Abstract
HCF-1 functions as a coactivator for herpes simplex virus VP16 and a number of mammalian transcription factors. Mature HCF-1 is composed of two subunits generated by proteolytic cleavage of a larger precursor at six centrally-located HCFPRO repeats. The resulting N-and C-terminal subunits remain tightly associated via two complementary pairs of self-association domains: termed SAS1N-SAS1C and SAS2N-SAS2C. Additional HCF proteins have been identified in mammals (HCF-2) and Caenorhabditis elegans (CeHCF). Both contain well-conserved SAS1 domains but do not undergo proteolytic processing. Thus, the significance of the cleavage and self-association of HCF-1 remains enigmatic. Here, we describe the isolation of the Drosophila HCF homologue (dHCF) using a genetic screen based on conservation of the SAS1 interaction. The N-terminal β-propeller domain of dHCF supports VP16-induced complex formation and is more similar to mammalian HCF-1 than other homologues. We show that full-length dHCF expressed in Drosophila cells undergoes proteolytic cleavage giving rise to tightly associated N-and C-terminal subunits. As with HCF-1, the SAS1N and SAS1C elements of dHCF are separated by a large central region, however, this sequence lacks obvious homology to the HCFPRO repeats required for HCF-1 cleavage. The conservation of HCF processing in insect cells argues that formation of separate N- and C-terminal subunits is important for HCF function.
HCF-1, also known as C1 factor, is a broadly-expressed nuclear protein involved in the regulation of cellular and viral gene expression. HCF-1 was first identified through its role in transcriptional activation by the herpes simplex virus (HSV) VP16 protein (Gerster and Roeder, 1988; Kristie et al., 1989). Association of HCF-1 with VP16 (also known as α-TIF or Vmw65) leads to the assembly of a multiprotein-DNA complex that includes the POU protein Oct-1 and the TAATGARAT element, a VP16-responsive DNA sequence present in all viral immediate-early (IE) gene promoters (reviewed in (Herr, 1998)). Nuclear localization of VP16 is dependent on association with HCF-1, which contributes a nuclear localization signal (NLS) located at its C-terminus (LaBoissière et al., 1999; Scarr et al., 2000; Wilson et al., 2000). In addition, HCF-1 contains its own activation domain and this is required for optimal transactivation by the VP16-induced complex (Luciano and Wilson, 2002). Thus, HCF-1 functions as a transcriptional coactivator in addition to its roles as an assembly and transport factor. Association with HCF-1 is, therefore, critical for VP16 to function as a transcription factor and VP16 mutants that cannot bind HCF-1 are unable to stimulate TAATGARAT-containing promoters in vivo (Lai and Herr, 1997).
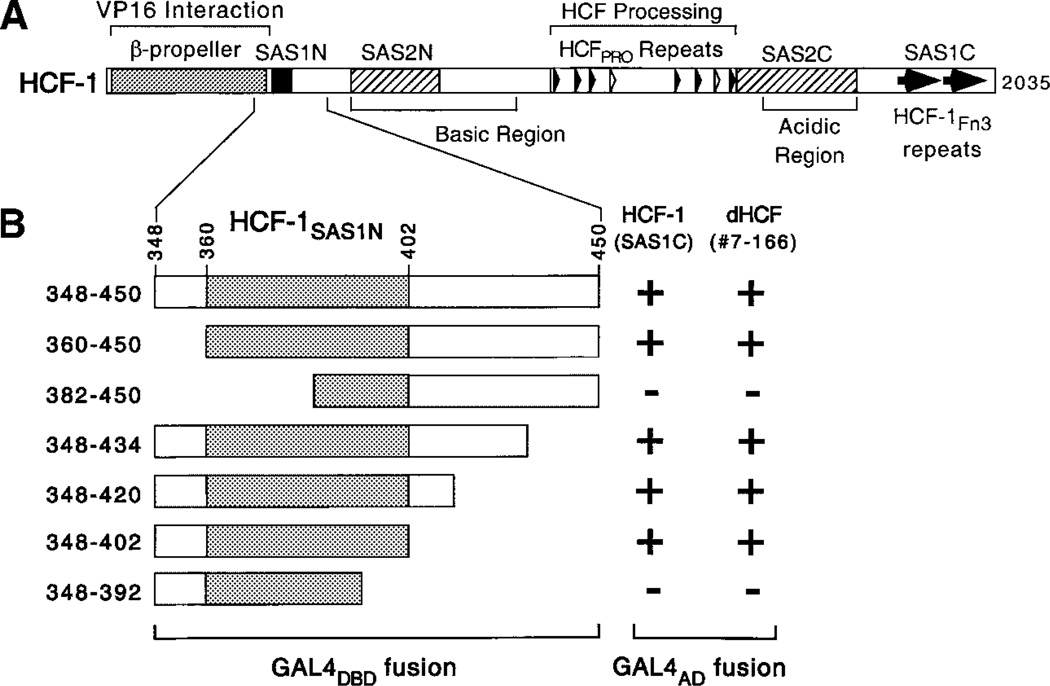
The arrangement of functional domains in human HCF-1 is shown in Figure 1A. HCF-1 is translated as a 230–300-kDa precursor and processed into N- and C-terminal subunits through autocatalytic proteolytic cleavage at a series of six HCFPRO repeats located near the center of the precursor polypeptide (Wilson et al., 1993b; Kristie et al., 1995; Wilson et al., 1995b; Vogel and Kristie, 2000). The resulting subunits remain stably associated via two matched pairs of interaction domains: termed SAS1N-SAS1C and SAS2N-SAS2C (Wilson et al., 2000). The SAS1C domain corresponds to a pair of fibronectin type 3 (Fn3) repeats, which interact with a 43-residue sequence (SAS1N) that is located in the N-terminus. The SAS2 self-association domains flank the HCFPRO repeat region and have been mapped with less precision. The majority of HCF-1 molecules in actively proliferating cells have undergone processing suggesting this is the active form of the protein (Wilson et al., 1993b; Kristie et al., 1995; Wilson et al., 1995b).
Fig. 1.
A: Structural features of human HCF-1. The two matched pairs of self-association sequences are shown as filled (SAS1) and hatched (SAS2) boxes. Tandem Fn3 repeats of SAS1C are indicated by filled arrows. The amino-terminal β-propeller domain involved in association with VP16 is represented as a shaded box. The six near-perfect HCF-1PRO repeats (filled arrowheads) and two degenerate nonfunctional repeats (open arrowheads) are indicated. B: Summary of the yeast two-hybrid analysis. Human HCF-1 SAS1C fragment (residues 1,578–2,014) was compared with dHCF clone 7–166 (residues 912–1,500) for interaction with a panel of HCF-1SAS1N fragments fused to the Gal4 DNA-binding domain. Positive interaction (+) was determined by β-galactosidase expression resulting in formation of blue yeast colonies when grown on X-gal containing media.
VP16 interacts with an N-terminal domain of HCF-1 (HCFVIC) composed of six kelch (HCFKEL) repeats that fold as a six-bladed β-propeller (LaBoissière et al., 1997; Simmen et al., 1997; Wilson et al., 1997; Hughes et al., 1999; Mahajan and Wilson, 2000). This association is mediated by a tetrapeptide sequence in VP16 (361-Glu-His-Ala-Tyr-364) known as the HCF-binding motif or HBM (Freiman and Herr, 1997). HBMs also occur in cellular transcription factors, including the basic-leucine zipper transcription factors LZIP (or Luman) and Zangfei (Freiman and Herr, 1997; Lu et al., 1997; Lu and Misra, 2000).
