Abstract
The plasma membrane is a highly complex, organized structure where the lateral organization of signaling proteins is tightly regulated. In the case of Ras proteins, it has been suggested that the differential activity of the various isoforms is due to protein localization in separate membrane compartments. To date, direct visualization of such compartmentalization has been achieved only by electron microscopy on membrane sheets. Here, we combine photoactivated light microscopy with quantitative statistical analysis to visualize protein distribution in intact cells. In particular, we focus on the localization of HRas and its minimal anchoring domain, CAAX. We demonstrate the existence of a complex partitioning behavior, where small domains coexist with larger ones. The protein content in these domains varied from two molecules to tens of molecules. We found that 40% of CAAX and 60% of HRas were localized in domains. Subsequently, we were able to manipulate protein distributions by inducing coalescence of supposedly cholesterol-enriched domains. Clustering resulted in an increase of the localized fraction by 15%.
Introduction
The currently accepted model of the plasma membrane is a heterogeneous bilayer organized into microdomains that results from molecular lipid-lipid and lipid-protein interactions and also reflects the interconnections between membrane components and the actin cytoskeleton (1). Although the first suggestions of protein clustering and membrane heterogeneity were highlighted in the 1970s in the classical article by Singer and Nicholson (2), it is only in the past decade that strong evidence of their existence has emerged. Domains ranging in size from a few tens of nanometers to ∼100–200 nm in diameter have been found in the past decade, both through indirect biochemical assays (3) and using sophisticated microscopy techniques including fluorescence recovery after photobleaching (4,5), fluorescence resonance energy transfer (6), fluorescence correlation spectroscopy (7–9), single-particle tracking (10–12), single-molecule microscopy (13–15), and electron microscopy (16). However, a common view on the nature and biological role of such membrane domains is still lacking. This uncertainty in part arises from the inability of the above techniques to directly identify domains in live cells. Earlier techniques identified domains indirectly by analysis of their effect on such processes as lateral diffusion of proteins and lipids (10–12,17). To date, direct observation of domains has been achieved in membrane sheets through cryo-immunogold electron microscopy (16). In the approach described here, the first of its kind to our knowledge, the advantages of the earlier approaches are combined to allow on one hand direct visualization of proteins and analysis of their organization and on the other experimentation with intact (though so far only fixed) cells. A further limitation of most studies of plasma membrane structure and function is that they have focused on the outer leaflet of the plasma membrane, in particular either labeled lipids or antibody-tagged outer-membrane components, which are more easily accessible experimentally. Less is known about the presence of domains in the inner leaflet of the plasma membrane or about possible coupling of inner-leaflet to outer-leaflet domains.
It is believed that domains in the inner leaflet would act as platforms where membrane-bound signaling proteins are segregated (1,18). This would add an additional layer to the regulation of signaling cascades. Interleaflet coupling could add an extra layer of control of signaling pathways, facilitating the interactions between transmembrane proteins and their effectors on the cytoplasmic leaflet (19,20).
The lipid anchored Ras proteins are involved in a multitude of pathways regulating cell growth, differentiation, and apoptosis (21). These proteins are small GTPases that function as molecular switches between an active, GTP-bound and an inactive, GDP-bound state in many intracellular signaling pathways. Ras proteins are localized in the cytosolic leaflet of the plasma membrane (22,23), although they can also be found in the Golgi apparatus and in the endoplasmic reticulum (24). The Ras-protein family comprises various highly homologous isoforms that significantly differ only in their C-terminal ends, the so-called hypervariable region (HVR) (25). The HVR contains the sequence that controls how the proteins are anchored to the plasma membrane. Three of the Ras isoforms, H-, N- and KRas, are ubiquitously expressed in mammalian cells. Different isoforms yield different signaling outputs, although each interacts with the same set of effector proteins. It has been suggested that this seeming controversy might be explained by differences in Ras localization and partitioning within the membrane. Given that the different isoforms use different membrane anchoring motifs, it is likely that their specificity to membrane domains might be dictated by different lipid content or structuring by the membrane skeleton (21,26,27).
Here, we applied stochastic superresolution imaging by photoactivation localization microscopy (PALM) (28–30) to study the partitioning of HRas and of its 10 C-terminal amino acids, which include the CAAX motif (hereafter CAAX) in mouse fibroblast cells. By detailed analysis of the surface distribution of the proteins with 35 nm positional accuracy, we were able to identify 100-nm-sized domains of increased HRas occupancy in the inner leaflet of the plasma membrane. Further, we were able to manipulate the partitioning of HRas by clustering of GM1 gangliosides on the extracellular leaflet induced by cholera toxin subunit B (CTX). HRas reorganization upon CTB administration indicates that cholesterol-rich domains on the outside are coupled to nanodomains on the inside of a cell.
