Abstract
The fungal kingdom is replete with unique adaptive capacities that allow fungi to colonize a wide variety of habitats, ranging from marine habitats to freshwater and terrestrial habitats. The diversity, importance, and ecological roles of marine fungi have recently been highlighted in deep-subsurface sediments using molecular methods. Fungi in the deep-marine subsurface may be specifically adapted to life in the deep biosphere, but this can be demonstrated only using culture-based analyses. In this study, we investigated culturable fungal communities from a record-depth sediment core sampled from the Canterbury Basin (New Zealand) with the aim to reveal endemic or ubiquist adapted isolates playing a significant ecological role(s). About 200 filamentous fungi (68%) and yeasts (32%) were isolated. Fungal isolates were affiliated with the phyla Ascomycota and Basidiomycota, including 21 genera. Screening for genes involved in secondary metabolite synthesis also revealed their bioactive compound synthesis potential. Our results provide evidence that deep-subsurface fungal communities are able to survive, adapt, grow, and interact with other microbial communities and highlight that the deep-sediment habitat is another ecological niche for fungi.
INTRODUCTION
Marine environments encompass coastal and open-ocean water columns and sediments, including the deep-marine subsurface. These environments harbor a broad diversity of microorganisms involved in biogeochemical cycling. There has recently been increasing interest in the diversity of microbial eukaryotes in these habitats, particularly in the subsurface biosphere. Hydrothermal vent sediments (1–4) as well as sediments from centimeters to hundreds of meters below the seafloor (5–13) have been investigated using both molecular and culture-dependent methods. Such integrated approaches have revealed the occurrence and activity of complex fungal communities in these extreme environments. Fungal DNA and RNA signatures as well as cultured isolates provide direct evidence that fungi persist in these marine sediments, although their role in these ecosystems is not yet clearly understood. Most of the fungal molecular signatures and cultured isolates from those habitats were affiliated with the phylum Ascomycota, and a few of them belonged to the phyla Basidiomycota and Chytridiomycota (14). While some fungal sequences are from potentially novel organisms, many sequences are from fungi with close relatives from terrestrial, freshwater, and marine environments (8).
To date, within subseafloor sediments, fungi have been identified from a few centimeters below the seafloor (5) down to 1,740 m below the seafloor (mbsf) (15), but we are still eager to assess whether they persist only as vegetative spores or play an active role(s) in biogeochemical cycling. Recent metatranscriptomic analyses suggest that fungi, while occurring in lower abundance than Bacteria and Archaea, possess the ability to degrade a variety of organic substrates in deep-subseafloor sediments (13).
Limitations on habitability in deep-subseafloor sediments are set by a variety of physical and chemical properties acting both individually and in combination, including low nutrient availability, salinity, hydrostatic and lithostatic pressures, pH, porosity, and temperature. The deep subsurface is defined by microbial ecologists as sediment layers with distinct microbial communities that lack the microbial imprint of water column communities (16). As many molecular signatures representing terrestrial fungi have been detected in deep-marine subsurface sediments, it is now necessary to determine whether such terrestrial microorganisms occurring in this ecosystem are specifically adapted to the extreme in situ conditions.
Microbial interactions between fungi, Bacteria, and Archaea may occur in subsurface sediments, including those resulting from the synthesis and release of bioactive compounds (from antimicrobials to cell cycle blockers). Marine fungi from coastal waters are known to synthesize a broad range of bioactive natural compounds with interesting bioactivities (17), including polyketides, peptides, alkaloids, and terpenoids. Consequently, the deep subseafloor may be an untapped reservoir of microorganisms producing biomolecules that may be useful for biotechnological applications.
Here we present the first culture-based analysis of fungi isolated from deep-subsurface sediments. The purpose of this survey was (i) to determine the distribution pattern of culturable fungal communities along a sediment core from the Canterbury Basin near New Zealand, (ii) to characterize their ecophysiological profiles and, consequently, to assess their ability to withstand harsh in situ conditions that include elevated hydrostatic pressure, high salinity, and a range of temperatures, and (iii) to evaluate their genetic potential for the synthesis of bioactive compounds.
MATERIALS AND METHODS
Site description and sediment sampling.
Sediment samples were obtained from a core collected in the Canterbury Basin on the eastern margin of the South Island of New Zealand at site U1352 (44°56′26.62″S, 172°1′36.30″E). This core reached a depth of 1,927.5 mbsf and was drilled during the Integrated Ocean Drilling Program (IODP) Leg 317 Expedition on DS JOIDES Resolution in a water depth of 344 m. Fluorescent microspheres were used as tracers for contamination during drilling. Samples were processed under strict contamination control conditions onboard and onshore, and only samples with undetectable contamination were used for this study (18). Core subsampling was performed onboard under sterile conditions, and only the center parts of unconsolidated sediments and intact pieces of rocks were kept for microbiological analyses, as reported elsewhere (18). Subsamples were immediately transferred into a sterile Schott bottle sealed with a butyl rubber stopper and were stored at 4°C under a nitrogen atmosphere for later cultivation. A recent culture-independent study of the same core did not detect contamination of those sediment samples (15, 19). For this study, 11 samples of sediment and sedimentary rock collected at depths ranging from 4 to 1,884 mbsf were analyzed (Table 1).
TABLE 1.
Culture collection of fungi from deep-subseafloor sediments
| Sample identifier | Core depth (mbsf) | No. of genera/no. of strains isolated: |
||
|---|---|---|---|---|
| In total | With enrichment under hydrostatic pressure | Under atmospheric pressure | ||
| 1H3 | 4 | 9/26 | 7/16 | 4/10 |
| 2H5 | 12 | 7/17 | 5/12 | 3/5 |
| 3H5 | 21 | 5/13 | 5/12 | 1/1 |
| 4H1 | 25 | 7/25 | 7/25 | 0 |
| 5H1 | 34 | 14/38 | 11/31 | 4/7 |
| 5H3 | 37 | 9/26 | 5/15 | 4/11 |
| 15H4 | 137 | 6/13 | 6/13 | 0 |
| 48X3 | 403 | 5/12 | 4/11 | 1/1 |
| 88X1 | 765 | 3/12 | 2/11 | 1/1 |
| 99R1 | 1,478 | 0 | 0 | 0 |
| 144R3 | 1,884 | 1/1 | 0 | 1/1 |
| Total | 21a/183 | 13/146 | 12/37 | |
Twenty-one genera plus one uncultured Agaricomycetes isolate.
Enrichment conditions mimicking the in situ natural environment.
Five different media were used for isolation of filamentous fungi and yeasts under atmospheric pressure: malt extract agar, potato dextrose agar, cornmeal agar, Sabouraud dextrose agar, and Czapek Dox agar. The media were diluted 1:5 to simulate the low nutrient conditions of deep sediments, as described elsewhere (5), and were formulated with and without 3% (wt/vol) sea salts and with and without antibiotics (500 mg/liter chloramphenicol, 200 mg/liter penicillin). Samples from 4, 12, 21, 25, 34, 37, and 137 mbsf were incubated at 5°C, 15°C, and 25°C; samples from 403 and 765 mbsf were incubated at 15°C, 25°C, and 30°C; and samples from 1,478 and 1,884 mbsf were incubated at 35°C, 45°C, and 55°C.
Three different media were used for the isolation of fungi under elevated hydrostatic pressure, i.e., Sabouraud dextrose broth, marine broth, and potato dextrose broth (PDB), diluted 1:5 with or without sea salts and antibiotics, as described above. Enrichment cultures were performed under aerobic conditions in sterile syringes, and the media were saturated with dissolved oxygen. Each syringe contained 5 ml of medium and 200 μl of sediment sample slurry. The syringes were then transferred into a high-pressure incubation system and incubated at different temperatures. Samples from 4, 12, 21, 25, 34, and 37 mbsf were incubated at 25°C and 4 MPa; samples from 137, 403, and 765 mbsf were incubated at 30°C and 11 MPa; and samples from 1,478 and 1,884 mbsf were incubated at 45°C and 37 MPa. After 14 days of enrichment under hydrostatic pressure, 100 μl of culture was dispensed into agar plates and incubated at the same temperature used for the original cultivation until fungal growth. For each condition, pure cultures were obtained by streaking and central picking on different enrichment media for fungi showing a distinct morphology.
As a control, nutrient plates were exposed within a laminar flow hood during all isolation procedures. No fungal colonies were obtained on control plates, indicating the absence of aerial contamination during isolation.
DNA extraction.
DNA extraction from all cultures was performed using a FastDNA spin kit (MP Biomedicals) following the manufacturer's recommendations for fungi. DNA quality and quantity were assessed using a NanoDrop spectrophotometer (NanoDrop Technologies) prior to gene-specific PCR amplifications.
