Abstract
Background
Sterol glycosyltransferases (SGTs) are ubiquitous but one of the most diverse group of enzymes of glycosyltransferases family. Members of this family modulate physical and chemical properties of secondary plant products important for various physiological processes. The role of SGTs has been demonstrated in the biosynthesis of pharmaceutically important molecules of medicinal plants like Withania somnifera.
Results
Analysis suggested conserved behaviour and high similarity in active sites of WsSGTs with other plant GTs. Substrate specificity of WsSGTs were analysed through docking performance of WsSGTs with different substrates (sterols and withanolides). Best docking results of WsSGTL1 in the form of stable enzyme-substrate complex having lowest binding energies were obtained with brassicasterol, transandrosteron and WsSGTL4 with solasodine, stigmasterol and 24-methylene cholesterol.
Conclusion
This study reveals topological characters and conserved nature of two SGTs from W. somnifera (WsSGTs) i.e. WsSGTL1 and WsSGTL4. However, besides being ubiquitous in nature and with broad substrate specificity, difference between WsSGTL1 and WsSGTL4 is briefly described by difference in stability (binding energy) of enzyme-substrate complexes through comparative docking.
Electronic supplementary material
The online version of this article (doi:10.1186/s12859-015-0563-7) contains supplementary material, which is available to authorized users.
Keywords: Sterol glycosyltransferases, Withanolides, Sterols, Docking, Enzyme-substrate complex
Background
Glycosylation is an important step in biosynthesis of various natural products that modifies the physical and chemical properties of plant metabolites. Glycosylation of metabolites have been reported to enhance their solubility and stability as well as facilitates their accumulation and storage in plant cells which ultimately determine bioactivity and bioavailability of natural product [1]. Glycosylation reaction is catalyzed by the specific enzymes, glycosyltransferases, (GTs; EC 2.4.x.y) which belong to a multigene family and responsible for glycosidic bond formation by transfer of activated glycosyl group to a nucleophilic acceptor molecule. Prior to the reaction, substrate that acts as glycosyl group donor is activated as nucleoside diphosphate sugars [2]. Acceptor molecule for the GTs are oligosaccharides, polysaccharides and glyco-conjugates of the secondary metabolites including phenolics, terpenoids, cyanohydrins, thiohydroximates, flavonoids, sterols and alkaloids [3,4].
GTs have been classified on the basis of sequence similarities into 97 families as mentioned in Carbohydrate Active Enzyme Database (http://www.cazy.org) [5]. Members of each GT family having related consensus sequence, corollary fold along with catalytic specificity [6-8]. A comprehensive survey of the GTs demonstrated to adopt either GT-A or GT-B fold. Tightly adjoining β/α/β domain of the GT-A fold form continuous central sheet with at least eight β-strands, while two Rossmann-like less tightly associated β/α/β domains “face each other” with ligand binding displays in the GT-B fold and associated with the conformational changes in relative orientation [9]. Formation of the enzyme substrate complex requires two highly conserved domain of GTs, one of which binds to the UDP-sugar and another binds to sugar acceptor. UDP sugar binding domain is also called as plant secondary product GT consensus sequence (PSPG) box [3,10,11]. Formation of the enzyme-substrate complex by GTs and UDP-sugar interaction has been described briefly in various reports [1,2,12,13].
Functional role of sterol glycosyltransferase is of great importance in medicinal plants like Withania somnifera, Panax ginseng, Crocus sativus, chickpea, etc. [14-17]. Comparative analysis of sterol glucosyltransferase (SGT) activity towards sterols without side chain suggests relatively higher activity in W. somnifera as compared to Arabidopsis [14]. Another study mentioned substrate specificity of purified cytosolic and membrane-bound sterol glycosyltransferase of W. somnifera towards different sterol substrates [18]. This medicinal plant has been reported to various pharmaceutical activities because of various glycosylated molecules synthesized in this plant [19-22] and has been a target for the genomic characterization [15,23-31].
Present study demonstrates catalytic behaviour of two characterized members of Withania somnifera glycosyltransferases (WsSGTs), WsSGTL1 and WsSGTL4 among the large gene family [29]. The structural model of WsSGTL1 and WsSGTL4 is not available in database and is reported for the first time in this study. Analysis suggested specificity of these SGTs to specific molecules which might be important for synthesis of unique molecules with specific pharmaceutical activities. Several reports are available with detailed mechanism of UDP-glucose binding motif. But, involvement of the putative sterol binding motif in reaction mechanism has not been illustrated earlier. In the present study, along with structural model of WsSGTs, we also describe the sterols/withanolides-WsSGTs complexes, which require putative sterol binding domain (UDPGT) of the enzyme in order to catalyze reaction mechanism. Comparative analysis of protein followed by docking experiments performed to evaluate the comparative docking concert of proteins using AutoDock’s standard protocol.
Methods
Selection and sequence alignment of WsSGTs
Two WsSGT cDNA sequence of W. somnifera, WsSGTL1 and WsSGTL4 with accession number DQ356887.1 and EU342374, respectively, were selected from the work of Sharma et al. [14] and Chaturvedi et al. [15] and retrieved from NCBI nucleotide database (http://www.ncbi.nlm.nih.gov/nucleotide/). Sequence homology searches between selected WsSGT proteins i.e. WsSGTL1 and WsSGTL4 were carried out using BLAST algorithm against protein data bank (PDB). The deduced polypeptide sequence alignment was performed using ClustalW program (http://www.ebi.ac.uk/clustalW/) while shading was done with the Boxshade 3.21 program (http://www.ch.embnet.org/software/BOX_form.html/).