Materials and Methods
Plasmids
The pDendra2 plasmid encoding for Dendra2 protein (Evrogen, Moscow, Russia) was digested using the restriction sites AgeI and BSrgI. The pcDNA3.1 vector encoding the 10 C-terminal amino acids of human HRas, including the CAAX motif fused to yellow fluorescent protein (YFP) (12), was cut using the same restriction sites. Dendra2 was subsequently ligated into the pcDNA 3.1 vector. An analogous procedure was followed for pcDNA3.1 vector encoding for HRas-YFP. The integrity of the resulting reading frames was controlled by sequence analysis.
Cell culture and transfection
Mouse 3T3 fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% newborn serum (NBS), streptomycin (200 μg/mL) and penicillin (200 U/mL) in a 7% CO2 humidified atmosphere at 37°C (95% humidity). Cells were passed every 4 days. For microscopy, cells were cultured on 25 mm glass slides, pretreated with hydrofluoric acid. Adherent cells were transfected with 1 μg DNA and 6 μL FuGENE 6HD (Roche Diagnostic, Mannheim, Germany) per slide. The transfection efficiency, as determined by fluorescence microscopy, was ∼20%. At 1 to 3 days after transfection, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) (GIBCO, Invitrogen, Grand Island, NY) at 37°C for 15 min. For CTB measurements, cells were incubated with 5 μg/mL Alexa647-cholera toxin B (Invitrogen, Thermo Fischer Scientific, Waltham, MA) for 10 min before fixation.
Single-molecule fluorescence microscopy
PALM imaging (28,29) was performed on a modified version of the wide-field single-molecule imaging setup that has been described in detail previously (31). In brief, 3T3 cells adhered to a glass slide were mounted onto an inverted microscope (Axiovert100, Zeiss, Jena, Germany), equipped with a 100× oil immersion objective (NA 1.4, Zeiss), and kept in PBS at room temperature. The apical membrane of the cells was imaged. For imaging, the 514 nm line from an Ar/Kr laser (Spectra Physics, Newport Corp., Irvine, CA) was coupled via an optical fiber to the excitation path of the microscope. Light from a 405 nm diode laser (Crystal Laser, Reno, NV) was used to photoconvert Dendra2. Images were taken consecutively for up to 104 images per experiment. The time lag between images was set to 120 ms. Precise timing and intensity setting for the excitation light was achieved by an acoustooptic tunable filter (AA Electroptique, Orsay, France). The illumination intensity at 514 nm was set to 4 kW/cm2 and the illumination time per frame to 5 ms. Both settings ensured that most of the photoconverted Dendra2 molecules photobleached after one frame. The intensity of the 405 nm activation laser for photoconversion was increased from 0.2 μW/cm2 to 20 μW/cm2 during acquisition, to ensure sufficient photoconversion of fluorophores per frame. The density of photoconverted molecules was <0.1 molecules/μm2. The fluorescence signal of individual photoconverted Dendra2-labeled proteins was subsequently detected on a slow-scan back-illuminated CCD camera (SpectraMax, Photometrics, Tucson, AZ) with a pixel size of 20 μm, which translates into a pixel size of 200 nm in the objective plane. To correct for the drift of the setup, an immobilized 200-nm-sized fluorescent bead (Crimson Red FluoSpheres, Molecular Probes, Eugene, OR) was imaged simultaneously with the cells.
Control
As a control for experiments and analysis, purified enhanced YFP (eYFP) was immobilized in a 15% acrylamide gel. This involved dilution of 200 μL of 20 nM buffered solution of eYFP in 2 mL of polyacrylamide before polymerization. At this low final concentration of 2 nM, the eYFP molecules were assumed to be randomly distributed. To obtain clusters of molecules, the concentration of YFP was increased 100 times to a final concentration of 200 nM. The sample was photobleached before measurement with continuous 514 nm illumination for 30 s at an intensity of 4 kW/cm2. This photobleaching step assured that on average only one molecule per frame was visible. We acquired 1000 consecutive frames 10 × 10 μm in size in which we observed the stochastic recovery (32) and photobleaching of eYFP.
Image analysis
Image and statistical analysis were performed using MATLAB (The MathWorks, Natick, MA). The signals of individual Dendra2 molecules on the charge-coupled device were fitted to two-dimensional Gaussian profiles of mean full width at half-maximum of 370 ± 40 nm set by the diffraction limit. The mean signal detected from individual molecules was 420 ± 60 cnts. At this signal, the accuracy for single-molecule lateral localization was found to be 35 nm in both the x and y directions (33). Before further analysis, all positions within the entire image stack were corrected for experimental drift, as determined by simultaneously tracking fiducial marker beads within each image. In this way, the drift was corrected to an accuracy of 5 nm throughout the image stack. Dendra2 signals that lasted for longer than one frame or reappeared within the 100 following frames, i.e., within 12 s, and were found within the positional accuracy of the first detection, were assumed to arise from the same molecule and were discarded (for details, see Section S2 in the Supporting Material).