MSP-PCR.
In order to cluster the isolates, mini- and microsatellite-primed PCR (MSP-PCR) was applied. The primer used for MSP-PCR was the core sequence of phage M13 (GAGGGTGGCGGTTCT) (20). The PCR was performed in 25-μl reaction volumes containing 1× PCR buffer, 2 mM MgCl2, 0.4 mM each deoxynucleoside triphosphate (dNTP), 0.8 mM the primer, 1 U of Taq polymerase, and 15 ng of genomic DNA from each cultured isolate. Amplification consisted of an initial denaturation step at 95°C for 5 min, followed by 40 cycles of 45 s at 93°C, 60 s at 50°C, and 60 s at 72°C and a final extension step of 6 min at 72°C (21). A negative control where DNA was replaced by sterile distilled water was also included. Amplified DNA fragments in 1× Tris-borate-EDTA (TBE) buffer were separated by electrophoresis on a 1.4% (wt/vol) agarose gel (Promega) at 85 V for 3.5 h. On each gel, a 1-kb molecular size marker was used (Promega). DNA banding patterns were visualized under UV transillumination, and picture files were generated using a Quantum ST4 system (Vilbert Lourmat). Genetic profiles were analyzed using BioNumerics software (version 6.6; Applied Maths). All fingerprints were grouped by similarity using the Dice coefficient, and clustering was based on the unweighted-pair group method using average linkages methods. The cophenetic correlation coefficient was calculated. Clusters were generated using an arbitrary cutoff of 75% sequence similarity with minor adjustments (±5%). Some representative sequences were selected from each cluster for further sequencing.
PCR amplification of 18S, ITS, and partial 28S/26S rRNA.
For genetic identification of fungi isolated in our sediment samples, 18S rRNA gene sequences from cultured isolates were amplified with primers NS1 (5′-GTAGTCATATGCTTGTCTC-3′) and SR6 (5′-AAGTAGAAGTCGTAACAAGG-3′) (22). Internal transcribed spacer (ITS) and partial 28S rRNA gene sequences were amplified using primers ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and LR6 (5′-CGCCAGTTCTGCTTACC-3′), respectively (22, 23). Amplifications of the D1/D2 region of the 26S rRNA gene were carried out with primer sets ITS5 (5′-GGAAGTAAAAGTCGTAACAAG-3′)/NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) and LR6 (5′-CGCCAGTTCTGCTTACC-3′)/NL1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′), as described by Gadanho and Sampaio (1). All PCRs were performed in 25-μl reaction volumes containing 1× GoTaq buffer, 0.4 mM dNTPs (Promega), 0.36 μM each primer (Sigma), 2 mM MgCl2, 1 U of GoTaq polymerase (Promega), and 1 μl of genomic DNA. The PCR assay for 18S rRNA gene and ITS sequences included an initial denaturation step at 94°C for 2 min, followed by 30 cycles of 30 s at 94°C, 30 s at 54°C, and 2 min at 72°C and a final extension step of 2 min at 72°C, before holding at 4°C. The PCR assay for the 26S rRNA gene included an initial denaturation step at 94°C for 2 min, followed by 30 cycles of 15 s at 94°C, 30 s at 54°C, and 1 min at 72°C and a final extension step of 2 min at 72°C, before holding at 4°C. A negative control in which DNA was replaced by sterile water was included. PCR products were analyzed by electrophoresis in a 0.8% (wt/vol) agarose gel (Promega) in 1× TBE buffer at 90 V for 90 min and stained with GelRed. DNA banding patterns were visualized under UV transillumination, and picture files were generated using a Quantum ST4 system (Vilbert Lourmat).
Sequencing and phylogenetic analyses.
Fungal small-subunit (SSU) ribosomal DNA (rDNA) amplicons were sequenced with forward primers NS1 (5′-GTAGTCATATGCTTGTCTC-3′) and NS3 (5′-GCAAGTCTGGTGCCAGCAGCC-3′) and reverse primers NS6 (5′-AACTTAAAGGAATTGACGGAA-3′) and SR6 (5′-AAGTAGAAGTCGTAACAAGG-3′) (22). For ITS and long-subunit (LSU) rDNA, primers ITS1, ITS4 (22), and LR6 (23) were used. The D1/D2 region of 26S rDNA was sequenced using NL1 on the ITS5-NL4 fragments and NL4 on the NL1-LR6 fragments (1). Sequences were obtained by use of the BigDye Terminator technology (Applied Biosystems) on a Biogenouest sequencing platform at the Station Biologique de Roscoff (http://www.sb-roscoff.fr/plateformes-techniques/genomique-sbr.html). The chromatograms obtained were translated into nucleotide sequences with BioNumerics software (version 6.6; Applied Maths). After the sequences were cleaned, they were imported into MEGA (version 5.0) software (24). Each sequence was analyzed using the BLASTn program to determine the nearest relatives in GenBank (25). Similarities between sequences were assessed using pairwise distances calculated with the MEGA (version 5.0) program. The sequences were trimmed to ensure that all sequences had the same start point and endpoint and were aligned using ClustalW (version 1.83) software (26). Phylogenetic analyses were performed by the use of maximum likelihood criteria with 500 bootstrap replicates in the MEGA (version 5.0) program (24). Shannon indices were calculated on the basis of the number and abundance of species for each depth using the vegan package in R.
Ecophysiological characterization.
Ecophysiological characterization was performed as described by Joubert et al. (27). Briefly, suspensions of conidia from selected isolates and dilutions of those isolates were made using PDB to achieve a final concentration of suspended spores of between 1 × 105 and 5 × 105 per ml. A 96-well plate was filled with the calibrated suspensions (300 μl/well) and incubated in a Laser nephelometer (NEPHELOstar Plus; BMG Labtech) for at least 33 h at 15, 25, 30, and 35°C. All experiments were performed in triplicate. Growth rates (kilo-relative nephelometric units [RNU] per hour) were determined for each culture condition, which included 0, 1.5, 3, and 4.5% sea salts at each temperature for detection of adaptation of these fungal isolates to subsurface in situ conditions (3% salinity all along the core and increasing temperatures, i.e., about 30 to 50°C per kilometer).
Occurrence of type I and III PKS, NRPS, hybrid PKS-NRPS, and TPS genes.
The presence of genes involved in the biosynthetic pathways of secondary metabolites was investigated. Type I polyketide synthase (PKS I), type III polyketide synthase (PKS III), nonribosomal peptide synthetase (NRPS), PKS-NRPS hybrid, and terpene synthase (TPS) genes were amplified using the following degenerate primer pairs with d-inosine modifications (indicated by i): (i) primers KAF1 (GARKSiCAYGGiACiGGiAC) and KAR1 (CCAYTGiGCiCCRTGiCCiGARAA) (28) were used for amplification of type I PKS genes, (ii) primers XKS1 (TTYGAYGCiBCiTTYTTYRA) and XKS2 (CRTTiGYiCCiCYDAAiCCAAA) (27) were used for amplification of PKS-NRPS hybrid genes, (iii) primers AUG003 (CCGGCACCACCGGNAARCCHAA) and AUG007 (GCTGCATGGCGGTGATGSWRTSNCCBCC) (29) were used for amplification of NRPS genes, (iv) primers TPS1 (GCiTAYGAYACiGCiTGGGT) and TPS2 (RAAiGCATiGCiGTRTCRTC) (30) were used for amplification of TPS genes, and (v) primers CHS1 (GAYTGGGCiVTNCAYCCBGGiGGD) and CHS2 (YTCNAYNKTRAKVCCiGGVCCRAA) were used for amplification of type III PKS genes. Primers targeting type III PKS genes were specifically designed for this study using ClustalW (version 1.83) (26). We selected 18 fungal strains from the NCBI database known to have those genes. Each degenerate primer was designed against two conserved domains of the C terminus of the protein [DWALHPGG and FGPG(L/I)(N/T/S)(M/V/I)E]. All PCRs were performed in 25-μl reaction mixtures including 1× GoTaq buffer, 2 mM MgCl2, 0.4 mM each dNTP (Promega), 0.6 μM each primer (Eurogentec) (except for 1 μM each primers KAF1/KAR2 and XKS1/XKS2), 1 U of GoTaq polymerase (Promega), and 100 ng of genomic DNA. Amplification of type I PKS and PKS-NRPS hybrid genes included an initial denaturation step at 98°C for 2 min, followed by 5 cycles of 98°C for 1 min, 45°C for 1 min, and 72°C for 2 min, followed by 30 cycles of 98°C for 1 min, 50°C for 1 min, and 72°C for 2 min and a final extension step for 10 min at 72°C. Amplification of type III PKS and TPS genes included an initial denaturation step at 98°C for 2 min, followed by 5 cycles of 98°C for 1 min, 48°C for 1 min, and 72°C for 2 min, followed by 30 cycles of 98°C for 1 min, 51°C for 1 min, and 72°C for 2 min and a final extension step for 10 min at 72°C. Amplification of the NRPS gene included an initial denaturation step at 2 min at 98°C, followed by 35 cycles of 1 min at 98°C, 1 min at 50°C, and 2 min at 72°C and a final extension step for 10 min at 72°C. For each experiment, we included two positive controls and two negative controls. For positive controls, we used DNA extracted from Aspergillus flavus (UBOCC-A-101081) and Penicillium chrysogenum (CBS 306.48), both of which are known to have PKS I, PKS III, NRPS, PKS-NRPS hybrid, and TPS genes in their genomes. For negative controls, we used DNA extracted from Saccharomyces cerevisiae (UBOCC-A-201006), as no PKS I, PKS III, NRPS, PKS-NRPS hybrid, or TPS gene has ever been detected in its genome (31). The other negative control was sterile water instead of DNA.