Topology alignment
Relationship of WsSGTs (WsSGTL1 and WsSGTL4) with the known proteins in the PDB along with resemblance of functionally important binding regions of proteins were analysed through the structural similarity scores using ProBis tool (http://probis.cmm.ki.si/) [32] by inspecting their physiochemical properties. Active sites for both the proteins are determined by DogSiteScorer [http://dogsite.zbh.uni-hamburg.de/] and ProBis tool. Three dimensional structures of proteins were further used for structural topology alignment against non-redundant PDB (nr-PDB) database to check the structural similarity and conserved regions in WsSGTs structure using by means of de novo comparisons of proteins ProBis tool. Structural superimposition was also performed using Chimera tool to find conserve structural folds [33].
Proteins data and model preparation
Osmani et al. [13] mentioned about the few reports available with demonstration of crystal structure of any GT. Structures of WsSGTL1 and WsSGTL4 were modelled using GENO3D (http://geno3d-pbil.ibcp.fr) [34] server where both sequences were submitted to search template by using condition NPS@3D sequences at 95% identity in PDB database with expectation value of 1e-06 applying blosum62 matrix. From provided outputs, pdb Id 3H4T was selected as template for the structure modelling. Structural refinement of both the models was accomplished by Molecular dynamics simulation (MD) using GROMOS56 force field in GROMACS along with the SPC model for water. MDs were done using a time step of 1 fs at 300 K, under these conditions 1 ns MD was performed using GROMACS [35]. Structural modelling provided insight about mechanism of action of WsSGTL1 and WsSGTL4, the information of participating amino acids and clarifying the mechanism of action of interaction between WsSGTL1 and WsSGTL4. Maximum likelihood algorithm in MEGA4 was used to construct phylogenetic tree using neighbour joining method with 100 bootstrap values. Accession numbers of the organisms are provided in Additional file 1.
Ligands data and preparation
The models of WsSGTL1 and WsSGTL4 were used to study protein-ligand conformations by automatic docking for 14 substrates including 10 sterols (β-sitosterol, brassicasterol, deactyl-16-DPA, dehydro-epiandrosteron, epoxypregnenolone, ergosterol, pregnenolone, transandrosterone, solasodine, stigmasterol and 24-methylene cholesterol) and 4 withanolides (withaferin A, withanolide A, withanolide B and withanolide D). Structure Data File (SDF) of the selected ligands were downloaded from the Pubchem (http://pubchem.ncbi.nlm.nih.gov) and further converted into PDB format using OpenBabel Tool (http://openbabel.org/wiki/Main_Page) [36].
Docking simulations
Docking studies were performed to predict the putative modification of binding modes of group of sterols and withanolides with the structural model of WsSGTL1 and WsSGTL4. The grid size was set to cover both acting domains present in WsSGTL1 and WsSGTL4 protein with grid spacing of 0.375 Å. Genetic algorithm (GA) was applied as searching parameter with 10 number of GA runs and setting population size 150, maximum number of energy evaluations was set to 25,00,000 with considering the maximum number of generations to 27,000. Binding of WsSGTs (WsSGTL1 and WsSGTL4) with different ligands were performed using AutoDock 4.0 (http://autodock.scripps.edu) [37]. The lowest binding energy conformation with H-Bonds in cluster was considered as the most favourable docking pose. Protein-ligand complexes obtained from AUTODOCK 4 were further viewed in UCSF-Chimera molecule viewer tool for better analysis of interaction [33]. In each case, 10 different docking arrangements were produced. The conformations obtained as result of rigid body docking were sorted by total binding energy, hydrogen bonds formed, bond lengths and close contacts between enzyme active sites.
Results and discussion
Evolutionary conserved nature of WsSGTs
Structural similarities as well as conserved functional domains of WsSGTs have been detected from a large database (ProBiS database) of the protein structures. Structural annotations of WsSGT proteins showed some interesting features. The active site of protein is aligned with nrPDB using PROBIS tool, which aligns active site geometry with similar amino acids. The result showed that the structure of active site is very much similar to chimeric glycosyltransferase (3H4T) of Actinoplanes teichomyceticus, the bacterial GT with high similarity of physiochemical properties on the basis of structural equivalence for the both proteins. The structure comparison of WsSGTL1 and WsSGTL4 was also made for the flavonoid glycosyltransferase protein of Vitis vinifera (pdb 2C1X) and iso-flavonoid glycosyltransferase protein of Medicago truncatula (pdb 2PQ6), which demonstrates the similarity in structures as well as evolutionary conserved regions in the protein structure (Figure 1).
Figure 1.
