Abstract
Background
Obesity is strongly associated with major depressive disorder (MDD) and various other diseases. Genome-wide association studies have identified multiple risk loci robustly associated with body mass index (BMI). In this study, we aimed to investigate whether a genetic risk score (GRS) combining multiple BMI risk loci might have utility in prediction of obesity in patients with MDD.
Methods
Linear and logistic regression models were conducted to predict BMI and obesity, respectively, in three independent large case–control studies of major depression (Radiant, GSK-Munich, PsyCoLaus). The analyses were first performed in the whole sample and then separately in depressed cases and controls. An unweighted GRS was calculated by summation of the number of risk alleles. A weighted GRS was calculated as the sum of risk alleles at each locus multiplied by their effect sizes. Receiver operating characteristic (ROC) analysis was used to compare the discriminatory ability of predictors of obesity.
Results
In the discovery phase, a total of 2,521 participants (1,895 depressed patients and 626 controls) were included from the Radiant study. Both unweighted and weighted GRS were highly associated with BMI (P <0.001) but explained only a modest amount of variance. Adding ‘traditional’ risk factors to GRS significantly improved the predictive ability with the area under the curve (AUC) in the ROC analysis, increasing from 0.58 to 0.66 (95% CI, 0.62–0.68; χ2 = 27.68; P <0.0001). Although there was no formal evidence of interaction between depression status and GRS, there was further improvement in AUC in the ROC analysis when depression status was added to the model (AUC = 0.71; 95% CI, 0.68–0.73; χ2 = 28.64; P <0.0001). We further found that the GRS accounted for more variance of BMI in depressed patients than in healthy controls. Again, GRS discriminated obesity better in depressed patients compared to healthy controls. We later replicated these analyses in two independent samples (GSK-Munich and PsyCoLaus) and found similar results.
Conclusions
A GRS proved to be a highly significant predictor of obesity in people with MDD but accounted for only modest amount of variance. Nevertheless, as more risk loci are identified, combining a GRS approach with information on non-genetic risk factors could become a useful strategy in identifying MDD patients at higher risk of developing obesity.
Electronic supplementary material
The online version of this article (doi:10.1186/s12916-015-0334-3) contains supplementary material, which is available to authorized users.
Keywords: Body mass index, Genetic risk score, Major depressive disorder, Obesity
Background
Obesity is a serious public health problem associated with an increased risk of various chronic diseases such as hypertension, diabetes, and cardiovascular disease [1]. It is estimated that over one-third of adults in the US are obese, whereas another one-third are overweight [2]. Moreover, the prevalence rate of obesity or overweight in most countries has been rising steadily over the past decades, resulting in a huge health burden [3]. There is also evidence that people with major depressive disorder (MDD) are more likely to be overweight or obese compared to psychiatrically-healthy controls [4], particularly in individuals with atypical depression, in whom increased appetite and weight gain are more prevalent. In addition, depressed people have a higher risk for various medical diseases and most of them are obesity-related. A recent meta-analysis further suggested the bi-directional relationship between obesity and MDD [5]. Given the high prevalence rate of both obesity and MDD, understanding the nature of their relationship is a pressing clinical problem.
Dietary factors and a lack of exercise as well as genetic factors contribute to the development of obesity. Twin and family studies have suggested the heritability of body mass index (BMI) to be between 0.4 and 0.7 [6]. The advance of genome-wide association studies (GWAS) has successfully identified multiple polymorphisms associated with the risk of obesity and higher BMI [7-9]. Among them, the fat mass and obesity associated (FTO) gene was consistently and reliably replicated in different studies. Our team has found that several polymorphisms in the FTO gene, the locus conferring the highest genetic risk contribution to obesity, are associated with increased BMI in people with MDD. A disease history of depression further moderates the effect of FTO on BMI [10]. However, each risk variant only confers a modest effect on the risk, resulting in a limited ability for obesity prediction by applying single variants. It has been suggested that combining multiple loci into a genetic risk score (GRS) might improve prediction of obesity. Although several studies have examined the joint genetic effect using different numbers of genetic variants to discriminate obesity in the general population [11-13], no study, to date, has investigated the combined genetic effects on obesity in people with MDD.
In this study, we aimed to investigate whether a GRS incorporating a number of well-defined common single nucleotide polymorphisms (SNPs) might have utility in prediction of obesity in patients with MDD.
Methods
Subjects and phenotypes
Discovery phase–Radiant study
A total of 3,244 participants (2,434 depressed patients and 810 healthy controls) were recruited from the Radiant study, which included the Depression Network (DeNT) study [14], the Depression Case–Control (DeCC) study [15], and the Genome-Based Therapeutic Drugs for Depression (GENDEP) study [16]. The DeNT study is a family study which recruited sibling pairs affected with recurrent unipolar depression from eight clinical sites across Europe and one in the USA. Only one proband from each family was recruited in our analysis. The DeCC study is a case–control study which recruited unrelated patients from three sites in the UK. All participants in the DeNT and DeCC studies experienced two or more episodes of major depression of at least moderate severity. The GENDEP study recruited individuals with at least one episode of depression of at least moderate severity from nine European centres. People who had ever fulfilled criteria of intravenous drug dependence, substance-induced mood disorder, schizophrenia, or bipolar disorder were excluded. The diagnosis of MDD was ascertained using the Schedules for Clinical Assessment in Neuropsychiatry (SCAN) [17] interview in all three studies. The controls were screened for lifetime absence of any psychiatric disorder using a modified version of the Past History Schedule [18]. Participants were excluded if they, or a first-degree relative, ever fulfilled the criteria for depression, bipolar disorder, or schizophrenia.
