Abstract
NR4A1 (Nur77, TR3) is an orphan nuclear receptor that is overexpressed in pancreatic cancer and exhibits pro-oncogenic activity. RNAi interference of NR4A1 expression in Panc-1 cells induced apoptosis and subsequent proteomic analysis revealed the induction of several markers of endoplasmic reticulum (ER) stress including glucose-related protein 78 (GRP78), CCAAT/enhancer-binding protein-homologous protein (CHOP), and activating transcription factor-4 (ATF-4). Treatment of pancreatic cancer cells with the NR4A1 antagonist 1,1-bis(3′-indolyl)-1-(p-hydroxyphenyl)methane (DIM-C-pPhOH), gave similar results. Moreover, both NR4A1 knockdown and DIM-C-pPhOH induced reactive oxygen species (ROS), and induction of ROS and ER stress by these agents was attenuated after co-treatment with antioxidants. Manipulation of NR4A1 expression coupled with gene expression profiling identified a number of ROS metabolism transcripts regulated by NR4A1. Knockdown of one of these transcripts, thioredoxin domain containing 5 (TXNDC5), recapitulated the elevated ROS and ER stress; thus, demonstrating that NR4A1 regulates levels of ER stress and ROS in pancreatic cancer cells to facilitate cell proliferation and survival. Finally, inactivation of NR4A1 by knockdown or DIM-C-pPhOH decreased TXNDC5, resulting in activation of ROS/ER stress and pro-apoptotic pathways.
Keywords: NR4A1, TR3, Nur77, pancreatic cancer, oxidative stress, endoplasmic stress, apoptosis, TXNDC5
INTRODUCTION
Nuclear receptor (NR) superfamily genes are critical regulators of homeostasis at all stages of development (1), and the 48 human NRs include the endocrine receptors, adopted orphan receptors, and orphan receptors. The NR4A orphan receptor subfamily consists of three orphan receptors (2, 3), namely NR4A1, (Nur77, TR3, NGFI-B), NR4A2 (Nurr1) and NR4A3 (Nor1), that were initially identified as immediate-early response genes induced by nerve growth factor in PC12 cells (4). The well-conserved DNA binding and C-terminal ligand binding domains of the NR4A receptors exhibit ~91-95% and ~60% homology, respectively, whereas their N-terminal A/B domains are highly divergent (5-7).
NR4A1 and NR4A2 bind as monomers and homodimers to NGFI-B response elements (NRBE) and Nur-responsive elements (NuRE), respectively. NR4A1 and NR4A2 (but not NR4A3) also form heterodimers with RXR and bind a DRE motif (8-10). NR4A receptors are induced by multiple stimuli/stressors and play essential roles in metabolic processes, inflammation, vascular function, steroidgenesis, and in the central nervous system. NR4A1 and NR4A2 are also overexpressed in multiple tumors and cancer cell lines (11, 12). RNA interference (RNAi) has been used to investigate the role of NR4A1 in cancer cells, and most studies indicate pro-oncongenic function where NR4A1 enhances both cancer cell proliferation and survival (11-20).
Zhang has summarized studies on a novel drug-dependent pro-apoptotic pathway in some cancer cell lines that is dependent on the nuclear export of NR4A1 and the subsequent formation of a mitochondria-associated NR4A1/bcl-2 pro-apoptotic complex (21). This has also led to development of peptide mimics that convert bcl-2 into an apoptotic complex, and paclitaxel, the taxane-derived anticancer drug, also exhibits comparable activity (22, 23). Multiple anticancer drugs and other apoptosis-inducers activate nuclear export of NR4A1, and there are reports that cytosporone B (CsnB) and related analogs may activate nuclear NR4A1 and also induce nuclear export of this receptor (24).
Knockdown of NR4A1 in lung and pancreatic cancer cell lines inhibits cell growth and induces apoptosis which is due, in part, to downregulation of the anti-apoptotic gene survivin (19). Mechanistic studies showed that survivin expression is regulated by a p300/NR4A1/Sp1 complex bound to the proximal GC-rich sites of the survivin promoter (19). NR4A1 also binds and inactivates wild-type p53 (25), and NR4A1 silencing results in mTOR inhibition due to activation of p53 and p53-dependent induction of sestrin2 which in turn activates AMPK (20). 1,1-Bis(3′-indolyl)-1-(p-hydroxyphenyl)methane (DIM-C-pPhOH) has been characterized as an NR4A1 antagonist which mimics the effects of NR4A1 silencing. DIM-C-pPhOH decreased cancer cell and tumor growth, activated p53-dependent inhibition of mTOR signaling, and downregulated survivin expression (19, 20).
NR4A1 silencing or treatment with DIM-C-pPhOH also altered pancreatic cancer cell morphology and fragmented the endoplasmic reticulum which is typically observed in cells undergoing endoplasmic reticulum (ER) stress. Results of this study show that NR4A1 regulates reactive oxygen species (ROS) production and expression of genes such as thioredoxin domain containing 5 (TXNDC5) that maintain stress levels that are permissive for cancer cell growth and survival, and both chemical and RNAi-mediated inactivation of NR4A1 induce ER stress and death pathways in pancreatic cancer cells.
MATERIALS AND METHODS
Cell lines and plasmids
Panc-1 and L3.6pL human pancreatic cancer cell lines were obtained and maintained as previously described (19). The TXNDC5 promoter (−1444/+25) reporter construct (pSESN2-Luc) was purchased from Genecopoeia, Inc. All other reporter constructs have been previously described (19).
Antibodies, reagents, quantitative real-time PCR, western blot analysis, and immunoprecipitation
NR4A1 antibody was purchased from Abcam (Cambridge, MA), and TXNDC5, PGK1 and ATP5A1 antibodies were purchased from GeneTex (Irvine, CA). XBP-1s (spliced XBP-1) and phosphor-PERK (Ser713) antibodies were purchased from BioLengend (San Diego, CA, USA). ATF4, CHOP, GRP78, β-actin, and IDH1 antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX, USA), and ATF6 andibody was purchased from Abgent (Atlanta, GA, USA). All remaining antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). DIM-C-pPhOH was synthesized and purified in this laboratory as previously described (19). Reporter lysis buffer, luciferase reagent, and β-galactosidase (β-gal) reagent were supplied by Promega (Madison, WI, USA). Quantitative real-time PCR and western blot analysis were undertaken as previously described (20). The real-time PCR primers for NR4A1 were purchased from Qiagen (Valencia, CA, USA), and all other sequences of the primers used for real-time PCR were shown in Supplemental Table 1.