Nucleotide sequence accession numbers.
All fungal isolates have been deposited in the Université de Bretagne Occidentale Culture Collection (UBOCC) and can be found in GenBank under accession numbers KM222200 to KM222349 and KM232435 to KM232507.
RESULTS
Culturable fungi isolated.
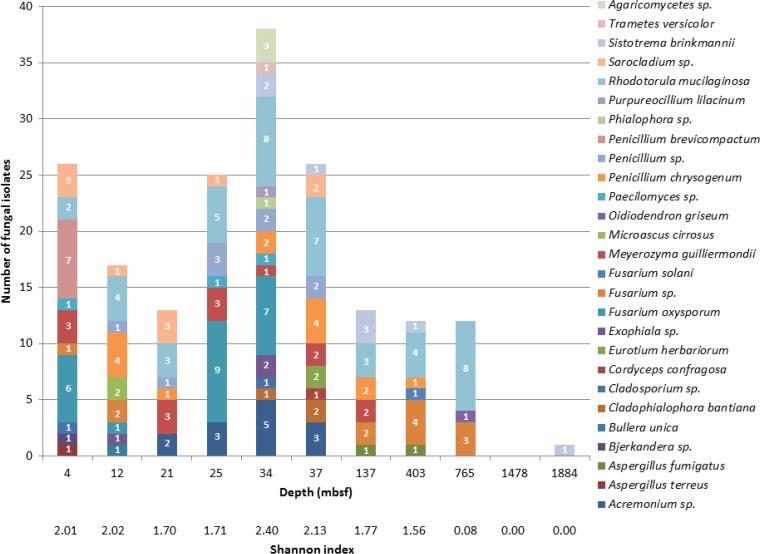
A total of 11 samples from 4 to 1,884 mbsf (Table 1) were tested for culturable fungi. Almost all samples (10 of 11, 91%) yielded fungal isolates (Fig. 1). Under the expected in situ hydrostatic pressures, 24 syringe cultures were processed per sample, with different conditions varying by temperature (1 temperature per sample according to the expected in situ temperature), salinity (0% and 3%), and nutrients (3 different media) with and without antibiotics being tested. Each combination of conditions was tested in duplicate. Under atmospheric pressure, 120 petri dishes were processed per sample, with different conditions varying by temperature (3 temperatures per sample according to the expected in situ temperature), salinity (0% and 3%), and nutrients (5 different media) with and without antibiotics being tested in duplicate. The final number of fungal isolates obtained was 183, and their distribution across the sediment core is shown in Table 1. The greatest number of isolates was obtained from sediments at 4 and 12 mbsf and from sediments at 25 to 37 mbsf. Only one isolate was obtained from the deepest layers, between 1,478 and 1,884 mbsf, the sedimentary rock (Fig. 1). Results were analyzed using two-way analysis of variance (ANOVA) to assess the effect of depth on isolation abundance. The effect of depth was highly significant (F = 3.48, P < 0.001). The Shannon index, which is a well-known parameter used to express the diversity of communities, was calculated for each depth and corroborates these findings, showing that the sample from 34 mbsf produced the most diverse culture collection. Thus, sediment depth clearly influenced the isolation rate and confirms that culturable fungi are present primarily at specific depths along the core. Additionally, it was more difficult to obtain positive cultures of samples from deeper layers. Indeed, only one isolate was obtained from 1,884 mbsf, and no fungus was ever isolated from a sample from 1,478 mbsf. This strong statistical evidence for the uniqueness of communities at different depths supports the idea that our isolates were not contaminants.
FIG 1.
Species richness along the core. Shannon diversity indices allow the identification of the relative complexity of fungal communities at different depths.
Morphological characteristics allowed us to separate filamentous fungal forms from unicellular yeasts. Observations revealed branching hyphae and spores for filamentous fungi (124 isolates) and budding cells for yeasts (59 isolates). Eighty-six isolates were obtained on culture media amended with 3% sea salts, and 97 were isolated on culture media without sea salts. Thirty-seven isolates were obtained under atmospheric pressure, and 146 were obtained after enrichment under hydrostatic pressure. Among the different conditions used to isolate deep-sea fungi, hydrostatic pressure appeared to be important for the retrieval of fungal isolates.
Clustering and phylogenetic diversity.
As a dereplication strategy, the MSP-PCR fingerprinting method was performed. We used similarity clusters to generate two dendrograms, one for filamentous fungi and one for yeasts. Using this strategy, 57 clusters, 48 for filamentous fungi and 9 for yeasts, were established as shown in Fig. S1 and S2 in the supplemental material. The minor fingerprint variability found within MSP-PCR clusters was defined as intraspecific heterogeneity. Several representatives from each cluster were selected for identification using SSU, ITS, and partial LSU rRNA sequencing; 72 filamentous fungi and 40 yeast isolates, or about 60% of the whole culture collection, were sequenced. Sequence analysis validated the MSP-PCR parameters used (cutoff of 75%).
Filamentous fungi.
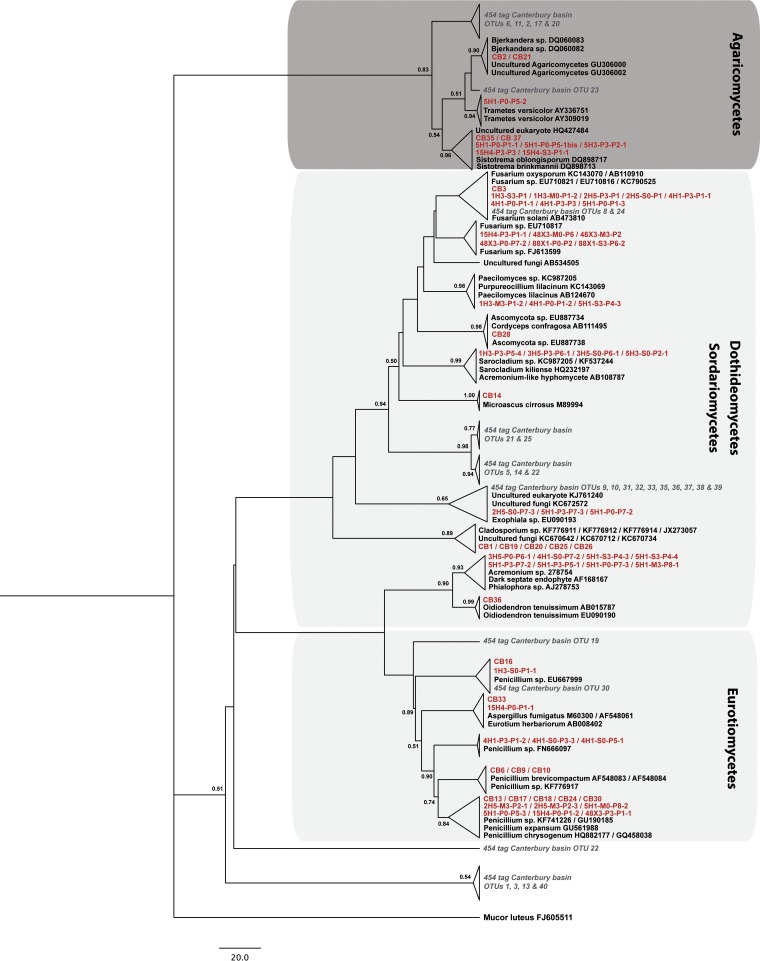
On the basis of the gene encoding SSU rRNA, representatives from each cluster were taxonomically assigned using the BLASTn program (25), and phylogenetic analyses integrating the closest relatives in GenBank were performed. When sequences from our culture collection could not be identified using 18S rRNA genes, complementary genetic analyses were performed using ITS and partial 28S rRNA genes. Filamentous fungal isolates were distributed among the Ascomycota and Basidiomycota, with the Ascomycota representing more than 89% of our culture collection. On the basis of 18S rRNA, ITS, and 28S rRNA gene sequence analysis, isolates belonging to the Ascomycota were clustered into 15 genera (Acremonium, Aspergillus, Cladophialophora, Cladosporium, Cordyceps, Eurotium, Exophiala, Fusarium, Microascus, Oidiodendron, Paecilomyces, Penicillium, Phialophora, Purpureocillium, and Sarocladium), isolates belonging to the Basidiomycota were clustered into 3 genera (Bjerkandera, Sistotrema, and Trametes), and 1 uncultured Agaricomycetes isolate was found (Fig. 2).