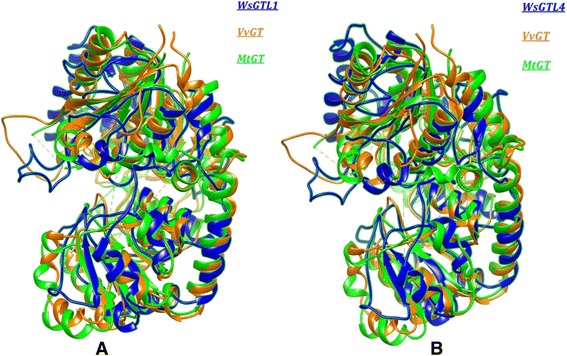
Topological alignment of WsSGTs. Local structural superimposition showing similarity of V. vinifera GT (yellow), M. truncatula GT (green), with (A) WsSGTL1 (blue) and (B) WsSGTL4 (blue).
On the basis of sequence similarity, more than 75% of GTs from prokaryotes to eukaryotes were grouped into three monophyletic super families named as GT-A, GT-B and GT-C. Among three, GT-A and GT-B evolved as most diverse and ubiquitous group of GTs as GT-A includes variety of organisms i.e. E. coli, Bacillus subtilis, Bos taurus, Oryctolagus cuniculus, Mus musculus, Neisseria meningitides, Homo sapiens, etc. [38]. Structure of WsSGTL1 and WsSGTL4 were modelled using homology modelling and are reported first time in this study. The proposed models show structural organization which contain 17 α-helices and 3 β-strands for the WsSGTL1 (Figure 2A) and 19 α-helix and 2 β-strands for the WsSGTL4 (Figure 2B). Previous studies indicated that the structure of these two domains of WsSGTs is crucial for the activity and therefore the domains in both proteins were evaluated [6,12,39]. Structures of both the proteins suggested that WsSGTL1 and WsSGTL4 belong to the GT-B family glycosyltransferase, as in members of GT-B family all β-sheets of protein are in parallel orientation (Figure 2).
Figure 2.
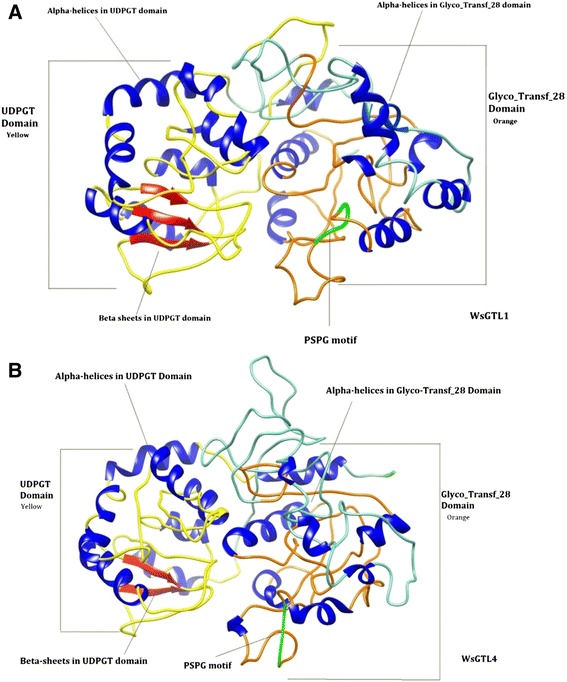
Ribbon diagrams of WsSGT proteins. (A) WsSGTL1 (B) WsSGTL4 showing Glyco_tranf_28 domain (orange), UDPGT domain (yellow), PSPG box (green), β-sheets (red) and α-helices (blue).
Differences between WsSGTL1 and WsSGTL4 proteins
WsSGTL1 (701aa) and WsSGTL4 (622 aa) have variation in sequence length as well as in their structure. The sequence alignment of these two sequences shows 55% identity with 72% conservative substitution (Figure 3A). Phylogenetic analysis of both the proteins suggested that WsSGTL1 was closer to S. lycopersicum GT, whereas WsSGTL4 have more similarity to M. truncatula GT (Figure 3B). Phylogenetic analysis of GTs performed by Coutinho et al. and Hashimoto et al. explained the ancient origin of GTs during evolution from the time of divergence of prokaryotes and eukaryotes [9,40]. Energy minimization results for both proteins reflects that WsSGTL4 seems more stable than the WsSGTL1, as WsSGTL1 stabilizes on −1.20 kJ/mol while WsSGTL4 stabilizes on −1.58 kJ/mol (Figure 3C),
Figure 3.
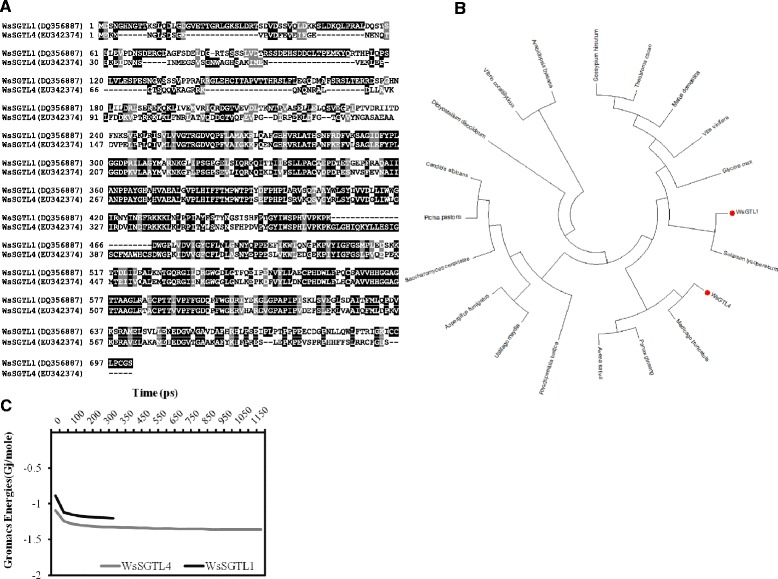
Similarity and difference between WsSGTL1 and WsSGTL4 proteins. (A) Sequence alignment of WsSGTL1, WsSGTL4 showing the close relationship between two enzymes. Shaded region showed highly conserved amino acids. (B) Gromacs energy plot of WsSGTL1 and WsSGTL4 proteins. (C) Phylogentic analysis of WsSGTL1 and WsSGTL4 with GTs of other organisms.