Self-reported weight and height were obtained during the SCAN interview for the individuals with depression and during telephone interview for controls. BMI was defined as weight in kilograms divided by height in meters squared. Obesity was defined as BMI ≥30 and normal weight was defined as BMI between 18.5 and 25. The reliability of self-report of height and weight was assessed in the GENDEP dataset (n = 811) where we also had measured height and weight. The correlations for measured versus self-reported height, weight, and BMI were 0.97, 0.95, and 0.95, respectively.
All participants were of white European ancestry. Approval was obtained from the local research ethics committees/institutional research boards of all of the participating sites. The full list of ethics committees can be seen in Additional file 1.
Replication phase – GSK-Munich study
Overall, 1,679 participants (822 cases and 857 controls) were recruited at the Max-Planck Institute of Psychiatry in Munich, Germany, and at two psychiatric hospitals in the Munich area (BKH Augsburg and Klinikum Ingolstadt). The same inclusion and exclusion criteria were applied in this study as the Radiant study. Patients had to fulfil the diagnosis of recurrent major depressive disorder of moderate or severe intensity using the SCAN interview. Controls were selected randomly from a Munich-based community and were screened for the presence of anxiety or mood disorders using the Composite International Diagnostic Screener (German version) [19]. Only individuals without mood and anxiety disorders were collected as controls. This study has been described in more detail elsewhere [20]. Anthropometric measures for patients and controls were taken at the Max Planck Institute and associated studies sites by trained technicians and study nurses [20].
This study was approved by the Ethics Committee of the Ludwig Maximilian University, Munich, Germany and written informed consent was obtained from all participants.
PsyCoLaus study
A total of 2,993 participants (1,296 cases and 1,697 controls) were recruited from a psychiatric sub-study (PsyCoLaus) of a community survey (CoLaus) carried out in Lausanne, Switzerland. A DSM-IV diagnosis of MDD was ascertained using the Diagnostic Interview for Genetics Studies [21]. The control subjects never fulfilled criteria for MDD. The PsyCoLaus study has been described in more detail elsewhere [22]. Weight and height were measured at the outpatient clinic at the Centre Hospitalier Universitaire Vaudois [23].
The Ethics committee of the Faculty of Biology and Medicine of the University of Lausanne approved the study and informed consent was obtained from all participants.
Selection of SNPs, genotyping, and quality control procedure
In the discovery phase, all the participants in Radiant were genotyped using the Illumina HumanHap610-Quad BeadChips (Illuminia, Inc., San Diego, CA, USA) by the Centre National de Génotypage as previously described [24]. All DNA samples underwent stringent quality control including exclusion if the sample genotype missing rate was >1%, or if abnormal heterozygosity or unmatched sex assignment were observed. SNPs with minor allele frequency <1% or showing departure from the Hardy-Weinberg equilibrium (P <1 × 10−5) were excluded. Quality control was described in detail elsewhere [24]. The risk alleles were defined as alleles associated with increased risk of BMI. We derived a 32-SNP additive GRS from the SNPs reported by Speliotes et al. [9] and Belsky et al. [25]. Of the 32 GRS SNPs, 14 were extracted from GWAS data after applying quality control, and 13 were extracted using proxy SNPs with r2 > 0.9. The remaining 5 SNPs, namely rs11847697, rs11083779, rs11165643, rs7640855, and rs1475219, were derived from the 1000 Genomes project imputed data. The quality measure of imputation for these SNPs was above 0.8. The call rate for most SNPs was more than 96% except for one SNP, rs1475219, which was approximately 91%. The detailed information of the 32 SNPs is shown in Table 1.
Table 1.
