium after xylose was exhausted by cells, with D2O added to 7%) and a sample using authentic glucose-6P in medium (gray contours, with D2O added to 7%). The molecules are distinguished because the presence of the phosphate group on d-glucose-6P promotes characteristic 1H and 13C chemical shift changes, compared to d-glucose, for the nuclei at positions nearer the site of glucose attachment (positions 6, 5, and 4, with smaller changes at 3, 2, and 1). Comparison with spectra of authentic d-glucose (not shown) confirms the identities. The data demonstrate that d-glucose is the fermentation product from xylose and that d-glucose does not form by extracellular hydrolysis of d-glucose-6P under the conditions of the experiments.
Batch process to accumulate glucose.
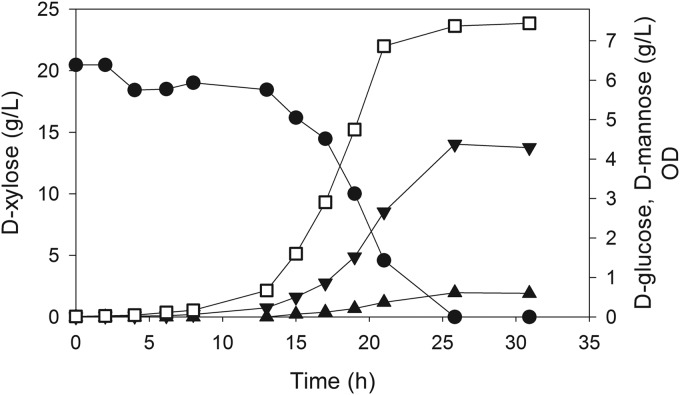
The previous experiments were all conducted in shake flasks using 5 g/liter l-arabinose or d-xylose. We next conducted an experiment using MEC143 in a controlled bioreactor with approximately 20 g/liter d-xylose to determine if a proportionate increase in d-glucose (and d-mannose) accumulation would be observed. In this batch run, 4.4 g/liter d-glucose and 0.61 g/liter d-mannose were formed in 25 h for an observed mass yield from d-xylose of 0.21 g d-glucose/g and 0.03 g d-mannose/g (Fig. 4). Furthermore, neither d-glucose nor d-mannose was reassimilated 5 h after d-xylose was exhausted. A nearly proportionate increase in product formation was observed in these controlled processes compared to the shake flask studies, suggesting that d-glucose formation is not inhibited or repressed by d-glucose accumulation. This result also demonstrates a potential for generating substantial quantities of 6-carbon hexoses from 5-carbon pentoses.
FIG 4.
Accumulation of d-glucose (▼) and d-mannose (▲) from 20 g/liter d-xylose (●) by E. coli MEC143 (MG1655 ptsG manZ glk pfkA zwf). Cell density is measured as optical density (□).
Continuous processes to accumulate glucose.
Chemostats are a convenient tool to study microbial growth and product formation under nutrient-limited conditions. During the batch process previously studied, the cells were grown under nutrient-excess conditions, and we reasoned that carbon flux might be maximal if the cells were grown under conditions for which growth was limited by a nutrient other than carbon. Furthermore, being at steady state and at a controlled growth rate, a chemostat would demonstrate whether the d-glucose observed is formed as a transient product or only during maximal cell growth. We therefore next grew MEC143 under nitrogen-limiting conditions by increasing the concentration of d-xylose and decreasing the concentration of the nitrogen source (see Materials and Methods). At four different dilution rates (0.08 h−1 to 0.15 h−1), the observed yields averaged 0.26 (±0.08 [standard deviation]) g d-glucose/g d-xylose and 0.23 (±0.03) g dry cells/g d-xylose, and these values did not vary with dilution rate. The mean carbon recovery was 108% (±11%), and 2.3 to 3.3 g/liter d-xylose and less than 0.5 mg/liter N were detected in the effluents. These results demonstrate that d-glucose formation is not a transient phenomenon. Since the yields in the nitrogen-limited chemostats were similar to yields observed in batch processes, d-glucose does appear to form as an overflow metabolite.