Active sites of both the protein involve UDPGT domain and a part of glyco_transf_28 domain. Comparison of active sites of these proteins indicated that cavity volume of WsSGTL4 was much bigger (2371.07 Å3) than WsSGTL1 (1481.16 Å3) (Table 1). Volume of cavities suggested more sensitive nature of WsSGTL4 for the reaction with substrates as compared to WsSGTL1. Number of H-bond donors being much more (66) in WsSGTL4 as compared to WsSGTL1 (38). Number of hydrophobic residues (116) is more in WsSGTL4 in comparison with WsSGTL1 (67 residues) as shown in Table 1.
Table 1.
Active site details of Ws SGTL1 and Ws SGTL4 proteins
| Active site descriptors | Ws SGTL1 | Ws SGTL4 |
|---|---|---|
| Size and shape | ||
| Volume [Å3] | 1481.16 | 2371.07 |
| Surface [Å2] | 1801.17 | 2989.3 |
| Lipophilic surface [Å2] | 1260.55 | 2211.65 |
| Depth [Å] | 22.86 | 39.6 |
| Ellipsoid main axis ratio c/a | 0.32 | 0.21 |
| Ellipsoid main axis ratio b/a | 0.7 | 0.26 |
| Enclosure | 0.05 | 0.07 |
| Functional groups | ||
| Hydrogen bond donors | 38 | 66 |
| Hydrogen bond acceptors | 100 | 159 |
| Metals | 0 | 0 |
| Hydrophobic interactions | 67 | 116 |
| Hydrophobicity ratio | 0.33 | 0.34 |
| Elements | ||
| Pocket atoms | 352 | 579 |
| Carbon (C) | 246 | 399 |
| Nitrogen (N) | 51 | 90 |
| Oxygen | 53 | 86 |
| Sulphur (S) | 2 | 4 |
| Other elements | 0 | 0 |
| Amino acid composition | ||
| Apolar amino acid ratio | 0.55 | 0.52 |
| Polar amino acid ratio | 0.31 | 0.32 |
| Positive amino acid ratio | 0.06 | 0.09 |
| Negative amino acid ratio | 0.08 | 0.08 |
| Amino acids | ||
| ALA | 10 | 14 |
| ARG | 1 | 5 |
| ASN | 4 | 6 |
| ASP | 2 | 6 |
| CYS | 2 | 2 |
| GLN | 2 | 4 |
| GLU | 4 | 3 |
| GLY | 7 | 13 |
| HIS | 3 | 4 |
| ILE | 1 | 10 |
| LEU | 9 | 8 |
| LYS | 1 | 2 |
| MET | 2 | 3 |
| PHE | 7 | 12 |
| PRO | 5 | 8 |
| SER | 3 | 4 |
| THR | 3 | 4 |
| TRP | 2 | 3 |
| TYR | 3 | 5 |
| VAL | 6 | 4 |
| Special amino acids | 0 | 0 |
Docking of WsSGTL1 and WsSGTL4 with different sterols
Protein-ligand conformations by automatic docking with chosen ligands (sterols and withanolides) have been analysed using proposed model of WsSGTs. For all ligands tested, the negative energies indicated a favourable interaction between the proteins and the ligands (Table 2). The obtained results revealed that the higher interaction energy was observed along with stable bonding for WsSGTL1 with brassicasterol, transandrosterone, 24-methylene cholesterol, ergosterol and β-sitosterol. Stigmasterol and solasodine have similar binding energies followed by pregnenolone (Table 2). The highest affinity energy of WsSGTL1 was −11.36, −9.95 and −9.75 kcal/mol for brassicasterol, transandrosterone and 24-methylene cholesterol, respectively. The best conformation of WsSGTL1 was found with brassicasterol having −11.36 kcal/mol of binding energy (Figure 4A). The model revealed that ASP535 is involved in formation of H-bond with the ‘3β-OH group’ of brassicasterol and the distance between the reactive functional group of protein and ligand was 1.863 Å. Another amino acid, PHE506 of WsSGTL1 formed H-Bond through –NH group reacting with oxygen of brassicasterol and the distance between the reactive functional groups of protein and ligand is 3.07 Å (Figure 4A). The second complex with the highest negative energy (−9.95 kcal/mol) of WsSGTL1 is with transandrosteron, which forms a stable complex by forming H-bond with ‘O’ of PRO55 (Figure 4B). Docking pose of 24-methylene cholesterol and ergosterol is represented in Figure 4C and D, respectively.