Two-dimensional gel electrophoresis and protein pathway analysis
Panc-1 cells were washed three times with ice-cold PBS. Cells were solubilized in a lysis buffer consisting of 5 mM EDTA, 9.5 M urea, 4% (v/v) CHAPS, 65 mM DTT, and protease inhibitors (Complete kit, Roche Diagnostics, Germany). Samples (310 μl) containing 450 μg of proteins were loaded onto IPG strips (17 cm, pH 3-10 nonlinear, pH 5-8, Bio-Rad). The second dimension was performed in 12% SDS-polyacrylamide gels that were then stained with Coomassie Brilliant Blue G-250 and scanned at 300-dpi resolution. Protein spots were analyzed with ImageMaster Platinum™ software (GE Healthcare, Pittsburgh, PA, USA) according to the manufacturer’s procedures. Proteins altered over 1.5-fold were considered to be significantly changed (p > 0.01 by Mann-Whitney test). The differentially expressed proteins were cut and digested, and dried sample were desalted with C18 Zip tips (Millipore, Billerica, MA, USA) and were resuspended in 100 μl of an aqueous solution containing 5% acetic acid and 5% acetonitrile for LC-MS/MS analysis. Each sample solution was loaded onto a 0.1 – 150-mm Magic C18AQ reverse phase column (Michrom Bioresources, Inc., Auburn, CA, USA) in line after a nanotrap column using the Paradigm MS4 HPLC system (Michrom Bioresources, Inc.). Separation of the peptides was performed at 500 nl/min and was coupled to on-line analysis by tandem mass spectrometry using an LTQ ion trap mass spectrometer (ThermoElectron, Waltham, MA, USA) equipped with a nanospray ion source (ThermoElectron). The peptides were detected in positive ion mode using a data-dependent method in which the seven most abundant ions detected in an initial survey scan were selected for MS/MS analysis. The MS/MS spectra of 2D samples were searched with Mascot Daemon (Matrix Science, Boston, MA, USA). The following MASCOT parameter settings were used: the peptide tolerance was 15 ppm and peptide charges and the MS/MS tolerance was 0.6 Da. One missed cleavages by trypsin were allowed, carbamidomethyl (C) was used as a fixed modification and oxidation (M) was used as variable modifications. When multiple proteins were identified in a single spot, the proteins with the highest number of peptides were considered as those corresponding to the spot, and the proteins with lower but significant scores were also recorded in the database.
Transmission electron microscopy
Cells were collected and fixed in 3% glutaraldehyde plus 2% paraformaldehyde in PBS for 2 hr, post-fixed with 1% OsO4 for 2 hr, and stained for 1 hr in 1% aqueous uranyl acetate. The samples were then dehydrated with increasing concentrations of ethanol, infiltrated, and embedded in Spurr’s low viscosity medium. Ultrathin sections were cut using a Leica Ultracut microtome (Leica, Deerfield, IL), counterstained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined under a JEM 1010 transmission electron microscope (JEOL USA Inc., Peabody MA).
Transfection, siRNA oligonucleotides, and reporter gene assay
Cells were transfected with 100 nM of each siRNA duplex for 6 hr using LipofectAMINE 2000 reagent (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s protocol. The sequences of siNR4A1 oligonucleotides used were 5′-CAG UCC AGC CAU GCU CCU C dTdT-3′. As a negative control, a nonspecific scrambled small inhibitory RNA (siScr) oligonucleotide was used (Qiagen) and all other siRNAs were purchased from Santa Cruz Biotechnology (Dallas, Texas). Reporter gene assays were performed as previously described (20).
Measurement of intracellular level of ROS
OxiSelect Intracellular ROS assay kit (Cell Biolabs, Inc., San Diego, CA) was used according to the manufacturer’s instructions. The treated cells were lysed and the amount of intracellular ROS was estimated from dichlorodihydrofluorescein (DCF) production measured by flow cytometry or a fluorometric plate reader (FLUOstar OPTIMA, BMG Labtechnologies, Cary, NC, USA) using excitation and emission wavelengths of 480 nm/530 nm.
DNA fragmentation assay
After incubation with siNR4A1 or DIM-C-pPhOH, cells were subjected to DNA fragmentation assay according to the manufacturer’s instructions (Cell Death Detection ELISAPLUS, Roche). Briefly, cells were lysed, cleared by centrifugation, and transferred into streptavidin-coated plate. Immunoreagent containing biotin-labeled anti-histone antibody and peroxidase conjugated anti-nucleosomal DNA-antibody were then added to each well, and the plate was incubated for 2 hr with gentle shaking. After incubation, 2,2′-azino-bis-[3-ethylbenzthiazoline-6-sulfonic acid] (ABTS) substrate solution was added to each well and the plate was incubated on a plate shaker at 250 rpm for 5 min. The absorbance was measured at 405 nm and 490 nm as test and reference wavelengths, respectively.
Intracellular Ca2+ assay
Intracellular cytosolic Ca2+ was measured using the Fluo-4 NW calcium assay kit (Molecular Probes, Invitrogen, USA). After incubation with siNR4A1 or DIM-C-pPhOH, cells were loaded with Fluo-4 according to the manufacturer’s instructions. The fluorescence of Fluo-4 was measured by a fluorometric plate reader (FLUOstar OPTIMA, BMG Labtechnologies, Cary, NC) using excitation at 485 nm and emission at 520 nm.
Protein pathway analysis
After protein identification, the accession numbers and fold changes of the differentially expressed proteins were tabulated in Microsoft Excel and imported into IPA (Ingenuity System, Mountain View, CA, USA). IPA is a software application that enables to identify the biological mechanisms, pathways and functions matching a particular dataset of proteins. IPA is based on a database obtained by abstracting and interconnecting a large fraction of the biomedical literature according to an algorithm integrating protein functions, cellular localization, small molecules, and disease inter-relationships. The networks are displayed graphically as nodes, representing individual proteins and edges representing the biological relation between nodes. Canonical pathway analysis within IPA utilizes well characterized metabolic and cell signaling pathways, which are generated prior to data input and on which identified proteins are overlaid.
Isolation of RNA and microarray analysis
Total RNA was extracted from Panc-1 cells by using a mirVanaTM miRNA isolation labeling kit (Ambion Inc., Austin, TX, USA). The total RNA was quantified by using a Nanodrop ND-1000 spectrophotometer (NanoDrop Technology, Wilmington, DE, USA). The total RNA samples with adequate RNA quality index (>7) were used for microarray analysis. Sample labeling was performed with an RNA amplification kit according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA, USA). We used 700 ng of total RNA for labeling and hybridization on HumanHT-12 v4 expression beadchip (Illumina Inc., San Diego, CA, USA) according to the manufacturer’s protocols (Illumina). After the bead chips were scanned with a BeadArray Reader (Illumina), the microarray data were normalized using the quantile normalization method in the Linear Models for Microarray Data (LIMMA) package in the R language (http://www.r-project.org). BRB-ArrayTools were primarily used for statistical analysis of gene expression data and the t-test was applied to identify the genes significantly different between two groups when compared. Cluster analysis was performed using the software programs Cluster and Heatmap was generated by Treeview.
