Abstract
Unlike zebrafish and newt hearts, mammalian hearts have limited capacity to regenerate. Upon injury or disease, the adult mammalian hearts form a fibrotic scar. Recently, it was shown that neonatal mouse hearts can regenerate similarly to adult zebrafish hearts. However, this capacity quickly decreases after postnatal day 7 (P7). Understanding the molecular mechanisms underlying neonatal heart regeneration might lead to therapeutic approaches for regenerating adult mammalian hearts. In this study, we utilized an inducible transgenic mouse model to determine the effects of FGF10 growth factor over expression on neonatal mouse heart regeneration/repair. Over expression of FGF10 in myocardium enhanced the expansion of Wt1 positive epicardial cells at 21 days after heart injury through increased proliferation. However, this expansion of epicardial cells did not lead to increased epithelial-to-mesenchymal transition or affect fibroblast formation or fibrosis, as seen by vimentin expression, after heart injury. Furthermore, neither continuous nor transient expression of FGF10 did not affect scar thickness or length after heart injury in neonatal hearts. Our results suggest that FGF10 can regulate epicardial cell expansion of neonatal mouse hearts after injury; however, FGF10 alone is not sufficient to cause beneficial effects on heart repair.
Keywords: Cardiomyocytes, Mesenchymal transition, Hypoplasia
Introduction
In contrast to newt and zebrafish, adult mammalian hearts have a very limited capacity to regenerate [1–3]. Ischemia in the mammalian heart causes significant tissue damage with the loss of many cardiac cells through apoptosis and necrosis, only to be replaced with nonfunctional fibrotic tissue and hypertrophy but not hyperplasia of the remaining cardiomyocytes. After an ischemic episode, the heart is permanently scarred and functionally impaired, leading to lifelong morbidity or ultimately death. Cardiovascular disease affects millions of people in the USA and is the leading cause of all mortality [4]. Determining the precise mechanisms and initiators for regeneration may lead to possible therapeutic agents for human patients, decreasing mortality and morbidity from ischemic heart diseases. Recently, neonatal mouse hearts were shown to regenerate after ventricular resection in a similar fashion to adult zebrafish [5]. Furthermore, it was demonstrated they can also regenerate after ischemia created by coronary artery ligation [6]. However, this regenerative capacity is lost by postnatal day seven [5]. Studying the molecular mechanisms of neonatal mouse heart regeneration might reveal important signalling pathways that can either extend the regenerative capacity past postnatal day 7 or lead to novel therapeutic approaches for adult heart regeneration and repair. Growth factors are ideal candidate molecules that can be utilized to improve heart regeneration. FGF (fibroblast growth factor) signalling is of particular interest and has shown promise in improving repair in the mammalian system in various processes that are required for regeneration. FGF ligands such as FGF1 and FGF2 contribute positively to repair after myocardial infarction in animal models [7,8]. Furthermore, inhibition of Fgf signaling during zebrafish heart regeneration decreases epicardial epithelial to mesenchymal transition (EMT) and neo vascularization [9]. Conversely, over expression of the constitutively activated FGF receptor in a mouse model after cardiac injury improved blood vessel formation after injury for an overall protective effect [10]. The functions of FGF in heart are mainly related to myocardium and epicardium interactions and are well studied in mouse embryos. The epicardium plays key roles during heart development, and likely during heart regeneration as well. During heart development, epicardial cells undergo an epithelial to mesenchymal transition (EMT) to form epicardium-derived cells (EPDCs) that reside within the mesenchyme to give rise to smooth muscle cells of blood vessels and fibroblasts that generate the heart’s connective tissue (reviewed in) [11]. Furthermore, the epicardium is a source of mitogens that can stimulate proliferation of cardiomyocytes during myocardial growth [12]. A retinoic acid-erythropoietin-IGF2 (insulin-like growth factor 2) axis between the liver, epicardium and myocardium [13] utilizes paracrine growth factor signaling to contribute to the proliferation of cardiomyocytes [14]. During late embryonic heart development, the FGF10-FGFR2b ligand-receptor axis is necessary for the EMT of epicardial cells that develop into fibroblasts, which in turn regulates cardiomyocyte proliferation [15]. Additionally, knockout of FGF receptors in cardiomyocytes but not endothelial cells, confirms that myocardium proliferation and vessel development depends on FGF signaling to the cardiomyocytes [16]. Although, the epicardium develops normally without FGF activity, cardiomyocyte proliferation decreases and leads to hypoplasia and lower vascularity of the heart [16,17]. In contrast to the ventricular resection or coronary artery ligation models, neonatal mouse hearts do not regenerate after severe cryoinjury. In this study, we specifically overexpressed FGF10 in neonatal heart with an inducible transgenic mouse model in order to test the hypothesis that FGF10 can promote heart repair and regeneration. We found that ectopic overexpression of FGF10 in myocardium can enhance expansion of epicardial cells after cryoinjury. However, the epicardial expansion does not affect EMT of epicardial cells or fibroblast formation after heart injury. Furthermore, overexpression of FGF10 in myocardium does not affect fibrosis and scar size. Our results suggest that overexpression FGF10 affects the epicardium but is not sufficient for beneficial effects on heart repair at the neonatal stage.
Materials and Methods
Maintenance, breeding and genotyping of transgenic mice and heart surgery
The animals were housed at Children’s Hospital Los Angeles (CHLA) animal facility. All protocols related to animal work were approved by CHLA IACUC. In order to create a tissue-specific and temporally controlled gene expression model, we crossed a tissue specific alpha-MHC driven rtTA (Myh6-rttA) (MMRRC) to the tet(o) Fgf10 mouse [18,19]. The tet-on (doxycycline inducible) transgene expression is limited to the cells expressing alpha-MHC and thus rtTA. Mice were genotyped at fourteen days of age using tail tissue, which was lysed in proteinase K (100 ug/ml) and used for PCR amplification. PCR primers for transgenes, alpha-MHC: rtTA and tet(o)Fgf10 are listed in the (table 1). Thirty-five cycles of 94°C for one minute, 58°C for thirty seconds to anneal, 72°C for one minute were performed to generate the amplicons. PCR products were separated on a 1.5% agarose/TAE gel. Cryo-probe injury and sham surgery of neonatal pups at 1 day of age (P1) were performed according to Porrello et al. [5]. The details for the cryoinjury procedure will be described by Darehzereshki et al. (in preparation).