Table 2.
Binding energy of selected substrates for Ws SGTL1 and Ws SGTL4 proteins
| Ligands | Binding energy (kcal/mol) | |
|---|---|---|
| Sterols | Ws SGTL1 | Ws SGTL4 |
| β-Sitosterol | −9.44 | −8.36 |
| Brassicasterol | −11.36 | −7.97 |
| Deactyl-16-DPA | −8.54 | −8.0 |
| Dehydroepiandrosteron | −8.28 | −7.57 |
| Epoxypregnenolone | −7.14 | −6.25 |
| Ergosterol | −9.71 | −8.21 |
| Pregnenolone | −8.97 | −7.88 |
| Solasodine | −9.42 | −9.4 |
| Stigmasterol | −9.42 | −9.4 |
| Transandrosterone | −9.95 | −8.44 |
| 24-methylene cholesterol | −9.75 | −8.65 |
| Withanolides | ||
| Withaferin A | −10.21 | −9.14 |
| Withanolide A | −9.28 | −10.51 |
| Withanolide B | −9.19 | −9.21 |
| Withanolide D | −9.28 | −8.96 |
Figure 4.
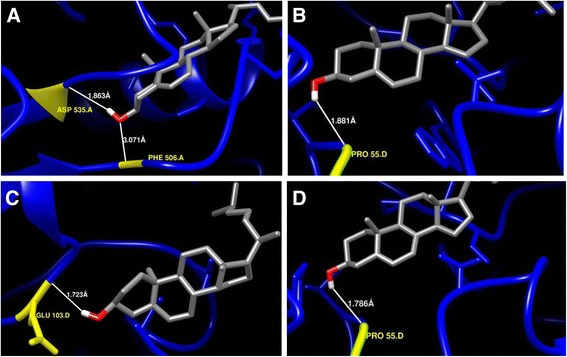
Enlarged view of interaction of WsSGTL1 protein with some selected ligand molecules. (A) Brassicasterol, (B) Transandrostrone, (C), 24-methylene cholesterol, and (D) Ergosterol. Interacting amino acid/s of protein, ‘H’ and ‘O’ atoms of hydroxyl group of interacting ligand are denoted by yellow, white and red colour, respectively.
For the WsSGTL4 protein, the best conformations were with solasodine and stigmasterol with affinity energy of −9.40 kcal/mol for both the ligands (Table 2). The best conformations observed in solasodine and stigmasterol reveals that these two substrates follow a similar way of interaction with WsSGTL proteins (Figure 5A,B). In WsSGTL4, both the ligands interact in a similar manner by stabilizing the complex with 2 H-bonds with same residues ASP11 with OD2 position and ALA350 with HN position maintaining the energy value of −9.4 kcal/mol. The second most stable confirmation is with 24-methylene cholesterol followed by transandrosterone with affinity energy of 8.65and −8.44 kcal/mol (Table 2; Figure 5C,D).
Figure 5.
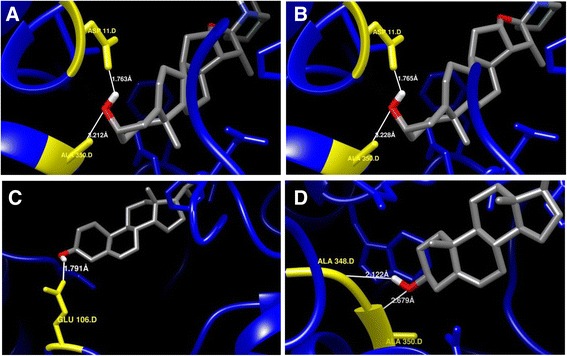
Enlarged view of interaction of WsSGTL4 protein with some selected ligand molecules. (A) Solasodine, (B) Stigmastrol, (C) 24-methylene cholesterol, and (D) Transandrosterone. Interacting amino acid/s of protein, ‘H’ and ‘O’ atoms of hydroxyl group of interacting ligand are denoted by yellow, white and red colour, respectively.
In all above interactions of sterols the main functional group interacting with WsSGTL proteins is ‘3β-OH group’ which indicates that it is the main active functional group in sterols. Sharma et al. and Madina et al. were also reported that the WsSGTLs have affinity for the interaction with ‘3β-OH group’ of sterols [14,18]. Docking of all these sterols is restricted to UDPGT as well as glyco_transf_28 domain. Gromacs energy plots of WsSGTL1 and WsSGTL4 for each ligand present in Table 2 were provided in Additional files 2 and 3. Most of the sterols prefer the nonpolar hydrophobic residues ALA, PRO and VAL where ‘O’ position of these amino acids is the preferential site for binding with the 3β-OH group of sterols. 3β-OH group also show affinity for negatively charged Aspartate at ‘O’ and OD1 position for their interaction. In case of positively charged ARG, HH11 and HN positions were preferred for binding. On the other hand, clear pattern of interaction was observed by overlooking for interactions of the WsSGTL4 with different sterols. Observation reveals that sterols show the tendency to bind with preferred residues with specificity for a position in some amino acids as ALA350, ALA348, ASP11, PRO200 and HIS146. WsSGTL4 prefer -HN position of ALA350 residue for 3β-OH group of sterols, where in case of WsSGTL1, sterols shows affinity towards the ‘O’ of ALA residue. The negatively charged Asp residue at 11th position consistently interacts with OD2 position. In all its interactions non polar cyclic residue PRO200 binds with sterols via ‘O’ position.