FIG 2.
Phylogenetic tree of deep-sea fungal isolates (red) and 454 pyrotags (gray) obtained by analysis of SSU rRNA genes. The tree topology is based on maximum likelihood criteria and a ClustalW (version 1.83) alignment. Bootstrap values greater than 50% are shown at the nodes. Phylogenetic data of Ciobanu et al. (15) were integrated as 454 pyrotags. Mucor luteus (GenBank accession number FJ605511), which belongs to the phylum Glomeromycota, was used as an outgroup. Dark and light gray boxes, Basidiomycota and Ascomycota, respectively. OTU, operational taxonomic unit.
Yeasts.
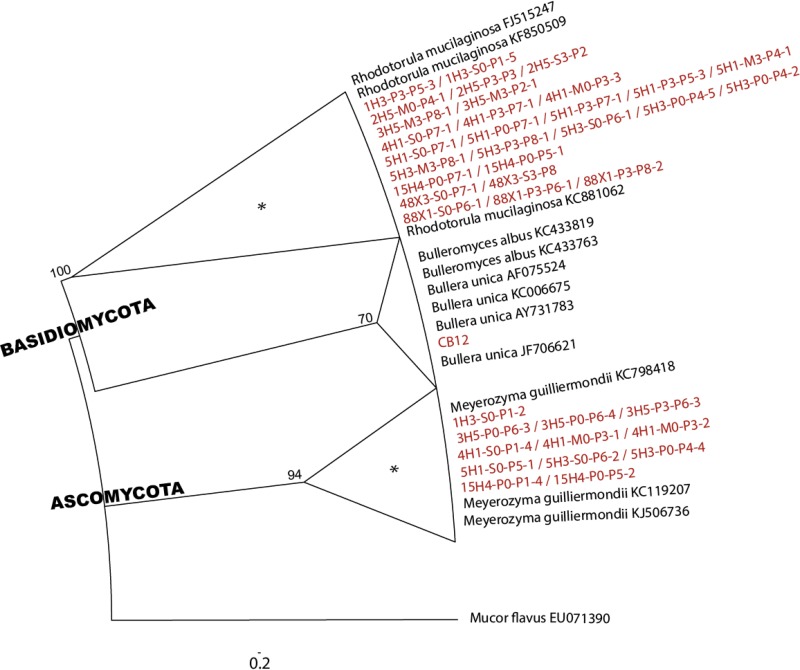
Phylogenetic analysis of the D1/D2 domain of the 26S rRNA gene revealed that cultured yeasts were distributed among the ascomycetous genus Meyerozyma and basidiomycetous genera Rhodotorula and Bullera. With 45 isolates, members of the Basidiomycota phylum represented more than 76% of our yeast culture collection (Fig. 3).
FIG 3.
Phylogenetic tree of deep-sea yeast isolates (red) obtained by analysis of the D1/D2 domain of the 26S rRNA gene. The topology is based on the maximum likelihood method and a ClustalW (version 1.83) alignment. Bootstrap values greater than 50% are shown at the nodes. Mucor flavus (GenBank accession number EU071390), which belongs to the phylum Zygomycota, was used as an outgroup. Clusters highlighted with an asterisk were also retrieved in molecular analysis data sets (15, 19).
Ecophysiological characterization.
Laser nephelometry was used as a high-throughput screening method for the ecophysiological characterization of fungal isolates obtained from deep-subsurface sediments. Different filamentous fungal isolates that were from the same cluster (on the basis of the 18S rRNA, ITS, and 28S rRNA gene sequences) but that were obtained from different depths were selected: 5 Fusarium spp. from 4 to 765 mbsf, 4 Penicillium spp. from 12 to 403 mbsf, 3 Paecilomyces spp. from 4 to 34 mbsf, 2 Exophiala spp. from 12 to 34 mbsf, and 1 Acremonium sp. at 25 mbsf. Maximum specific growth rates (μmax) were calculated for each isolate under all sets of conditions (4 temperatures and 4 salinities).
Most of the isolates were determined to be nonhalophiles (for which the maximal growth rate was observed in the absence of sea salts and decreasing growth rates were observed with increasing concentrations of sea salts in the media) or halotolerant (in which the isolates were able to grow in the absence as well as in the presence of sea salts) at 25°C and 30°C (Fig. 4). No useful information was obtained from incubations at 15°C, as most of the isolates were still in lag phase after 35 h. Only one Fusarium sp. collected at 4 mbsf was able to grow at 15°C in less than 35 h, and this organism displayed the same general trend at 15°C as it did at 25°C.
FIG 4.
Physiological analysis of filamentous fungi (A) and yeast (B) isolates. Growth was measured at different temperatures (25°C, 30°C, and 35°C) and different sea salt concentrations (0, 1.5, 3, and 4.5%), and the general growth trend is shown as 3-part bars representing minimum, medium, and maximum growth. Blank cells indicate that the results were not determined. Red, green, and blue dots represent, respectively, nonhalophilic, halotolerant, and halophilic fungal isolates.
Isolates affiliated with Fusarium spp. were defined as nonhalophiles at 25 and 30°C, regardless of the depth from which the sample was obtained. Isolates affiliated with Penicillium spp. were defined as halotolerant at 25°C, with better growth being observed in the presence of 3% sea salts. Isolates affiliated with Paecilomyces spp., Exophiala spp., and Acremonium spp. displayed the same nonhalophilic pattern at 25°C. Interestingly, at 30°C, these isolates were halotolerant and even halophilic in the case of the deepest Exophiala spp. and Acremonium spp. This ecophysiological shift was clearly observed at 35°C, where all isolates selected for ecophysiological characterization were defined as halophiles or halotolerant, in the case of the Paecilomyces spp. from 25 mbsf. This shift toward halophily was much more significant for Penicillium spp. recovered from deeper sediments (137 and 403 mbsf).
Different yeasts from the same cluster but from different sampling depths were also selected for ecophysiological analysis. Using the same methodology, their maximum specific growth rates at one temperature (30°C) and 4 salinities were calculated. All isolates affiliated with Rhodotorula spp. (n = 8) were defined as nonhalophiles. Indeed, growth rates decreased when the sea salt concentration increased. Isolates affiliated with Meyerozyma spp. were much more diverse, on the basis of their responses. Isolates obtained from between 4 and 25 mbsf were nonhalophiles, isolates obtained from between 37 and 137 mbsf were halotolerant, and one isolate obtained from 21 mbsf was a halophile (no growth without sea salts).
The screening strategy allowed us to assess ecophysiological differences between isolates using temperature and salinity as important in situ parameters. Our results suggest the adaptation of some fungal isolates, possibly of terrestrial origin, to deep-subsurface conditions.
Presence of genes involved in the synthesis of bioactive compounds.