Table 1.
Primer sequences for genotyping.
| Gene | Sense | Antisense | Reference |
|---|---|---|---|
| aMHC- rtTA | cac ctg ggg ttc cca ccc tta tgt | agc agc tcc agt gcgctg tta | |
| tet (o)fgf10 | gac gcc atc cac gct gtt ttg acc | att tgc ctg cca ttg tgc tgc cag | Clark et al. [19] |
Induction of gene expression by doxycycline
Pregnant female mice had a diet of normal feed and water as prescribed by CHLA animal facilities. On the day of surgery, the mother’s normal feed was replaced with a doxycycline-supplemented (0.0625%) rodent diet (Harlan Teklad). They remained on the doxycycline-supplemented diet and pups were allowed to nurse as usual until tissue was harvested for continuous induction that was used for all experiments unless noted otherwise. The short-term induction consisted of replacing the normal feed to doxycycline- supplemented diet at 3 dpc and returning to normal diet at 10 dpc.
RNA isolation and quantitative/RT PCR
Ventricle tissue was homogenized in Trizol® using beaddisruption and RNA isolated as indicated with Trizol® extraction. RNA was quantified using Nanodrop® spectrophotometer. Generation of cDNA was done in vitro using a First Strand cDNA synthesis kit® (Invitrogen©) using one microgram of starting RNA. Total RNA total quantity was assessed by qPCR amplification of the cDNA using primers and probe combinations designed by the Roche Assay Design Center. The program was forty cycles at 94°C for ten seconds, 60°C for thirty seconds to anneal. The software calculated the C.P. and target gene levels were compared to housekeeping genes to determine the relative fold change in expression. All the primer sequences for PCR analysis are listed in table 1.
Histological analysis
Harvested tissues were fixed in 4% paraformaldehyde overnight at 4°C. Fixed tissue was than dehydrated, cleared in toluene, and paraffinized. Paraffin embedded tissue was sectioned at 10 µm. Sections were stained with hematoxylin and eosin and examined by light microscopy for histological changes.
Immunostaining and quantification
Tissue sections of 10 microns on glass slides were processed for immunostaining as follows: paraffin was removed with toluene washes, and then rehydrated in PBS with graded ethanol. Heat and acid antigen retrieval was done with Vector© Antigen Retrieval solution and a microwave. Tissue sections were stained with Wilm’s Tumor 1 antibody (Santa Cruz) and Vimentin antibody (Sigma) followed with secondary antibody goat anti-rabbit conjugated to Alexa 594 (Molecular Probes) and goat anti-mouse Alexa 488 and co-stained with Vectashield© mounting media with propridium iodide. Confocal images were taken on Zeiss LSM 710® single photon confocal microscope. EMT marker staining was done with antibody to Snail and Slug (Abcam) but followed with goat anti-rabbit_HRP (Dako) and color developed with DAB (Sigma) until brown color was visible then counterstained with hematoxylin. The average ratio of the number of Wt-1 positive nuclei (double stained with antibody and DAPI) to total nuclei (DAPI- stained) ratio was quantified by at least three sections of each sample for Wt1-positive cells within the injury area (scar regions were identified by consecutive tissue slides stained by AFOG and by a lack of green autofluorescence from myocardium for reference) for each heart. The measurements were analyzed for statistical significance using the Wilcoxon rank sum test.
Scar measurement analysis
To identify collagen scars, AFOG staining was performed as described [20]. Briefly, 10 micron tissue sections on slides were deparaffinized and rehydrated to PBS. The tissue slides were placed in Bouin’s fixative for 2.5 hours at 56°C then one hour at room temperature. The slides were washed of Bouin’s fixative for 20 minutes in running water. The slides were treated with 1% phosphomolybdic acid for 5 minutes then washed briefly with deionized water. Slides were immersed in and stained with acid fuschin, Orange G, Aniline blue (AFOG) trichrome staining solution for 5 minutes and then briefly rinsed with deionized water. Slides were then quickly processed with alcohol to dehydrate, cleared in toluene, and mounted with Cytoseal®. Tissue sections were imaged with bright-field microscopy. Images of sections were calibrated for measurement on Image J software. Three to five measurements spanning the width of the tissue were taken and averaged for scar length. Left ventricle circumference and the ratio of the average scar length to average left ventricle circumference was calculated. Similarly, three to five measurements spanning the length of the tissue were taken and averaged for scar thickness and septum thickness, and the ratio of the average scar thickness to average septum thickness was calculated. The measurements were analyzed for statistical significance using the Wilcoxon rank sum test.
Imaging/ microscopy
Whole mount imaging was performed with a Leica dissecting microscope and using Spot Advanced® imaging software. Fluorescent imaging of sections was performed using a Zeiss LSM 710© single photon confocal microscope with Zen® imaging software.