Docking of WsSGTL1 and WsSGTL4 with withanolides
Withanolides might also be serving as a substrate for WsSGTs, as these enzymes catalyze the biosynthesis of glycol-withanolides. It was observed that WsSGTL1-withaferin-A complex generated with −10.21 kcal/mol of binding energy with the single hydrogen-bond via -HN of ALA327 with the bond length of 2.686 Å and −3.747 kcal/mol of the bond energy (Figure 6A). The distance between the interacting functional groups showed that this position is quite favourable for the stabilizing the complex through the H-bond. Binding of withanolide A reveals that it forms a significantly stable complex with WsSGTL1 protein by forming 3 H-bonds with energy value of −9.28 kcal/mol involving ALA325 at ‘O’ position, ASP11 at OD1 position and ALA327 at HN position. It also reflects that it prefers hydrophobic side chain for the interaction.
Figure 6.
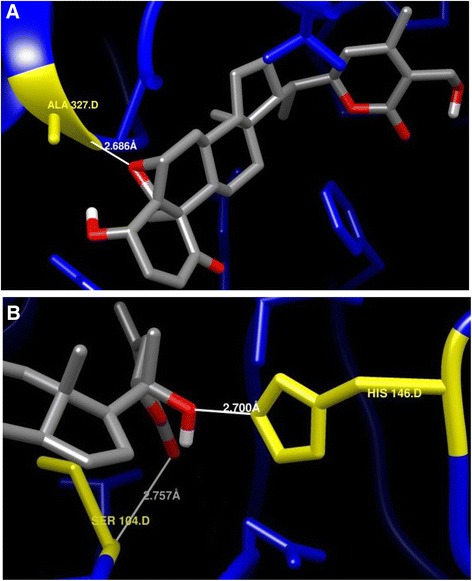
Enlarged view of interaction of WsSGT proteins with some selected withanolides. (A) WsSGTL1 with withaferin A, (B) WsSGTL4 with withanolide A. Interacting amino acid/s of protein, ‘H’ and ‘O’ atoms of hydroxyl group of interacting ligand are denoted by yellow, white and red colour, respectively.
Interactions of WsSGTL4 with withanolides shows its affinity towards withanolide A and withanolide B with higher (negative) binding energy forming the conformations of −10.51 and −9.21 kcal/mol, respectively. Complex of withanolide A with WsSGTL4 showed enzyme-substrate complex with −10.15 kcal/mol of binding energy and stabilized by 2 hydrogen-bonds (hydrogen of -NH group of SER104 and hydrogen of HIS146 residue) between withanolide A and WsSGTL4 enzyme (Figure 6B). Distance between the interacting functional groups are 2.757 Å (SER104 at HN position) and 2.70 Å (HIS146 at HE2 position), which are suitable distance for the supporting H-bonds. Other higher energy conformation is of witanolide B which forms single H-bond with SER74 at OG position with binding energy of −9.21 kcal/mol. Gromacs energy plots of the WsSGTL1 and WsSGTL4 is provided in Additional file 4.
Withanolides, the steroidal lactones, prefer ALA327, ALA325 and ASP11 residues of WsSGTL1 mainly for binding where -HN position for ALA327, ‘O’ position for ALA325 and OD1 position for ASP11 were observed for involvement in binding. In Withaferin A, the 5β- position ‘O’ molecule from steroid chain and 27th position hydroxyl group in lactone chain, participate in the reaction. In reaction of Withanolide A with WsSGTL1, again the same group participates in the interaction, whereas interaction of WsSGTL4 with the withanolide A involves participation of the 26th position ‘O’ as well as 22th position -OH group of the lactone chain in the complex formation. These results clearly reflect the tendency of the interaction of hydrophobic and negatively charged residues of WsSGTs while in sterols, the 3β-OH group is the main interacting chemical domain. In withanolides, no such common interacting group was observed as each withanolide dock with the residues with the different chemical moieties.
Conclusion
Glycosyltransferases are one of the largest families of enzymes which catalyze glycosylation of variety of acceptor molecules by the transfer of glycosyl moiety from activated nucleoside diphosphate sugar donar [41]. WsSGTL1 and WsSGTL4 differ in size as observed through sequence alignment and difference in their affinity towards different substrates. Observations collected in this study indicated that WsSGTL1 and WsSGTL4 interacts with different substrate and follow the different pattern of interaction. Hydrophobic amino acids as well as those with charged side chains play important role in the interaction with sterols. Results obtained in this study indicated that brassicasterol and withanolide A are the preferred substrates for WsSGTL1 and WsSGTL4, respectively. The interactions with different ligand molecules reveal that both proteins interact with all the mentioned ligands due to broad substrate specificity, however, have different affinity for the same substrate. The current study is predictive and needs to be confirmed experimentally using functional genomics approaches. This study shed light to understand glycosylation mechanism of sterol glycosides in plants.
Acknowledgements
The authors are thankful to the Council of Scientific and Industrial Research, New Delhi, Govt. of India, for providing the financial support to carry out this work under NMITLI scheme. VP and PG acknowledge CSIR for senior research fellowship.