Single nucleotide polymorphisms included in the genetic risk score in the RADIANT study
| Chr | Nearest gene | SNP name | Alleles | BMI-increasing allele | Frequency of BMI-increasing allele | GWAS effect-size for BMI | Call rate |
|---|---|---|---|---|---|---|---|
| 1 | NEGR1 | rs2568958 | A/G | A | 62.5% | 0.13 | 99.95% |
| TNNI3K | rs1514175 | A/G | A | 42.3% | 0.07 | 99.86% | |
| PTBP2 | rs11165643 | C/T | T | 58.8% | 0.06 | 99.07% | |
| SEC16B | rs10913469 | C/T | C | 19.2% | 0.22 | 100% | |
| 2 | TMEM18 | rs2867125 | C/T | C | 82.9% | 0.31 | 99.98% |
| ADCY3,RBJ | rs10182181 | A/G | G | 46.9% | 0.14 | 99.40% | |
| FANCL | rs759250 | A/G | A | 28.4% | 0.1 | 100% | |
| LRP1B | rs6714473 | C/T | T | 9.7% | 0.09 | 99.85% | |
| 3 | CADM2 | rs7640855 | A/G | A | 19.0% | 0.1 | 96.83% |
| ETV5 | rs7647305 | C/T | C | 79.0% | 0.14 | 99.93% | |
| 4 | GNPDA2 | rs12641981 | C/T | T | 44.1% | 0.18 | 100% |
| SLC39A8 | rs13107325 | C/T | T | 7.5% | 0.19 | 99.91% | |
| 5 | FLJ35779 | rs253414 | C/T | T | 66.4% | 0.1 | 99.93% |
| ZNF608 | rs6864049 | A/G | A | 47.2% | 0.07 | 100% | |
| 6 | TFAP2B | rs987237 | A/G | A | 18.2% | 0.13 | 100% |
| NUDT3 | rs206936 | A/G | G | 18.0% | 0.06 | 95.99% | |
| 9 | LRRN6C | rs2183825 | C/T | C | 32.9% | 0.11 | 99.98% |
| 11 | STK33, RPL27A | rs10840065 | A/G | A | 51.6% | 0.06 | 100% |
| BDNF | rs6265 | C/T | C | 79.8% | 0.19 | 100% | |
| MTCH2 | rs10838738 | A/G | G | 34.5% | 0.06 | 100% | |
| 12 | BCDIN3, FAIM2 | rs7138803 | A/G | A | 37.5% | 0.12 | 100% |
| 13 | MTIF3 | rs1475219 | C/T | C | 20.4% | 0.09 | 90.61% |
| 14 | PRKD1 | rs11847697 | C/T | T | 3.6% | 0.17 | 96.87% |
| NRXN3 | rs10146997 | A/G | G | 21.9% | 0.13 | 100% | |
| 15 | MAP2K5 | rs2241423 | A/G | G | 77.2% | 0.13 | 99.96% |
| 16 | GPRC5B | rs12446632 | A/G | G | 86.1% | 0.17 | 99.93% |
| SH2B1 | rs4788102 | A/G | A | 39.0% | 0.15 | 100% | |
| FTO | rs3751812 | G/T | T | 41.0% | 0.39 | 100% | |
| 18 | MC4R | rs921971 | C/T | C | 26.6% | 0.23 | 99.98% |
| 19 | KCTD15 | rs29941 | A/G | G | 68.3% | 0.06 | 100% |
| ZC3H4, TMEM160 | rs2303108 | C/T | C | 71.4% | 0.09 | 100% | |
| QPCTL | rs11083779 | C/T | T | 95.8% | 0.15 | 98.28% |
The GSK Munich study was used for replication. Genotyping was performed using the Illumina HumanHap550 SNP Chip arrays. All SNPs with a call frequency below 95% were excluded. The details were described elsewhere [26]. The same criteria to construct the GRSs was applied here; whenever possible, SNPs were extracted from the GWAS data after applying quality control, and the rest of the SNPs were extracted using proxy SNPs.
Participants in the PsyCoLaus study were genotyped using the Affymetrix 500 K SNP chip [22]. The genotype was obtained via the BRLMM algorithm. The SNPs were removed from the analysis based on gender inconsistency, call rate less than 90%, and inconsistent duplicate genotypes. The GRSs were constructed as in the discovery phase.
Construction of the unweighted and weighted GRS
To evaluate the combined effects of the 32 SNPs on BMI, an additive model was used to construct both unweighted and weighted GRSs. The unweighted GRS (uGRS) was calculated by summation of the number of risk alleles across the 32 variants. The weighted GRS (wGRS) was calculated by multiplying the number of risk alleles at each locus (0, 1, 2) for the corresponding effect sizes, in kg/m2 per allele, as reported by Speliotes et al. [9] and then summing the products. In order to reduce the bias caused by missing data, only the participants without any missing data were included in our GRS analysis.
Statistical analysis
Linear regression models using traditional risk factors (age, sex, and principal components of ancestry) and GRS were calculated to predict BMI. Since BMI did not follow a normal distribution, a natural log-transformed BMI was used for the analyses. The analyses were first performed in the whole sample and then separately in the depressive cases and controls.
Binary logistic regression adjusted by age, sex, depression status and ancestry was used to predict probabilities of obesity in each model. Receiver-operating characteristics (ROC) curve analysis was conducted to calculate the area under the curve (AUC) to evaluate the discriminatory ability of each model. We first compared the difference between AUCs from models incorporating traditional risk factors (age, sex, and ancestry) with and without GRS. Then we compared the models comprising GRS only and the models incorporating other risk factors. To correct for the possible presence of population stratification, all analyses were adjusted for the first five principal components of ancestry, which were calculated with EIGENSOFT [27].
The analyses were performed first in the whole sample, and then separately in depressed patients and controls. All data were analyzed using STATA version 12.1 (STATA Corp, Texas). Two-tailed value of P <0.05 were considered significant.
Results
Discovery phase – Radiant study
Demographic characteristics
After excluding people with any missing genotypes, a total of 2,521 participants (2,086 non-obese and 435 obese) were included in the analysis. There were no differences in sex, age, and depression status between included and excluded people (all P >0.05). The mean age ± SD of participants was 43.9 ± 12.8 years (non-obese 43.2 ± 13.1, obese 47.3 ± 10.7, t = −6.08, P <0.0001) and 67.7% were female (72.9% female in obese and 66.6% female in non-obese, χ2 = 6.50, P = 0.011). Obese people were more likely to be depressed (90.3% vs. 72.0%, χ2 = 64.87, P <0.001).