DISCUSSION
During growth on d-xylose or l-arabinose, wild-type E. coli generates 2 mol of d-fructose-6P and 1 mol d-glyceraldehyde-3P from 3 mol of either pentose (equation 1). If the glycolytic pathway is complete, the 2 mol of d-fructose-6P formed via equation 1 readily generates 4 mol of d-glyceraldehyde-3P. Indeed, because the conversion of d-fructose-6P to d-glyceraldehyde-3P is readily accomplished in widely studied microorganisms, d-fructose-6P is typically not thought of as an intermediate of pentose metabolism. However, our results demonstrate that E. coli can direct this metabolic intermediate d-fructose-6P into other 6-carbon (i.e., hexose) end products such as d-glucose when three conditions are met.
A first condition for the accumulation of products derived from pentoses via d-fructose-6P is that glycolysis must be disrupted between d-fructose-6P and d-glyceraldehyde-3P. By blocking glycolysis, the pentose phosphate pathway essentially becomes a branched pathway during the metabolism of d-xylose or l-arabinose with two separate products, d-fructose-6P and d-glyceraldehyde-3P (Fig. 1). That is, when d-fructose-6P cannot enter glycolysis, it becomes available for the formation of other 6-carbon products, while the d-glyceraldehyde-3P remains available for the generation of ATP, NADH, and the precursors that exist metabolically “below” d-glyceraldehyde-3P via the terminal steps of glycolysis and the tricarboxylic acid cycle. In E. coli, the entry of d-fructose-6P into glycolysis can be blocked by a deletion in the pfkA gene.
A second condition to facilitate the accumulation of hexoses from pentoses is that metabolites should be prevented from reentering the PP pathway, for example, from d-glucose-6P. In E. coli the reentry of d-glucose-6P into the PP pathway can be prevented by a knockout of the zwf gene (Fig. 1). Finally, as a third condition, the ultimate product must be excreted and should not be remetabolized. Our results demonstrate that the knockouts in the ptsG, manZ, and glk genes effectively block d-glucose metabolism and at least curtail its reassimilation once generated.
E. coli MEC143, which met these three conditions, accumulated significant d-glucose from either of two pentoses, l-arabinose or d-xylose. Interestingly, d-glucose was also observed, but to a much lesser extent, when glycerol or d-fructose was the sole carbon source in the same strain, probably as a result of the formation of a small quantity of d-fructose-6P generated via the nonoxidative PP pathway. Another sugar derived from d-fructose-6P, d-mannose, was also consistently observed as a by-product of d-glucose formation. d-Mannose likely accumulated as a result of the manZ gene deletion in all the strains studied, which prevented the uptake of not only d-glucose but also this sugar.
Of the several knockouts examined to clarify the route for d-glucose formation from the intermediate d-fructose-6P, only a deletion of the pgi gene, encoding d-glucose-6P isomerase, eliminated d-glucose formation. Although this result implicates d-glucose-6P as the direct precursor to extracellular d-glucose, we do not establish how d-glucose-6P itself is hydrolyzed. Unfortunately, E. coli has numerous candidate enzymes that could hydrolyze d-glucose-6P: a periplasmic acid phosphatase (25), an alkaline phosphatase (26), and eight different haloacid dehalogenase-like hydrolases (27) have all been observed to hydrolyze d-glucose-6P under various environmental conditions. Each of these enzymes might mediate the final step to d-glucose during growth on d-xylose or l-arabinose.
All strains in this study had knockouts in the ptsG, manZ, and glk genes, encoding proteins involved in the principal means for d-glucose uptake in E. coli (28). There is no report of these proteins being involved in d-glucose export, and our results provide no guidance to this process. E. coli has several known porins and permeases that can translocate d-glucose through the outer and cytoplasmic membranes (though previous studies have invariably focused on sugar import). For example, porins OmpF and OmpC transport d-glucose by passive diffusion across the outer membrane (29, 30). LamB functions as a broad-specificity glycoporin which transports d-glucose among other mono- and polysaccharides (31). Galactose permease (GalP) readily transports d-glucose across the cytoplasmic membrane (32). In fact, for strains deficient in the phosphotransferase system (PTS) uptake system (ptsH, ptsI, and crr knockouts), GalP very effectively replaces the transport functions of the IICBGlc PTS protein (33). Similarly, the periplasmic d-glucose/d-galactose binding receptor protein encoded by mglB binds d-glucose (34, 35) and contributes significantly to the growth and transport affinity for d-glucose at low extracellular d-glucose concentrations (36). Additionally, E. coli responds to knockouts in transport genes: the expression of the galP, mglB, and lamB permease genes increased as a result of a ptsHI crr knockout (37). The mechanisms of d-glucose uptake under d-glucose-limiting and -excess conditions have been reviewed (38), and the various proteins involved in d-glucose uptake have recently been examined collectively in E. coli in the context of overflow metabolism and vaccine production (39). Interestingly, GluP has been implicated in d-glucose export in Bacillus subtilis (40), but we found no similar E. coli protein. In our current study, any of these or other proteins could also be involved in d-glucose excretion.