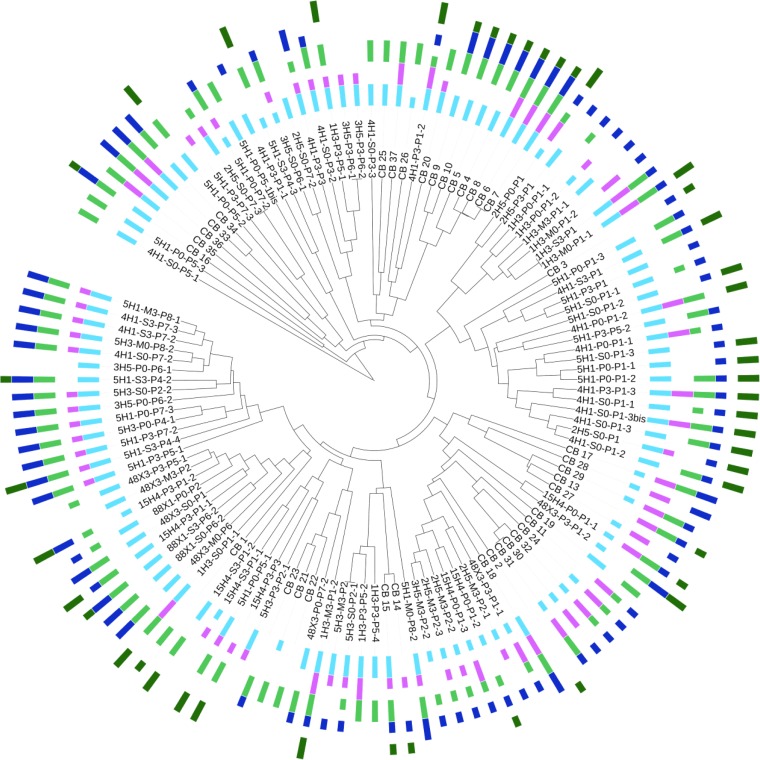
All fungal isolates were investigated for genes coding for type I and type III PKSs, NRPSs, PKS-NRPS hybrids, and TPS (Fig. 5 and 6). Among the 124 filamentous fungi isolated, all harbored at least one of those genes for bioactive compounds: 6 isolates (4.8%) had 1 gene, 23 (18.5%) had 2 genes, 30 (24.2%) had 3 genes, 53 (42.7%) had 4 genes, and 12 (9.7%) had all targeted genes (Fig. 5). Among the 59 yeasts isolated, 52 harbored at least one of those genes: 16 isolates (27.1%) had 1 gene, 7 (11.8%) had 2 genes, 12 (20.3%) had 3 genes, and 17 (28.8%) had 4 genes. Seven yeasts did not show evidence of having any of the targeted genes (Fig. 6). Contrary to the results from filamentous fungi, genes coding for PKS-NRPS hybrids were not detected in any of our yeast isolates. This result appears to be consistent with a previous report indicating the absence of PKS-NRPS hybrid genes in yeast genomes (31). Filamentous fungal isolates from deep-marine subsurface sediments appear to have a greater potential for synthesis of bioactive compounds than yeasts.
FIG 5.
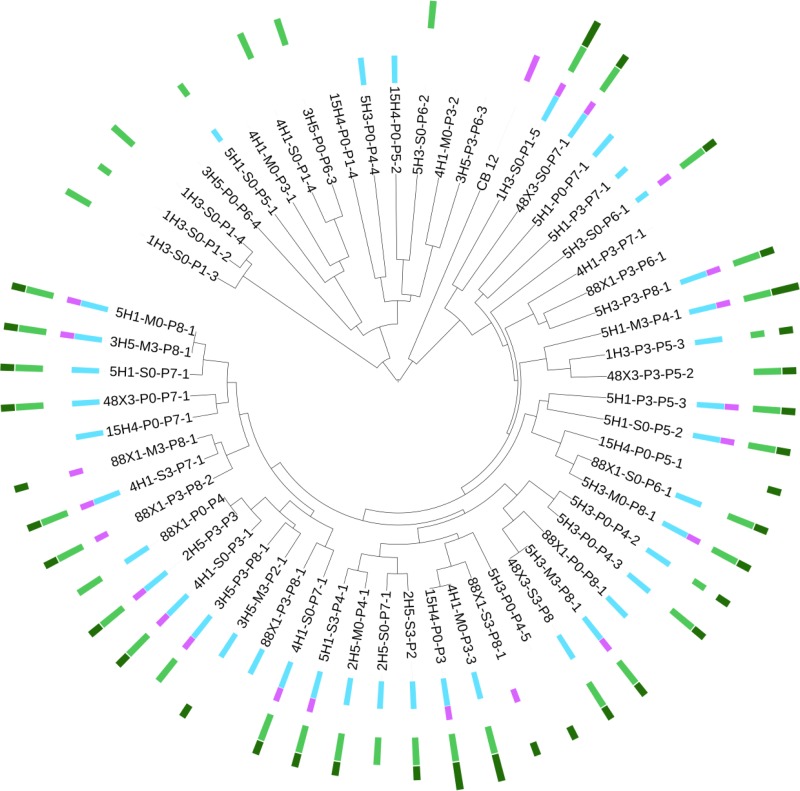
Presence/absence of genes encoding type I and III PKSs, NRPSs, PKS-NRPS hybrids, and TPS in filamentous fungi. Filamentous fungal MSP-PCR fingerprints coupled with type I PKS gene (light blue), type III PKS gene (pink), NRPS gene (light green), PKS-NRPS hybrid gene (dark blue), and TPS gene (dark green) occurrences are presented using an aligned multivalue bar chart (short bar, only one gene; long bar, several genes). This figure was generated using Interactive Tree of Life (version 2) (59).
FIG 6.
Presence/absence of genes coding type I and III PKSs, NRPSs, PKS-NRPS hybrids, and TPS in yeasts. Yeast MSP-PCR fingerprints coupled with type I PKS gene (light blue), type III PKS gene (pink), NRPS gene (light green), PKS-NRPS hybrid gene (dark blue), and TPS gene (dark green) occurrences are presented using an aligned multivalue bar chart (short bar, only one gene; long bar, several genes). This figure was generated using the Interactive Tree of Life (version 2) (59).
The relative occurrence of PKS I, PKS III, NRPS, PKS-NRPS hybrid, and TPS genes at depths ranging from 4 to 1,884 mbsf was analyzed (see Table S2 in the supplemental material). No trend was evident for yeasts, but filamentous fungal isolates from deeper layers seemed to harbor fewer genes for bioactive compounds than isolates from shallower depths.
DISCUSSION
The aim of this study was to assess deep-marine subsurface sediments for the presence of culturable fungi and to determine their genetic potential for bioactive compound synthesis. We examined subsurface sediments collected from the Canterbury Basin at 11 depths ranging from 4 mbsf to 1,884 mbsf. The culture collection of filamentous fungi and yeasts obtained provides insights into the culturable fraction of in situ fungal communities colonizing the subseafloor at this site.
Controlling contamination.
The potential for microbiological contamination of samples was monitored carefully during the IODP Leg 317 Expedition and during subsequent laboratory procedures (described above). The sampling procedures and contamination controls used during this expedition have been previously described (15). One of our samples (from 1,478 mbsf) did not produce cultivable fungi (Fig. 1), in spite of the fact that the same methods were applied for sampling and organism isolation by culture for samples from all depths. Although we cannot definitively rule out the possibility that one or more of our cultured isolates represents a contaminant, the fact that one of our samples did not produce cultivable fungi is further support for the absence of contamination during our sampling and culturing efforts. Our ANOVA analysis and Shannon diversity indices also clearly indicate that the distribution of our isolates was dependent on the sampling depth. This strongly supports the idea that the recovered fungi were not contaminants. The high rate of recovery of fungal isolates using enrichments under elevated hydrostatic pressure conditions further indicates that these fungi are able to grow under in situ pressure and suggests that our established culture collection consists of indigenous taxa.
Distribution pattern of fungal communities.
The phylogenetic diversity of fungi revealed in this study is relatively low compared to that in well-characterized organic matter-rich terrestrial or marine habitats (i.e., soils, plants, mangroves, etc.). Despite that fact, there was a greater diversity from the upper sediment layers down to 37 mbsf, with 25 different species representing more than 92% of the whole diversity within our fungal culture collection being recovered. In samples from greater depths from 137 to 1,884 mbsf, only 8 different species, representing about 30% of the recovered diversity, were observed in our culture collection (Fig. 1). This could be explained by a reduction in accessible organic matter with depth. Indeed, the organic matter quality changed from relatively labile material in shallow sediments to more stable protokerogen at greater sediment depths (15). Among the 27 species recovered in our cultures, some were isolated from a unique depth and several species were shared between 2 or more depths. The highest rate of occurrence of a species along the core was reported for Rhodotorula mucilaginosa, isolated at 9 depths ranging from 4 to 765 mbsf. A maximal overlap between 2 depths was observed for samples from 34 and 37 mbsf, with 7 shared species, i.e., an Acremonium sp., Cladophialophora bantiana, Penicillium chrysogenum, a Penicillium sp., Sistotrema brinkmannii, and the yeasts Meyerozyma guilliermondii and Rhodotorula mucilaginosa. Interestingly, a Phialophora sp., Purpureocillium lilacinum, Trametes versicolor, and some species of the Agaricomycetes were isolated only from the sample from 34 mbsf and seemed to be depth dependent. Beyond 765 mbsf, the diversity within our culture collection decreased considerably, with only 4 different species being recovered, including isolates belonging to the Fusarium genus, one isolate identified as Oidiodendron griseum that was isolated only from 765 mbsf, and the red yeast Rhodotorula mucilaginosa. One isolate, Sistotrema brinkmannii, was obtained from 1,884 mbsf, and it was also retrieved from samples recovered at 34 and 403 mbsf.