The frequencies of uGRS and wGRS were approximately within normal distribution (Figure 1). The mean uGRS, the total number of risk alleles of 32 SNPs, was 29.5 ± 3.5 in obese and 28.6 ± 3.5 in non-obese participants (t = −4.47, P <0.0001), whereas the mean wGRS was slightly higher in obese compared to non-obese participants (4.14 ± 0.50 vs. 4.03 ± 0.53, t = −4.18, P <0.0001).
Figure 1.
Distribution of weighted genetic risk score in RADIANT study.
Principal component analysis was used to control for population stratification. The top five principal component scores were used to discriminate the subpopulation of white Europeans. Principal component 1 (distinguishes southeast Europe from northwest European ancestry) and principal component 2 (distinguishes east Europe from west Europe) were significantly associated with BMI and were included as covariates.
Linear regression analyses with BMI as the outcome variable
A base linear regression model including age, sex, depression status, ancestry, and significant interaction between ancestry and age accounted for 8.29% of the variance in log-transformed BMI. After adding weighted GRS to the base model, there was improvement of fit and an additional 1.27% of phenotypic variance of BMI explained giving a total of 9.56% (Table 2). Using either weighted or unweighted GRS made little difference for the explained variance of BMI (9.56% vs. 9.58%). No interaction between traditional covariates or between GRS and traditional covariates were found (data not shown). Although the interaction between depression and GRS on BMI did not meet the conventional 5% level of significance (ß = 0.27, s.e. = 0.02, P = 0.078), stratifying by depression status with GRS incorporated in the model explained an extra 1.63% of variance of BMI in depressed patients but only explained an extra 0.34% of variance of BMI in healthy controls.
Table 2.
Linear regression models with BMI as the outcome variable
| Study/sample | Model | F | Adj. R 2 | Additional variance explained by GRS |
|---|---|---|---|---|
| Radiant | ||||
| Total | Model 1: adjusted by age, sex, and depression | 38.98 | 0.0829 | 1.27% |
| Model 2: model 1 + wGRS | 39.16 | 0.0956 | ||
| Depressed cases | Model 1: adjusted by age and sex | 17.85 | 0.0426 | 1.63% |
| Model 2: model 1 + wGRS | 20.75 | 0.0589 | ||
| Controls | Model 1: adjusted by age and sex | 11.71 | 0.0789 | 0.34% |
| Model 2: model 1 + wGRS | 10.34 | 0.0823 | ||
| GSK-Munich | ||||
| Total | Model 1: adjusted by age, sex, and depression | 34.02 | 0.1056 | 0.53% |
| Model 2: model 1 + wGRS | 29.80 | 0.1109 | ||
| Depressed cases | Model 1: adjusted by age and sex | 8.02 | 0.0372 | 1.32% |
| Model 2: model 1 + wGRS | 7.13 | 0.0504 | ||
| Controls | Model 1: adjusted by age and sex | 25.66 | 0.1306 | 0.23% |
| Model 2: model 1 + wGRS | 21.98 | 0.1329 | ||
| PsyCoLaus | ||||
| Total | Model 1: adjusted by age, sex, and depression | 40.20 | 0.0843 | 0.93% |
| Model 2: model 1 + wGRS | 39.47 | 0.0936 | ||
| Depressed cases | Model 1: adjusted by age and sex | 14.84 | 0.0605 | 1.09% |
| Model 2: model 1 + wGRS | 15.15 | 0.0714 | ||
| Controls | Model 1: adjusted by age and sex | 31.25 | 0.0970 | 0.77% |
| Model 2: model 1 + wGRS | 29.21 | 0.1047 |
Prediction of obesity
Logistic regression models were used to examine the relationship between GRS and obesity in addition to age, sex, ancestry, and depression status. The discriminative power of the regression model was measured by the AUC. The AUC was significantly higher in the model combining all non-genetic risk factors (age, sex, ancestry, and depression status) and genetic factors compared to the model only applying non-genetic risk factors (AUC increased from 0.69 to 0.71, χ2 = 9.83, P = 0.0017). We further investigated whether GRS alone is able to discriminate obesity or not. The AUC was only 0.58 (95% CI, 0.55–0.61) while only including genetic risk score and ancestry into the base regression model. However, the AUC increased to 0.65 (95% CI, 0.62–0.68) after adding traditional risk factors such as age and sex (χ2 = 21.46, P <0.0001). The AUC further increased to 0.71 (95% CI, 0.68–0.73) on incorporating depression status into the above model (χ2 = 32.33, P <0.0001; Figure 2). Again, the unweighted GRS produced similar results as the wGRS when incorporated into our regression model (AUC increased from 0.58 to 0.65 to 0.70).
Figure 2.
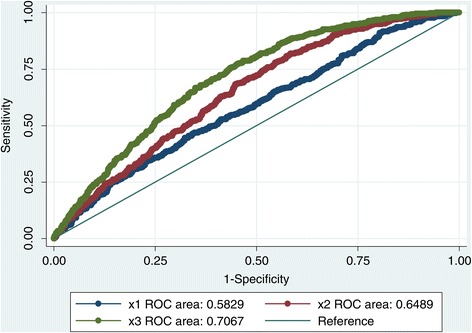
Receiver operating characteristic curves for models predicting obesity in the discovery phase. The AUC for the full model combining depression status, age, sex, and GRS (×3) is significantly greater than AUC for the model combining age, sex, and GRS (×2), which in turn is significantly greater than AUC for the base model with only GRS (×1).