The drift-corrected single-molecule position data were subsequently used to reconstruct a two-dimensional image of the cell membrane. Each position was thereby represented by a two-dimensional Gaussian of width given by the mean positional accuracy, . For each image, >104 positions were used.
Statistical analysis
Ripley’s analysis
For statistical analysis of potential nonrandom distributions of proteins on the plasma membrane, we used Ripley’s L(r) − r function (34),
| (1) |
with Ripley’s K-function that counts the mean number of neighbors in a distance of radius r for N points in an area A:
| (2) |
dij is the Euclidean distance between two points and the counting measure is Ir(dij) = 1 for distance dij ≤ r and Ir(dij) = 0 otherwise. wij ≥ 1 is a weighting factor that accounts for edge effects (34). It corresponds to the inverse of the proportion of the circumference of a circle with its center in i and passing through j, which lies within the area A.
For a spatially randomly arranged point pattern, K(r) = πr2, and thus L(r) − r approaches 0. For a nonrandom pattern with well-separated clusters of typical size R, L(r) − r displays a characteristic maximum at R and decays to zero for larger radii (35).
The superresolution images of the size of a cell area (commonly 7 × 7 μm2) typically were split into 20 square tiles of 1 × 1 μm2. Each of the tiles contained at least 70 positions. Ripley’s L(r) − r function was subsequently applied to each tile separately. From the analysis of the individual tiles, the mean <L(r) − r> was calculated, as was a 1σ-confidence interval as defined by the quantil.
Density-based spatial cluster analysis with noise
Following the approach of Nan et al. (36), we used another density-based algorithm, density-based spatial cluster analysis with noise (DBSCAN), to complement the information obtained with Ripley’s analysis. DBSCAN is a widely used algorithm in spatial pattern analysis and data mining (37,38). It generates a list of clusters and assigns each point in the image within a certain distance (here 40 nm) to a cluster. Points that do not belong to any cluster are classified as noise (randomly distributed points). The minimal number of points required to form a cluster was set to 2 (corresponding to protein dimers).
Simulations
Point patterns were simulated using MATLAB. The simulations were used to validate our statistical analysis and to determine the parameter values needed to reproduce the experimental results. As an initial guess for the parameters, we used values found in the experiments. The radius of maximum clustering, R, obtained from Ripley’s analysis was used as an initial estimate for the simulations (35) (see Section S2 in the Supporting Material)). However, to reproduce the results more closely, we subsequently assumed a distribution of cluster sizes between 40 and 150 nm. The range of concentrations, 5000 < c < 25,000, corresponded to the number of molecules detected in the experiments. Other parameters included the fraction of randomly distributed molecules, α (0.4 < α < 0.6 (12)), the number of clusters, Nc, the number of molecules per cluster, pc = c × (1 − α)/Nc, (i.e., 2 < pc < 15), and the cluster radius, R (40 nm < R < 500 nm). On top of the randomly distributed positions, Nc clusters were randomly distributed over the area, with each cluster containing pc positions randomly distributed in a circular area of radius R around the center of mass. L(r) − r was calculated as in Eq. 1 for each realization, and the results were compared to the experimental data. For a purely random distribution of positions (α = 1), L(r) − r fluctuates around zero. Further, we found that for the case of clustering, the position of the maximum of L(r) − r is located close to clustering radius R, as was reported in Kiskowski et al. (35), and the maximal value of L(r) − r increases with the number of positions within a cluster, pc (for a summary of those simulations, see the Supporting Material).
Results
YFP-acrylamide gel
The strategies for experiment and analysis were validated by examining the spatial distribution of eYFP molecules immobilized in an acrylamide gel (see Materials and Methods).