Results
Fgf10 is strongly expressed in the hearts in an inducible transgenic mouse model
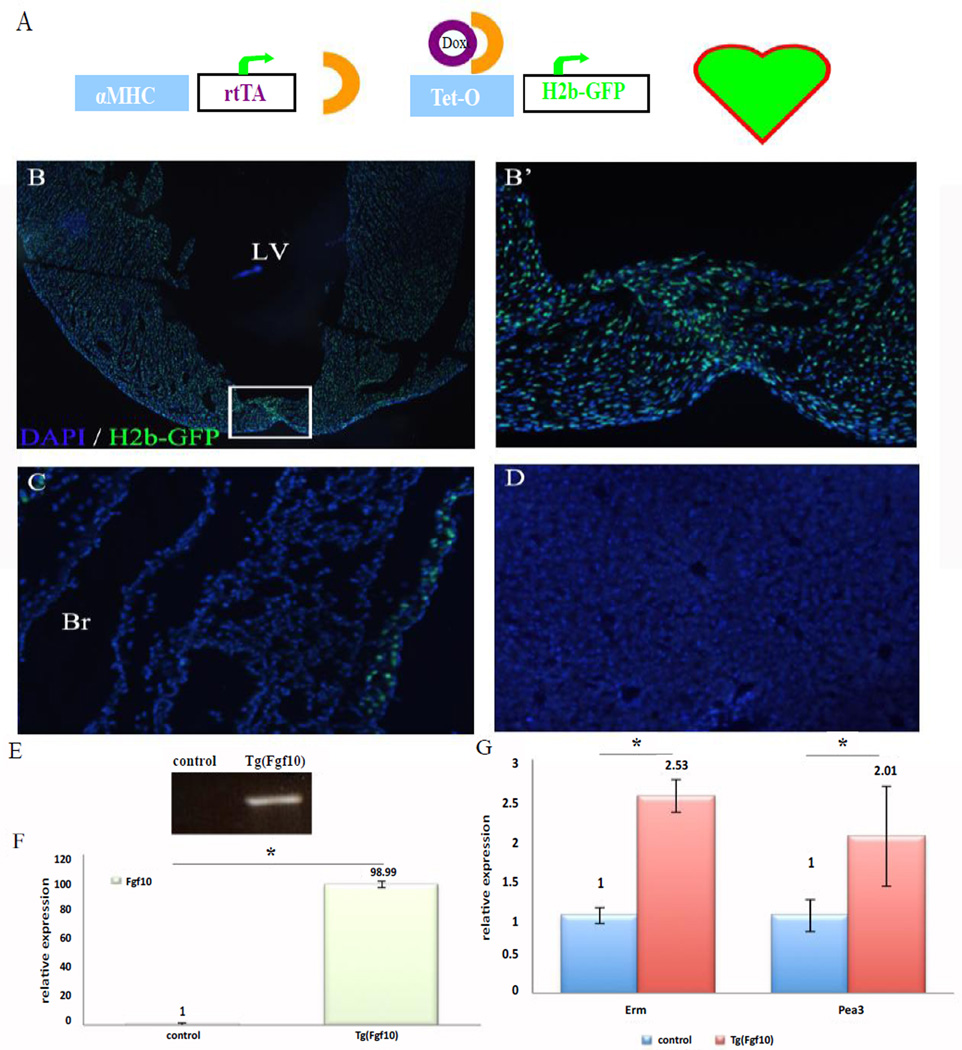
To achieve overexpression of Fgf10 in the myocardium, we crossed tet-O-Fgf10 mice to the α-myosin heavy chain (MHC) promoter driven rtTA mouse line (α-MHCrtTA) to generate double transgenic mice [Tg(Fgf10)] [19]. We utilized another tet- inducible reporter, tet-OH2b-GFP, which has GFP labeled histones that mark nuclei of double transgenic cells to validate tissue specificity of this double transgenic model [21]. The expression of the H2b-GFP or Fgf10 transgenes was controlled temporally via the addition of doxycycline (Figure 1A). α-MHC-rtTA; tet-O-H2b-GFP double transgenic mice showed myocardial specific expression (Figures 1B and 1B’). Other tissues examined, such as lung (Figure 1C) and liver (Figure 1D) did not show H2B-GFP expression except epithelium of the bronchi. When Tg(Fgf10) double transgenic mice were induced for 24 hr (from P9–P10), we detected a ~99 fold increase of total Fgf10 mRNA expression over control littermates (p<0.05) with qPCR analysis (Figures 1E and 1F; Table 2), indicating that strong Fgf10 expression can be induced in this double transgenic model. We further confirmed the induction of the FGF downstream targets Pea3 and Erm in the double transgenic mice. Real time PCR showed a 2.53- fold and 2.01-fold increase for Erm and Pea3, respectively (p<0.05) (Figure 1G).
Figure 1. Fgf10 is strongly expressed in the hearts in an inducible transgenic mouse model.
(A) Schematic of DNA constructs of the two transgenes that compose the tetinducible, heart- specific expression model of GFP- tagged histone (H2b-GFP) for assaying inducibility and tissue specificity using the alpha-MHC: rtTA in neonatal mice. Induction with doxycycline in neonatal mice up to the tenth day of age was used to label cells with both transgenes expressed. (B) Fluorescent microscopy images of heart. LV, left ventricle. The box in B indicates magnified area (B’). H2b-GFP expression was identified in only a few cells of the lung (C) bronchi (Br) but not in the liver (D). DAPI: blue; H2B-GFP: green. (E)RT -PCR with primers that specifically detect the Fgf10 transgene. Strong transgene expression after 24 hr doxycycline-induction was only detected in the hearts of Tg(Fgf10) double transgenic mice, but not in control littermates. (F) A ~99 fold increase of total Fgf10 RNA was determined by qPCR analysis. (G). The FGF target genes Erm and Pea3,showed a 2.53- and 2.01- fold increase in expression, respectively, in double transgenic mice compared to control littermates. Asterisk: P<0.05. n=3 for all the groups.
Table 2.
qPCR analysis.