We used the same analysis stratifying by depression status and found that, in depressed patients, the AUC increased from 0.58 (95% CI, 0.55–0.61) to 0.61 (95% CI, 0.58–0.64; χ2 = 5.65, P = 0.0175) while in healthy controls it remained at 0.67 (95% CI, 0.60–0.73; χ2 = 0.00, P = 0.98). No interaction was found between depression, GRS, and obesity (OR = 1.08, s.e. = 0.36, P = 0.81).
Replication phase – GSK Munich study
Demographic characteristics
A total of 1,679 participants (244 obese and 1,435 non-obese) were included in this study. The mean age ± SD was 51.49 ± 13.50 years (53.29 ± 11.51 for obese and 51.19 ± 13.80 for non-obese, P = 0.01). There was no sex difference between obese and non-obese patients (64.75% obese and 67.24% non-obese patients were female, P = 0.44). Obese people were more likely to be depressed (64.75% vs. 46.27%, P <0.001).
Linear regression analyses with BMI as the outcome variable
Linear regression models to predict BMI suggested the wGRS accounts for 0.63% of the variance in log-transformed BMI. While stratifying by depression status, we found wGRS explained an extra 1.32% of phenotypic variance of BMI in depressed patients but only accounted for 0.23% of variance in healthy controls (Table 2).
No significant interaction was found between depression and GRS on BMI (ß = 0.25, s.e. = 0.01, P = 0.18).
Prediction of obesity
Logistic regression models were used to examine the relationship between GRS and obesity in addition to age, sex, ancestry, and depression status. The AUC was approximately 0.59 (95% CI, 0.55–0.63) while only including genetic risk score and ancestry into the base regression model. The AUC increased to 0.64 (95% CI, 0.60–0.68) while adding traditional risk factors such as age and sex (χ2 = 8.21, P = 0.004). The AUC further increased to 0.69 (95% CI, 0.66–0.73) while incorporating depression status into the above model (χ2 = 10.67, P = 0.001). Stratified analyses by depression status showed that using wGRS to discriminate obesity was statistically significant in depressed patients (AUC increased from 0.53 (95% CI, 0.48–0.58) to 0.58 (95% CI, 0.53–0.63), χ2 = 4.19, P = 0.041) but not in healthy controls (AUC remained at 0.66 (95% CI, 0.60–0.72), χ2 = 0.34, P = 0.56).
No significant interaction was found between depression and GRS on obesity (OR = 1.38, s.e. = 0.39, P = 0.26).
PsyCoLaus study
Demographic characteristics
Overall, 2,993 subjects (409 obese and 2,584 non-obese) were included in PsyCoLaus study. The mean age ± SD was 50.19 ± 8.84 years (52.94 ± 8.80 for obese and 49.76 ± 8.77 for non-obese, P <0.0001). There were no sex differences between obese and non-obese patients (49.87% of obese and 53.44% of non-obese people were female, P = 0.18). Obese people and non-obese people had equal depression rates (40.83% vs. 43.69%, P = 0.28).
Linear regression analyses with BMI as the outcome variable
Linear regression analysis to predict BMI suggested the wGRS accounts for 0.90% of the variance in log-transformed BMI. While stratifying by depression status, we found that wGRS explained an extra 1.09% of phenotypic variance of BMI in depressed patients but only accounted for 0.77% of variance of BMI in healthy controls (Table 2).
No significant interaction was found between depression and GRS on BMI (ß = 0.09, s.e. = 0.01, P = 0.52).
Prediction of obesity
Again, logistic regression models were used to examine the relationship between GRS and obesity in addition to age, sex, ancestry, and depression status. The AUC was approximately 0.56 (95% CI, 0.53–0.58) while only including GRS and ancestry into the base regression model. The AUC increased to 0.62 (95% CI, 0.59–0.65) while adding traditional risk factors such as age and sex (χ2 = 14.61, P = 0.0001). The AUC remained at 0.62 (95% CI, 0.59–0.65) while incorporating depression status into the above model (χ2 = 0.11, P = 0.74). Stratified analyses by depression status showed that using wGRS to discriminate obesity was not statistically significant neither in depressed patients (AUC increased from 0.61 (95% CI, 0.56–0.66) to 0.63 (95% CI, 0.58–0.67), χ2 = 3.66, P = 0.0558) nor in healthy controls (AUC increased from 0.61 (95% CI, 0.57–0.65) to 0.62 (95% CI, 0.59–0.66), χ2 = 2.66, P = 0.1).
No significant interaction was found between depression and GRS on obesity (OR = 0.98, s.e. = 0.21, P = 0.94).
Discussion
In this study, we developed both weighted and unweighted GRS, including 32 well-established risk loci from a recent meta-analysis of GWAS on BMI [9]. We aimed to investigate whether these GRSs are associated with BMI and predict obesity.
Prediction of BMI
Both uGRS and wGRS were associated with BMI (P <0.0001) and accounted for 1.27%, 0.63%, and 0.90% of phenotypic variance of BMI in Radiant, GSK Munich, and PsyCoLaus studies, respectively, and there was little difference in explained variance of BMI in each study. For each unit increase in uGRS, which is equal to one additional risk allele, BMI increased by approximately 0.175 kg/m2. Our overall result was thus in keeping with a previous study [9] using the same method to construct a GRS for BMI, but which did not take into account the relationship between BMI and depression.