As shown by Dickson at al. (39), eYFP can be reactivated multiple times from the dark state and thus is a suitable fluorophore for superresolution imaging by PALM (32). The positions of the molecules in each frame were determined by fitting the intensity distribution from individual eYFP molecules to a two-dimensional Gaussian (see Materials and Methods). Images of eight separate areas of the gel showed an average of 13 eYFP molecules/μm2. To reconstruct a high-resolution image, each molecule was plotted as a two-dimensional Gaussian with a standard deviation that equals the positional accuracy, <σ1D> = 35 nm. The resulting image shows a random distribution of molecules throughout the region of interest (Fig. 1 A). An analysis based on both the DBSCAN algorithm and Ripley’s K-function was performed to confirm the visual inspection. The observed area was divided into nonoverlapping tiles of 1 × 1 μm in size, and L(r) − r was evaluated in each box up to a radius of r = 0.5 μm. The DBSCAN algorithm identified 84% of the molecules as being unclustered, and the remaining 16% were assigned to small clusters containing two or three molecules (see Fig. 1 B). The mean Ripley’s analysis of the eight independent measurements is shown in Fig. 1 C. For comparison, the figure shows the mean results obtained from 10 simulated random point patterns using the same number of data points (Fig. 1 C, black dashed line). To demonstrate the capability of the algorithm to distinguish random distributed fluorophores from clusters, we performed the same analysis on gel containing a 100× higher concentration of fluorophore. The resulting image shows large aggregates (see Fig. 1 D). DBSCAN identifies only 9% of these as randomly distributed molecules, whereas the remaining 91% belong to clusters that vary in content from a few tens to hundreds of molecules, as shown in Fig. 1 E. Finally, Ripley’s K-function confirmed this observation (Fig. 1 F), showing a marked deviation from the results obtained at the lower concentration of fluorophores. Hence, these control experiments validate the approach chosen here to detect a random distribution by means of superresolution microscopy in conjunction with Ripley’s analysis.
Figure 1.
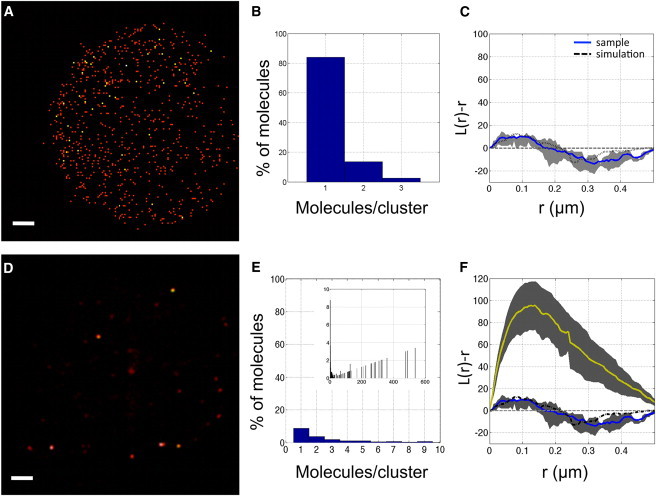
(A) PALM image of eYFP gel. Each position is represented as a two-dimensional Gaussian. Scale bar, 1 μm. (B) Distribution of molecules in the gel according to the DBSCAN algorithm, showing that 84% of molecules are unclustered. (C) Plot of L(r) − r calculated for the eYFP gel (blue solid line). The function fluctuates around zero, as expected for a randomly distributed sample. Also plotted are the experimental confidence interval (gray area) and the L(r) − r for a set of 10 simulations (black dashed line). (D) PALM image of a 200 nM eYFP gel. Clusters of molecules are clearly visible. (E) Distribution of molecules in (D) according to the DBSCAN algorithm. Only 9% of the molecules are randomly distributed. (Inset) Full histogram. Clusters containing up to 600 molecules are detected. (F) Plot of L(r) − r calculated for the 200 nM eYFP gel (yellow solid line). The function has a maximum at 120 ± 15 nm. The gray area indicates the confidence interval. To see this figure in color, go online.
CAAX clustering
Initially, we studied the membrane distribution of the minimal anchoring motif of the HRas protein CAAX. Association of CAAX to membrane domains between 20 and 200 nm in size has been suggested previously based on results from studies that used various indirect methods (9,12). Here, we performed superresolution experiments to directly visualize those potential domains in fixed cells. In these and all subsequent experiments, we chose to use the photoconvertible genetically encoded fluorophore Dendra2. Dendra2 offers active regulation of the photoconversion and does not require a high-fluency photobleaching step, which may damage the sample. Wide-field and confocal images verified that the construct was faithfully located at the membrane (not shown). The localization of CAAX was studied in 11 cells. Before superresolution imaging, a bright-field image was taken to select a cell and define a region of interest on its apical membrane. High-resolution fluorescence images were taken for up to 104 frames per sequence. On average, 2–10 molecules were activated per frame. For the analysis, single fluorophore positions were localized and selected as described in Materials and Methods. Upon visual inspection, PALM images show a nonhomogenous distribution of CAAX molecules. The density map showed irregular patches containing a higher density of molecules alternating with regions of sparsely distributed molecules (Fig. 2, A and B).
Figure 2.