In addition to its interaction with viral and cellular transcription factors, HCF-1 is also known to be required for cell proliferation. This is most clearly illustrated by a cell line (tsBN67) that carries a missense mutation in the HCF-1 β-propeller domain that changes proline-134 to serine. When shifted to the non-permissive temperature, these cells undergo a stable but reversible G1/G0 cell cycle arrest (Goto et al., 1997; Wilson et al., 1997). It has also been shown that in resting peripheral blood mononucleocytes and serum-arrested human primary fibroblasts, HCF-1 is inactivated by proteolysis in a manner unrelated to the cleavage producing N- and C-terminal subunits (Scarr et al., 2000). Presumably, inactivation of HCF-1 promotes entry into G0 by modulating the activity of transcription factors required for cell cycle progression, either by actively sequestering these factors or by limiting their ability to activate target genes.
Mammalian cells also express a second HCF protein termed HCF-2, which is composed of the N-terminal β-propeller domain and pair of SAS1 elements separated by a short central region lacking HCFPRO repeats (Johnson et al., 1999). In terms of function, HCF-2 differs from HCF-1 in five important respects: (i) HCF-2 does not undergo proteolytic processing, (ii) requires domains outside of the β-propeller domain for efficient VP16-induced complex formation, (iii) does not support transactivation by full-length VP16, (iv) associates poorly with LZIP and lastly, (v) cannot complement the tsBN67 deficiency (Johnson et al., 1999; Lee and Herr, 2001). These properties suggest that HCF-2 may function as an antagonist to HCF-1, perhaps by interacting with a subset of cellular target proteins to mute or modify their function.
Although HSV infection is restricted to mammalian cells, extracts from invertebrate animals such as nematodes (Caenorhabditis elegans), moths (Spodoptera frugiperda), and fruitflies (Drosophila melanogaster), also support VP16-induced complex formation (Kristie et al., 1989; Wilson et al., 1993a; Liu et al., 1999). Each VP16-induced complex has a unique gel-mobility, consistent with the incorporation of HCF proteins of distinct size or shape (Wilson et al., 1993a). The C. elegans genome encodes a single HCF protein (CeHCF), composed of an N-terminal β-propeller domain, and potential SAS1N and SAS1C domains separated by a short unique region (LaBoissière et al., 1997; Hughes et al., 1999; Liu et al., 1999). As with HCF-2, there is no evidence that CeHCF undergoes proteolytic processing and the uncleaved polypeptide is sufficient for VP16-induced complex formation (Liu et al., 1999; Lee and Herr, 2001; Wysocka et al., 2001). The functional conservation of the HCF activity in distantly-related metazoan organisms suggests that VP16 has evolved to exploit a preexisting interaction between cellular transcription factors (Freiman and Herr, 1997; Goto et al., 1997; Wilson et al., 1997).
Here we describe the Drosophila HCF homologue (termed dHCF), isolated through a functional screen that exploits the self-association properties of mammalian HCF-1. The predicted dHCF protein resembles HCF-1 in having a large (819-residue) intervening region that separates the SAS1N and SAS1C domains. We show that the dHCF β-propeller domain supports VP16-induced complex formation in the presence of bacterially-expressed VP16 and Oct-1 proteins. In striking contrast to HCF-2 and CeHCF, full-length dHCF undergoes proteolytic cleavage near the center of the polypeptide, generating N- and C-terminal subunits which remain associated via the conserved SAS1N and SAS1C elements. Processing of the HCF precursor provides an elegant mechanism for generating paired sub-units with equal stoichiometry and the conservation of HCF processing in insect cells suggests that this unusual form of post-translational processing plays a fundamental role in HCF function.
MATERIALS AND METHODS
Yeast two-hybrid screen
1 × 106 clones from a Drosophila embryonic (12–24 h) cDNA library (gift of Grace Gill, Harvard Medical School) were screened using residues 348–450 of HCF-1 fused to the Gal4 DNA-binding domain (pYDBT-HCF-1N348–450) as bait. The reporter strain Y190 contained Gal4-responsive his3 and lacZ genes, allowing selection by dual selection. Constitutive activation by the bait protein was suppressed by including 20 mM 3-amino-triazole in the media.
Library screening and sequence analysis
An oligo(dT)-primed Drosophila embryonic (3–12 h) cDNA library in λgt 10 ((Poole et al., 1985), a kind gift of Grace Gill, Harvard Medical School) was screened by nucleic acid hybridization using standard methodology. DNA was prepared from positive phages using Lamb-daSorb Phage Absorbent (Promega). Searches of the protein databases were performed using BLASTP and TBLASTN and the searches of FlyBase by BLASTN. The sequence of the composite Drosophila HCF cDNA has been deposited in the GenBank™ database under the accession number AF251006.
Northern and RT-PCR analysis
Total RNA was isolated from Schneider line 2 cells (gift of J. Treisman, New York University School of Medicine) using a thiocyanate/phenol mixture (Tri Reagent , Sigma-Aldrich, Inc.). 25 µg of RNA was resolved by denaturing gel electrophoresis and blotted onto a Hybond-N+ membrane (Amersham). Hybridization was carried out at 42°C in Rapid-hyb buffer (Amersham) and The probe corresponds to the entire dHCF open reading frame and was labeled with P 32 by random priming. Temporal expression of dHCF mRNA was determined by RT-PCR using a commercial first-strand cDNA panel (Rapid-Scan™ Gene Expression Panel, Origene Technologies, Rockville, MD). The annealing reactions was performed at 55°C and subject to 35 PCR cycles. To detect dHCF transcripts, a 691-bp fragment spanning exons 8–11 was amplified using the following oligonucleotide primers: 5′ -CTGCAATCG-TAAGTGGTGAT-3′ and 5′-CGAAAACGGTAAGCCGT-TCC-3′. Normalization of the template cDNA pools was confirmed using primers complementary to the Drosophila ribsomal protein 49 gene and yield a 433 bp fragment. Products were resolved by electrophoresis through a 1.5% agarose gel and visualized by staining with ethidium bromide.
Expression plasmids
Fragments derived from the dHCF open reading frame were generated by high-fidelity PCR amplification (Expand High Fidelity PCR System, Boehringer Mannheim) and subcloned into the mammalian expression vectors pCGN, pCGNGal4(1–94) and pCGT or the in vitro transcription/translation vector pNCITE (Wilson et al., 1997). During amplification of the β-propeller domain (amino acids 2–420 or 51–420), a G to C substitution was incorporated at nucleotide 430, inactivating a naturally occurring BamH I site. Likewise, to facilitate subcloning of the isolated SAS1C domain, site-directed mutagenesis (Quik Change protocol, Stratagene) was used to create a silent G to A substitution at nucleotide 4,768, also inactivating a natural BamH I site. The sequences of all PCR-generated fragments were verified by DNA sequence analysis.
To express full-length dHCF, two overlapping fragments spanning the complete ORF were generated by high-fidelity PCR amplification from library clones 10F7 and 3E1. To facilitate joining of the amplified fragments, a synthetic NgoM IV site was engineered using silent nucleotide substitutions within codons 526 and 527. The resulting complete ORF was subcloned into the CMV enhancer-driven vector pCGT, for expression in mammalian cells or the Drosophila actin 5C promoter-driven vector pACXT, for expression in Drosophila cells. Both vectors introduce a T7 epitope tag at the dHCF N-terminus. A FLAG-epitope tag was inserted between codon 1,500 and the stop codon during the PCR amplification. DNA sequencing revealed a single T to K substitution at codon 695. This residue lies in the central non-conserved region and its relevance to dHCF function is unknown.