Our results suggest that GRS explained more phenotypic variance of BMI in depressed patients than in healthy controls, although the interaction analyses were suggestive (Radiant) but not significant (GSK Munich and PsyCoLaus), this could reflect the fact that conventional levels of significance for interaction are often difficult to detect when the outcome variable has been log transformed. Interestingly, the case/control difference in the effect of GRS was more prominent when depression was diagnosed in clinical settings (RADIANT and GSK Munich studies) than in a community study (PsyCoLaus study).
Prediction of obesity
We further explored the utility of a GRS approach using ROC analysis to compare the discriminatory ability of predictors of obesity. Conventionally, it is accepted that the AUC in a ROC analysis should be >0.8 to be of clinical value for screening. During the discovery phase, AUC fell short of this threshold but combining genetic factors and non-genetic factors proved better than using GRS alone in the prediction of obesity (with the AUC increasing from 0.69 to 0.71). In the replication phase, findings were similar except that depression had a small and non-significant association with obesity in the PsyCoLaus study, which could reflect the fact that PsyCoLaus was a community-based study with less severe cases of MDD than the clinically ascertained RADIANT and Munich GSK studies. Our results suggest that GRS might improve obesity prediction in depressed patients compared to controls.
In other respects, the results were similar to previous studies, which used only genome wide significant genetic variants to construct a GRS [11], in finding that the optimum AUC was obtained by combining GRS and non-genetic risk factors. A significant novel feature of the present study was that combining these factors with depression status further improves the prediction of obesity. This is in keeping with the association between obesity and MDD that has been found in either the general population or clinical settings [4,5,28]. Although the relationship between these two diseases may be bi-directional [5], our own recent analyses using a Mendelian Randomization approach [29] do not support a direction of cause from high BMI to depression. In addition, the fact that GRS has a larger effect on BMI and obesity in depressed patients, especially clinically severe depression, might reflect the importance of genetic effects on the association between obesity and clinically significant depression.
Limitations
There are certainly some limitations that should be mentioned. First, we only selected the risk loci that reached genome-wide levels of significance. It is highly probable that there are additional as yet to be identified loci that will emerge when even larger sample sizes are included in GWAS. Second, since the established common variants from GWAS explain only a small proportion of the variation in BMI, future studies should include rare variants with larger effects and copy number variants to construct future GRS. In addition, gene-gene interactions and gene-environment interactions should be taken into account as well to maximize the obesity prediction ability of GRS. For example, our group [10] has found that depression status moderates the effect of FTO gene on BMI (although we did not find evidence of interaction between depression and GRS in the current study). Third, the 32 BMI loci used to construct the GRS were identified in GWAS of white European origin. The allele frequencies and their effect size may be different from non-European populations and the results should probably not be generalized to other ethnicities. Furthermore, the present study is a cross sectional study and cannot therefore take into account BMI fluctuations across the life span.
A further minor drawback is that PsyCoLaus is a subset of the CoLaus study, which was one of the 46 studies from which the GRS was derived [9], and therefore cannot, on its own, provide independent estimation of the risk score effect.
Conclusions
In summary, we found that either a wGRS or a uGRS based on 32 well-established risk loci were significantly associated with BMI. Although GRS on its own explained only a small amount of variance of BMI, a significant novel feature of this study is that including non-genetic risk factors together with GRS and depression came close to the conventional threshold for clinical utility used in ROC analysis and improves the prediction of obesity.
Our results suggest that the GRS might predict obesity better in depressed patients than in healthy controls. This has potential clinical implications as well as implications for future research directions in exploring the links between depression and obesity-associated disorders.
While it is likely that future genome-wide studies with very large samples will detect variants other than the common ones, it seems probable that a combination of non-genetic information will still be needed to optimize the prediction of obesity.
Acknowledgements
The authors would like to thank those who agreed to participate in the studies and the many colleagues who contributed to collection and phenotypic characterization of the clinical samples, as well as to genotyping and statistical analyses.
Abbreviations
- AUC
Area under the curve
- BMI
Body mass index
- DeCC
Depression case–control study
- DeNT
Depression network study
- FTO
Fat mass and obesity associated gene
- GENDEP
Genome-based therapeutic drugs for depression
- GRS
Genetic risk score
- GWAS
Genome-wide association studies
- MDD
Major depressive disorder
- ROC
Receiver operating characteristic
- SCAN
Schedules for Clinical Assessment in Neuropsychiatry
- SNP
Single nucleotide polymorphism
- uGRS
Unweighted genetic risk score
- wGRS
Weighted genetic risk score
Additional file
List of institutions where the ethical committees gave approval for the Radiant study.