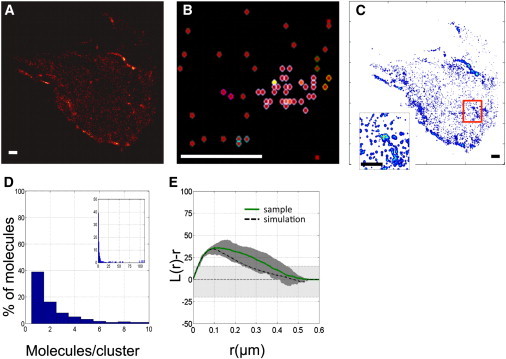
(A) PALM image of CAAX. Scale bar, 1 μm. (B) DBSCAN output for a selected region of the cell image in (A). Scale bar, 500 nm. (C) Density map of molecule distribution along the membrane. Scale bar, 1 μm. (Inset) Zoom of the red-outlined region in the larger image. A region with higher density of molecules is visible. Scale bar, 1 μm. (D) Cluster-size distribution for the cell in (A) as determined by DBSCAN. In this cell, ∼40% of the molecules are randomly distributed. Clustered molecules are mainly distributed in small clusters (containing up to 10 molecules.) (Inset) The full distribution of cluster sizes shows that a few molecules are in bigger clusters with up to 100 molecules. (E) Plot of the mean L(r) − r calculated for CAAX in 11 cells (green solid line). The function has a maximum at r = 112 ± 13 nm. Also plotted are the confidence interval (gray area) and the mean L(r) − r of 10 simulations (black dashed line). The light gray shaded area represents the confidence interval of the random sample. To see this figure in color, go online.
To quantitatively estimate the observed clustering, we performed both the density-based spatial clustering analysis with noise (DBSCAN) and Ripley’s K-function analysis (34–38). The first method estimates the percentage of clustered molecules and the protein content of such clusters, whereas the latter method provides information on cluster size.
DBSCAN analysis classified 60 ± 3% of the CAAX molecules as randomly distributed monomers. The remaining 40% are distributed in clusters with varying protein content. Small clusters of two to three proteins account for around 50% of the clustered fraction. Another 25% of molecules are found in clusters with a maximum of 10 proteins. An example of one such cell distribution is shown in Fig. 2 D. The remaining 25% of molecules are distributed in bigger clusters, which can contain up to hundreds of proteins (see Fig. 2 D, inset).
Ripley’s analysis was implemented iteratively on 1 × 1 μm tiles, covering the entire cell. The density of molecules in each tile varied between 40 and 170 molecules/μm2. L(r) − r was calculated for each tile. The mean over all tiles, <L(r) − r>, was assumed to be representative of the spatial distribution of CAAX in a single cell. The average and confidence intervals for all 11 cells were calculated to include potential biological variability of the data set and were assumed to be representative of CAAX distribution in 3T3 cells. (Fig. 2 E). The <L(r) − r> plot displays maximal clustering at 112 ± 13 nm. The errors were calculated as the mean ± SEM of the different single-cell measurements. To quantitatively interpret Ripley’s K test, we performed a set of simulations to verify the parameter values that were able to reproduce the experimental pattern. We assumed that α = 60% of molecules are randomly distributed, as determined by DBSCAN. The remaining 40% were distributed in clusters of two to five molecules. Bigger clusters (as identified by DBSCAN) were taken into account by including clusters of 15 molecules. L(r) − r was calculated on each point pattern. The average curve for 10 point patterns is shown in Fig. 2 E (black dashed line) overlaid with the experimental data. We assumed the distribution of cluster sizes to be between 40 and 150 nm. For details of the simulations, see the Supporting Material.
HRas spatial distribution
After assessing the distribution of the minimal anchoring motif, CAAX, we studied the full-length HRas protein. The concentration of HRas-Dendra2 in the range 50–180 molecules/μm2 was comparable to that determined for CAAX-Dendra2. This allows a direct comparison of the resulting curves. Superresolution images were acquired and analyzed as described above. The density map of HRas exhibits large aggregates, connoting a nonrandom distribution of HRas on the plasma membrane (Fig. 3 A). DBSCAN analysis showed a decrease in the fraction of randomly distributed monomers to 38 ± 5% compared to the 60% observed for CAAX. The majority of clustered molecules appear to be concentrated in clusters of two to three molecules, as seen for the membrane anchor, CAAX (see Fig. 3 B). Ripley’s analysis displayed a maximum at 160 ± 22 nm. The simulation parameters that could reproduce the results of Ripley’s analysis were α = 33% of randomly distributed molecules, cluster sizes varying in the range 80–180 nm, and protein concentration, c, in the range 2–15 molecules. The resulting curve is shown in Fig. 3 C overlaid with the curve obtained from the experimental data.
Figure 3.