DNA-binding assay and chromatin immunoprecipitation (ChIP) assay
GC-rich DNA binding of NR4A1 was measured using a Universal EZ-TFA transcription factor assay Chemiluminescent kit (Upstate Biotechnology, Inc., Lake Placid, NY, USA) according to the manufacturer’s protocol. A biotinylated double-stranded oligouncleotide containing the putative NBRE sequence of TXNDC5 was used as a capture probe and an unlabeled oligonucleotide containing the identical consensus sequence as the capture probe was used as a competitor. A negative control without the capture probe was also used in each assay. The ChIP assay was performed using ChiP-IT Express Magnetic Chromatin Immunoprecipitation kit (Active Motif, Carlsbad, CA, USA) as previously described (20). The TXNDC5 primers which contain the NBRE were 5′-CTA ATT CAG GTG CAA ACC CCA CC −3′ (sense) and 5′-AGC GGC GGT GAA CAC ACG AAG TA −3′ (antisense).
Statistical analysis
Statistical significance of differences between groups were analyzed using Student’s t-test. The results are expressed as means with error bars representing 95% confidence intervals for three experiments for each group unless otherwise indicated, and a P value of less than 0.05 was considered statistically significant. All statistical tests were two-sided.
RESULTS
1. Inactivation of NR4A1 induces ER stress and apoptosis
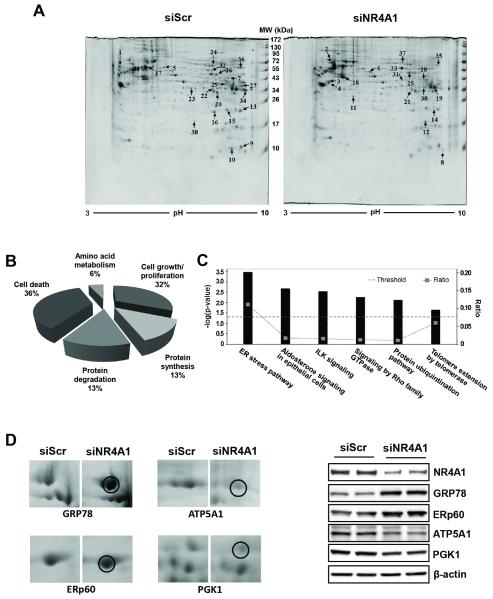
Silencing NR4A1 or inactivation of the receptor by DIM-C-pPhOH inhibited pancreatic cancer cell and tumor growth through decreased expression of some NR4A1-regulated pro-survival genes such as survivin (19). In this study, we have used a proteomic and gene array approach to identify other key genes regulated by this receptor. Silencing of NR4A1 by RNA interference (RNAi, siNR4A1) followed by two-dimensional gel electrophoresis (Fig. 1A) and mass spectrometry analysis of individual spots, identified 38 proteins induced or suppressed by >2-fold (Suppl. Table 2). The functional distribution (Fig. 1B) and canonical pathways (Fig. 1C) resulting from NR4A1 knockdown demonstrate that this receptor plays a key role in regulating ER stress in Panc-1 cells and this represents a hitherto unknown function for this receptor. Figure 1D illustrates that after knockdown of NR4A1 in Panc-1 cells, there is increased intensity of gel spots identified by mass spectrometry as GRP78 and ERp60, two ER chaperone proteins, and decreased expression of the mitochondrial ATP5A1 and cytosolic PGK1 proteins. In a separate experiment, western blot analysis of whole cell lysates from Panc-1 cells transfected with siNR4A1 or siScr (non-specific oligonucleotide) confirmed induction of the ER stress markers (GRP78 and ERp60) and downregulation of ATP5A1 and PGK1 (Fig. 1D).
Figure 1. 2D-PAGE gels showing differentially expressed proteins in Panc-1 cells transfected with siNR4A1 and functional classification of the proteins.
Cells were transfected with either siScr or siNR4A1 for 60 hr, and whole cell lysates (400 μg of protein) were prepared, and the pH gradient in the first dimension was from 3 to 10. The second dimension was a 12% acrylamide SDS-PAGE. Gels were stained by colloidal staining with Coomassie blue G250 (A). Arrows point to the proteins of interest, and the numbers assigned to the spots correspond to the numbers listed in Supplemental Table 2. Functional distribution (B) and canonical pathway (C) of the 38 identified proteins. Assignments were made based on information from the NCBI, the Swiss-Prot/TrEMBL Protein Knowledge Base, and the Ingenuity Pathways Knowledge Base. Enlarged images of protein spots for which image analysis software reported ≥2-fold differences in accumulation in siNR4A1-transfected cells are shown (D, left panel). Panc-1 cells were transfected with either siScr or siNR4A1 for 60 hr, and whole cell lysates were analyzed by western blot analysis (D, right panel).
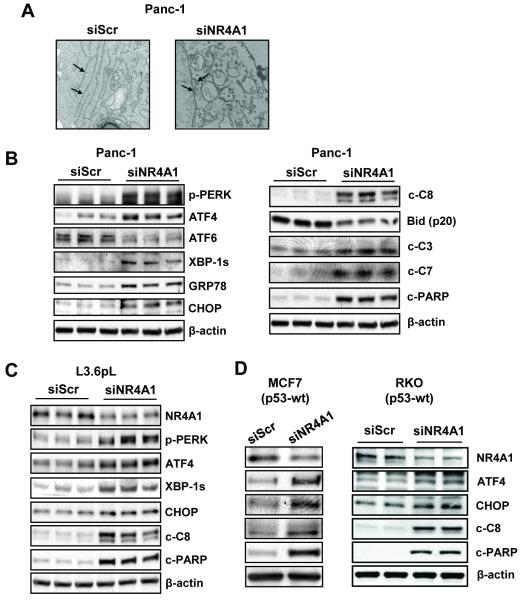
The novel observation that NR4A1 regulates stress in pancreatic cancer cells was further confirmed in Panc-1 cells transfected with siNR4A1 and examined by transmission electron microscopy (Fig. 2A). The results show that typical structural features of the ER are disrupted and fragmented. Western blot analysis of Panc-1 cell lysates after NR4A1 silencing confirmed the induction of several ER stress markers including ATF4, XBP-1s, GRP78 and CHOP (Fig. 2B). Moreover, NR4A1 silencing also resulted in induction of apoptosis as evidenced by induction of cleaved caspases 8, 3 and 7 and PARP and decreased expression of Bid (p20) (Fig. 2B), and these responses are typically observed after induction of ER stress (26-28). NR4A1 silencing in L3.6pL pancreatic cancer cells also induced ER stress (p-PERK, ATF4, XBP-1s and CHOP) and apoptosis (cleaved caspase-8 and PARP) (Fig. 2C), and NR4A1 silencing in MCF-7 breast and RKO colon cancer cells (Fig. 2D) (also MDA-MB-231 and Jurkat cells, data not shown) confirmed that this receptor also regulated ER stress in several cancer cell lines. Moreover, tumor lysates from mice treated with corn oil (control) or DIM-C-PhOH (30 mg/kg/d) (19) also showed enhanced expression of ER stress markers in the latter group (Suppl. Fig. S1A).
Figure 2. Knockdown of NR4A1 induces ER stress and apoptosis in multiple cancer cell lines.
(A) Panc-1 cells were transfected with either siScr or siNR4A1, and TEM analysis was performed at 60 hr after transfection. The structure of the ER (arrows) in cells transfected with siNR4A1 showed morphological disorders. (B, C, and D) Panc-1, L3.6pL, MCF-7/RKO cells, respectively, were transfected with either siScr or siNR4A1 for 72 hr, and whole cell lysates were analyzed by western blot analysis. β-Actin was used as a loading control.