Footnotes
Competing interests
AEF and PM have received consultancy fees and honoraria for participating in expert panels for pharmaceutical companies including GlaxoSmithKline. PM has received speaker’s fees from Pfizer. FH is cofounder of the biotech company HolsboerMaschmeyerNeuroChemie GmbH (HMNC GmbH) in Germany. WM is member of the Advisory Board or has received speaker fees from Eli Lilly and Lundbeck. MP is part of the advisory boards for Eli Lilly and Lundbeck. All other authors declare no competing interests. This study was funded by the Medical Research Council, UK. GlaxoSmithKline (G0701420) funded the DeNT study and were co-funders with the Medical Research Centre for the GWAS of the whole sample. The GENDEP study was funded by a European Commission Framework 6 grant, EC Contract Ref.: LSHB-CT-2003-503428. This study presents independent research [part-] funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. The CoLaus/PsyCoLaus was funded by four grants from the Swiss National Science Foundation (#32003B-105993, #32003B-118308, #33CSC0-122661, and #139468), the Faculty of Biology and Medicine of Lausanne, and two grants from GlaxoSmithKline Clinical Genetics.
Authors’ contributions
PM, MR, and CH participated in the design of the study. CH performed statistical analyses and wrote the first draft of the manuscript. DC, TC, and CW helped with the statistical analyses and extraction of the SNPs from the GWAS data. SB, ZK, and BM contributed to the analyses. PM and MR lead the study and made critical revisions to the report. GB, PM, and MR supervised the statistical analyses. SK, NC, MG, FH, LJ, IJ, AK, SL, WM, OM, MO, JR, MR, RU, PV, GW, AF, MP, and PM participated in the collection of clinical and phenotype data. IC contributed to the coordination of genotyping for the Radiant study. All authors were involved in drafting the manuscript or revising it critically for important intellectual content, and approved the final version of the manuscript to be published.
Contributor Information
Chi-Fa Hung, Email: chifa.hung@gmail.com.
Gerome Breen, Email: gerome.breen@gmail.com.
Darina Czamara, Email: darina@mpipsykl.mpg.de.
Tanguy Corre, Email: tanguy.corre@unil.ch.
Christiane Wolf, Email: cwolf@mpipsykl.mpg.de.
Stefan Kloiber, Email: stkloiber@mpipsykl.mpg.de.
Sven Bergmann, Email: sven.bergmann@unil.ch.
Nick Craddock, Email: craddockn@Cardiff.ac.uk.
Michael Gill, Email: mgill@tcd.ie.
Florian Holsboer, Email: holsboer@mpipsykl.mpg.de.
Lisa Jones, Email: l.a.jones@bham.ac.uk.
Ian Jones, Email: jonesir1@cf.ac.uk.
Ania Korszun, Email: a.korszun@qmul.ac.uk.
Zoltan Kutalik, Email: zoltan.kutalik@unil.ch.
Susanne Lucae, Email: lucae@mpipsykl.mpg.de.
Wolfgang Maier, Email: wolfgang.maier@ukb.uni-bonn.de.
Ole Mors, Email: nielmors@rm.dk.
Michael J Owen, Email: owenmj@cardiff.ac.uk.
John Rice, Email: jrice@wustl.edu.
Marcella Rietschel, Email: marcella.rietschel@zi-mannheim.de.
Rudolf Uher, Email: rudolf.uher@cdha.nshealth.ca.
Peter Vollenweider, Email: peter.vollenweider@chuv.ch.
Gerard Waeber, Email: gerard.waeber@chuv.hospvd.ch.
Ian W Craig, Email: ian.craig@kcl.ac.uk.
Anne E Farmer, Email: anne.farmer@kcl.ac.uk.
Cathryn M Lewis, Email: cathryn.lewis@kcl.ac.uk.
Bertram Müller-Myhsok, Email: bmm@mpipsykl.mpg.de.
Martin Preisig, Email: martin.preisig@chuv.ch.
Peter McGuffin, Email: peter.mcguffin@kcl.ac.uk.
Margarita Rivera, Email: margarita.rivera_sanchez@kcl.ac.uk.
References
- 1.Nguyen NT, Magno CP, Lane KT, Hinojosa MW, Lane JS. Association of hypertension, diabetes, dyslipidemia, and metabolic syndrome with obesity: findings from the National Health and Nutrition Examination Survey, 1999 to 2004. J Am Coll Surg. 2008;207:928–34. doi: 10.1016/j.jamcollsurg.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999–2008. JAMA. 2010;303:235–41. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 3.Wang YC, McPherson K, Marsh T, Gortmaker SL, Brown M. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet. 2011;378:815–25. doi: 10.1016/S0140-6736(11)60814-3. [DOI] [PubMed] [Google Scholar]
- 4.Farmer A, Korszun A, Owen MJ, Craddock N, Jones L, Jones I, et al. Medical disorders in people with recurrent depression. Br J Psychiatry. 2008;192:351–5. doi: 10.1192/bjp.bp.107.038380. [DOI] [PubMed] [Google Scholar]
- 5.Luppino FS, de Wit LM, Bouvy PF, Stijnen T, Cuijpers P, Penninx BWJH, et al. Overweight, obesity, and depression: a systematic review and meta-analysis of longitudinal studies. Arch Gen Psychiatry. 2010;67:220. doi: 10.1001/archgenpsychiatry.2010.2. [DOI] [PubMed] [Google Scholar]
- 6.Maes HHM, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27:325–51. doi: 10.1023/A:1025635913927. [DOI] [PubMed] [Google Scholar]