A consistent result was that a greater yield of d-glucose was attained from l-arabinose than from d-xylose, and this difference must result from differences in the metabolism of these two pentoses by E. coli. Interestingly, d-xylose and l-arabinose are transported and enter the PP pathway through different routes in E. coli. d-Xylose is transported by several routes: a d-xylose/proton symporter (41), an ATP-binding-dependent system (42), and promiscuous transporter activity (43). The ATP-dependent system appears to predominate under normal growth conditions (44), indicating that d-xylose uptake generally demands energy directly in the form of ATP. Cellular options for the transport of l-arabinose similarly include a high-affinity ATP-dependent system and a low-affinity proton symport (45), as well as promiscuous transport (46). For this pentose, the low-affinity, ATP-independent system appears to predominate when both systems are present and the l-arabinose concentration is relatively high (45), although this process presumably affects availability of ATP also. After cellular uptake, both of these pentoses are ultimately converted to the common intermediate d-xylulose-5P (Fig. 1), through steps which require ATP for phosphorylation via d-xylulokinase or l-ribulokinase, respectively, for d-xylose or l-arabinose. Since the metabolism of these two pentoses would appear identical after d-xylulose-5P, one might speculate that the difference in d-glucose yield between the two pentoses might be due to the difference in sugar transport mechanisms. Additional studies will have to clarify this difference.
If 2 mol of d-fructose-6P generated from 3 mol of d-xylose or l-arabinose (equation 1) is available for d-glucose formation (corresponding to 0.67 mol/mol), the theoretical d-glucose mass yield from either pentose is 0.80 g/g. This calculation considers the d-glyceraldehyde-3P generated from the flux-balanced PP pathway to be unavailable for d-glucose formation because additional d-glyceraldehyde-3P cannot reenter the PP pathway without consuming d-fructose-6P. On the other hand, inclusion of the hypothetical conversion of d-glyceraldehyde-3P through the reverse Embden-Meyerhof-Parnas pathway to d-fructose-6P would result in a theoretical maximum yield 1.0 g/g. The greatest yield observed in the current study was 0.3 g/g, a result probably due to the assimilation of some of the intermediate monosaccharides by other enzymes present in E. coli and not deleted in this study, and by the reversible enzymes in the PP pathway (transaldolase and transketolase), which would limit d-fructose-6P formation if these reactions approached equilibrium. Although not likely to serve as a process for generating the specific hexose d-glucose, this work demonstrates an approach to convert 5-carbon saccharides into 6-carbon saccharides, which could thereby both build carbon length and generate hexoses derived from d-fructose-6P not possible under typical conditions during growth on d-glucose.
Our results highlight two other aspects of metabolism in strains that have deletions in d-glucose uptake and other genes in upper metabolism. First, we detected no d-fructose as a product in any of the experiments, and our shake flask study with MEC143 growing on d-fructose yielded only a low concentration of d-glucose. The absence of d-fructose in the (extracellular) medium is likely because E. coli uses a d-fructose-specific phosphotransferase system (fruA and fruB genes) and d-fructose-1P kinase (fruK) to metabolize d-fructose to d-fructose-1,6P2, bypassing d-fructose-6P. In other words, assimilation of d-fructose via this route bypasses d-fructose-6P and would also prevent d-fructose accumulation in the strains studied. In contrast, once d-glucose is transported out of the cell, deletions of the ptsG and manZ genes prevent its uptake. A second noteworthy result from the mutants studied lies in the absence of d-glucose repression. For example, previous research has demonstrated that xylose isomerase is repressed in the presence of d-glucose (47). This effect appears to be caused specifically by d-glucose catabolite repression (48), an occurrence requiring the active catabolism of d-glucose and which therefore is avoided in a strain unable to metabolize d-glucose. In our batch process accumulating nearly 5 g/liter d-glucose (Fig. 3), we did not observe any deceleration of d-xylose utilization: the cells acted as though d-glucose were not present. Thus, E. coli is able to metabolize d-xylose in the presence of d-glucose when the d-glucose is not being metabolized.