| Gene | Sense | Antisense | Probe number (Roche universal probe library) |
|---|---|---|---|
| Fgf10 | cgg gac caa gaa tga aga ct | gca aca act ccg att tcc ac | 80 |
| Erm | tcc tac atg aga ggc gggta | gta cag cct ggg gtc ctt ct | 66 |
| Pea3 | cag act tcg cct acg act ca | gcc ata acc cat cac tcc at | 80 |
| 18s | aaa tca gtt atg gtt cct ttg gtc | gct cta gaa tta cca cag tta tcc aa | 55 |
| Fgfr2 | cct acc tca agg tcc tga agc | cat cca tct ccg tca cat tg | 21 |
Exogenous FGF10 overexpression did not affect the epicardium in the absence of injury
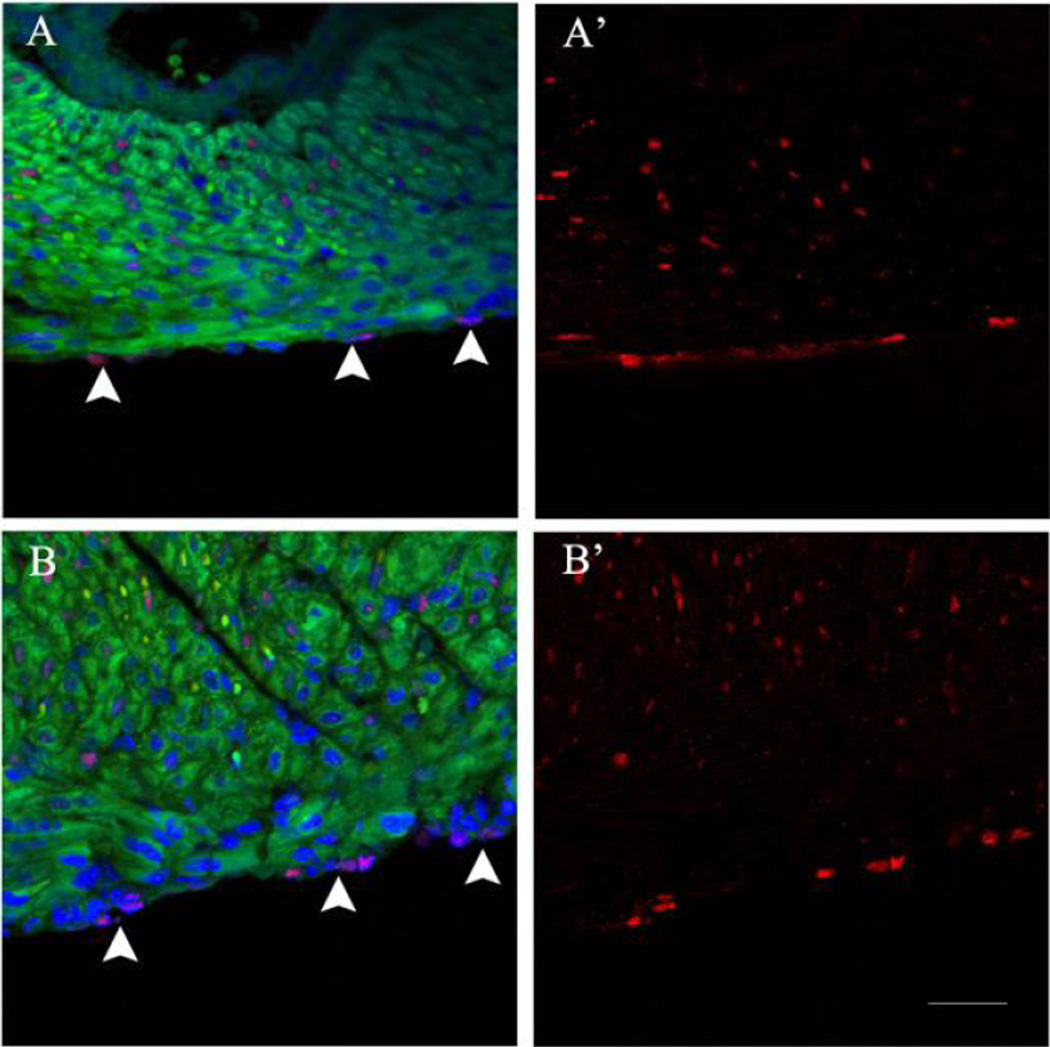
We employed the Tg(Fgf10) double transgenic mouse model to determine the effect of FGF10 overexpression on the Wt1-positive epicardial cells without heart injury. FGF10 was overexpressed from birth until collection at fourteen days of age (P14). Heart sections from the Tg(Fgf10) double transgenic mice and control littermates were stained with Wt1 antibody to detect changes to the thickness of the epicardium (layers of Wt1- positive cells). Control littermates (Figures 2A and 2A’) and Tg(Fgf10) mice (Figures 2B and 2B’) both had a single layer of Wt1 positive cells in the epicardium. Control littermates (Figures 2A and 2A’) and Tg(Fgf10) (Figures 2B and 2B’) mice both exhibited apparently normal histology of the epicardium with a single squamous epithelial morphology and positive nuclear staining for Wt1. These results suggest that overexpression of FGF10 had no effect on the morphology of epicardial cells in neonatal hearts in the absence of injury.
Figure 2. FGF10 overexpression had no effect on the morphology of the epicardium (Wt1-positive cells) in uninjured hearts.
Fluorescent microscopy images of hearts from the Tg(Fgf10) double transgenic mice and control littermate were stained with Wt1 antibody to identify the epicardium (layers of Wt1-positive cells). Control littermates (A and A’) and Tg(Fgf10) (B and B’) both demonstrated a single layer of Wt-1-positive cells in the epicardium (arrowheads) with no remarkable histological changes. Wt1: red; myocardium autofluorescence: green; DAPI: blue. Scale bar = 20 µm. n=3 for all the groups.