Protein expression, gel-mobility shift assays, and immunoblotting
Human 293T cells were transiently transfected using Lipofectamine 2000 (Life Technologies, Inc.). Drosophila SL2 cells (2–5 × 106 cells) were transfected with 10 µg of expression plasmids encoding T7 and/or FLAG epitope tagged versions of full-length dHCF or VP16ΔC using a modified calcium phosphate procedure (Chen and Okayama, 1987). Whole cell lysates were prepared by direct lysis in sample buffer containing 2% SDS, briefly sonicated to disrupt high molecular weight DNA and then boiled for 10 min. Immunoblotting was performed as previously described (Mahajan and Wilson, 2000), except that Supersignal Femto chemiluminescence reagent (Pierce) was required to detect recombinant dHCFFL in SL2 cell lysates. For immuno-precipitation, extracts were prepared from 1 × 107 cells using 300 ml of high salt extraction buffer (420 mM KCl, 20 mM Tris-HCl [pH 7.9], 0.25% NP-40, 10% glycerol) supplemented with fresh 5 mM PMSF and mixed with αT7- or αFLAG-coupled beads. Immunoprecipitates were washed three times in the 1 ml of the same buffer.
For cell-free expression, dHCF-encoding fragments were subcloned into pNCITE, a derivative of pCITE2a+ (Novagen, Inc.), that includes the HA epitope at the N-terminus of the expressed protein. In vitro transcription and translation reactions were performed in the presence of [35S] methionine using the TNT Quick Coupled Transcription/Translation System (Promega, Inc.). Electrophoretic mobility shift assays were performed as described previously (Johnson et al., 1999; Mahajan and Wilson, 2000), except that binding reactions were assembled at varying temperatures. Electrophoresis was carried out at room temperature.
RESULTS
Isolation of cDNAs encoding dHCF
To identify proteins that interact with the SAS1N domain of HCF-1, we performed a yeast two-hybrid screen using a Drosophila embryonic cDNA library and a bait protein corresponding to residues 348–450 of human HCF-1 fused to the GAL4 DNA-binding domain (Gal4-HCF-1N348–450). From screening 1 ×106 clones, we isolated ten positives that interacted specifically with SAS1N. Five of these clones encoded an undescribed Drosophila protein containing two Fn3 repeats located at the C-terminus of the predicted polypeptide, strongly reminiscent of the tandem Fn3 repeats in mammalian and nematode HCF. The paired Fn3 repeats of the Drosophila protein were 57% identical and 70% similar to HCFFn31 and HCFFn32 repeats of mammalian HCF-1, suggesting we had isolated a functional SAS1C domain. Consistent with this idea, the smallest interacting clone (9–124) corresponded to little more than the paired Fn3 repeats.
To confirm that the two-hybrid interaction represented a genuine SAS1N-SAS1C interaction, we transformed yeast with a panel of HCF-1SAS1N truncations fused to the Gal4 DNA-binding domain and assayed for interaction with the longest Drosophila cDNA (clone 7–166). As a control, we also assayed the SAS1C domain of HCF-1 (residues 1,758–2,014). This experiment is summarized in Figure 1B. The association of clone 7–166 with the HCF-1SAS1N truncations paralleled that of HCF-1SAS1C indicating the recapitulation of a genuine SAS1N-SAS1C interaction. We, therefore, identified the new protein as Drosophila HCF (dHCF).
Structure of the dHCF protein
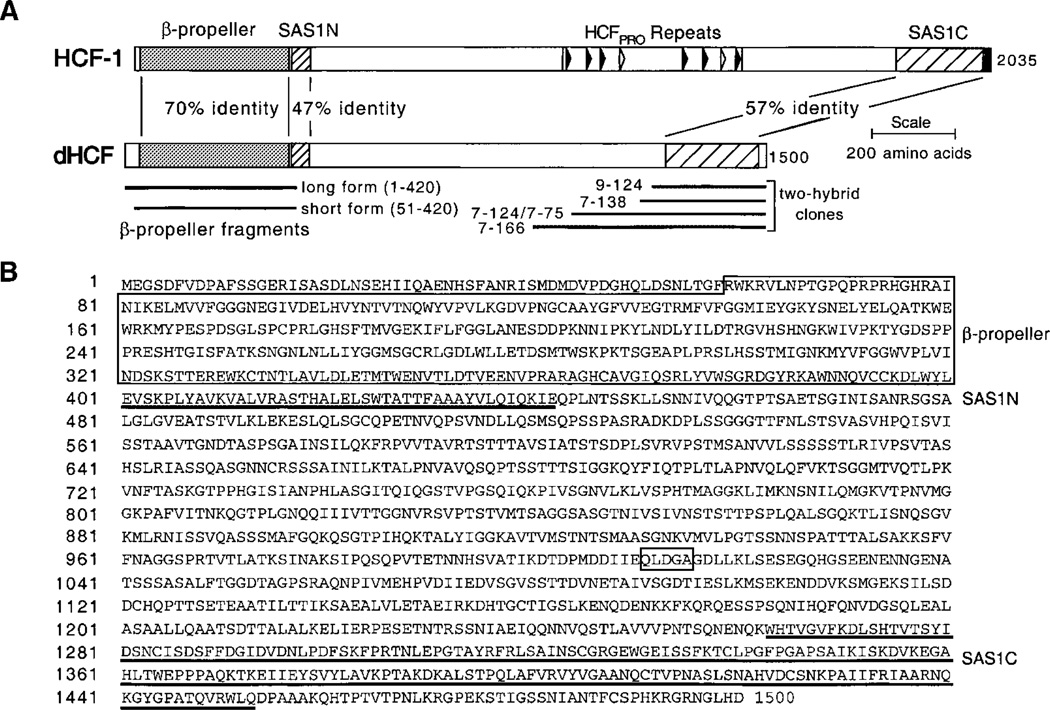
The dHCF clones isolated in the yeast screen lacked amino-terminal sequences. To extend the open reading frame, we screened two separate Drosophila cDNA libraries by hybridization using a probe derived from the 5′ end of clone 7–166. Multiple rounds of screening were necessary to obtain the entire open reading frame (ORF) that included a putative initiation codon and in-frame stop codon. The corresponding genomic locus (gene identifier CG1710) located at 102B5 on chromosome 4, was identified by a BLAST search of the near-complete Drosophila genome (Berkeley Drosophila Genome Project). The composite ORF determined from overlapping cDNAs contains exactly 1,500 codons and encodes a poly-peptide with a theoretical molecular mass of 160 kDa. The amino acid sequence and predicted domain organization of dHCF is shown in Figure 2. The dHCF protein possesses the characteristic features of the other HCF proteins, namely a β-propeller domain and HCFSAS1N element towards the N-terminus and a HCFSAS1C element composed of two Fn3 repeats at the C-terminus (indicated in Fig. 2A). The six HCFKEL repeats that make up the b-propeller domain (residues 59–400 in dHCF) are 70% identical and 84% similar to the β-propeller domain of HCF-1. The degree of homolgy was slightly lower when the dHCF b-propeller sequence was compared to HCF-2 (58% identical and 75% similar) and C. elegans HCF (49% identical, 69% similar).
Fig. 2.
Sequence of the dHCF protein. A: Conserved organization of function domains within HCF-1 and Drosophila HCF is shown schematically together with the degree of sequence identity between the β-propeller and SAS1 domains. The coding regions of each of dHCF clone isolated by the two-hybrid screen (clone 7–166, residues 913– 1,500; clones 7–214/7–75, residues 991–1,500; clone 7–138, residues 1,155–1,500; clone 9–124, residues 1,185–1,500) and the β-propeller constructs used in Figure 3 are indicated. B: Predicted amino acid sequence of dHCF. The β-propeller domain (boxed) comprising six HCFKEL repeats, the 43 amino acid SAS1N domain (underlined) and the tandem Fn3 repeats of the SAS1C domain (underlined) are indicated. A sequence motif found in dTFIIA-L is also boxed. The nucleotide and predicted amino acid sequence have been submitted to GenBank™ with accession number AF251006.