Additional files
Accession numbers of organisms used in construction of phylogenetic tree.
Gromacs energy plots of Ws SGTL1 and Ws SGTL4 with sterols.
Gromacs energy plots of Ws SGTL1 and Ws SGTL4 with withanolides.
Chemical structures of sterols and withanolides.
Footnotes
Vibha Pandey, Yogeshwar Vikram Dhar and Parul Gupta contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
VP, YVD and PG performed the experiments. VP and PG wrote the manuscript. YVD and PG performed data analysis. PKT and MHA designed the study, gave experimental suggestions and helped in the writing of the manuscript. SKB, NA and PM helped in troubleshooting during the analysis. All authors read and approved the final manuscript.
Contributor Information
Vibha Pandey, Email: vibhashekharmishra@gmail.com.
Yogeshwar Vikram Dhar, Email: yogi.bioinfo@gmail.com.
Parul Gupta, Email: gupta.parulnbri@gmail.com.
Sumit K Bag, Email: sumit.bag@nbri.res.in.
Neelam Atri, Email: neelam.bhu@gmail.com.
Mehar Hasan Asif, Email: mh.asif@nbri.res.in.
Prabodh Kumar Trivedi, Email: prabodht@nbri.res.in.
Pratibha Misra, Email: pratibhaflora@yahoo.com.
References
- 1.Wang X. Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett. 2009;583:3303–9. doi: 10.1016/j.febslet.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 2.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–55. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 3.Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 2000;5:380–6. doi: 10.1016/S1360-1385(00)01720-9. [DOI] [PubMed] [Google Scholar]
- 4.Jones P, Vogt T. Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta. 2001;213:164–74. doi: 10.1007/s004250000492. [DOI] [PubMed] [Google Scholar]
- 5.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database CAZy in 2013. Nucleic Acids Res. 2014;42:D490–5. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross J, Li Y, Lim E, Bowles DJ. Higher plant glycosyltransferases. Genome Biol. 2001;2:3004. doi: 10.1186/gb-2001-2-2-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y, Walker S. Remarkable structural similarities between diverse glycosyltransferases. Chem Biol. 2002;9:1287–96. doi: 10.1016/S1074-5521(02)00295-8. [DOI] [PubMed] [Google Scholar]
- 8.Breton C, Snajdrová L, Jeanneau C, Koca J, Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16:29R–37. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- 9.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–17. doi: 10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 10.Paquette S, Møller BL, Bak S. On the origin of family 1 plant glycosyltransferases. Phytochemistry. 2003;62:399–413. doi: 10.1016/S0031-9422(02)00558-7. [DOI] [PubMed] [Google Scholar]
- 11.Mulichak AM, Losey HC, Walsh CT, Garavito RM. Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure. 2001;9:547–57. doi: 10.1016/S0969-2126(01)00616-5. [DOI] [PubMed] [Google Scholar]
- 12.Chaturvedi P, Misra P, Tuli R. Sterol glycosyltransferases-the enzymes that modify sterols. Appl Biochem Biotechnol. 2011;165:47–68. doi: 10.1007/s12010-011-9232-0. [DOI] [PubMed] [Google Scholar]
- 13.Osmani SA, Bak S, Møller BL. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modelling. Phytochemistry. 2009;70:325–47. doi: 10.1016/j.phytochem.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Sharma LK, Madina BR, Chaturvedi P, Sangwan RS, Tuli R. Molecular cloning and characterization of one member of 3b-hydroxy sterol glucosyltransferase gene family in Withania somnifera. Arch Biochim Biophys. 2007;460:48–55. doi: 10.1016/j.abb.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 15.Chaturvedi P, Mishra M, Akhtar N, Gupta P, Mishra P, Tuli R. Sterol glycosyltransferases-identification of members of gene family and their role in stress in Withania somnifera. Mol Biol Rep. 2012;39:9755–64. doi: 10.1007/s11033-012-1841-3. [DOI] [PubMed] [Google Scholar]
- 16.Sharma R, Rawat V, Suresh CG. Genome-Wide Identification and Tissue-specific expression analysis of UDP-glycosyltransferases genes confirm their abundance in Cicer arietinum chickpea genome. PLoS One. 2014;9:e109715. doi: 10.1371/journal.pone.0109715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapero A, Ahrazem O, Rubio-Moraga A, Jimeno ML, Gómez MD, Gómez Gómez L. Characterization of a glucosyltransferase enzyme involved in the formation of kaempferol and quercetin sophorosides in Crocus sativus. Plant Physiol. 2012;159:1335–54. doi: 10.1104/pp.112.198069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madina BR, Sharma LK, Chaturvedi P, Sangwan RS, Tuli R. Purification and physico-kinetic characterization of 3β-hydroxy specific sterol glucosyltransferase from Withania somnifera L. and its stress response. Biochim Biophys Acta. 2007;1774:392–402. doi: 10.1016/j.bbapap.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Ahlawat P, Khajuria A, Bhagwat DP, Kalia B. Therapeutic benefits of Withania somnifera: An exhaustive review. Int J Pharma Chem Sci. 2012;1:491–6. [Google Scholar]