- 7.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2008;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 8.Willer CJ, Speliotes EK, Loos RJF, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2008;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivera M, Cohen-Woods S, Kapur K, Breen G, Ng M, Butler AW, et al. Depressive disorder moderates the effect of the FTO gene on body mass index. Mol Psychiatry. 2012;17:604–11. doi: 10.1038/mp.2011.45. [DOI] [PubMed] [Google Scholar]
- 11.Sandholt CH, Sparsø T, Grarup N, Albrechtsen A, Almind K, Hansen L, et al. Combined analyses of 20 common obesity susceptibility variants. Diabetes. 2010;59:1667–73. doi: 10.2337/db09-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson RE, Maes HH, Holmans P, Sanders AR, Levinson DF, Shi J, et al. Genetic risk sum score comprised of common polygenic variation is associated with body mass index. Hum Genet. 2011;129:221–30. doi: 10.1007/s00439-010-0917-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Zhao JH, Luan J, Luben RN, Rodwell SA, Khaw KT, et al. Cumulative effects and predictive value of common obesity-susceptibility variants identified by genome-wide association studies. Am J Clin Nutr. 2010;91:184–90. doi: 10.3945/ajcn.2009.28403. [DOI] [PubMed] [Google Scholar]
- 14.Farmer A, Breen G, Brewster S, Craddock N, Gill M, Korszun A, et al. The Depression Network (DeNT) Study: methodology and sociodemographic characteristics of the first 470 affected sibling pairs from a large multi-site linkage genetic study. BMC Psychiatry. 2004;4:42. doi: 10.1186/1471-244X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen-Woods S, Gaysina D, Craddock N, Farmer A, Gray J, Gunasinghe C, et al. Depression Case Control (DeCC) Study fails to support involvement of the muscarinic acetylcholine receptor M2 (CHRM2) gene in recurrent major depressive disorder. Hum Mol Genet. 2009;18:1504–9. doi: 10.1093/hmg/ddp051. [DOI] [PubMed] [Google Scholar]
- 16.Uher R, Huezo-Diaz P, Perroud N, Smith R, Rietschel M, Mors O, et al. Genetic predictors of response to antidepressants in the GENDEP project. Pharmacogenomics J. 2009;9:225–33. doi: 10.1038/tpj.2009.12. [DOI] [PubMed] [Google Scholar]
- 17.Wing JK, Babor T, Brugha T, Burke J, Cooper J, Giel R, et al. SCAN: schedules for clinical assessment in neuropsychiatry. Arch Gen Psychiatry. 1990;47:589. doi: 10.1001/archpsyc.1990.01810180089012. [DOI] [PubMed] [Google Scholar]
- 18.McGuffin P, Katz R, Aldrich J. Past and present state examination: the assessment of ‘lifetime ever’ psychopathology. Psychol Med. 1986;16:461–5. doi: 10.1017/S0033291700009302. [DOI] [PubMed] [Google Scholar]
- 19.Wittchen HU, Höfler M, Gander F, Pfister H, Storz S, Üstün B, et al. Screening for mental disorders: performance of the Composite International Diagnostic–Screener (CID–S) Int J Methods Psychiatr Res. 1999;8:59–70. doi: 10.1002/mpr.57. [DOI] [Google Scholar]
- 20.Lucae S, Salyakina D, Barden N, Harvey M, Gagné B, Labbé M, et al. P2RX7, a gene coding for a purinergic ligand-gated ion channel, is associated with major depressive disorder. Hum Mol Genet. 2006;15:2438–45. doi: 10.1093/hmg/ddl166. [DOI] [PubMed] [Google Scholar]
- 21.Nurnberger JI, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, et al. Diagnostic interview for genetic studies: rationale, unique features, and training. Arch Gen Psychiatry. 1994;51:849–59. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 22.Preisig M, Waeber G, Vollenweider P, Bovet P, Rothen S, Vandeleur C, et al. The PsyCoLaus study: methodology and characteristics of the sample of a population-based survey on psychiatric disorders and their association with genetic and cardiovascular risk factors. BMC Psychiatry. 2009;9:9. doi: 10.1186/1471-244X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Firmann M, Mayor V, Vidal PM, Bochud M, Pécoud A, Hayoz D, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K, et al. Genome-wide association study of major recurrent depression in the UK population. Am J Psychiatr. 2010;167:949–57. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- 25.Belsky DW, Moffitt TE, Houts R, Bennett GG, Biddle AK, Blumenthal JA, et al. Polygenic risk, rapid childhood growth, and the development of obesity evidence from a 4-decade longitudinal study. Arch Pediatr Adolesc Med. 2012;166:515–21. doi: 10.1001/archpediatrics.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muglia P, Tozzi F, Galwey N, Francks C, Upmanyu R, Kong X, et al. Genome-wide association study of recurrent major depressive disorder in two European case–control cohorts. Mol Psychiatry. 2010;15:589–601. doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Zhao G, Ford E, Dhingra S, Li C, Strine T, Mokdad A. Depression and anxiety among US adults: associations with body mass index. Int J Obes (Lond) 2009;33:257–66. doi: 10.1038/ijo.2008.268. [DOI] [PubMed] [Google Scholar]
- 29.Hung C-F, Rivera M, Craddock N, Owen MJ, Gill M, Korszun A, et al. Relationship between obesity and the risk of clinically significant depression: Mendelian randomisation study. Br J Psychiatr. 2014;205:24–8. doi: 10.1192/bjp.bp.113.130419. [DOI] [PMC free article] [PubMed] [Google Scholar]