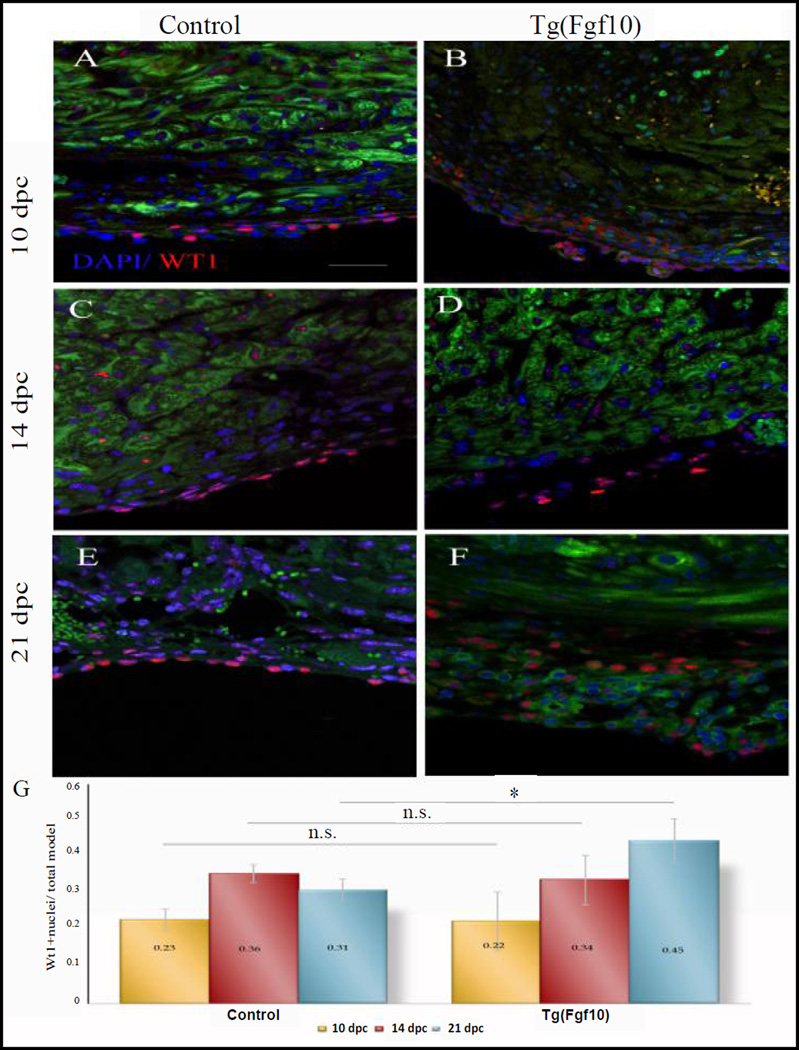
Epicardial cells respond to injury with an expansion Wt1- positive epicardial cells that were proliferative in response to FGF10
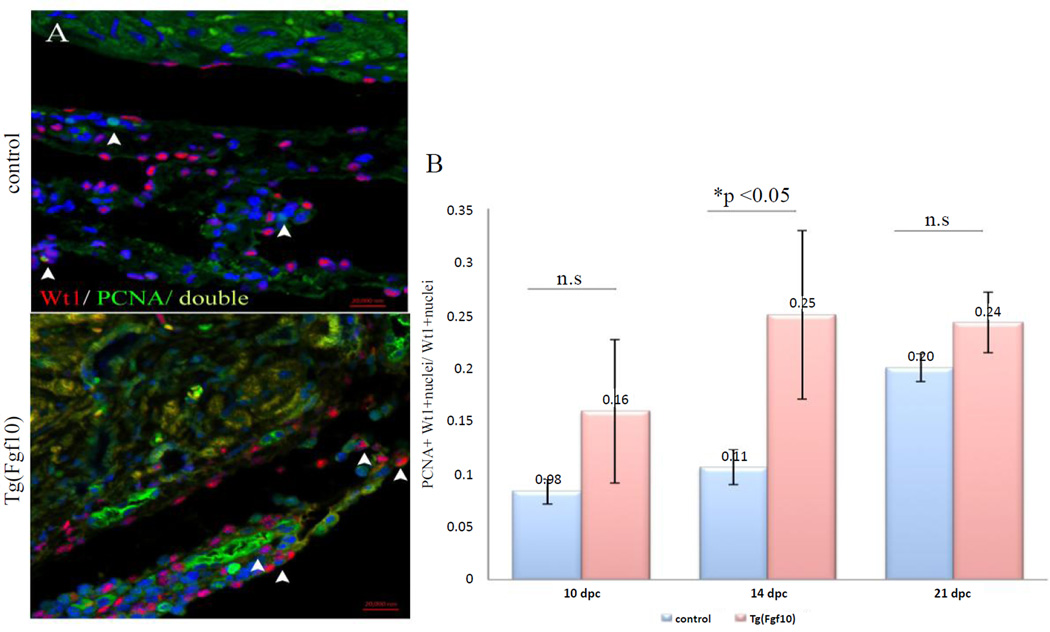
Similar to what was observed after ventricular resection, cryoinjury of neonatal mouse hearts also leads to an expansion of epicardial cells, which are usually a single layer thick to multiple layers of Wt1-positive epicardial cells and Darehzereshki et al. (unpublished) [5]. Analysis of the epicardial expansion in the injury area by Wt1 immunostaining showed that Wt1- positive cells increased to more than a single layer. Quantification of the ratio of Wt1- positive nuclei to total nuclei in the injury area demonstrated that a significant increase (p<0.05) of Wt1- positive cells was observed in Tg(Fgf10) double transgenic mice by 21 days post cryoinjury (dpc) (Figures 3E–3G), but not at the earlier time points of 10 dpc (Figures 3A and 3B) and 14 dpc (Figures 3C and 3D). To determine if the expansion of Wt1-positive epicardium resulted from epicardial cell proliferation, we performed double immunostaining of Wt1 and proliferating cell nuclear antigen (PCNA) to analyze Wt1- positive cells in the injury area at 10, 14 and 21 dpc. The PCNA-positive epicardial cells were low at all time points for both control (0.08 at 10 dpc, 0.11 at 14 dpc, 0.20 at 21 dpc) and Tg(Fgf10) mice (0.16 at 10 dpc, 0.25 at 14 dpc, and 0.24 at 21 dpc) (Figure 4A). Only Tg(Fgf10) double transgenic mice had a greater number of double-stained nuclei compared to controls at 14 dpc (p<0.05) (Figure 4B). The increased proliferation of Wt1-positive cells detected at 14 dpc contributes to an overall increase in this population, as seen at 21 dpc.
Figure 3. FGF10 enhances cryoinjury induced expansion of Wt1-positive cells in neonatal mouse heart at 21 dpc.
Wt1 staining of the neonatal heart of double transgenic Tg(Fgf10) mice (B, D, F) and control littermates (A, C, E) at 10 dpc (A, B), 14 dpc (C, D) and 21 dpc (E, F). Wt1: red; myocardium: green; DAPI: blue. Scale bar=20 µm. F. Quantification of Wt1 positive nuclei in Tg(Fgf10) double transgenic mice and control mice at 10, 14 and 21 dpc. Tg(Fgf10) double transgenic mice had a greater number (p<0.05) (asterisk) of Wt1-positive cells compared to the control littermates at 21 dpc. N=3 for all the groups.
Figure 4. FGF10 induces proliferation of Wt1-positive cells in neonatal mouse heart at 14 dpc.