Diversity of fungal communities.
The Ascomycota dominated our filamentous fungal culture collection, whereas the Basidiomycota dominated our yeast collection. This result appears to be consistent with the findings of another phylogenetic analysis of culturable fungi from deep-sea sediments of the Central Indian Basin (32). In our study, among the yeasts, Rhodotorula mucilaginosa was the most abundant species isolated from samples from depths ranging from 4 to 765 mbsf. The red yeast Rhodotorula mucilaginosa is widely distributed and has already been isolated from deep-sea sediments and basaltic crust (32–34). Bullera unica, isolated from 12 mbsf in our study, was the only isolate belonging to the Tremellomycetes. Interestingly, Bullera unica was previously isolated from New Zealand seawater (35). Meyerozyma guilliermondii was the only ascomycetous yeast in our culture collection isolated from 4 to 137 mbsf. Culturable representatives of this opportunistic pathogen have been detected in marine environments, including hydrothermal vents (2) and deep-sea volcanoes (36), and from Canterbury Basin sediments by culture-independent methods (19). Yeasts were previously suggested to be the dominant marine fungal forms on the basis of fungus-specific clone libraries from marine water column and sediment environments (37). Here, yeasts appeared to be able to adapt to and colonize the deep subseafloor, but our results suggest that filamentous fungi are more diverse in marine habitats than previously suggested (37). In agreement with the findings of molecular studies, the classes Eurotiomycetes and Sordariomycetes are the fungal taxa most frequently detected from deep-sea environments (14).
Among the Sordariomycetes, Fusarium oxysporum is the most abundant taxon. F. oxysporum has already been shown to grow under marine conditions (38) and was described to be a denitrifying fungus in marine sediments (39). The other well-represented members of the Sordariomycetes were (i) Fusarium solani, recently detected from Canterbury Basin sediments using culture-independent methods (19), (ii) representatives of Acremonium spp. already reported from several marine environments, including deep-sea sediments (10, 32, 38), (iii) Sarocladium spp. previously isolated from brine sediments (40), (iv) representatives of Paecilomyces spp. formerly isolated from deep-sea coral (41) and from the first centimeter of coastal marine sediments off Goa, India (39), and (v) Microascus spp. that may be opportunistic pathogens of deep-sea animals, such as corals (42). Phialophora spp., Purpureocillium lilacinum, Cordyceps confragosa, and Oidiodendron griseum were less represented in our culture collection.
Among the Eurotiomycetes, Penicillium was the most frequently isolated genus. Molecular signatures of this genus were previously found in the Canterbury Basin deep sediments (19). Fourteen fungal isolates of Penicillium chrysogenum were obtained. This species was described to be the dominant phylotype among the Ascomycota in a DNA-based clone library prepared from deep-sea methane cold-seep sediments (9). The other well-represented Eurotiomycetes were (i) Aspergillus fumigatus and Aspergillus terreus, known to be able to germinate under hydrostatic pressure (5), and (ii) Eurotium herbariorum, previously isolated from other extreme environments, including hypersaline waters of solar salterns (43), the hypersaline Dead Sea (44), hydrothermal sediments (2), and deep-sea sediments (11).
The class Dothideomycetes was represented in our culture collection by 3 isolates belonging to the genus Cladosporium. Among the Basidiomycota, Agaricomycetes were the most frequently recovered, with isolates being affiliated with Bjerkandera spp., Sistotrema brinkmannii, Trametes versicolor, and uncultured Agaricomycetes. Although the cultivable representatives of Agaricomycetes appear to be rare in marine environments (36), the molecular signatures of Agaricomycetes have been detected in deep-sea environments (3, 9, 11, 45) and Agaricomycetes have been identified to be the dominant fungal class in mangrove habitats (46).
All of the fungal taxa isolated from Canterbury Basin are well-known in terrestrial environments, raising intriguing ecological questions regarding the abilities of terrestrial fungi to adapt to deep-subsurface conditions that include confinement, pressure, temperature, access to only refractory organic matter, etc. The fungal taxa detected here using culture-dependent methods can be compared with fungal 454 pyrotags previously obtained from the same sediment core (15, 19). The two methods provided different pictures of diversity. Only fungal signatures and isolates belonging to the genera Exophiala, Fusarium, Penicillium, Rhodotorula, and Meyerozyma were detected by both culture-independent and culture-dependent methods. Culture-based and molecular diversity survey approaches appear here to provide two complementary sources of information. The fact that some isolates have not been detected by molecular methods could be explained by several reasons, including the fact that fungal spores are lysis resistant, bias in universal primers, and the use of a limited (or insufficient) sequencing depth. Efficient nucleic acid extraction from vegetative spores may be an important factor to consider. Indeed, previous results obtained by molecular analysis based on RNA suggest that few persistent subsurface fungal taxa are able to colonize the deep subsurface of the Canterbury Basin (19). Our results strongly support the importance of applying a polyphasic approach using culture-dependent and culture-independent methods, which together generate a general and comprehensive view of deep-subsurface fungal diversity. One of the main advantages of handling fungal cultures is that the specific adaptations that they use to cope with different stresses can be assessed. Ecophysiological analyses provide concrete and relevant data that may be used to estimate their adaptations and understand the role of fungal communities in the deep subsurface.
Adaptation.
In our study, we decided to examine the effect of elevated hydrostatic pressure, since this is likely to be one of the most significant physical challenges for microbiota living in the abyss (47). We specifically examined whether cultivation under increased pressure enhanced culture efficiency. We also performed ecophysiological analyses to examine the impacts of temperature and salinity.
Hydrostatic pressure enhanced the growth of a number of fungal isolates in our culture collection. About 80% of our isolates were obtained with a preliminary enrichment under elevated hydrostatic pressure. Thus, the elevated hydrostatic and lithostatic pressures in deep sediments do not curb the germination and growth of all fungi in the deep subseafloor. Previous studies have demonstrated efficient culturing of filamentous fungi and yeasts under elevated hydrostatic pressure (5, 11, 32, 48, 49). Although no true piezophilic fungi have been reported to date, some fungi are able to develop mechanisms of adaptation. For example, even the well-known yeast Saccharomyces cerevisiae is able to modify its membrane composition to tolerate high hydrostatic pressure (50), revealing the potential of fungi, including terrestrial ones, to adapt to deep-sea conditions.
Temperature and salinity are two additional significant parameters used to examine the adaptation of fungi to deep-subsurface conditions. Indeed, the temperature increases along the sediment core at a rate of 30 to 50°C per kilometer. The temperature at the bottom of the core hole (at 1,922 mbsf) was estimated to be in the range of from 60 to 100°C (15). Salinities in sediment samples near the seafloor are slightly lower than the salinity of normal seawater at 3.3%, and salinity rapidly declines to 3.0% at 28 mbsf and remains relatively constant at 2.9 to 3.1% over the rest of the core section analyzed to 1,400 mbsf (18). Our ecophysiological analysis revealed shifts along the core. Most of the filamentous fungal isolates analyzed shifted from nonhalophilic to halotolerant when incubation temperatures increased from 25 to 35°C. Only the deepest Fusarium and Penicillium isolates demonstrated a complete shift to halophily when the temperature increased. Such a shift from terrestrially adapted to marine-adapted lifestyles along the core may indicate a transition from shallow layers, where fungi are not specifically adapted but are able to survive, to deeper layers, where fungi are more adapted to higher temperatures and higher salinities. Ecophysiological studies on deep-subseafloor prokaryotes are scarce. Biddle et al. (51) used a low-salt medium (R2A) to culture bacterial strains from deep sediments of the Peru Margin and obtained colonies only from shallow depth layers. This may indicate the occurrence of nonmarine strains in the first layers and endemic or adapted strains at deeper layers, even if the possibility of a decrease in isolation success with depth cannot be ruled out. However, another study described some generalist subseafloor sediment bacteria to be more adapted to different environmental conditions than their surface counterparts (52). Such results strongly support our conclusions.
Fungi detected by culture-based and molecular approaches in the deep-subseafloor biosphere may range from those that are buried alive and are simply dormant to those that adapt to subsurface conditions. The same trend is observed for bacterial endospores that are as abundant as vegetative prokaryotic cells in the deep biosphere (53). Recently, Jørgensen (54) proposed that most deep-subseafloor microbial cells are on physiological standby and that only a few cells may be active at any given time. The metabolic activities of long-buried microbes could be activated due to tectonic activity or if subsurface fluid flows deliver an infusion of new electron donors and acceptors. This hypothesis fits well with our finding that some fungi may possess a range of physiological adaptations for handling different physical and chemical stressors, while others might be microbial zombies (55). Complementary genomic, transcriptomic, and proteomic analyses are needed to better understand the metabolic capabilities of deep-subseafloor fungi.