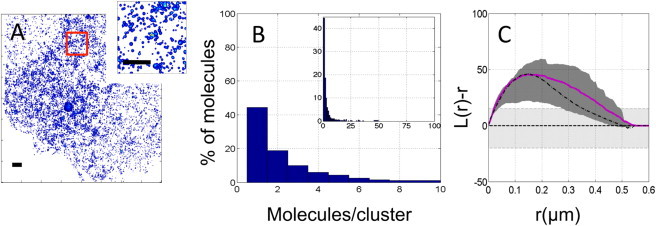
(A) Density map of HRas distribution in the membrane. (Inset) Zoom of the red-outlined region in the larger figure, where clusters are visible. (B) Cluster-size distribution as determined from DBSCAN for the cell in (A). In this cell, ∼45% of the molecules are randomly distributed. Clustered molecules are mainly distributed in small clusters (containing up to 10 molecules.) (Inset) The full distribution of cluster sizes shows that a few molecules are clustered in bigger clusters with up to 100 molecules. (C) Plot of the mean L(r) − r calculated for HRas in 11 cells (magenta solid line). The function has a maximum at r = 160 ± 22 nm. Also plotted are the confidence interval (gray area) and the mean L(r) − r of 10 simulations (black dashed line). The light gray shaded area represents the confidence interval of the random sample. To see this figure in color, go online.
CTB-induced protein reorganization
After showing the existence of membrane-bound protein clusters, we investigated whether we could manipulate these structures by modifying the organization of lipids in the outer leaflet of the plasma membrane. To induce cholesterol-dependent coalescence of the membrane, cells were treated with CTB. Cells were imaged and analyzed as described above. Upon CTB incubation, the organization of CAAX-Dendra2 exhibited a significant change in <L(r) − r>, as shown in Fig. 4. An additional 20% of molecules formed clusters (thus reducing the percentage of randomly distributed molecules from 60% to 40%), as shown in Fig. 4, B and C. This increased clustering corresponded also to an apparent increase in the radius of maximal clustering to 156 ± 13 nm (Fig. 4, A (left), B, and D).
Figure 4.
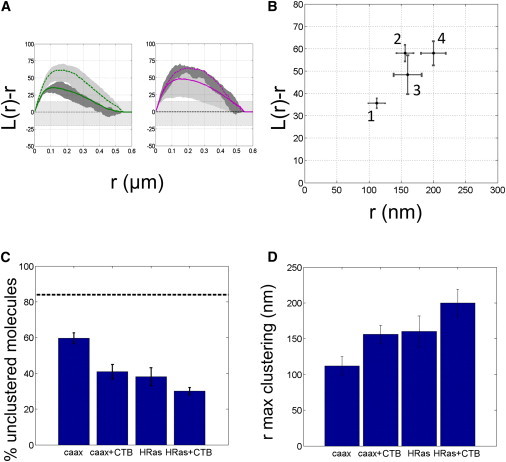
(A) Ripley’s analysis of CAAX distribution (left) and HRas distribution (right) before (solid line) and after (dashed line) treatment with CTB. (B) Position and value of the maximum of Ripley’s function in the four experimental conditions: CAAX alone, CAAX+CTB, HRas alone, and HRas+CTB. (C) Fraction of randomly distributed molecules in the different experiments. A decrease is observed upon CTB treatment. The dashed line indicates the unclustered fraction in the control experiment. (D) Radius of maximal clustering (as determined by Ripley’s analysis) in the four experimental conditions. To see this figure in color, go online.
For the full-length HRas protein, the changes were not as striking, although they were statistically significant. The unclustered fraction decreased to 30 ± 2%, as determined by DBSCAN. The radius of maximum clustering increased to 200 ± 19 nm, a change from the untreated cells at the limit of our confidence interval (Fig. 4, A (right), B, and D).
Discussion
The differential activity of the various isoforms of Ras proteins has been suggested to result from protein localization in separate membrane compartments (21). This partitioning is supposed to be mediated by the different C-terminal membrane anchoring sequences (23,27). Interestingly, it has been found that partitioning is connected to the activation state of Ras proteins (12), thus hinting at a functional role of membrane-bound protein clusters. However, a complete understanding of the mechanism underlying the spatial distribution of those proteins is still lacking. Here, we applied superresolution imaging to directly visualize the organization of HRas and its minimal anchoring motif in the inner leaflet of the plasma membrane of 3T3 fibroblasts. Data on CAAX distribution support that ∼40% of the molecules were organized into clusters. In our previous study, using single-molecule tracking, we observed that 40% of CAAX in live cells were undergoing confined diffusion, whereas the remaining 60% were freely diffusing in the plane of the membrane (40). We suggested that this was related to membrane confinement of the membrane anchor, CAAX. Here, we provide direct evidence of the presence of molecular clusters, and we suggest that the 60% of randomly distributed molecules correspond to the fast-diffusing fraction in tracking experiments. The wide distributions found for the spatial correlation indicate a wide distribution of domain sizes, ranging from 40 to 150 nm, as estimated from our simulations. The lower bound of 40 nm is in excellent agreement with the domain size obtained by immuno-electron microscopy when taking into account our limited positional accuracy of σ = 35 nm. The limited positional accuracy will increase any real domain size, rreal, by a factor of . Thus, from the experimentally determined lower bound of 40 nm, we infer a real domain size of 10 nm. This finding is comparable to the value of ∼12 nm found using electron microscopy (16,41). However, the combination of direct imaging by superresolution microscopy with spatial distribution analysis allowed us to unravel more complex partitioning behavior, where bigger domains coexist with smaller domains, potentially in a dynamic equilibrium. It is probably impossible to unravel such data using the earlier approaches by single-molecule tracking due to the spatial averaging required in the latter experiments. Using DBSCAN, we were also able to determine the protein content of each cluster, which varies from two to tens of proteins, but with the majority of observed clusters formed of two to three molecules.