(A) Double staining of Wt1 and proliferating cell nuclear antigen (PCNA) (arrowheads) to analyze proliferation of Wt1-positive cells in the injury area at 10, 14 and 21 dpc. PCNA: green; Wt1: red; DAPI: blue. PCNA-positive epicardial cells were infrequent at all time points for both control and Tg(Fgf10). (B) Quantification of PCNA positive epicardial cells (Wt1 positive) as a ratio of total epicardial cells. Tg(Fgf10) had a statistically greater number of double-stained nuclei compared to controls only at 14dpc (B). Scale bar =20 µm. n =3 for all the groups.
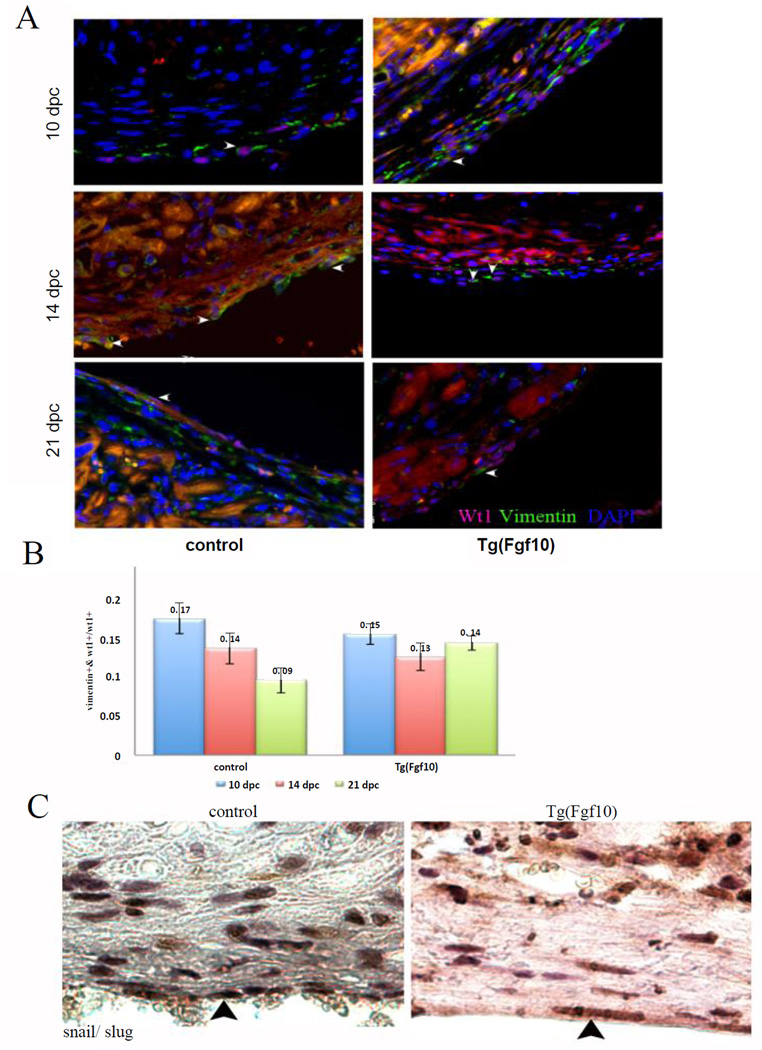
Fgf10 did not increase vimentin expression in epicardial cells (Wt1-positive cells) in cardiac injury
Cardiac injury alone leads to expansion of epicardial cells in neonatal mouse hearts [5]. Although Fgf10 overexpression increased the number of Wt1-positive cells in the injury by proliferation, it did not increase the expression of vimentin in Wt1- positive cells. Double staining of Wt1 and vimentin in hearts of Tg(Fgf10) and control littermates at 10 dpc, 14 dpc and 21 dpc showed similar expression in the injury area (Figure 5A). The ratio of vimentin-positive epicardial cells to total Wt1-positive cells was not significantly different between the Tg(Fgf10) mice (n=3 for all time points) and control mice (n=3 for all time points) at any of the time points (Figure 5B). Immunohistochemical staining for Snail and Slug indicated that epicardial cells at the injury site underwent EMT even at 21 dpc, but there was not a marked difference in expression of the EMT markers between Tg(Fgf10) mice and control (Figure 5C). These results indicate that FGF10 overexpression did not enhance EMT or vimentin expression in Wt1-positive epicardial cells after injury.
Figure 5. FGF10 overexpression does not increase vimentin-expressing epicardial cells in the injury area.
Double staining of Wt1 (magenta) and vimentin (green) (arrowheads) to determine fibroblast differentiation in Tg(Fgd10) double transgenic mice and control littermates at 10 dpc, 14 dpc and 21 dpc. (B). Quantification of cells expressing vimentin (green) in the cytoplasm and Wt1 (red) in the nuclei as ratio of total Wt1-positive nuclei, in Tg(Fgf10) double transgenic mice (n=3 for all time points) and control littermates (n=3 for all time points). The ratio of vimentin-positive/ Wt1-positive cells was not significantly different between the Tg(Fgf10) mice and controls at any of the time points. Scale bar=20µm. (C) Light microscopy image of 21 dpc hearts stained for Snail/ Slug indicate positive staining (arrowheads) on the epicardium at the of the injury site in both control and Tg(Fgf10).