Beyond the HCFKEL repeats, the sequence homology to the HCF proteins was limited to the SAS1N and SAS1C domains: 47 and 57% identical to their counterparts in HCF-1. Interestingly, the SAS1N and SAS1C elements are separated by large intervening region (residues 444–1,262) rich (26.1%) in serine and threo-nine residues. This is highly reminiscent of HCF-1 in which SAS1N and SAS1C are separated by a 1,411 amino acid segment that includes the central HCFPRO repeats that specify proteolytic processing (Wilson et al., 2000). Careful examination of the unique central sequence from dHCF did not reveal any convincing matches to the HCFPRO repeats.
The dHCF gene is transcribed throughout development
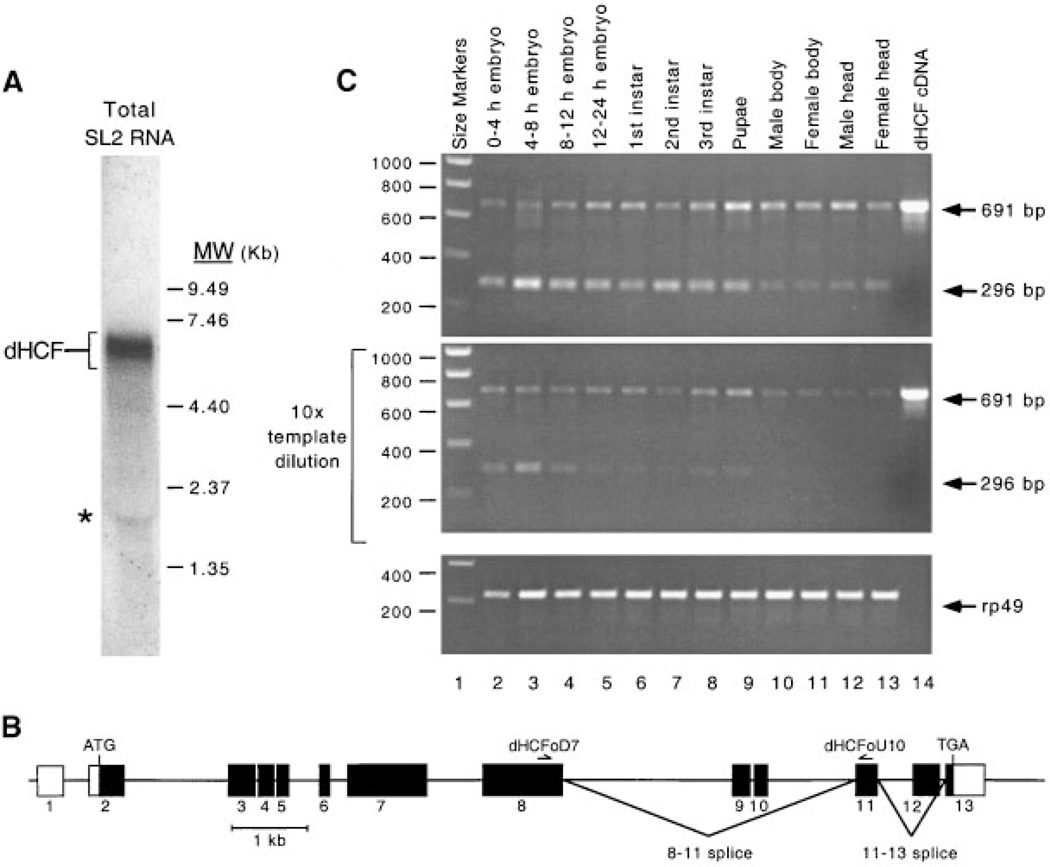
Northern analysis using total RNA prepared from SL2 cells and probed with the complete coding sequence of dHCF, identified an abundant transcript of 6.95–7.0-kb (Fig. 3A). To determine when the dHCF gene is expressed during development, we performed RT-PCR analysis using first-strand cDNA prepared from polyA+ RNA isolated from various embryonic and larval stages as well as from adult tissues. Amplification using oli-gonucleotide primers (dHCFoD7 and dHCFoU10) complementary to sequences in exons 8 and 11, gave rise to the expected 691 bp fragment in all cDNA samples (Fig. 3B). There appeared to be only minor variation in the amount of amplification product. These results indicate that dHCF mRNA is present throughout development and is also present in adults. We also observed a smaller amplification product (296 bp) in all samples, although this product was more abundant in embryonic and larval tissues than in adults. Sequencing of the smaller amplification product showed that it corresponds to an alternative splice variant lacking exons 9 and 10 (data not shown). To confirm that equal quantities of cDNA were used, parallel amplification reactions were performed using primers derived from the broadly expressed ribosomal protein 49 gene (Fig. 3B, lower part).
Fig. 3.
Drosophila HCF mRNA is present throughout development. A: Northern blot analysis of total RNA (25 µg) prepared from Drosophila SL2 tissue culture cells. RNA was resolved on a 1% denaturing gel and probed with a P32-labeled DNA fragment corresponding to the entire dHCF open reading frame. Sizes of RNA molecular weight standards (Gibco-BRL) are given in thousands of nucleotides. An asterisk denotes a non-specific band. B: RT-PCR analysis using oligonucleotide primers derived from dHCF (upper and middle panels) and ribosomal protein 49 (lower part). In the middle panel, cDNA templates were diluted 10-fold prior to amplification. Sources of mRNA were as follows: 0–4 h embryos (lane 2), 4–8 h embryos (lane 3), 8–12 h embryos (lane 4), 12–24 h embryos (lane 5), 1st instar larvae (lane 6), 2nd instar larvae (lane 7), 3rd instar larvae (lane 8), pupae (lane 9), male head (lane 10), female head (lane 11), male body (lane 12), female body (lane 13), and cloned dHCF cDNA as a positive control (lane 14). Sizes of DNA markers (lane 1) are given in bp. C: The predicted exonintron structure of the dHCF gene. Coding sequences are indicated by filled boxes. Oligonucleotide primers used for RT-PCR analysis are shown above the appropriate exon. Two alternative mRNA splice variants are indicated (labeled 8–11 and 11– 13 splice). The splice variant which deletes exon 12 is based on a single cDNA clone isolated from the embryo library.