Biotechnological potential of deep-sediment fungal isolates.
Many marine microorganisms have been screened for novel bioactive compounds (56, 57), and some research has been specifically dedicated to marine fungal secondary metabolites (17). In this study, we examined the secondary metabolite synthesis potential of deep-subsurface fungal isolates under the assumption that deep-sea sediments represent an untapped reservoir of bio- and chemodiversity.
Almost all fungal isolates (96%) had at least one gene involved in a secondary metabolite biosynthesis pathway. This clearly indicates their genetic potential for the synthesis of numerous secondary metabolites. Many of these genes might be responsible for the biosynthesis of natural compounds that may help these fungi to compete with other microorganisms for available substrates (28). Isolates affiliated with the genera Penicillium, Fusarium, Paecilomyces, and Rhodotorula have genetic differences depending on sampling depth. Taxa from shallow depths (from 4 to 25 mbsf) harbored type III PKS genes, while taxa from deeper layers (from 25 to 700 mbsf) did not. These subseafloor fungi may have lost or gained some of these genes in order to adapt to their environment. Indeed, as previously demonstrated, secondary metabolism gene clusters evolve rapidly through multiple rearrangements, duplications, or losses (58). It may confirm our conclusion that some of these isolates exhibit evidence of ecophysiological adaptation. More detailed genome-based and/or phylogenetic analyses should be performed to determine their evolutionary origin.
Our results (i) demonstrate our success in isolating some representatives of fungal taxa occurring within deep-subseafloor communities, (ii) give insights into their adaptation to subsurface in situ conditions, with fungi from deeper layers appearing to be more adapted to deep-subseafloor conditions, and (iii) illustrate the potential for their synthesis of genes for secondary metabolites of possible biotechnological importance. While new molecular methods have recently allowed the genetic capacities of uncultured microorganisms to be studied using single-cell genomics and metatranscriptomics, many important physiological features remain culture dependent. Our established culture collection offers the possibility for future applied biotechnological investigations specifically dedicated to the expression of selected genes of interest.
Supplementary Material
ACKNOWLEDGMENTS
This study received grants from the Ministère de l'Enseignement Supérieur et de la Recherche (Paris, France), Région Bretagne, Rennes, France, and EU project MaCuMBA (Marine Microorganisms: Cultivation Methods for Improving Their Biotechnological Applications, FP7, grant agreement 311975, Brussels, Belgium).
We thank Virginia Edgcomb and Maria G. Pachiadaki for proofreading of the manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04064-14.
REFERENCES
- 1.Gadanho M, Sampaio JP. 2005. Occurrence and diversity of yeasts in the mid-Atlantic ridge hydrothermal fields near the Azores Archipelago. Microb Ecol 50:408–417. doi: 10.1007/s00248-005-0195-y. [DOI] [PubMed] [Google Scholar]
- 2.Burgaud G, Le Calvez T, Arzur D, Vandenkoornhuyse P, Barbier G. 2009. Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environ Microbiol 11:1588–1600. doi: 10.1111/j.1462-2920.2009.01886.x. [DOI] [PubMed] [Google Scholar]
- 3.Le Calvez T, Burgaud G, Mahé S, Barbier G, Vandenkoornhuyse P. 2009. Fungal diversity in deep-sea hydrothermal ecosystems. Appl Environ Microbiol 75:6415–6421. doi: 10.1128/AEM.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgaud G, Arzur D, Durand L, Cambon-Bonavita MA, Barbier G. 2010. Marine culturable yeasts in deep-sea hydrothermal vents: species richness and association with fauna. FEMS Microbiol Ecol 73:121–133. doi: 10.1111/j.1574-6941.2010.00881.x. [DOI] [PubMed] [Google Scholar]
- 5.Damare S, Raghukumar C, Raghukumar S. 2006. Fungi in deep-sea sediments of the Central Indian Basin. Deep Sea Res Part I Oceanogr Res Pap 53:14–27. doi: 10.1016/j.dsr.2005.09.005. [DOI] [Google Scholar]
- 6.Lai X, Cao L, Tan H, Fang S, Huang Y, Zhou S. 2007. Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J 1:756–762. doi: 10.1038/ismej.2007.51. [DOI] [PubMed] [Google Scholar]
- 7.Nagano Y, Nagahama T, Hatada Y, Nunoura T, Takami H, Miyazaki J, Takai K, Horikoshi K. 2010. Fungal diversity in deep-sea sediments—the presence of novel fungal groups. Fungal Ecol 3:316–325. doi: 10.1016/j.funeco.2010.01.002. [DOI] [Google Scholar]
- 8.Edgcomb VP, Beaudoin D, Gast R, Biddle JF, Teske A. 2011. Marine subsurface eukaryotes: the fungal majority. Environ Microbiol 13:172–183. doi: 10.1111/j.1462-2920.2010.02318.x. [DOI] [PubMed] [Google Scholar]
- 9.Nagahama T, Takahashi E, Nagano Y, Abdel-Wahab MA, Miyazaki M. 2011. Molecular evidence that deep-branching fungi are major fungal components in deep-sea methane coldseep sediments. Environ Microbiol 13:2359–2370. doi: 10.1111/j.1462-2920.2011.02507.x. [DOI] [PubMed] [Google Scholar]
- 10.Mouton M, Postma F, Wilsenach J, Botha A. 2012. Diversity and characterization of culturable fungi from marine sediment collected from St. Helena Bay, South Africa. Microb Ecol 64:311–319. doi: 10.1007/s00248-012-0035-9. [DOI] [PubMed] [Google Scholar]
- 11.Singh P, Raghukumar C, Meena RM, Verma P, Shouche Y. 2012. Fungal diversity in deep-sea sediments revealed by culture-dependent and culture-independent approaches. Fungal Ecol 5:543–553. doi: 10.1016/j.funeco.2012.01.001. [DOI] [Google Scholar]
- 12.Orsi W, Biddle JF, Edgcomb V. 2013. Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PLoS One 8:e56335. doi: 10.1371/journal.pone.0056335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orsi WD, Edgcomb VP, Christman GD, Biddle JF. 2013. Gene expression in the deep biosphere. Nature 499:205–208. doi: 10.1038/nature12230. [DOI] [PubMed] [Google Scholar]
- 14.Nagano Y, Nagahama T. 2012. Fungal diversity in deep-sea extreme environments. Fungal Ecol 5:463–471. doi: 10.1016/j.funeco.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Ciobanu MC, Burgaud G, Dufresne A, Breuker A, Rédou V, BenMaamar S, Gaboyer F, Vandenabeele-Trambouze O, Lipp JS, Schippers A, Vandenkoornhuyse P, Barbier G, Jebbar M, Godfroy A, Alain K. 2014. Microorganisms persist at record depths in the subseafloor of the Canterbury Basin. ISME J 8:1370–1380. doi: 10.1038/ismej.2013.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teske A, Sørensen KB. 2008. Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J 2:3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- 17.Rateb ME, Ebel R. 2011. Secondary metabolites of fungi from marine habitats. Nat Prod Rep 28:290–344. doi: 10.1039/c0np00061b. [DOI] [PubMed] [Google Scholar]
- 18.Fulthorpe CS, Hoyanagi K, Blum P, the Expedition 317 Scientists. 2011. Proceedings of the Integrated Ocean Drilling Program, vol 317 Integrated Ocean Drilling Program Management International, Inc, Tokyo, Japan. doi: 10.2204/iodp.proc.317.104.2011. [DOI] [Google Scholar]
- 19.Rédou V, Ciobanu MC, Pachiadaki MG, Edgcomb V, Alain K, Barbier G, Burgaud G. 2014. In-depth analyses of deep subsurface sediments using 454-pyrosequencing reveals a reservoir of buried fungal communities at record-breaking depths. FEMS Microbiol Ecol 90:908–921. doi: 10.1111/1574-6941.12447. [DOI] [PubMed] [Google Scholar]
- 20.Meyer W, Mitchell TG, Freedman EZ, Vilgalys R. 1993. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J Clin Microbiol 31:2274–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampaio JP, Gadanho M, Santos S, Duarte FL, Pais C, Fonseca A, Fell JW. 2001. Polyphasic taxonomy of the basidiomycetous yeast genus Rhodosporidium: Rhodosporidium kratochvilovae and related anamorphic species. Int J Syst Evol Microbiol 51:687–697. [DOI] [PubMed] [Google Scholar]