Fgf10 overexpression did not affect infarct size after cryoinjury
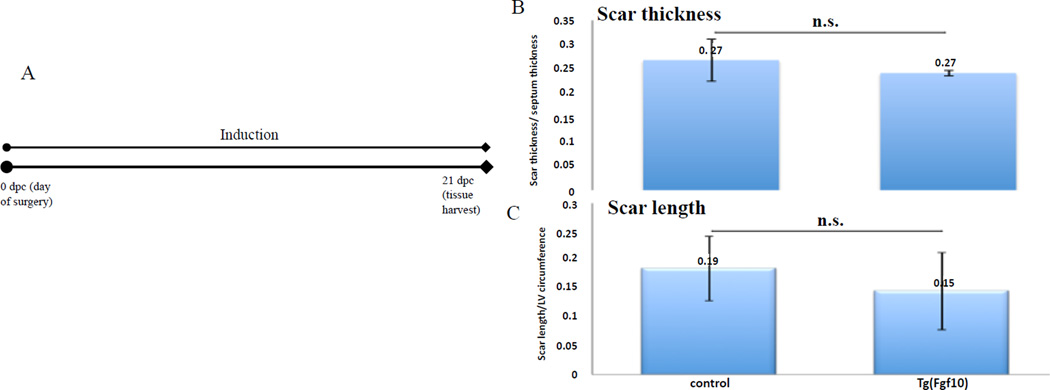
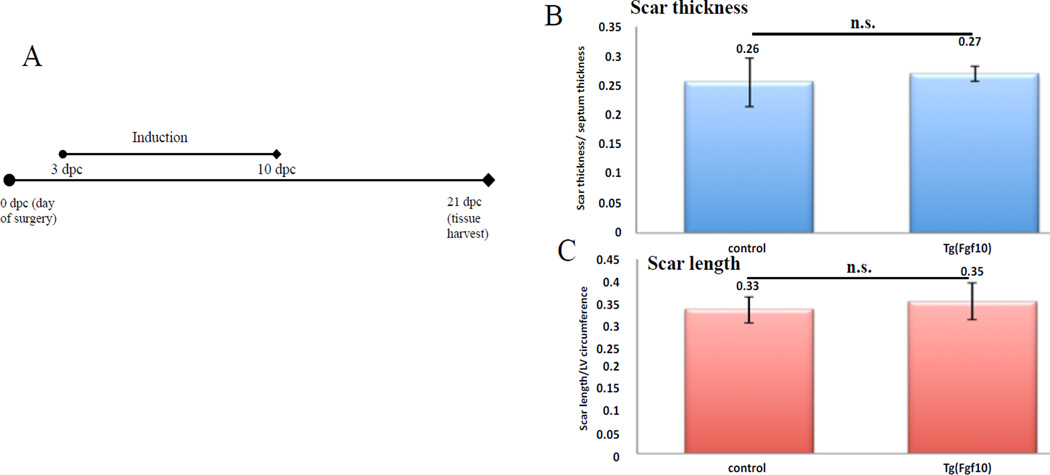
Neonatal mouse hearts do not regenerate after cryoinjury (Darehzereshki et al., in preparation). To determine if Fgf10 overexpression affects the amount of fibrosis in the heart after cryoinjury, we tested two time periods of Fgf10 induction in cryoinjured mice (Figures 6A and 7A). First, a continuous induction period beginning from the day of surgery and lasting until tissue procurement on day 21 post surgery was performed (Figure 6A). AFOG (acid fuschin, orange G, aniline blue) staining was used to analyze the scar; scar length and scar thickness were evaluated for changes in scar size. The length of the scar was measured and normalized to the circumference of the left ventricle (LV). The thickness of the scar area was also measured and normalized to the septum between the left and right ventricle. Continuous Fgf10 overexpression resulted in an average scar thickness ratio of 0.24 for Tg(Fgf10) hearts, which was not significantly different than control hearts (0.27) (Figure 6B). The average scar length to left ventricle circumference ratio was 0.15 in Tg(Fgf10) heart, which was also not significantly different from control hearts (0.19) (Figure 6C). To avoid the possibility that overexpression of FGF10 might affect inflammation, short-term overexpression of FGF10 from 3 dpc to 10 dpc was done (Figure 7A). Similar to the continuous overexpression, the average scar thickness ratio for Tg(Fgf10) hearts was 0.27 and not significantly different than control hearts (0.26) (Figure 7B). The average scar length to left ventricle circumference ratio in Tg(Fgf10) was 0.35 and the average for the control was 0.33, which were also not significantly different (Figure 7C). Overexpression of exogenous Fgf10 after the inflammatory phase of the cardiac ischemia injury, did not lead to a reduction in fibrosis.
Figure 6. Continuous overexpression of FGF10 does not alter scar size after cryoinjury.
(A) Doxycycline induction time period (immediately after surgery to 21 dpc). Transverse sections of the heart at 21 days post surgery were stained for AFOG. (B) The average scar thickness ratio for Tg(Fgf10) hearts was 0.24 and not significantly different than control hearts (0.27). (C) The average scar length to left ventricle circumference ratio in Tg(Fgf10) was 0.15 and the average for the control was 0.19, which were not significantly different. (n =3 for all the groups).
Figure 7. Transient overexpression of FGF10 does not alter scar size after cryoinjury.
(A) Doxycycline induction time period (3 to 7 dpc). Transverse sections of the heart at 21 days post surgery were stained for AFOG. (B) The average scar thickness ratio for Tg(Fgf10) hearts was 0.27 and not significantly different than control hearts (0.26). (C). The average scar length to left ventricle circumference ratio in Tg(Fgf10) was 0.35 and the average for the control was 0.33, which were not significantly different. (n=3 for all the groups).
Discussion
In this study, we utilized an inducible transgenic mouse model to study functions of Fgf10 in neonatal mouse heart repair. We demonstrated that Fgf10 could be strongly induced in myocardium in a temporally controlled manner. Overexpressing Fgf10 in the myocardium for paracrine signaling to the epicardium replicates the endogenous pattern seen in the late embryo [14]. However, the downstream targets of FGF signaling (such as Erm or Pea3) were only slightly induced. It is likely that the diffusion of Fgf10 is extremely limited by the extracellular matrix. During late embryonic heart development (E17.5), Fgf10 is expressed in the compact myocardium and the FGFR2 is mainly located in epicardium [15]. It is likely that FGFR2 has a similar expression pattern in the epicardium of neonatal mouse hearts. Therefore, even though we strongly overexpressed Fgf10 in the entire myocardium, only a small fraction of ectopically expressed Fgf10 might reach and signal to FGFR2b in the epicardium. During late embryonic heart growth (E17.5), myocardial Fgf10 signals to FGFR2b in epicardium and induces migration of epicardium derived cells (EPDC) into the myocardium. These migratory EPDCs then differentiate into vimentin expressing fibroblasts, which are important for the development of the myocardial extracellular matrix and correlates with cardiomyocyte proliferation during myocardial growth [15]. In our study at the neonatal stage, we observed expansion of Wt1 positive epicardial cells. In contrast to the observation at late embryonic stage Fgf10 treatment did not increase the number of vimentin-positive, Wt1-positive fibroblasts after heart injury. Heart injury can induce epicardial EMT; Fgf10 increases proliferation of epicardial cells [15]. However, our data show that Fgf10 does not seem to enhance EMT. During heart development, the epicardium can undergo EMT and contribute to fibroblasts and perivascular cells including smooth muscle [22–25]. During zebrafish heart regeneration, epicardium is reactivated and proliferates to contribute to perivascular cells and fibroblasts [2,9,26,27]. This regenerative process recapitulates what happens during embryonic heart development. Interestingly, the epicardium is also activated in adult mouse hearts after ischemic injury [28]. These EPDCs also express fibroblast and perivascular cell markers but remain a thickened layer at the surface of the heart and secrete paracrine factors that promote angiogenesis [28]. Similarly, activation and expansion of epicardial cells were also observed in neonatal mouse heart regeneration [5]. It remains to be determined if the expansion of epicardial cells plays beneficial or detrimental roles for neonatal heart regeneration and repair.