In conclusion, d-glucose formation from either l-arabinose or d-xylose occurs as a result of the PP pathway leading to d-fructose-6P, which, unable to proceed into the glycolytic pathway due to a knockout in pfkA, equilibrates to d-glucose-6P. d-Glucose-6P likely hydrolyzes by one of several possible enzymes to d-glucose, which then accumulates when the cells are unable to metabolize it. We envision that an analogous route could be used to generate similar sugars or sugar-containing compounds.
ACKNOWLEDGMENTS
We acknowledge the National Science Foundation (CBET-0929893) for financial support of portions of this work.
We thank Sarah Lee and Janet Fisher for technical assistance.
REFERENCES
- 1.Eiteman MA, Lee SA, Altman E. 2008. A co-fermentation strategy to consume sugar mixtures effectively. J Biol Eng 2:3. doi: 10.1186/1754-1611-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eiteman MA, Lee SA, Altman R, Altman E. 2009. A substrate-selective co-fermentation strategy with Escherichia coli produces lactate by simultaneously consuming xylose and glucose. Biotechnol Bioeng 102:822–827. doi: 10.1002/bit.22103. [DOI] [PubMed] [Google Scholar]
- 3.Xia T, Eiteman MA, Altman E. 2012. Simultaneous utilization of glucose, xylose and arabinose in the presence of acetate by a consortium of Escherichia coli strains. Microb Cell Fact 11:77. doi: 10.1186/1475-2859-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba T, Ara T, Hasegawa M, Takaki Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battley EH. 1991. Calculation of the heat of growth of Escherichia coli K-12 on succinic acid. Biotechnol Bioeng 37:334–343. doi: 10.1002/bit.260370407. [DOI] [PubMed] [Google Scholar]
- 7.Eiteman MA, Chastain MJ. 1997. Optimization of the ion-exchange analysis of organic acids from fermentation. Anal Chim Acta 338:69–75. doi: 10.1016/S0003-2670(96)00426-6. [DOI] [Google Scholar]
- 8.Bertrand A, Barbe J-C. 2002. Formation of γ-gluconolactone in a wine-like model system. J Sci Food Agric 82:1571–1573. doi: 10.1002/jsfa.1217. [DOI] [Google Scholar]
- 9.U.S. EPA. 1983. Nitrogen, ammonia. Method 250.1 (colorimetric), p 350-1.1–350-1.4. In Methods for chemical analysis of water and wastes. EPA-600/4-79-020 U.S. EPA, Cincinnati, Ohio, USA. [Google Scholar]
- 10.Bagno A, Rastrelli F, Saielli G. 2007. Prediction of the 1H and 13C NMR spectra of α-d-glucose in water by DFT methods and MD simulations. J Org Chem 72:7373–7381. doi: 10.1021/jo071129v. [DOI] [PubMed] [Google Scholar]
- 11.Curatolo W, Neuringer LJ, Ruben D, Haberkorn R. 1983. Two-dimensional J-resolved 1H-nuclear magnetic resonance spectroscopy of α,β-d-glucose at 500 MHz. Carbohydr Res 112:197–300. [Google Scholar]
- 12.Peng F, Ren JL, Xu F, Bian J, Peng P, Sun RC. 2010. Fractionation of alkali-solubilized hemicelluloses from delignified Populus gansuensis: structure and properties. J Agric Food Chem 58:5743–5750. doi: 10.1021/jf1003368. [DOI] [PubMed] [Google Scholar]
- 13.Pfeffer PE, Valentine KM, Parrish FW. 1979. Deuterium-induced differential isotope shift 13C NMR. 1. Resonance reassignments of mono- and disaccharides. J Am Chem Soc 101:1265–1274. [Google Scholar]
- 14.Rossi C, Marchettini N, Donati A, Medaglini D, Valassina M, Bastianoni S, Cresta E. 1995. 13C-NMR determination of simultaneous xylose and glucose fermentation by a newly isolated strain (G11) of Klebsiella planticola. Biomass Bioenergy 8:197–202. doi: 10.1016/0961-9534(95)00005-R. [DOI] [Google Scholar]
- 15.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao H, Markley JL. 2008. BioMagResBank. Nucleic Acids Res 36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6:135–140. [DOI] [PubMed] [Google Scholar]
- 17.Sebastian J, Asensio C. 1972. Purification and properties of the mannokinase from Escherichia coli. Arch Biochem Biophys 151:227–233. doi: 10.1016/0003-9861(72)90492-4. [DOI] [PubMed] [Google Scholar]