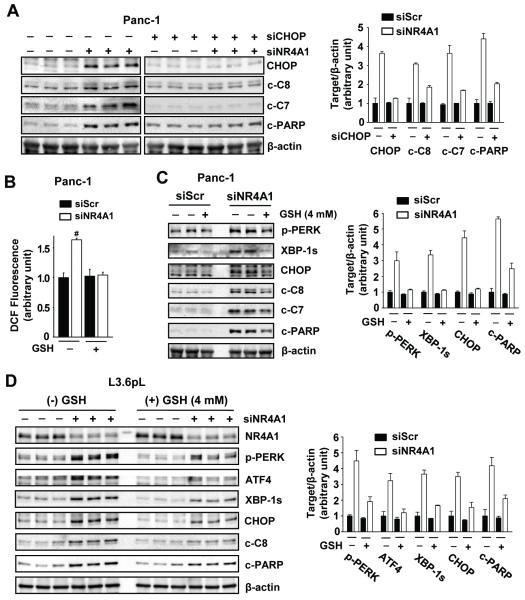
Since CHOP is a key stress-induced pro-apoptotic protein, we further investigated the role of CHOP in mediating the effects of NR4A1 silencing in Panc-1 cells (Fig. 3A). Knockdown of NR4A1 enhanced activation of caspases 8 and 7 (cleaved) and PARP cleavage and, in cells transfected with siNR4A1, the pro-apoptotic responses were attenuated in cells cotransfected with siCHOP. These results confirm a role for CHOP and ER stress in mediating siNR4A1-induced apoptosis. We also observed that both siNR4A1 and DIM-C-pPhOH induce LC3-II, a marker of autophagy and siCHOP attenuated this response (Suppl. Figs. S1B and S1C), indicating that CHOP plays a role in ER stress-induced autophagy as previously reported (29).
Figure 3. Knockdown of NR4A1 induces ER stress-mediated apoptosis by increasing ROS production in pancreatic cancer cells.
(A) Panc-1 cells were transfected with each indicated siRNA for 72 hr, and whole cell lysates were analyzed by western blot analysis. (B-D) Cells were transfected with siScr or siNR4A1 for 6 hr. At 60 hr after transfection, ROS production was measured by the oxidation of non-fluorescent DCFH-DA to fluorescent DCF using a fluorescence plate reader (B). #P<0.001 vs. siScr without reduced glutathione (GSH). At 24 hr after transfection, the cells were treated with GSH for an additional 48 hr and whole cell lysates were analyzed by western blot analysis (C and D). β-Actin was used as a loading control. The multiple lanes represent lysates from different experiments
2. Inactivation of NR4A1 induces ROS
ROS is an important inducer of ER stress, and therefore we examined the effects of NR4A1 silencing on induction of ROS using the cell permeant dichloroflurescein diacetate (DFC-DA) probe and observed that cells transfected with siNR4A1 exhibited significantly induced ROS which was inhibited after cotreatment with glutathione (GSH) (Fig. 3B). Moreover, in Panc-1 cells transfected with siScr or siNR4A1, the induction of ER stress (p-PERK, XBP-1s and CHOP) and apoptosis (cleaved caspases 8 and 7 and PARP) markers after NR4A1 silencing were partially reversed after cotreatment with GSH (Fig. 3C). The role of NR4A1 silencing in inducing ROS-dependent ER stress was also confirmed in L3.6pL pancreatic cancer cells in which transfection with siNR4A1 increased markers of ER stress (p-PERK, XBP-1s, ATF4 and CHOP) and apoptosis (cleaved caspase 8 and PARP) and these responses were attenuated by cotreatment with GSH (Fig. 3D).
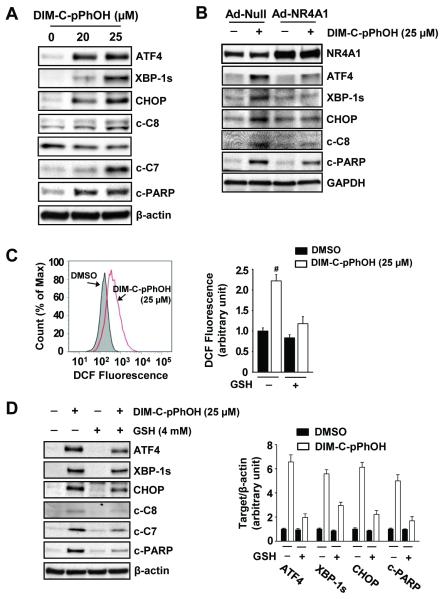
Previous studies have shown that DIM-C-PhOH acts as an NR4A1 antagonist and mimics the effects of NR4A1 silencing (19, 20), and Figure 4A shows DIM-C-pPhOH significantly increased expression of markers of ER stress (ATF4, XBP-1s and CHOP) and apoptosis (decreased Bid and increased cleavage of caspase 8 and 7 and PARP) (Fig. 4A) which was comparable to the effects of siNR4A1 (Fig. 2B). Moreover, using a quantitative DNA fragmentation assay (Suppl. Fig. S2A), we showed that both siNR4A1 and DIM-C-PhOH significantly induced DNA damage, and both treatments also increased ER calcium flux (Suppl. Fig. S2B) associated with ER stress. DIM-C-pPhOH-induced ATF4, XBP-1s, CHOP, cleaved caspase 8 and PARP was attenuated in Panc-1 cells infected with adenovirus overexpressing NR4A1 (Fig. 4B). Moreover, like NR4A1 silencing, inactivation of this receptor by DIM-C-pPhOH also induced ROS which was attenuated after cotreatment with GSH (Fig. 4C), and DIM-C-pPhOH-induced markers of ER stress (CHOP, ATF4, XBP-1s and CHOP) and apoptosis (cleaved caspases 8 and 7 and PARP) were also decreased after cotreatment with GSH (Fig. 4D).
Figure 4. DIM-C-pPhOH, the NR4A1 inactivator, induces ER stress-mediated apoptosis by increasing ROS production in Panc-1 cells.
(A) Cells were treated with either DMSO or DIM-C-pPhOH for 24 hr. (B) Cells were transfected with either Ad-Null or Ad-NR4A1 for 12 hr, and treated with DIM-C-pPhOH (25 μM) for another 24 hr. Whole cell lysates were analyzed by western blot analysis. (NR4A1 protein levels were induced 4-fold by Ad-NR4A1). (C) Cells were treated with DMSO or DIM-C-pPhOH for 18 hr, and ROS production was measured by the oxidation of non-fluorescent DCFH-DA to fluorescent DCF using either flow cytometric analysis (left panel) or a fluorescence plate reader (right panel). (D) Cells were treated with DMSO or DIM-C-pPhOH in the presence or absence of GSH for 24 hr and whole cell lysates were analyzed by western blot analysis. β-Actin was used as a loading control.