References
- 1.Kikuchi K, Poss KD. Cardiac regenerative capacity and mechanisms. Annu Rev Cell Dev Biol. 2012;28:719–741. doi: 10.1146/annurev-cellbio-101011-155739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lien CL, Harrison MR, Tuan TL, Starnes VA. Heart repair and regeneration: Recent insights from zebrafish studies. Wound Repair Regen. 2012;20:638–646. doi: 10.1111/j.1524-475X.2012.00814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poss KD. Getting to the heart of regeneration in zebrafish. Semin Cell Dev Biol. 2007;18:36–45. doi: 10.1016/j.semcdb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Roger Véronique L., Go Alan S., Lloyd-Jones Donald M., Adams Robert J., Berry Jarett D., et al. Executive Summary: Heart Disease and Stroke Statistics-- 2011 Update: A Report from the American Heart Association. Circulation. 2011;123:459–463. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porrelloa Enzo R., Mahmoudc Ahmed I., Simpsona Emma, Johnsona Brett A., Grinsfelderc David, et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. PNAS. 2012;110:1, 187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engel Felix B., Hsieh Patrick C. H, Lee Richard T., Keating Mark T. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc Natl Acad Sci U S A. 2006;103:15546–15551. doi: 10.1073/pnas.0607382103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheikh F, Sontag DP, Fandrich RR, Kardami E, Cattini PA. Overexpression of FGF-2 increases cardiac myocyte viability after injury in isolated mouse hearts. Am J Physiol Heart Circ Physiol. 2001;280:1039–1050. doi: 10.1152/ajpheart.2001.280.3.H1039. [DOI] [PubMed] [Google Scholar]
- 9.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 10.Matsunaga S, Okigaki M, Takeda M, Matsui A, Honsho S, et al. Endothelium-targeted overexpression of constitutively active FGF receptor induces cardioprotection in mice myocardial infarction. J Mol Cell Cardiol. 2009;46:663–673. doi: 10.1016/j.yjmcc.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Winter EM, Gittenberger-de Groot AC. Epicardium-derived cells in cardiogenesis and cardiac regeneration. Cell Mol Life Sci. 2007;64:692–703. doi: 10.1007/s00018-007-6522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sucov HM, Gu Y, Thomas S, Li P, Pashmforoush M. Epicardial control of myocardial proliferation and morphogenesis. Pediatr Cardiol. 2009;30:617–625. doi: 10.1007/s00246-009-9391-8. [DOI] [PubMed] [Google Scholar]
- 13.Brade T, Kumar S, Cunningham TJ, Chatzi C, Zhao X, et al. Retinoic acid stimulates myocardial expansion by induction of hepatic erythropoietin which activates epicardial Igf2. Development. 2011;138:139–148. doi: 10.1242/dev.054239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li P, Cavallero S, Gu Y, Chen TH, Hughes J, et al. IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development. 2011;138:1795–1805. doi: 10.1242/dev.054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vega-Hernández M, Kovacs A, De Langhe S, Ornitz DM. FGF10/FGFR2b signaling is essential for cardiac fibroblast development and growth of the myocardium. Development. 2011;138:3331–3340. doi: 10.1242/dev.064410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavine KJ, White AC, Park C, Smith CS, Choi K, et al. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavine KJ, Yu K, White AC, Zhang X, Smith C, et al. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Valencik ML, McDonald JA. Codon optimization markedly improves doxycycline regulated gene expression in the mouse heart. Transgenic Res. 2001;10:269–275. doi: 10.1023/a:1016601928465. [DOI] [PubMed] [Google Scholar]
- 19.Clark JC, Tichelaar JW, Wert SE, Itoh N, Perl AK, et al. FGF-10 disrupts lung morphogenesis and causes pulmonary adenomas in vivo. Am J Physiol Lung Cell Mol Physiol. 2001;280:705–715. doi: 10.1152/ajplung.2001.280.4.L705. [DOI] [PubMed] [Google Scholar]
- 20.Poss Kenneth D., Wilson Lindsay G., Keating Mark T. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 21.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merki E, Zamora M, Raya A, Kawakami Y, Wang J, et al. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci USA. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikawa T, Fischman DA. Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proc Natl Acad Sci USA. 1992;89:9504–9508. doi: 10.1073/pnas.89.20.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 25.Olivey HE, Compton LA, Barnett JV. Coronary vessel development: the epicardium delivers. Trends Cardiovasc Med. 2004;14:247–251. doi: 10.1016/j.tcm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Rubin N, Huang Y, Tuan TL, Lien CL. In vitro culture of epicardial cells from adult zebrafish heart on a fibrin matrix. Nat Protoc. 2012;7:247–255. doi: 10.1038/nprot.2011.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Wu Q, Zhang Y, Wiens KM, Huang Y, et al. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc Natl Acad Sci USA. 2010;107:17206–17210. doi: 10.1073/pnas.0915016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou B, Honor LB, He H, Ma Q, Oh JH, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest. 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]