- 18.Bhosale SH, Rao MB, Deshpande VV. 1996. Molecular and industrial aspects of glucose isomerase. Microbiol Rev 60:280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epting KL, Vieille C, Zeikus JG, Kelly RM. 2005. Influence of divalent cations on the structural thermostability and thermal inactivation kinetics of class II xylose isomerases. FEBS J 272:1454–1464. doi: 10.1111/j.1742-4658.2005.04577.x. [DOI] [PubMed] [Google Scholar]
- 20.Voronovsky AY, Ryabova OB, Verba OV, Ishchuk OP, Dmytruk KV, Sibirny AA. 2005. Expression of xylA genes encoding xylose isomerases from Escherichia coli and Streptomyces coelicolor in the methylotrophic yeast Hansenula polymorpha. FEMS Yeast Res 5:1055–1062. doi: 10.1016/j.femsyr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Sapunova LI, Lobanok AG, Kazakevich IO, Shlyakhotko EA, Evtushenkov AN. 2006. Biosynthetic features and properties of xylose isomerases from Arthrobacter nicotianae, Escherichia coli, and Erwinia carotovora subsp. atroseptica. Appl Biochem Microbiol (Russ) 42:246–251. doi: 10.1134/S0003683806030045. [DOI] [PubMed] [Google Scholar]
- 22.Liu SY, Wiegel J, Gherardini FC. 1996. Purification and cloning of a thermostable xylose (glucose) isomerase with an acidic pH optimum from Thermoanaerobacterium strain JW/SL-YS 489. J Bacteriol 178:5938–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Schie BJ, Hellingwerf KJ, van Kijken JP, Elferink MGL, van Dijl JM, Kuenen JG, Konings WN. 1985. Energy transduction by electron transfer via a pyrrolo-quinoline quinone-dependent glucose dehydrogenase in Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter calcoaceticus (var. lwoffi). J Bacteriol 163:493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsushita K, Arents JC, Bader R, Yamada M, Adachi O, Postma PW. 1997. Escherichia coli is unable to produce pyrroloquinoline quinone (PQQ). Microbiol 143:3149–3156. doi: 10.1099/00221287-143-10-3149. [DOI] [PubMed] [Google Scholar]
- 25.Passariello C, Forleo C, Micheli V, Schippa S, Leone R, Mangani S, Thaller MC, Rossonlini GM. 2006. Biochemical characterization of the class B acid phosphatase (AphA) of Escherichia coli MG1655. Biochim Biophys Acta 1764:13–19. doi: 10.1016/j.bbapap.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 26.Heppel LA, Harkness DR, Hilmoe RJ. 1962. A study of the substrate specificity and other properties of the alkaline phosphatase of Escherichia coli. J Biol Chem 237:841–846. [PubMed] [Google Scholar]
- 27.Kuznetsova E, Proudfoot M, Gonzalez CF, Brown G, Omelchenko MV, Borozan I, Carmel L, Wolf YI, Mori H, Savchenko AV, Arrowsmith CH, Koonin EV, Edwards AM, Yakunin AF. 2006. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehydrogenase-like phosphatase family. J Biol Chem 281:36149–36161. doi: 10.1074/jbc.M605449200. [DOI] [PubMed] [Google Scholar]
- 28.Curtis SJ, Epstein W. 1975. Phosphorylation of d-glucose in Escherichia coli mutants defective in glucosephosphotransferase, mannosephosphotransferase, and glucokinase. J Bacteriol 122:1189–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikaido H, Nakae T. 1979. The outer membrane of Gram-negative bacteria. Adv Microbial Physiol 20:163–250. [DOI] [PubMed] [Google Scholar]
- 30.Nikaido H, Rosenberg EY. 1983. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J Bacteriol 153:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luckey M, Nikaido H. 1980. Specificity of diffusion channels produced by l-phase receptor protein of Escherichia coli. Proc Natl Acad Sci U S A 77:167–171. doi: 10.1073/pnas.77.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald TP, Walmsley AR, Henderson PJF. 1997. Asparagine 394 in putative helix 11 of the galactose-H+ symport protein (GalP) from Escherichia coli is associated with the internal binding site for cytochalasin B and sugar. J Biol Chem 272:15189–15199. doi: 10.1074/jbc.272.24.15189. [DOI] [PubMed] [Google Scholar]