- 22.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In PCR protocols: a guide to methods and applications. Academic Press, Orlando, FL. [Google Scholar]
- 23.Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul S, Gish W, Miller W, Myers E, Lipman D. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joubert A, Calmes B, Berruyer R, Pihet M, Bouchara JP, Simoneau P, Guillemette T. 2010. Laser nephelometry applied in an automated microplate system to study filamentous fungus growth. Biotechniques 48:399–404. doi: 10.2144/000113399. [DOI] [PubMed] [Google Scholar]
- 28.Amnuaykanjanasin A, Punya J, Paungmoung P, Rungrod A, Tachaleat A, Pongpattanakitshote S, Cheevadhanarak S, Tanticharoen M. 2005. Diversity of type I polyketide synthase genes in the wood-decay fungus Xylaria sp. BCC 1067. FEMS Microbiol Lett 194:125–136. doi: 10.1016/j.femsle.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 29.Slightom JL, Metzger BP, Luu HT, Elhammer AP. 2009. Cloning and molecular characterization of the gene encoding the aureobasidin A biosynthesis complex in Aureobasidium pullulans BP-1938. Gene 43:67–79. doi: 10.1016/j.gene.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Kawaide H, Imai R, Sassa T, Kamiya Y. 1997. Ent-kaurene synthase from the fungus Phaeosphaeria sp. L487. cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. J Biol Chem 272:21706–21712. [DOI] [PubMed] [Google Scholar]
- 31.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. 2010. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh P, Raghukumar C, Verma P, Shouche Y. 2010. Phylogenetic diversity of culturable fungi from the deep-sea sediments of the Central Indian Basin and their growth characteristics. Fungal Divers 40:89–102. doi: 10.1007/s13225-009-0009-5. [DOI] [Google Scholar]
- 33.Nagahama T, Hamamoto M, Nakase T, Takami H, Horikoshi K. 2001. Distribution and identification of red yeasts in deep-sea environments around the northwest Pacific Ocean. Antonie Van Leeuwenhoek 80:101–110. doi: 10.1023/A:1012270503751. [DOI] [PubMed] [Google Scholar]
- 34.Smith A, Popa R, Fisk M, Nielsen M, Wheat CG, Jannasch HW, Fisher AT, Becker K, Sievert SM, Flores G. 2011. In situ enrichment of ocean crust microbes on igneous minerals and glasses using an osmotic flow-through device. Geochem Geophys Geosyst 12:Q06007. doi: 10.1029/2010GC003424. [DOI] [Google Scholar]
- 35.Francis M. 2013. A preliminary investigation of marine yeast biodiversity in New Zealand waters. M.S. thesis Victoria University of Wellington, Wellington, New Zealand. [Google Scholar]
- 36.Connell L, Barrett A, Templeton A, Staudigel H. 2009. Fungal diversity associated with an active deep sea volcano: Vailulu'u Seamount, Samoa. Geomicrobiol J 26:597–605. doi: 10.1080/01490450903316174. [DOI] [Google Scholar]
- 37.Bass D, Howe A, Brown N, Barton H, Demidova M, Michelle H, Li L, Sanders H, Watkinson SC, Willcock S, Richards TA. 2007. Yeast forms dominate fungal diversity in the deep oceans. Proc Biol Sci 274:3069–3077. doi: 10.1098/rspb.2007.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burgaud G, Woehlke S, Rédou V, Orsi W, Beaudoin D, Barbier G, Biddle FJ, Edgcomb VP. 2013. Deciphering the presence and activity of fungal communities in marine sediments using a model estuarine system. Aquat Microb Ecol 70:45–62. doi: 10.3354/ame01638. [DOI] [Google Scholar]
- 39.Cathrine SJ, Raghukumar C. 2009. Anaerobic denitrification in fungi from the coastal marine sediments off Goa, India. Mycol Res 113:100–109. doi: 10.1016/j.mycres.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Zhang WP, Cao HL, Shek CS, Tian RM, Wong YH, Batang Z, Al-Suwailem A, Qian PY. 2014. Diversity and distribution of eukaryotic microbes in and around a brine pool adjacent to the Thuwal cold seeps in the Red Sea. Front Microbiol 5:37. doi: 10.3389/fmicb.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galkiewicz JP, Stellick SH, Gray MA, Kellogg CA. 2012. Cultured fungal associates from the deep-sea coral Lophelia pertusa. Deep Sea Res Part I Oceanogr Res Pap 67:12–20. doi: 10.1016/j.dsr.2012.05.001. [DOI] [Google Scholar]
- 42.Höller U, Wright AD, Matthee GF, Konig GM, Draeger S, Aust HJ, Schulz B. 2000. Fungi from marine sponges: diversity, biological activity and secondary metabolites. Mycol Res 104:1354–1365. doi: 10.1017/S0953756200003117. [DOI] [Google Scholar]
- 43.Butinar L, Zalar P, Frisvad JC, Gunde-Cimerman N. 2005. The genus Eurotium—members of indigenous fungal community in hypersaline waters of salterns. FEMS Microbiol Ecol 51:155–166. doi: 10.1016/j.femsec.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Kis-Papo T, Grishkan I, Oren A, Wasser SP, Nevo E. 2001. Spatiotemporal diversity of filamentous fungi in the hypersaline Dead Sea. Mycol Res 105:749–756. doi: 10.1017/S0953756201004129. [DOI] [Google Scholar]
- 45.Xu W, Pang KL, Luo ZH. 2014. High fungal diversity and abundance recovered in the deep-sea sediments of the Pacific Ocean. Microb Ecol 68:688–698. doi: 10.1007/s00248-014-0448-8. [DOI] [PubMed] [Google Scholar]
- 46.Arfi Y, Marchand C, Wartel M, Record E. 2012. Fungal diversity in anoxic-sulfidic sediments in a mangrove soil. Fungal Ecol 5:282–285. doi: 10.1016/j.funeco.2011.09.004. [DOI] [Google Scholar]
- 47.Lauro FM, Bartlett DH. 2008. Prokaryotic lifestyles in deep sea habitats. Extremophiles 12:15–25. doi: 10.1007/s00792-006-0059-5. [DOI] [PubMed] [Google Scholar]
- 48.Lorenz R, Molitoris HP. 1997. Cultivation of fungi under simulated deep-sea conditions. Mycol Res 101:1355–1365. doi: 10.1017/S095375629700405X. [DOI] [Google Scholar]
- 49.Raghukumar C, Raghukumar S. 1998. Barotolerance of fungi isolated from deep-sea sediments of the Indian Ocean. Aquat Microb Ecol 15:153–163. doi: 10.3354/ame015153. [DOI] [Google Scholar]
- 50.Simonato F, Campanaro S, Lauro FM, Vezzi A, D'Angelo M, Vitulo N, Giorgio V, Bartlett DH. 2006. Piezophilic adaptation: a genomic point of view. J Biotechnol 126:11–25. doi: 10.1016/j.jbiotec.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 51.Biddle JF, House CH, Brenchley JE. 2005. Microbial stratification in deeply buried marine sediment reflects changes in sulfate/methane profiles. Geobiology 3:287–295. doi: 10.1111/j.1472-4669.2006.00062.x. [DOI] [Google Scholar]
- 52.Sass H, Parkes RJ. 2011. Sub-seafloor sediments: an extreme but globally significant prokaryotic habitat (taxonomy, diversity, ecology), p 1016–1036. In Horikoshi K, Antranikian G, Bull AT, Robb FT, Stetter KO (ed), Extremophiles handbook. Springer, Tokyo, Japan. [Google Scholar]
- 53.Lomstein BA, Langerhuus AT, D'Hondt S, Jørgensen BB, Spivack AJ. 2012. Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment. Nature 484:101–104. doi: 10.1038/nature10905. [DOI] [PubMed] [Google Scholar]
- 54.Jørgensen BB. 2011. Deep subseafloor microbial cells on physiological standby. Proc Natl Acad Sci U S A 108:18193–18194. doi: 10.1073/pnas.1115421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colwell FS, D'Hondt S. 2013. Nature and extent of the deep biosphere. Rev Mineral Geochem 75:547–574. doi: 10.2138/rmg.2013.75.17. [DOI] [Google Scholar]
- 56.Weber T. 2014. In silico tools for the analysis of antibiotic biosynthetic pathways. Int J Med Microbiol 304:230–235. doi: 10.1016/j.ijmm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 57.Keller NP, Turner G, Bennett JW. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol 3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 58.Khaldi N, Collemare J, Lebrun MH, Wolfe KH. 2008. Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol 9:R18. doi: 10.1186/gb-2008-9-1-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Letunic I, Bork P. 2011. Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 23:127–128. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.