The β-propeller domain of dHCF supports VP16-induced complex formation
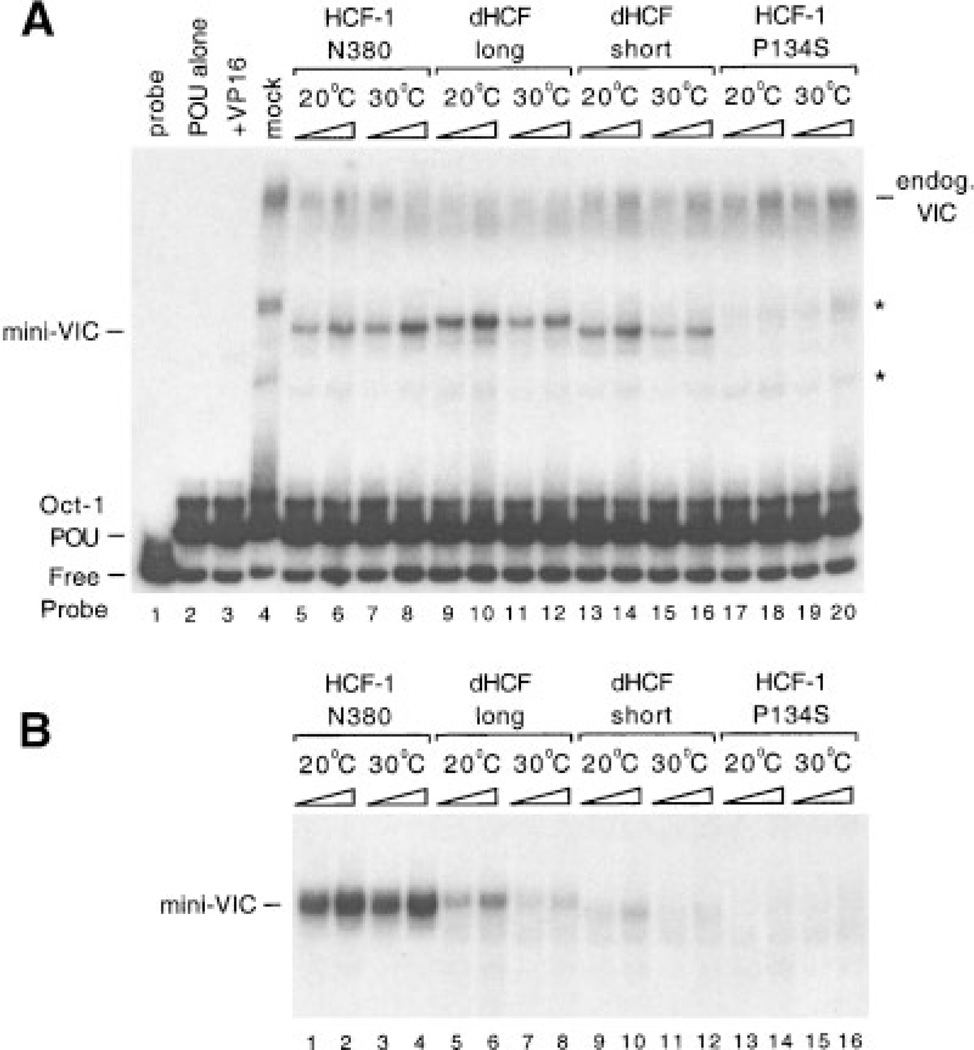
To determine whether dHCF was capable of supporting VP16-induced complex formation, we expressed the b-propeller domain by in vitro translation and assayed for VP16-induced complex formation using a labeled TAATGARAT motif containing probe and recombinant VP16 and the Oct-1 POU domain proteins purified from bacteria (Fig. 4A). Two versions of the dHCF β-propeller were tested; a ‘long form’ comprising the -propeller and fifty amino acid N-terminal tail (residues 1–421, lanes 9–12) and ‘short form’ corresponding to the six HCFKEL repeats alone (residues 51–421, lanes 13–16). Both dHCF fragments gave rise to a specific complex (labeled mini-VIC) with similar mobility to the complex formed by the human HCF-1 β-propeller (lanes 5–8). As expected, complex formation was not detected using an inactivating mutant (P134S, see reference (Wilson et al., 1997)) of HCF-1 β-propeller (lanes 17–20). This result shows that the protein encoded by the dHCF cDNA is capable of promoting VP16-induced complex formation and is thus likely to account for the activity previously identified in Drosophila cell extracts (Wilson et al., 1993a).
Fig. 4.
The dHCF β-propeller domain supports VP16-induced complex formation. A: Proteins were expressed by in vitro translation at 20°C and tested for VP16-induced complex forming activity by electrophoretic mobility shift assay. The first four lanes are controls showing probe alone (lane 1), Oct-1 POU domain protein alone (lane 2), Oct-1 POU mixed with bacterially produced GST-VP16DC (lane 3), or unprogrammed reticulocyte lysate mixed with Oct-1 POU and GST-VP16DC (lane 4). In the remaining lanes, lysates expressing HCF-1N380 (lanes 5–8), dHCFN420 (lanes 9–12), dHCFN51–420 (lanes 13–16), or HCF-1N380P134S (lanes 17–20) polypeptides were mixed with GST-VP16DC and Oct-1 POU. Binding reactions were assembled on ice and then incubated for 30 min at 20 or 308C as indicated. Positions of the free probe, Oct-1 POU domain complex, and VIC containing rabbit HCF-1 from the lysate (labeled endog. VIC) or truncated recombinant HCF (mini-VIC) are indicated. Non-specific complexes are indicated with an asterisk. B: Equivalent to A except that in vitro translation reactions were performed at 30°C. Only VP16-induced complexes containing recombinant HCF (mini-VIC) are shown.
Activity of the dHCF β-propeller is temperature-sensitive
To identify optimal conditions for VP16-induced complex assembly, dHCF was translated in vitro at 20°C and the binding reactions performed at 20 or 30°C (Fig. 4A). For both long and short forms of dHCF, the efficiency of complex formation was greater at the lower temperature (compare lanes 10 and 12 and also lanes 14 and 16). In contrast, the HCF-1 β-propeller was more effective at the higher temperature (compare lanes 6 and 8). This result suggests that the activity of the dHCF β-propeller is temperature-dependent and that the optimum temperature is lower for the insect protein than its mammalian counterpart. To determine whether temperature influences the intrinsic activity of the β-propeller or some aspect of the assembly process, we repeated the experiment using recombinant dHCF and HCF-1 proteins translated at 30°C (Fig. 4B). Relative to HCF-1 (lanes 1–4), both long and short forms of the dHCF β-propellers were significantly impaired for complex formation (lanes 5–12), indicating that the temperature at which polypeptide is translated has a lasting effect on activity. The dHCF β-propeller shows maximal activity at a lower temperature than human HCF-1, consistent with the physiological differences between insects and mammals.
Functional conservation of the SAS1N and SAS1C pairing
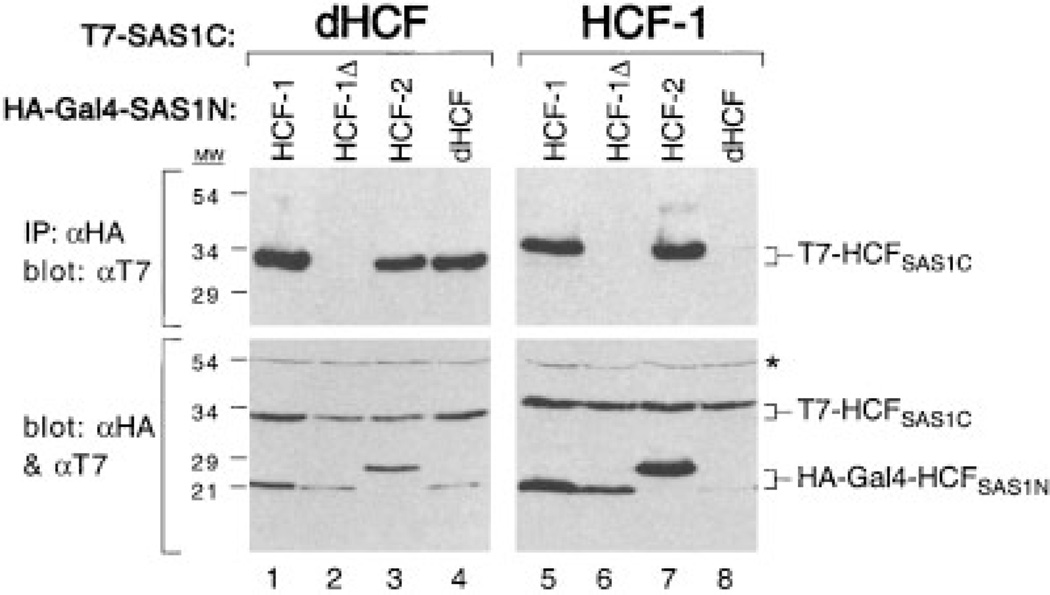
The two-hybrid analysis described in Figure 1, showed that the SAS1C domain of dHCF can associate effectively with the SAS1N domain of HCF-1. To determine whether there is an interaction between the SAS1N and SAS1C modules of dHCF, we assayed self-association by coimmunoprecipitation from transfected cell extracts (Fig. 5). Human 293T cells were transfected with expression plasmids encoding the SAS1N elements of HCF-1 (residues 348–402 and 348–392), HCF-2 (residues 345–416) and dHCF (residues 389–443) together with the SAS1C element from either dHCF (residues 1,209–1,500) or HCF-1 (residues 1,758– 2,035). The SAS1N fragments were tagged with a flu HA-epitope and the SAS1C elements were tagged with the phage T7 gene 10 epitope. Note that the relatively short SAS1N peptides were expressed as fusions to Gal4 (residues 1–94) to ensure stable expression (Wilson et al., 2000). The HA-tagged Gal4-SAS1N fusion proteins were recovered by immunoprecipitation using an aHA monoclonal antibody, resolved by SDS–PAGE and immunoblotted with an aT7 antibody. Consistent with the two-hybrid analysis (Fig. 1B), the dHCFSAS1C fragment (dHCFSAS1C) could be coimmunoprecipitated by the SAS1N domain of HCF-1 (Gal4-HCF-1N348–402, lane 1) but not with a truncated form (Gal4-HCF-1N348–392, lane 2). The dHCFSAS1C fragment was also recovered using the SAS1N fragment so fHCF-2 (lane 3) and dHCF (lane 4). This result indicates that dHCF contains a functional pair of SAS1 elements that are capable of self-associating when expressed separately in mammalian cells. The result also shows that the tandem Fn3 repeats of dHCFSAS1C can interact with the SAS1N domains of both HCF-1 and HCF-2.