Further, we studied the full-length HRas protein. For the experiment, the inactive, GDP-bound form of the protein was used. The presence of the highly conserved G-domain did not influence greatly the lateral segregation of HRas. We found an increase in the clustered fraction, with ∼60% of the molecules grouped into clusters varying in size from 80 to 180 nm. This finding suggests that the membrane anchor is only partially responsible for the partitioning of the protein, and that further interactions (either between the proteins or between proteins and other membrane components such as the cytoskeleton) play a significant role. It has been proposed that HRas distribution can play a physiological role in signaling (42).
Previously published studies have suggested that HRas is localized in cholesterol-enriched raft domains. Thus, to investigate whether we could manipulate such a distribution, we incubated cells with CTB, which induces coalescence of cholesterol-enriched domains (43). Indeed, we observed a significant increase in the percentage of clustered molecules for both CAAX and HRas, associated with an increase in cluster size.
In conclusion, we demonstrate the superiority of superresolution techniques to visualize membrane structuring in cells. Our data indicate that inner-leaflet HRas-related domains exist in a broad size range between 40 and 150 nm, each containing 2–15 Ras molecules. From these data, we suggest a model for inner-membrane partitioning into dynamic domains varying in size from a few tens to hundreds of nanometers in diameter. HRas partitions dynamically into and out of those domains, and its localization is determined primarily by the anchoring motif CAAX. CTB-induced reorganization of CAAX and HRas clusters suggests that this differential localization is mediated, at least partially, by the lipid composition and organization in both the inner and outer leaflets. Membrane reorganization at the extracellular side induced by coalescence of cholesterol-enriched domains resulted in protein enrichment of microdomains in the inside, supporting the theory of coupling between the leaflets.
Acknowledgments
The authors thank Sylvie Olthuis for support with cell cultures and plasmid design. The authors further thank the reviewers for their helpful comments and constructive criticisms.
Supporting Material
References
- 1.Lommerse P.H., Spaink H.P., Schmidt T. In vivo plasma membrane organization: results of biophysical approaches. Biochim. Biophys. Acta. 2004;1664:119–131. doi: 10.1016/j.bbamem.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Singer S.J., Nicolson G.L. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 3.Brown D.A., Rose J.K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 4.Yechiel E., Edidin M. Micrometer-scale domains in fibroblast plasma membranes. J. Cell Biol. 1987;105:755–760. doi: 10.1083/jcb.105.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotblat B., Prior I.A., Hancock J.F. Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Mol. Cell. Biol. 2004;24:6799–6810. doi: 10.1128/MCB.24.15.6799-6810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenworthy A.K., Edidin M. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 Å using imaging fluorescence resonance energy transfer. J. Cell Biol. 1998;142:69–84. doi: 10.1083/jcb.142.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwille P., Korlach J., Webb W.W. Fluorescence correlation spectroscopy with single-molecule sensitivity on cell and model membranes. Cytometry. 1999;36:176–182. doi: 10.1002/(sici)1097-0320(19990701)36:3<176::aid-cyto5>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 8.Schwille P., Haupts U., Webb W.W. Molecular dynamics in living cells observed by fluorescence correlation spectroscopy with one- and two-photon excitation. Biophys. J. 1999;77:2251–2265. doi: 10.1016/S0006-3495(99)77065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggeling C., Ringemann C., Hell S.W. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 10.Kusumi A., Sako Y., Yamamoto M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 1993;65:2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pralle A., Keller P., Hörber J.K. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 2000;148:997–1008. doi: 10.1083/jcb.148.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lommerse P.H.M., Snaar-Jagalska B.E., Schmidt T. Single-molecule diffusion measurements of H-Ras at the plasma membrane of live cells reveal microdomain localization upon activation. J. Cell Sci. 2005;118:1799–1809. doi: 10.1242/jcs.02300. [DOI] [PubMed] [Google Scholar]