3. Inactivation of NR4A1 induces genes involved in ROS metabolism
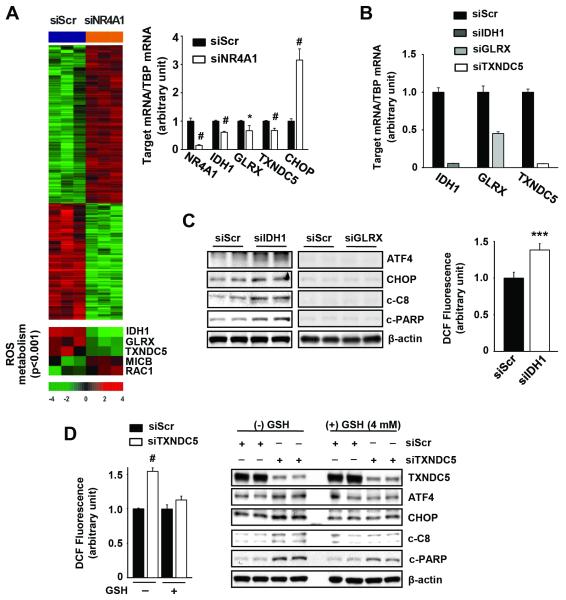
Thus, siNR4A1 or inactivation of NR4A1 by DIM-C-pPhOH induced ROS-dependent ER stress, and identification of NR4A1-regulated genes involved in ROS metabolism was further investigated by RNAi using Illumina beadchip arrays (Fig. 5A and Suppl. Table 3) and several canonical pathways were also affected (Suppl. Fig. S3). Only three potential NR4A1-regulated genes involved in ROS metabolism were decreased in Panc-1 cells, namely, IDH1, GLRX and TXNDC5, and in a separate experiment, we confirmed by real time PCR that these genes are regulated by NR4A1 (Fig. 5A). The potential roles of the NR4A1-regulated IDH1, GLRX and TXNDC5 in mediating ER stress were investigated by RNAi (Fig. 5B). GLRX protein expression was minimal and ER stress genes were not induced by siGLRX; however, knockdown of IDH1 induced CHOP and cleaved caspase 8 and PARP and this was accompanied by induction ROS (Fig. 5C). We also observed that silencing of TXNDC5 by RNAi (siTXNDC5) resulted in the induction of ROS which was attenuated after cotreatment with GSH; transfection of Panc-1 cells with siTXNDC5 also induced expression of ATF4, CHOP, cleaved caspase 8 and PARP and these responses were attenuated in cells transfected with siScr or siTXNDC5 and cotreated with 4 mM GSH (Fig. 5D). Thus, both TXNDC5 and IDH1 are NR4A1-regulated genes that maintain low ROS and ER stress levels in Panc-1 cells, and the former gene was consistently more active in repetitive studies.
Figure 5. TXNDC5 is a novel NR4A1-regulated gene involved in ER stress-mediated apoptosis.
(A) Heat map of genes including ROS metabolism genes regulated by siNR4A1 in Panc-1 cells (left panel). Each cell in the matrix represents the expression level of a gene feature. Red and green reflect relatively high and low expression levels of genes, respectively, as indicated in the scale bar (a log2-transformed scale). (A, right panel and B) Panc-1 cells were transfected with each indicated siRNA for 48 hr and mRNA levels were determined by real-time PCR, as described in the Materials and Methods. TATA-binding protein (TBP) was used as an internal control and mRNA levels are presented as means with s.d. of three experiments. *P<0.05 and #P<0.001 vs. siScr. (C) Panc-1 cells were transfected with each indicated siRNA for 72 hr (left panel) for western blot analysis or 60 hr (right panel) for measurement of ROS. (D) Panc-1 cells were transfected with siScr or siTXNDC5 for 6 hr. At 60 hr after transfection, ROS production was measured by the oxidation of non-fluorescent DCFH-DA to fluorescent DCF using a fluorescence plate reader (left panel). #P<0.001 vs. siScr without GSH. At 24 hr after transfection, the cells were treated with GSH for an additional 48 hr and whole cell lysates were analyzed by western blot analysis (right panel). β-Actin was used as a loading control and the two lanes for each siRNA represent different experiments.
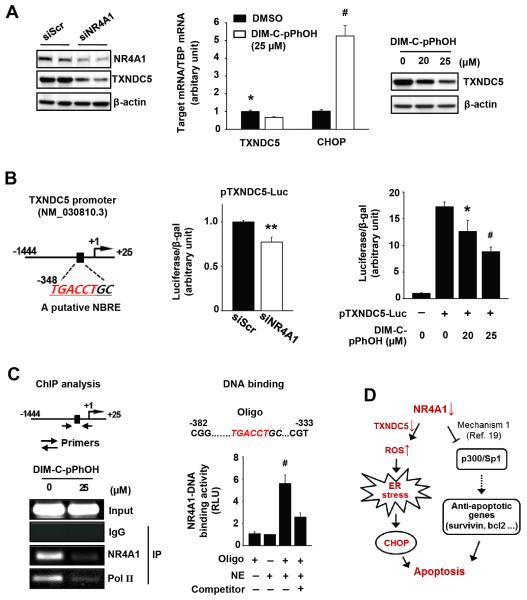
Figure 6A shows that siNR4A1i and DIM-C-pPhOH decreased TXNDC5 expression in Panc-1 cells, and we also observed induction of CHOP mRNA by DIM-C-pPhOH (Fig. 6A). Moreover, DIM-C-pPhOH also decreased expression of TXNDC5 and induced markers of ER stress in the presence of leptomycin B, a nuclear export inhibitor (Suppl Fig. S4A); DIM-C-pPhOH also did not induce nuclear export of NR4A1 (Suppl. Fig. S4B), confirming that these responses were nuclear NR4A1-dependent. The TXNDC5 promoter has a putative NBRE at −348 and, in Panc-1 cells transfected with the pTXNDC5 luciferase reporter gene construct containing a −1444 to +25 TXNDC5 promoter insert, both siNR4A1 and DIM-C-pPhOH decreased luciferase activity (Fig. 6B). Results of a ChIP assay demonstrated that NR4A1 bound the region containing the NBRE and treatment with DIM-C-pPhOH resulted in loss of NR4A1 binding to this site. This was accompanied by decreased binding of pol II (Fig. 6C, left panel) which is consistent with the decreased expression of TXNDC5 after treatment of Panc-1 cells with DIM-C-pPhOH (Fig. 6A). We also investigated NR4A1 binding to the NBRE of the TXNDC5 promoter using a nonradioactive ZZ transcription factor DNA binding assay, and the results (Fig. 6C, right panel) show that NR4A1 bound to the promoter region of TXNDC5 which contains the NBRE. Panc-1 cells express minimal levels of NR4A3 (data not shown), but NR4A2 protein/mRNA are expressed (Suppl. Figs. S5A and S5B). Knockdown of NR4A2 by RNAi did not induce apoptosis (Suppl. Fig. S5A), affect expression of TXNDC5 and CHOP mRNA and protein (Suppl. Figs. S5B and S5C), or affect reporter gene activity in cells transfected with the TXNDC5 promoter (Suppl. Fig. S5D). Thus, NR4A2 does not play a role in the observed stress/ROS responses in Panc-1 cells. Thus, induction of ROS, ER stress and apoptosis by siNR4A1 or inactivation of NR4A1 by DIM-C-pPhOH in Panc-1 cells is primarily due to decreased expression of TXDNC5 and to a lesser extent IDH1, suggesting that NR4A1 antagonists that also decrease TXNDC5 expression represent a novel pathway for activating stress-dependent pancreatic cancer cell death (Fig. 6D).