Fig. 5.
Cross-species pairing of SAS1N and SAS1C domains. T7-tagged dHCFSAS1C (lanes 1–4) or HCF-1SAS1C (lanes 5–8) polypep-tides were expressed in transfected 293T cells together with HA-tagged fragments (fused to GAL4 residues 1–94) corresponding to residues 348–450 of HCF-1 (lanes 1 and 5), 382–450 of HCF-1 (lanes 2 and 6), 341–394 of HCF-2 (lanes 3 and 7), and 359–421 of dHCF (lanes 4 and 8). HCFSAS1N and HCFSAS1C association was assayed by coimmunoprecipitation (IP) with the αHA antibody followed by immunoblotting with the αT7 antibody (upper part). Protein expression was confirmed by direct immunoblotting of extracts with a mixture of αT7 and αHA antibodies (lower part). An asterisk indicates a non-specific protein recognized by the αHA antibody.
Equivalent coimmunoprecipitations were also performed using HCF-1SAS1C (lanes 5–8). As shown previously, the HCF-1SAS1C polypeptide can be coimmunoprecipitated by the SAS1N fragments from HCF-1 (lane 5) and HCF-2 (lane 7) but not by the nonfunctional HCF-1N348–392, (lane 6). In striking contrast, dHCFSAS1N was unable to coprecipitate the human HCF-1SAS1C fragment (lane 8). It is unlikely that the relatively poor expression of Gal4-dHCFSAS1N was responsible for lack of association because the slightly higher levels seen in lane 4 resulted in very efficient recovery of dHCFSAS1C. Thus, although interaction between SAS1N and SAS1C has been conserved through metazoan evolution, there are subtle differences between the SAS1N domains of mammalian and Drosophila HCF.
Proteolytic processing of dHCF
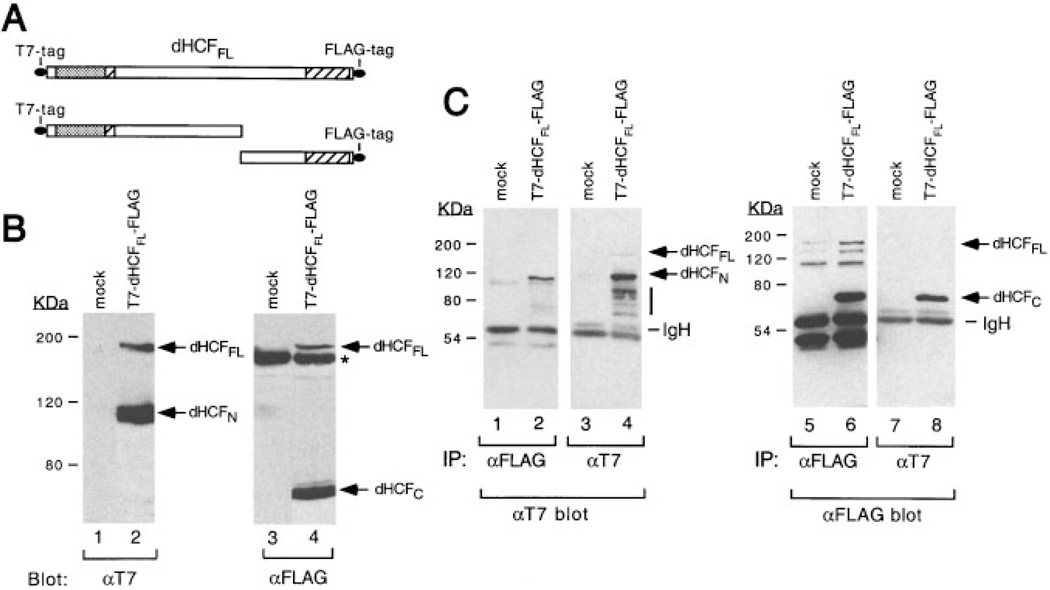
Drosophila HCF resembles mammalian HCF-1 in having a large (819-residue) spacer region separating the SAS1N and SAS1C elements. One function of the spacer region in HCF-1 is to specify proteolytic processing (Wilson et al., 1993b; Kristie et al., 1995; Wilson et al., 1995b; Vogel and Kristie, 2000). To determine if the dHCF protein is also processed, we transiently transfected Drosophila SL2 cells with an expression vector encoding full-length dHCF tagged at the N-terminus with the T7 epitope and at the C-terminus with a FLAG epitope (illustrated schematically in Fig. 6A). Cell lysates were prepared under denaturing conditions, resolved by SDS/PAGE and immunoblotted with the aT7 antibody (Fig. 6B). Two T7-tagged polypeptides were detected (lane 2), a prominent species with an apparent molecular weight of 120 kDa and a less abundant species of ~180 kDa. Neither species were seen in mock-transfected cells (lane 1). Full-length dHCF has a predicted molecular weight of 160 kDa, suggesting that the larger polypeptide corresponds to the full-length translation product and the smaller polypeptide corresponds to an N-terminal fragment. To confirm this, the same extracts were probed with an antibody against the FLAG epitope (lanes 3 and 4). This recognized the same 180-kDa species as well as a smaller and more abundant 72-kDa polypeptide (lane 4) that corresponds to the C-terminus of dHCF. Thus, dHCFFL undergoes proteolytic cleavage at one or more sites within the central non-conserved region, giving rise to discrete N- and C-terminal subunits.
Fig. 6.
Drosophila HCF undergoes proteolytic processing. A: Schematic showing the full-length dHCF polypeptide tagged at the N-terminus with a synthetic T7-epitope and at the C-terminus with the FLAG epitope (T7-dHCFFL-FLAG). B:Drosophila SL2 cells (2 × 106 cells) were transfected with 10 µg of an empty expression vector (lanes 1 and 3) or plasmid encoding T7-dHCFFL-FLAG (lanes 2 and 4). Live cells were lysed in sample buffer containing 2% SDS, incubated at 90°C for 10 min and briefly sonicated to shear high molecular weight DNA Proteins were resolved by SDS–8% polyacrylamide gel electrophoresis and blotted with an αT7 monoclonal antibody (lanes 1 and 2) or αFLAG antibody (lanes 3 and 4). An irrelevant band that cross-reacts with the aFLAG antibody is indicated with an asterisk. The sizes of prestained protein markers are given in kilodaltons. C: The N-and C-terminal subunits of dHCF remain associated after processing. Drosophila SL2 cells were transfected with an empty expression vector (lanes 1, 3, 5, and 7) or plasmid encoding T7-dHCFFL-FLAG (lanes 2, 4, 6, and 8). Whole cell extracts were prepared under non-denaturing conditions and immunoprecipitated using αFLAG (lanes 1, 2, 5, and 6) or αT7 (lanes 3, 4, 7, and 8) conjugated beads. Immunoprecipitates were resolved by SDS–8% polyacrylamide gel electro-phoresis and probed by immunoblotting with αT7 (lanes 1–4) or αFLAG (lanes 5–8) antibodies. Bands corresponding to the T7-tagged N-terminus or FLAG-tagged C-terminus are indicated.