- 33.Flores S, Gosset G, Flores N, Graaf AA, Bolívar F. 2002. Analysis of carbon metabolism in Escherichia coli strains with an inactive phosphotransferase system by 13C labeling and NMR spectroscopy. Metabol Eng 4:124–137. doi: 10.1006/mben.2001.0209. [DOI] [PubMed] [Google Scholar]
- 34.Kalckar HM. 1971. The periplasmic galactose-binding protein of Escherichia coli. Science 174:557–565. doi: 10.1126/science.174.4009.557. [DOI] [PubMed] [Google Scholar]
- 35.Miller DM III, Olson JS, Quiocho FA. 1980. The mechanism of sugar binding to the periplasmic receptor for galactose chemotaxis and transport in Escherichia coli. J Biol Chem 255:2465–2471. [PubMed] [Google Scholar]
- 36.Hua Q, Yang C, Oshima T, Mori H, Shimizu K. 2004. Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl Environ Microbiol 70:2354–2366. doi: 10.1128/AEM.70.4.2354-2366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flores S, Flores N, Anda R, González A, Escalante A, Sigala JC, Gosset G, Bolívar F. 2005. Nutrient-scavenging stress response in an Escherichia coli strain lacking the phosphoenolpyruvate:carbohydrate phosphotransferase system, as explored by gene expression profile analysis. J Mol Microbiol Biotechnol 10:51–63. doi: 10.1159/000090348. [DOI] [PubMed] [Google Scholar]
- 38.Ferenci T. 1996. Adaptation to life at micromolar nutrient levels: the regulation of Escherichia coli glucose transport by endoinduction and cAMP. FEMS Microbiol Rev 18:301–317. doi: 10.1111/j.1574-6976.1996.tb00246.x. [DOI] [PubMed] [Google Scholar]
- 39.Fuentes LG, Lara AR, Martínez LM, Ramírez OT, Martínez A, Bolívar R, Gosset G. 2013. Modification of glucose import capacity in Escherichia coli: physiological consequences and utility for improving DNA vaccine production. Microb Cell Fact 12:42. doi: 10.1186/1475-2859-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mesak LR, Mesak FM, Dahl MK. 2004. Expression of a novel gene, gluP, is essential for normal Bacillus subtilis cell division and contributes to glucose export. BMC Microbiol 4:13. doi: 10.1186/1471-2180-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam VM, Daruwalla KR, Henderson PJ, Jones-Mortimer MC. 1980. Proton-linked d-xylose transport in Escherichia coli. J Bacteriol 143:396–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahlem C, Huisman W, Heslund G, Dahms AS. 1982. Purification and properties of a periplasmic d-xylose-binding protein from Escherichia coli K-12. J Biol Chem 257:2926–2931. [PubMed] [Google Scholar]
- 43.Khankal R, Chin JW, Cirino PC. 2008. Role of xylose transporters in xylitol production from engineered Escherichia coli. J Biotechnol 134:246–252. doi: 10.1016/j.jbiotec.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Hasona A, Kim T, Healy FG, Ingram LO, Shanmugam KT. 2004. Pyruvate formate lyase and acetate kinase are essential for anaerobic growth of Escherichia coli on xylose. J Bacteriol 186:7593–7600. doi: 10.1128/JB.186.22.7593-7600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daruwalla KR, Paxton AT, Henderson PJF. 1981. Energization of the transport systems for arabinose and comparison with galactose transport in Escherichia coli. Biochem J 200:611–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desai TA, Rao CV. 2010. Regulation of arabinose and xylose metabolism in Escherichia coli. Appl Environ Microbiol 76:1524–1532. doi: 10.1128/AEM.01970-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batt CA, Bodis MS, Picataggio SK, Claps MC, Jamas S, Sinskey AJ. 1985. Analysis of xylose operon regulation by Mud (Apr, lac) fusion: trans effect of plasmid coded xylose operon. Can J Microbiol 31:930–933. doi: 10.1139/m85-174. [DOI] [Google Scholar]
- 48.Briggs K, Lancashire W, Hartley B. 1984. Molecular cloning, DNA structure and expression of the Escherichia coli d-xylose isomerase. EMBO J 3:611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sprenger GA. 1995. Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch Microbiol 164:324–330. [DOI] [PubMed] [Google Scholar]