Figure 6. Regulation of TXNDC5 transcriptional activity by NR4A1.
(A) Panc-1 cell swere transfected with siNR4A1 for 72 hr (left panel) or treated with DIM-C-pPhOH for 24 hr (middle and right panel), and TXNDC5 protein and mRNA levels were determined by western blot analysis and real-time PCR, respectively. (B, left panel) A putative NBRE in the TXNDC5 promoter. (B, middle panel) Cells were cotransfected with each siRNA and pTXVDC5-Luc (−1444/+25), and luciferase activity (relative to β-galactosidase activity) was determined. (B, right panel) Panc-1 cells were transfected with pTXVDC5-Luc (−1444/+25) for 4 hr and treated with DIM-C-pPhOH for another 18 hr. Luciferase activity (relative to β-galactosidase activity) was determined, and the corresponding empty vector was used as a control. (C, left panel) ChIP assay. Panc-1 cells were treated with DIM-C-pPhOH for 6 hr, and the ChIP assay was performed as described in the Materials and Methods. (C, right panel) DNA binding assay. Nuclear extracts of Panc-1 cells were tested for NR4A1-DNA binding activity as described in Materials and Methods. (D) Schematic diagram illustrating induction of apoptosis by inactivation of NR4A1.
DISCUSSION
NR4A1 is a unique nuclear receptor that is overexpressed in cancer cells and tumors, and results of RNAi demonstrate the pro-oncogenic activity of this receptor (12). Some cancer cell lines treated with phorbol esters, cyclic retinoids and other apoptosis-inducing agents induce nuclear export of NR4A1 which forms a pro-apoptotic complex with bcl-2 (21-24). This unusual NR4A1-dependent drug-induced apoptotic pathway does not require the DNA binding domain of NR4A1 and is blocked by nuclear export inhibitors such as leptomycin B (LMB). Studies in this laboratory have investigated the effects of 1,1-bis(3′-indolyl)-1-(p-substituted phenyl)methane (C-DIM) derivatives on NR4A1-mediated-transactivation and DIM-C-pPhOH was identified as an NR4A1 antagonist that inhibited nuclear NR4A1-mediated responses and mimicked the effects of NR4A1 knockdown by RNAi (19, 20). DIM-C-pPhOH and siNR4A1 inhibited pancreatic and lung cancer cell proliferation, and DIM-C-pPhOH also inhibited tumor cell growth. Mechanistic studies showed that NR4A1 regulated expression of pro-survival and growth promoting genes through p300-NR4A1-Sp1 complex interactions with GC-rich gene promoter sequences (e.g. survivin) (19), and NR4A1 also activated mTOR(20) by inhibiting the function of p53 (25). Knockdown of NR4A1 by RNAi or inactivation of the receptor by DIM-C-pPhOH reversed these responses, resulting in inhibition of cell and tumor growth, induction of apoptosis and inhibition of mTOR (19, 20). However, microscopic examination of Panc-1 cells transfected with siNR4A1 or treated with DIM-C-pPhOH showed that NR4A1 was important for regulating stress levels since loss of this receptor resulted in significant loss and fragmentation of ER structure (Fig. 2A), indicative of ER stress.
We initially used a proteomics approach to identify stress-related gene products that are modulated after silencing NR4A1 (Figs. 1B and 1C), and both siNR4A1 and DIM-C-pPhOH induced several prototypical ER stress-related genes. Analysis of the induced and repressed proteins by mass spectrometry demonstrated that NR4A1 silencing in Panc-1 cells resulted in induction of the stress proteins GRP78 and ERp60 and several other classes of proteins including the mitochondrial and cytosolic ATP5A and PGK1 proteins, respectively (Fig. 1D). NR4A1 silencing or treatment with DIM-C-pPhOH induced several other ER stress-related genes including ATF4, XBP-1s and CHOP as well as several markers of apoptosis that are associated with activation of ER stress (26-28) (Figs. 2B and 4A). Moreover, many of the same markers of ER stress and apoptosis were also induced in L3.6pL pancreatic, MCF-7 breast, and RKO colon cancer cells transfected with siNR4A1 (Figs. 2C and 2D), suggesting that NR4A1 serves as an important regulator (inhibitor) of ER stress in cancer cells.
Based on results of CHOP silencing (Fig. 3A), it was also evident that this gene product plays a critical role in siNR4A1 and DIM-C-pPhOH-induced ER stress and subsequent induction of cleaved caspases and PARP. Studies with several anticancer agents have demonstrated a direct linkage between drug-induced ROS and ROS-dependent induction of CHOP and downstream responses including apoptosis (30-32). For example, the polyherbal formulation zyflamend induces ROS and ER stress (including CHOP) in colon and pancreatic cancer cells (33) and, using HCT116 colon cancer cells as a model, it was shown that cotreatment with antioxidants reverse zyflamend-induced CHOP and apoptosis. The observations are consistent with our data showing that both siNR4A1 and DIM-C-pPhOH induced ROS in Panc-1 cells (Figs. 3B and 4C) and the antioxidant GSH significantly inhibited induction of CHOP and other markers of ER stress and apoptosis (Figs. 3D and 4D). These results suggest that NR4A1 attenuates stress levels in pancreatic cancer cells by regulating ROS metabolism and production. GSH only partially protected against NR4A1-dependent stress induced by DIM-C-pPhOH since C-DIM compounds induce stress via direct perturbation of mitochondria (34, 35).
Results of gene array studies showed that only three ROS metabolism genes were suppressed in Panc-1 cells transfected with siNR4A1 and these include IDH1, GLRX and TXNDC5 (Fig. 5A). IDH1 plays a critical role in the citric acid cycle and the generation of reducing equivalents, and the relative expression of wild-type and mutant IDH1 have both functional and prognostic significance in cancer (36, 37). Glutaredoxin (GLRX) is a thioredoxin that contributes to the cellular redox status and some GLRX genes are overexpressed in human lung and breast tumors (38, 39). TXNDC5 is a member of the thioredoxin family of ER proteins that contain a disulfide isomerase-like (PDI) domain (40-42). Transfection of Panc-1 cells with siNR4A1 decreases expression of all three genes; however, GLRX protein levels are low and knockdown of GLRX by RNAi does not activate ER stress or apoptosis, whereas silencing IDH1 and TXNDC5 induces ER stress and apoptosis (Figs. 6A and 6B). Thus, NR4A1 regulation of IDH1 and TXNDC5 serves to maintain homeostatic levels of ROS in Panc-1 cells, and silencing of IDH1 or TXNDC5 by RNAi induces markers of ER stress and apoptosis (Figs. 5C and 5D) and mimics the effects of siNR4A1 or treatment of these cells with DIM-C-pPhOH (Figs. 4A and 4D). Induction of ER stress and apoptosis markers were more pronounced in Panc-1 cells transfected with siTXNDC5 vs. siIDH1 (Fig. 5C and 5D), and results of ChIP assays and transfection with the TXNDC5 promoter construct confirmed that TXNDC5 is directly regulated by NR4A1. The parallel results observed after knockdown of NR4A1 or TXNDC5 or after treatment with the NR4A1 antagonist (DIM-C-pPhOH) suggest that regulation of TXNDC5 expression by NR4A1 is an important pro-oncogenic function of this receptor which serves to maintain sufficiently low stress levels that facilitate cancer cell growth and survival.