The processed subunits of dHCF remain associated
To determine whether the N-and C-terminal subunits remain associated after processing, we prepared extracts from SL2 cells expressing T7- and FLAG-tagged dHCFFL under non-denaturing conditions and performed immunoprecipitations using either αT7 or αFLAG antibody beads (Fig. 6C). Immunoprecipitates were fractionated by SDS/PAGE and probed separately with each antibody. The T7-tagged N-terminus was recovered efficiently by immunoprecipitation with the αFLAG antibody (Fig. 6C, lane 2) and conversely, the FLAG-tagged C-terminus was recovered using the αT7 antibody (lane 8). Immunoprecipitation of an extract from mock-transfected cells serves to identify a number of relevant cross-reacting bands (lanes 1, 3, 5, and 7). The less abundant full-length dHCFFL polypeptide (just visible in lane 4) was barely detectable in this experiment. Denaturation of the extracts prior to immunoprecipitation abolished coimmunoprecipitation of the untagged subunit confirming that association is non-covalent (data not shown). In summary, these results show that proteolytic cleavage of the dHCF precursor gives rise to two independent subunits, which remain stably associated.
DISCUSSION
Exploiting the self-association properties of human HCF-1, we have isolated cDNAs encoding an HCF protein from the fruit fly Drosophila melanogaster. In terms of general architecture, the predicted dHCF protein resembles its mammalian and nematode counterparts in having a well-conserved β-propeller domain at its N-terminus, followed by a matched pair of SAS1 self-association elements. As with HCF-1, the SAS1N and SAS1C domains are separated by a large unique region that shares little obvious sequence homology with other proteins. The dHCF β-propeller was sufficient for VP16-induced complex assembly in the presence of bacterially expressed Oct-1 and VP16, supporting the view that dHCF corresponds to the functional activity previously detected in crude Drosophila cell extracts (Wilson et al., 1993a). Searches of the Drosophila genome (Berkeley Drosophila Genome Project) and expressed sequence tag (EST) databases provide no evidence for a second HCF-like gene.
In several respects, dHCF appears more similar to HCF-1 than to HCF-2 or C. elegans HCF (CeHCF). The dHCF β-propeller domain shares 70% amino acid sequence identity to the β-propeller of HCF-1 but only 58% identity to HCF-2 and 49% identity to CeHCF. Likewise, the tandem Fn3 repeats of SAS1C domain are 57% identical to HCF-1 compared to 50% for HCF-2 and CeHCF. More significant perhaps is the size of the central region separating the SAS1N and SAS1C domains. Both dHCF and HCF-1 have a comparatively large central region (819 and 1,411 residues respectively) whereas HCF-2 and C. elegans HCF have a much shorter spacer (205 and 137 residues respectively) (Johnson et al., 1999; Liu et al., 1999; Lee and Herr, 2001). The presence of a large central region may be related to the fact that dHCF and HCF-1 undergoes proteolytic cleavage and self-association. There is currently no evidence that CeHCF or HCF-2 undergo a comparable form of proteolytic processing, although it is possible this could occur in an undiscovered subset of cells (Johnson et al., 1999; Liu et al., 1999; Lee and Herr, 2001). Full-length HCF-2 and CeHCF can support VP16-induced complex formation but interestingly, the resulting complexes activate transcription poorly compared to the complex formed by HCF-1 (Lee and Herr, 2001). Experiments to determine the transcriptional capacity of dHCF are underway. The existence of both processed (HCF-1, dHCF) and unprocessed (HCF-2, CeHCF) forms of HCF suggests that the ancestors of multicellular organisms may have encoded multiple HCF genes, only one of which is retained in the two invertebrate lineages examined to date.
The dHCF locus
A search of the near-complete Drosophila genomic sequence showed that the dHCF gene lies on chromo-some 4, within a 1.2 Mb region (cytogenetic bands 101E-102F) that contains the majority of the chromosome’s transcribed genes. The exonintron structure (Fig. 3C) was derived by comparison of our composite cDNA with the genomic sequence. Chromosome 4 is extremely small (5 Mb total) and displays a number of unusual properties, including diffuse appearance in salivary gland preparations, very limited recombination, and variegated expression of P-element transgenes. These peculiar features have severely hampered analysis and relatively few genetic screens have been designed to search for genes on this chromosome (Locke et al., 1999). Further mapping analysis will determine whether dHCF corresponds to one of the twenty-five or so lethal alleles that have been mapped to chromosome 4 but not yet assigned to a specific gene.
Conservation of HCF processing and self-association during metazoan evolution
By expressing epitope-tagged versions of dHCF in Drosophila tissue culture cells, we have shown that the primary translation product undergoes proteolytic cleavage to generate N- and C-terminal fragments. As with mammalian HCF-1, the processed dHCF fragments remain stably, but non-covalently associated. Presumably, this self-association is mediated by the functional SAS1N and SAS1C elements. Previously, we proposed that conservation of the SAS1 elements in HCF-1 and HCF-2 might be due to interactions with other cellular proteins and imagined that the SAS1N and SAS1C domains might transiently dissociate to allow these interactions (Wilson et al., 2000). Because the N- and C-terminal subunits of HCF-1 are not covalently linked, one function of the SAS2 elements might be to maintain the integrity of the HCF-1 complex. If correct, this model predicts that dHCF will also contain a second set of self-association elements, although not necessarily homologous to the mammalian SAS2 elements.
Based on the relative sizes of the N- and C-terminal fragments, we estimate that cleavage of dHCF occurs between amino acids 900 and 1,100 within the non-conserved central region. In mammalian HCF-1, processing occurs within each of the six active HCFPRO repeats and also within an unrelated sequence directly C-terminal to the last HCFPRO repeat (Kristie et al., 1995; Wilson et al., 1995a). For the HCFPRO repeats, the cleavage event is highly specific; alanine substitutions at 11 of the 26 residues flanking the cleavage point in HCFPRO repeat 2 abolishes cleavage activity and implies a protease activity that requires an unusually large recognition sequence or a precise secondary structure (Wilson et al., 1995a). Careful examination of the dHCF sequence revealed no convincing blocks of similarity to the HCFPRO repeat sequence leaving us to conclude that either a variety of unrelated sequences can serve as the substrate for the autocatalytic processing activity or that dHCF is processed through an entirely different mechanism. The 20- and 30-kDa subunits of Drosophila TFIIA are also generated by proteolytic cleavage of a larger precursor, TFIIA-L (Yokomori et al., 1993). The precise cleavage point is not known but can be approximated from the sizes of the processed products. Within this region there is a pentapeptide sequence (257-QLDGA-261, indicated in Fig. 2B) that is also found within dHCF (1011-QLDGA-1015) raising the intriguing possibility that that dHCF and dTFIIA-L might be processed through a similar mechanism.
Consistent with a role in regulation of cell proliferation, dHCF is expressed throughout Drosophila development and the promoter contains at least one binding site for DREF, a transcription factor that coordinates the expression of genes involved in DNA replication and proliferation (SSM and ACW, unpublished results). By analogy to HCF-1, we predict that a variety of Droso-phila transcription factors interact with the β-propeller domain dHCF. Freiman and Herr have shown that the basic-leucine zipper protein BBF2/dCREB-A contains a functional HBM located in the N-terminus of the protein (Freiman and Herr, 1997) and association with dHCF may be required for proper regulation of BBF2/dCREB-A target genes. Although fly strains carrying mutations in the dHCF gene have not been identified, loss of BBF2/ dCREB-A function results in an embryonic lethal pheno-type suggesting that dHCF mutants would similarly die during development (Rose et al., 1997). Given these technical limitations, transient ablation of dHCF expression may offer the best prospects for studying dHCF function in vivo. Initial attempts to silence dHCF expression in cultured cells using RNA interference have been hampered by the abundance and stability of dHCF protein in these cells (SSM and ACW, unpublished results). Similar interference experiments in transgenic or microinjected flies may prove more illuminating.
