Main Text
In 1901, Alfred Russel Wallace felt confident to assert that: “natural selection is not the all-powerful, all-sufficient and only cause of the development of organic forms.” In the article by Kurz et al. (1) in this issue of the Biophysical Journal, cutting-edge culmination of more than a century’s research has led to the realization that it is the organizing power of energy flow within the nonequilibrium network systems of living organisms that provides the accumulating information that takes life to ever increasing levels of complexity (2).
Living systems are characterized by coherence (coordinated occurrence of events in space and time) and robustness (coordination resilient to external or internal perturbations). This is achieved through oscillations that are essential for synchronous operation and behavior and intra- and intercellular cooperation, not only in the heart (3,4), but even in unicellular organisms (5,6), and their ubiquity indicates evolutionary conservation over billions of years (7). Oscillator synchronization even in the simplest physical systems is incompletely understood. In a huge population of oscillators it involves Kuramoto’s rule of coupling strength (or strength of interaction) and sudden cohesion of a small but growing cluster as the spread of natural frequencies is decreased below a threshold. This represents a specific example of Prigogine’s theory of time order in which spontaneous transitions give spatiotemporal patterns in nonequilibrium open systems.
From this (1), the latest in a series of contributions (8,9) over almost two decades from the cardiology group at Johns Hopkins University, we learn much about the rich dynamics of the process of healthy cardiac performance. In humans at rest, this requires the daily pumping of ∼7200 L of blood and the turnover of ∼6 Kg of ATP. Central to this are tightly coupled ensembles of individual cardiomyocyte mitochondria working as oscillating clusters. The measured variable, the inner trans-membrane electrochemical potential (Ψm), is measured by the fluorescence of two-photon excited preparations. This oscillates in cells subjected to metabolic or oxidative stress, as reactive oxygen species (ROS)-activated inner membrane channels release ROS, initiate propagating waves of depolarization, and synchronize limit-cycle oscillations of (Ψm) in the mitochondrial network. These events were observed both in isolated cardiac cells and in intact perfused hearts. Coupling constants, quantitatively determined using the Kuramoto model, varied so as to alter cluster size, oscillatory frequencies, and respiratory substrate selection. Fig. 1 summarizes the hierarchical levels of organization found in the human cardiovascular system.
Figure 1.
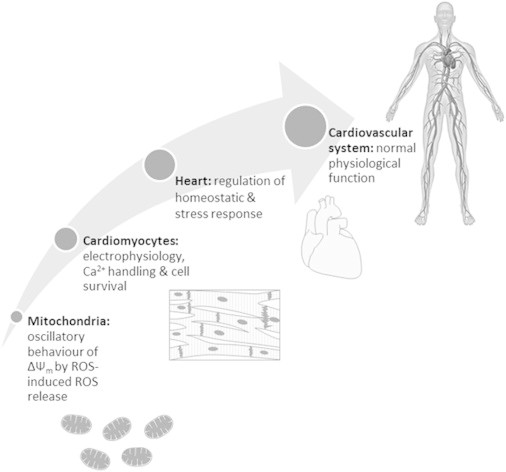
Hierarchical organization of the human cardiovascular system
Pathological changes during declining organ function may well result from molecular and microscopic events resulting in decreased intermitochondrial coupling and homeodynamic instability.
Time-series analyses utilized a recently-developed wavelet method, and a major challenge in the future is to use similar and more advanced modifications of this approach to measure and analyze ROS, especially if the frequencies of ROS oscillations (like those of Ψm) vary inversely with cluster size. This process of modulation (exquisitely and specifically balanced by ROS production and scavenging, e.g., by the interplay between Cu,Zn SOD and MnSOD levels) has been demonstrated in pathophysiological conditions, but whether a similar signaling process also functions under normal states is not known (10). It is established that substrate selection differs markedly from normal in many important and widespread disease states (e.g., in diabetes and dilated cardiomyopathy).
This research continues to open many possible new avenues toward future clinical applications. The new “tunes” thereby being uncovered remind us that mighty antiphonic choruses can arise from huge numbers of organized solo voices, but in biology, it is network dynamics that provide the synchrony and coordination, not the maestro conductor.
References
- 1.Kurz F.T., Derungs T., Armoundas A.A. Mitochondrial networks in cardiac myocytes reveal dynamic coupling behavior. Biophys. J. 2015;108:1922–1933. doi: 10.1016/j.bpj.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace D.C. Bioenergetics in human evolution and disease: implications for the origins of biological complexity and the missing genetic variation of common diseases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120267. doi: 10.1098/rstb.2012.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodsky V.Y. Circahoralian (ultradian) metabolic rhythms. Biochem. Mosc. 2014;79:483–495. doi: 10.1134/S0006297914060017. [DOI] [PubMed] [Google Scholar]
- 4.Porat-Shliom N., Chen Y., Weigert R. In vivo tissue-wide synchronization of mitochondrial metabolic oscillations. Cell Rep. 2014;9:514–521. doi: 10.1016/j.celrep.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashford C.L., Chance B., Poole R.K. Oscillations of redox states in synchronously dividing cultures of Acanthamoeba castellanii and Schizosaccharomyces pombe. Biophys. J. 1980;29:1–11. doi: 10.1016/S0006-3495(80)85114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aon M.A., Cortassa S., Lloyd D. Single and cell population respiratory oscillations in yeast: a 2-photon scanning laser microscopy study. FEBS Lett. 2007;581:8–14. doi: 10.1016/j.febslet.2006.11.068. [DOI] [PubMed] [Google Scholar]
- 7.Lloyd D., Cortassa S., Aon M.A. What yeast and cardiomyocytes share: ultradian oscillatory redox mechanisms of cellular coherence and survival. Integr. Biol. (Camb) 2012;4:65–74. doi: 10.1039/c1ib00124h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aon M.A., Cortassa S., O’Rourke B. The fundamental organization of cardiac mitochondria as a network of coupled oscillators. Biophys. J. 2006;91:4317–4327. doi: 10.1529/biophysj.106.087817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortassa S., Caceres V., Aon M.A. From metabolomics to fluxomics: a computational procedure to translate metabolite profiles into metabolic fluxes. Biophys. J. 2015;108:163–172. doi: 10.1016/j.bpj.2014.11.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kembro J.M., Aon M.A., Cortassa S. Integrating mitochondrial energetics, redox and ROS metabolic networks: a two-compartment model. Biophys. J. 2013;104:332–343. doi: 10.1016/j.bpj.2012.11.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]