- 20.Alam N, Hossain M, Mottalib MA, Sulaiman SA, Gan SH, Khalil MI. Methanolic extracts of Withania somnifera leaves, fruits and roots possess antioxidant properties and antibacterial activities. BMC Complement Altern Med. 2012;12:175. doi: 10.1186/1472-6882-12-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kataria H, Wadhwa R, Kaul SC, Kaur G. Withania somnifera water extract as a potential candidate for differentiation based therapy of human neuroblastomas. PLoS One. 2013;8:e55316. doi: 10.1371/journal.pone.0055316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khedgikar V, Kushwaha P, Gautam J, Verma A, Changkija B, Kumar A, et al. Withaferin A: a proteasomal inhibitor promotes healing after injury and exerts anabolic effect on osteoporotic bone. Cell Death Dis. 2013;4:e778. doi: 10.1038/cddis.2013.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandey V, Misra P, Chaturvedi P, Mishra MK, Trivedi PK, Tuli R. Agrobacterium tumefaciens mediated transformation of Withania somnifera L. Dunal: an important medicinal plant. Plant Cell Rep. 2010;29:133–41. doi: 10.1007/s00299-009-0805-0. [DOI] [PubMed] [Google Scholar]
- 24.Pandey V, Niranjan A, Atri N, Chandrashekhar K, Mishra MK, Trivedi PK, et al. WsSGTL1 gene from Withania somnifera, modulates glycosylation profile, antioxidant system and confers biotic and salt stress tolerance in transgenic tobacco. Planta. 2014;239:1217–31. doi: 10.1007/s00425-014-2046-x. [DOI] [PubMed] [Google Scholar]
- 25.Akhtar N, Gupta P, Sangwan NS, Sangwan RS, Trivedi PK. Cloning and functional characterization of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Withania somnifera: an important medicinal plant. Protoplasma. 2013;250:613–22. doi: 10.1007/s00709-012-0450-2. [DOI] [PubMed] [Google Scholar]
- 26.Gupta N, Sharma P, Santosh Kumar RJ, Vishwakarma RK, Khan BM. Functional characterization and differential expression studies of squalene synthase from Withania somnifera. Mol Biol Rep. 2012;39:8803–12. doi: 10.1007/s11033-012-1743-4. [DOI] [PubMed] [Google Scholar]
- 27.Gupta P, Akhtar N, Tewari SK, Sangwan RS, Trivedi PK. Differential expression of farnesyl diphosphate synthase gene from Withania somnifera in different chemotypes and in response to elicitors. Plant Growth Regul. 2011;65:93–100. doi: 10.1007/s10725-011-9578-x. [DOI] [Google Scholar]
- 28.Gupta P, Agarwal AV, Akhtar N, Sangwan RS, Singh SP, Trivedi PK. Cloning and characterization of 2-C-methyl-D-erythritol-4-phosphate pathway genes for isoprenoid biosynthesis from Indian ginseng, Withania somnifera. Protoplasma. 2013;250:285–95. doi: 10.1007/s00709-012-0410-x. [DOI] [PubMed] [Google Scholar]
- 29.Gupta P, Goel R, Pathak S, Srivastava A, Singh SP, Sangwan RS, et al. De novo assembly, functional annotation and comparative analysis of Withania somnifera leaf and root transcriptomes to identify putative genes involved in the withanolides biosynthesis. PLoS One. 2013;8:e62714. doi: 10.1371/journal.pone.0062714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishra MK, Chaturvedi P, Singh R, Singh G, Sharma LK, Pandey V, et al. Overexpression of WsSGTL1 gene of Withania somnifera enhances salt tolerance, heat tolerance and cold acclimation ability in transgenic Arabidopsis plants. PLoS One. 2013;8:e63064. doi: 10.1371/journal.pone.0063064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Razdan S, Bhat WW, Rana S, Dhar N, Lattoo SK, Dhar RS, et al. Molecular characterization and promoter analysis of squalene epoxidase gene from Withania somnifera L. Dunal Mol Biol Rep. 2013;40:905–16. doi: 10.1007/s11033-012-2131-9. [DOI] [PubMed] [Google Scholar]
- 32.Konc J, Janezic D. ProBiS-2012: web server and web services for detection of structurally similar binding sites in proteins. Nucleic Acids Res. 2012;40:W214–21. doi: 10.1093/nar/gks435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;2513:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Combet C, Jambon M, Deléage G, Geourjon C. Geno3D: automatic comparative molecular modelling of protein. Bioinformatics. 2002;18:213–4. doi: 10.1093/bioinformatics/18.1.213. [DOI] [PubMed] [Google Scholar]
- 35.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–18. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 36.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open babel: an open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Mushegian A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003;12:1418–31. doi: 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yonekura-Sakakibara K, Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011;66:182–93. doi: 10.1111/j.1365-313X.2011.04493.x. [DOI] [PubMed] [Google Scholar]
- 40.Hashimoto K, Madej T, Bryant SH, Panchenko AR. Functional states of homooligomers: insights from the evolution of glycosyltransferases. J Mol Biol. 2010;399:196–206. doi: 10.1016/j.jmb.2010.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paquette SM, Jensen K, Bak S. A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases http://www.P450.kvl.dk . Phytochem. 2009;70:1940–7. doi: 10.1016/j.phytochem.2009.08.024. [DOI] [PubMed] [Google Scholar]