Although the functional consequences of HCF processing remain a mystery, the observation that the Drosophila protein undergoes a similar pattern of cleavage followed by stable association, strengthens the view that the processing is important to some aspect of HCF function. Cleavage may provide a novel mechanism for regulation through the controlled dissociation of the two subunits or alternatively allow for greater flexibility in HCF’s association with other proteins. As we have noted previously, the sequences of the SAS1N and SAS1C domains are well-conserved irrespective of whether the parent protein is processed or not (Wilson et al., 1997). To our minds, this conservation implies a selective pressure beyond the need to tether one subunit to the other. Distinguishing between these and other possibilities presents an exciting challenge for the future.
ACKNOWLEDGMENTS
We are grateful to Winship Herr and Robert Tjian for their initial encouragement and continued enthusiasm for this project. We also wish to thank Grace Gill, Michael Garabedian, Stavros Giannakopoulos, Randy Luciano, Helen Sink, Jessica Treisman, and Mark van Doren for generously providing reagents, assistance and much needed advice. Naoko Tanese, Richard Freiman, and Winship Herr provided valuable comments on the manuscript. This work was supported by funds from the Kaplan Comprehensive Cancer Center, the National Science Foundation (MCB-98–16856) and the National Institutes of Health (R01-GM61139).
LITERATURE CITED
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman RN, Herr W. Viral mimicry: Common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerster T, Roeder RG. A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc Natl Acad Sci USA. 1988;85:6347–6351. doi: 10.1073/pnas.85.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- Herr W. The herpes simplex virus VP16-induced complex: Mechanisms of combinatorial transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:599–607. doi: 10.1101/sqb.1998.63.599. [DOI] [PubMed] [Google Scholar]
- Hughes TA, La Boissiere S, O’Hare P. Analysis of functional domains of the host cell factor involved in VP16 complex formation. J Biol Chem. 1999;274:16437–16443. doi: 10.1074/jbc.274.23.16437. [DOI] [PubMed] [Google Scholar]
- Johnson KM, Mahajan SS, Wilson AC. Herpes simplex virus transactivator VP16 discriminates between HCF-1 and a novel family member, HCF-2. J Virol. 1999;73:3930–3940. doi: 10.1128/jvi.73.5.3930-3940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristie TM, LeBowitz JH, Sharp PA. The octamer-binding proteins form multi-protein–DNA complexes with the HSV alpha TIF regulatory protein. Embo J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristie TM, Pomerantz JL, Twomey TC, Parent SA, Sharp PA. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- LaBoissière S, Walker S, O’Hare P. Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol Cell Biol. 1997;17:7108–7118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBoissière S, Hughes T, O’Hare P. HCF-dependent nuclear import of VP16. EMBO J. 1999;18:480–489. doi: 10.1093/emboj/18.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JS, Herr W. Interdigitated residues within a small region of VP16 interact with Oct-1, HCF, and DNA. Mol Cell Biol. 1997;17:3937–3946. doi: 10.1128/mcb.17.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Herr W. Stabilization but not the transcriptional activity of herpes simplex virus VP16-induced complexes is evolutionarily conserved among HCF family members. J Virol. 2001;75:12402–12411. doi: 10.1128/JVI.75.24.12402-12411.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hengartner MO, Herr W. Selected elements of herpes simplex virus accessory factor HCF are highly conserved in Caenorhabditis elegans . Mol Cell Biol. 1999;19:909–915. doi: 10.1128/mcb.19.1.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J, Podemski L, Roy K, Pilgrim D, Hodgetts R. Analysis of two cosmid clones from chromosome 4 of Drosophila melanogaster reveals two new genes amid an unusual arrangement of repeated sequences. Genome Res. 1999;9:137–149. [PMC free article] [PubMed] [Google Scholar]
- Lu R, Misra V. Zhangfei: A second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res. 2000;28:2446–2454. doi: 10.1093/nar/28.12.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Yang P, O’Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano RL, Wilson AC. An activation domain in the C-terminal subunit of HCF-1 is important for transactivation by VP16 and LZIP. Proc Natl Acad Sci USA in press. 2002 doi: 10.1073/pnas.202200399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan SS, Wilson AC. Mutations in host cell factor 1 separate its role in cell proliferation from recruitment of VP16 and LZIP. Mol Cell Biol. 2000;20:919–928. doi: 10.1128/mcb.20.3.919-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole SJ, Kauvar LM, Drees B, Kornberg T. The engrailed locus of Drosophila: Structural analysis of an embryonic transcript. Cell. 1985;40:37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Rose RE, Gallaher NM, Andrew DJ, Goodman RH, Smolik SM. The CRE-binding protein dCREB-A is required for Drosophila embryonic development. Genetics. 1997;146:595–606. doi: 10.1093/genetics/146.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr RB, Smith MR, Beddall M, Sharp PA. A novel 50-kilodalton fragment of host cell factor 1 (C1) in G(0) cells. Mol Cell Biol. 2000;20:3568–3575. doi: 10.1128/mcb.20.10.3568-3575.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen KA, Newell A, Robinson M, Mills JS, Canning G, Handa R, Parkes K, Borkakoti N, Jupp R. Protein interactions in the herpes simplex virus type 1 VP16-induced complex: VP16 peptide inhibition and mutational analysis of host cell factor requirements. J Virol. 1997;71:3886–3894. doi: 10.1128/jvi.71.5.3886-3894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JL, Kristie TM. Autocatalytic proteolysis of the transcription factor-coactivator C1 (HCF): A potential role for proteolytic regulation of coactivator function. Proc Natl Acad Sci USA. 2000;97:9425–9430. doi: 10.1073/pnas.160266697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Cleary MA, Lai J-S, LaMarco K, Peterson MG, Herr W. Combinatorial control of transcription: The herpes simplex virus VP16-induced complex. Cold Spring Harbor Symp Quant Biol. 1993a;58:167–178. doi: 10.1101/sqb.1993.058.01.021. [DOI] [PubMed] [Google Scholar]
- Wilson AC, LaMarco K, Peterson MG, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993b;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Parrish JE, Massa HF, Nelson DL, Trask BJ, Herr W. The gene encoding the VP16-accessory protein HCF (HCFC1) resides in human Xq28 and is highly expressed in fetal tissues and the adult kidney. Genomics. 1995a;25:462–468. doi: 10.1016/0888-7543(95)80046-o. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Peterson MG, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 1995b;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Freiman RN, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Boutros M, Johnson KM, Herr W. HCF-1 amino- and carboxy-terminal subunit association through two separate sets of interaction modules: Involvement of fibronectin type 3 repeats. Mol Cell Biol. 2000;20:6721–6730. doi: 10.1128/mcb.20.18.6721-6730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Liu Y, Kobayashi R, Herr W. Developmental and cell-cycle regulation of Caenorhabditis elegans HCF phosphorylation. Biochemistry. 2001;40:5786–5794. doi: 10.1021/bi010086o. [DOI] [PubMed] [Google Scholar]
- Yokomori K, Admon A, Goodrich JA, Chen JL, Tjian R. Drosophila TFIIA-L is processed into two subunits that are associated with the TBP/TAF complex. Genes Dev. 1993;7:2235–2245. doi: 10.1101/gad.7.11.2235. [DOI] [PubMed] [Google Scholar]