- 13.van Zanten T.S., Gómez J., Garcia-Parajo M.F. Direct mapping of nanoscale compositional connectivity on intact cell membranes. Proc. Natl. Acad. Sci. USA. 2010;107:15437–15442. doi: 10.1073/pnas.1003876107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schütz G.J., Kada G., Schindler H. Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. EMBO J. 2000;19:892–901. doi: 10.1093/emboj/19.5.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakoshi H., Iino R., Kusumi A. Single-molecule imaging analysis of Ras activation in living cells. Proc. Natl. Acad. Sci. USA. 2004;101:7317–7322. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prior I.A., Muncke C., Hancock J.F. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez J.B., Segura J.M., Vogel H. Monitoring the diffusion of single heterotrimeric G proteins in supported cell-membrane sheets reveals their partitioning into microdomains. J. Mol. Biol. 2006;363:918–930. doi: 10.1016/j.jmb.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 18.Eisenberg S., Beckett A.J., Henis Y.I. Raft protein clustering alters N-Ras membrane interactions and activation pattern. Mol. Cell. Biol. 2011;31:3938–3952. doi: 10.1128/MCB.05570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Field K.A., Holowka D., Baird B. FcεRI-mediated recruitment of p53/56lyn to detergent-resistant membrane domains accompanies cellular signaling. Proc. Natl. Acad. Sci. USA. 1995;92:9201–9205. doi: 10.1073/pnas.92.20.9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheets E.D., Holowka D., Baird B. Membrane organization in immunoglobulin E receptor signaling. Curr. Opin. Chem. Biol. 1999;3:95–99. doi: 10.1016/s1367-5931(99)80017-9. [DOI] [PubMed] [Google Scholar]
- 21.Prior I.A., Hancock J.F. Ras trafficking, localization and compartmentalized signalling. Semin. Cell Dev. Biol. 2012;23:145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock J.F., Paterson H., Marshall C.J. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 23.Roy S., Luetterforst R., Parton R.G. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat. Cell Biol. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- 24.Chiu V.K., Bivona T., Philips M.R. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 25.Willumsen B.M., Christensen A., Lowy D.R. The p21 ras C-terminus is required for transformation and membrane association. Nature. 1984;310:583–586. doi: 10.1038/310583a0. [DOI] [PubMed] [Google Scholar]
- 26.Abankwa D., Gorfe A.A., Hancock J.F. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc. Natl. Acad. Sci. USA. 2010;107:1130–1135. doi: 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niv H., Gutman O., Henis Y.I. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J. Cell Biol. 2002;157:865–872. doi: 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hess S.T., Girirajan T.P.K., Mason M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 30.Bates M., Huang B., Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harms G.S., Cognet L., Schmidt T. Single-molecule imaging of l-type Ca2+ channels in live cells. Biophys. J. 2001;81:2639–2646. doi: 10.1016/S0006-3495(01)75907-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biteen J.S., Thompson M.A., Moerner W.E. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat. Methods. 2008;5:947–949. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt T., Schütz G.J., Schindler H. Characterization of photophysics and mobility of single molecules in a fluid lipid membrane. J. Phys. Chem. 1995;99:17662–17668. [Google Scholar]
- 34.Ripley B.D. Tests of randomness for spatial point patterns. J. R. Stat. Soc. Series B Stat. Methodol. 1979;41:368–374. [Google Scholar]
- 35.Kiskowski M.A., Hancock J.F., Kenworthy A.K. On the use of Ripley’s K-function and its derivatives to analyze domain size. Biophys. J. 2009;97:1095–1103. doi: 10.1016/j.bpj.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nan X., Collisson E.A., Chu S. Single-molecule superresolution imaging allows quantitative analysis of RAF multimer formation and signaling. Proc. Natl. Acad. Sci. USA. 2013;110:18519–18524. doi: 10.1073/pnas.1318188110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ester, M., H. P. Kriegel, …, X. Xu. 1996. A density-based algorithm for discovering clusters in large spatial databases with noise. Proc. Int. Conf. Knowl. Discov. Data Mining (KDD Ô96), 2nd, Portland, OR. 226–231.
- 38.Daszykowski M., Walczak B., Massart D.L. Density-based clustering for exploration of analytical data. Anal. Bioanal. Chem. 2004;380:370–372. doi: 10.1007/s00216-004-2582-5. [DOI] [PubMed] [Google Scholar]
- 39.Dickson R.M., Cubitt A.B., Moerner W.E. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 40.Lommerse P.H., Blab G.A., Schmidt T. Single-molecule imaging of the H-ras membrane-anchor reveals domains in the cytoplasmic leaflet of the cell membrane. Biophys. J. 2004;86:609–616. doi: 10.1016/S0006-3495(04)74139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plowman S.J., Muncke C., Hancock J.F. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hancock J.F., Parton R.G. Ras plasma membrane signalling platforms. Biochem. J. 2005;389:1–11. doi: 10.1042/BJ20050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lingwood D., Ries J., Simons K. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc. Natl. Acad. Sci. USA. 2008;105:10005–10010. doi: 10.1073/pnas.0804374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