TXNDC5 expression is upregulated in non-small cell lung cancer, colorectal adenomas and tumors of the cervix, uterus and stomach (compared to non-tumor tissue) (41, 43-45), and overexpression of TXNDC5 in gastric cancer cells increased proliferation and migration and decrease apoptosis (46). Although data mining of pancreatic cancer patient arrays did not detect higher TXNDC5 mRNA expression in tumor vs. non-tumor tissue (data not shown), results of this study demonstrate that TXNDC5 is an important NR4A1-regulated gene that significantly contributes to the pancreatic cancer cell phenotype. Thus, NR4A1 inactivates p53 to activate mTOR signaling (20), regulates expression of pro-survival genes with GC-rich promoters (e.g. survivin and bcl-2) (19), and regulates expression of genes such as TXNDC5 to maintain low levels of ROS-induced stress in cancer cells. These NR4A1-regulated genes/pathways highlight the pro-oncogenic activity of this receptor and the importance of this gene as a target for DIM-C-pPhOH and related compounds that are currently being developed as NR4A1 antagonists for cancer chemotherapy.
Supplementary Material
IMPLICATIONS.
The NR4A1 receptor is pro-oncogenic, regulates ROS/ER stress pathways, and inactivation of the receptor represents a novel pathway for inducing cell death in pancreatic cancer.
Acknowledgements
The financial support of the National Institutes of Health (R01-CA124998) and Texas AgriLife Research is greatly appreciated.
Funding: Funded by NIH (R01-CA124998) and Texas AgriLife.
Footnotes
Disclosure of Conflicts of Interest: None
REFERENCES
- 1.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–99. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearen MA, Muscat GE. Minireview: Nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol. 2010;24:1891–903. doi: 10.1210/me.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milbrandt J. Nerve growth factor induces a gene homologous to the glucocorticoid receptor gene. Neuron. 1988;1:183–8. doi: 10.1016/0896-6273(88)90138-9. [DOI] [PubMed] [Google Scholar]
- 5.Murphy EP, Dobson AD, Keller C, Conneely OM. Differential regulation of transcription by the NURR1/NUR77 subfamily of nuclear transcription factors. Gene Expr. 1996;5:169–79. [PMC free article] [PubMed] [Google Scholar]
- 6.Paulsen RF, Granas K, Johnsen H, Rolseth V, Sterri S. Three related brain nuclear receptors, NGFI-B, Nurr1, and NOR-1, as transcriptional activators. J Mol Neurosci. 1995;6:249–55. doi: 10.1007/BF02736784. [DOI] [PubMed] [Google Scholar]
- 7.Saucedo-Cardenas O, Kardon R, Ediger TR, Lydon JP, Conneely OM. Cloning and structural organization of the gene encoding the murine nuclear receptor transcription factor, NURR1. Gene. 1997;187:135–9. doi: 10.1016/s0378-1119(96)00736-6. [DOI] [PubMed] [Google Scholar]
- 8.Maira M, Martens C, Philips A, Drouin J. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol Cell Biol. 1999;19:7549–57. doi: 10.1128/mcb.19.11.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perlmann T, Jansson L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev. 1995;9:769–82. doi: 10.1101/gad.9.7.769. [DOI] [PubMed] [Google Scholar]
- 10.Zetterstrom RH, Solomin L, Mitsiadis T, Olson L, Perlmann T. Retinoid X receptor heterodimerization and developmental expression distinguish the orphan nuclear receptors NGFI-B, Nurr1, and Nor1. Mol Endocrinol. 1996;10:1656–66. doi: 10.1210/mend.10.12.8961274. [DOI] [PubMed] [Google Scholar]
- 11.Lee SO, Li X, Khan S, Safe S. Targeting NR4A1 (TR3) in cancer cells and tumors. Expert Opin Ther Targets. 2011;15:195–206. doi: 10.1517/14728222.2011.547481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Safe S, Kim K, Li X, Lee SO. NR4A orphan receptors and cancer. Nucl Recept Signal. 2011;9:e002. [Google Scholar]
- 13.Wu H, Lin Y, Li W, Sun Z, Gao W, Zhang H, et al. Regulation of Nur77 expression by beta-catenin and its mitogenic effect in colon cancer cells. FASEB J. 2011;25:192–205. doi: 10.1096/fj.10-166462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke N, Claassen G, Yu DH, Albers A, Fan W, Tan P, et al. Nuclear hormone receptor NR4A2 is involved in cell transformation and apoptosis. Cancer Res. 2004;64:8208–12. doi: 10.1158/0008-5472.CAN-04-2134. [DOI] [PubMed] [Google Scholar]
- 15.Zeng JZ, Sun DF, Wang L, Cao X, Qi JB, Yang T, et al. Hypericum sampsonii induces apoptosis and nuclear export of retinoid X receptor-alpha. Carcinogenesis. 2006;27:1991–2000. doi: 10.1093/carcin/bgl046. [DOI] [PubMed] [Google Scholar]
- 16.Bras A, Albar JP, Leonardo E, de Buitrago GG, Martinez AC. Ceramide-induced cell death is independent of the Fas/Fas ligand pathway and is prevented by Nur77 overexpression in A20 B cells. Cell Death Differ. 2000;7:262–71. doi: 10.1038/sj.cdd.4400653. [DOI] [PubMed] [Google Scholar]
- 17.Kolluri SK, Bruey-Sedano N, Cao X, Lin B, Lin F, Han YH, et al. Mitogenic effect of orphan receptor TR3 and its regulation by MEKK1 in lung cancer cells. Mol Cell Biol. 2003;23:8651–67. doi: 10.1128/MCB.23.23.8651-8667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uemura H, Chang C. Antisense TR3 orphan receptor can increase prostate cancer cell viability with etoposide treatment. Endocrinology. 1998;139:2329–34. doi: 10.1210/endo.139.5.5969. [DOI] [PubMed] [Google Scholar]
- 19.Lee SO, Abdelrahim M, Yoon K, Chintharlapalli S, Papineni S, Kim K, et al. Inactivation of the orphan nuclear receptor TR3/Nur77 inhibits pancreatic cancer cell and tumor growth. Cancer Res. 2010;70:6824–36. doi: 10.1158/0008-5472.CAN-10-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SO, Andey T, Jin UH, Kim K, Singh M, Safe S. The nuclear receptor TR3 regulates mTORC1 signaling in lung cancer cells expressing wild-type p53. Oncogene. 2012;31:3265–76. doi: 10.1038/onc.2011.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang XK. Targeting Nur77 translocation. Expert Opin Ther Targets. 2007;11:69–79. doi: 10.1517/14728222.11.1.69. [DOI] [PubMed] [Google Scholar]
- 22.Kolluri SK, Zhu X, Zhou X, Lin B, Chen Y, Sun K, et al. A short Nur77-derived peptide converts Bcl-2 from a protector to a killer. Cancer Cell. 2008;14:285–98. doi: 10.1016/j.ccr.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferlini C, Cicchillitti L, Raspaglio G, Bartollino S, Cimitan S, Bertucci C, et al. Paclitaxel directly binds to Bcl-2 and functionally mimics activity of Nur77. Cancer Res. 2009;69:6906–14. doi: 10.1158/0008-5472.CAN-09-0540. [DOI] [PubMed] [Google Scholar]
- 24.Zhan Y, Du X, Chen H, Liu J, Zhao B, Huang D, et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat Chem Biol. 2008;4:548–56. doi: 10.1038/nchembio.106. [DOI] [PubMed] [Google Scholar]
- 25.Zhao BX, Chen HZ, Lei NZ, Li GD, Zhao WX, Zhan YY, et al. p53 mediates the negative regulation of MDM2 by orphan receptor TR3. EMBO J. 2006;25:5703–15. doi: 10.1038/sj.emboj.7601435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breckenridge DG, Germain M, Mathai JP, Nguyen M, Shore GC. Regulation of apoptosis by endoplasmic reticulum pathways. Oncogene. 2003;22:8608–18. doi: 10.1038/sj.onc.1207108. [DOI] [PubMed] [Google Scholar]
- 27.Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004;11:372–80. doi: 10.1038/sj.cdd.4401378. [DOI] [PubMed] [Google Scholar]
- 28.Madeo F, Kroemer G. Intricate links between ER stress and apoptosis. Mol Cell. 2009;33:669–70. doi: 10.1016/j.molcel.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008;15:1460–71. doi: 10.1038/cdd.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prasad S, Yadav VR, Kannappan R, Aggarwal BB. Ursolic acid, a pentacyclin triterpene, potentiates TRAIL-induced apoptosis through p53-independent up-regulation of death receptors: evidence for the role of reactive oxygen species and JNK. J Biol Chem. 2011;286:5546–57. doi: 10.1074/jbc.M110.183699. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Prasad S, Yadav VR, Ravindran J, Aggarwal BB. ROS and CHOP are critical for dibenzylideneacetone to sensitize tumor cells to TRAIL through induction of death receptors and downregulation of cell survival proteins. Cancer Res. 2011;71:538–49. doi: 10.1158/0008-5472.CAN-10-3121. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Sheveleva EV, Landowski TH, Samulitis BK, Bartholomeusz G, Powis G, Dorr RT. Imexon induces an oxidative endoplasmic reticulum stress response in pancreatic cancer cells. Mol Cancer Res. 2012;10:392–400. doi: 10.1158/1541-7786.MCR-11-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JH, Park B, Gupta SC, Kannappan R, Sung B, Aggarwal BB. Zyflamend sensitizes tumor cells to TRAIL-induced apoptosis through up-regulation of death receptors and down-regulation of survival proteins: role of ROS-dependent CCAAT/enhancer-binding protein-homologous protein pathway. Antioxid Redox Signal. 2012;16:413–27. doi: 10.1089/ars.2011.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong J, Samudio I, Chintharlapalli S, Safe S. 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes decrease mitochondrial membrane potential and induce apoptosis in endometrial and other cancer cell lines. Mol Carcinog. 2008;47:492–507. doi: 10.1002/mc.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei P, Abdelrahim M, Cho SD, Liu X, Safe S. Structure-dependent activation of endoplasmic reticulum stress-mediated apoptosis in pancreatic cancer by 1,1-bis(3′-indoly)-1-(p-substituted phenyl)methanes. Mol Cancer Ther. 2008;7:3363–72. doi: 10.1158/1535-7163.MCT-08-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102:932–41. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin G, Reitman ZJ, Duncan CG, Spasojevic I, Gooden DM, Rasheed BA, et al. Disruption of wild-type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res. 2013;73:496–501. doi: 10.1158/0008-5472.CAN-12-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cha MK, Kim IH. Preferential overexpression of glutaredoxin3 in human colon and lung carcinoma. Cancer Epidemiol. 2009;33:281–7. doi: 10.1016/j.canep.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Song JJ, Rhee JG, Suntharalingam M, Walsh SA, Spitz DR, Lee YJ. Role of glutaredoxin in metabolic oxidative stress. Glutaredoxin as a sensor of oxidative stress mediated by H2O2. J Biol Chem. 2002;277:46566–75. doi: 10.1074/jbc.M206826200. [DOI] [PubMed] [Google Scholar]
- 40.Knoblach B, Keller BO, Groenendyk J, Aldred S, Zheng J, Lemire BD, et al. ERp19 and ERp46, new members of the thioredoxin family of endoplasmic reticulum proteins. Mol Cell Proteomics. 2003;2:1104–19. doi: 10.1074/mcp.M300053-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan DC, Huminiecki L, Moore JW, Boyle JJ, Poulsom R, Creamer D, et al. EndoPDI, a novel protein-disulfide isomerase-like protein that is preferentially expressed in endothelial cells acts as a stress survival factor. J Biol Chem. 2003;278:47079–88. doi: 10.1074/jbc.M308124200. [DOI] [PubMed] [Google Scholar]
- 42.Wrammert J, Kallberg E, Leanderson T. Identification of a novel thioredoxin-related protein, PC-TRP, which is preferentially expressed in plasma cells. Eur J Immunol. 2004;34:137–46. doi: 10.1002/eji.200324225. [DOI] [PubMed] [Google Scholar]
- 43.Vincent EE, Elder DJ, Phillips L, Heesom KJ, Pawade J, Luckett M, et al. Overexpression of the TXNDC5 protein in non-small cell lung carcinoma. Anticancer Res. 2011;31:1577–82. [PubMed] [Google Scholar]
- 44.Nissom PM, Lo SL, Lo JC, Ong PF, Lim JW, Ou K, et al. Hcc-2, a novel mammalian ER thioredoxin that is differentially expressed in hepatocellular carcinoma. FEBS Lett. 2006;580:2216–26. doi: 10.1016/j.febslet.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Ma Y, Lu B, Xu E, Huang Q, Lai M. Differential expression of mimecan and thioredoxin domain-containing protein 5 in colorectal adenoma and cancer: a proteomic study. Exp Biol Med (Maywood) 2007;232:1152–9. doi: 10.3181/0701-RM-8. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Hou Y, Li N, Wu K, Zhai J. The influence of TXNDC5 gene on gastric cancer cell. J Cancer Res Clin Oncol. 2010;136:1497–505. doi: 10.1007/s00432-010-0807-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.