Abstract
Interleukin-22 (IL-22) is a recently described IL-10 family cytokine that is produced by T-helper (Th)-17 cells, γδ T cells, NKT cells and newly described innate lymphoid cells (ILCs). Knowledge of IL-22 biology has rapidly evolved since its discovery in 2000, and a role for IL-22 has been identified in numerous tissues including the intestines, lung, liver, kidney, thymus, pancreas and skin. IL-22 primarily targets non-hematopoietic epithelial and stromal cells where it can promote proliferation and play a role in tissue regeneration. In addition, IL-22 regulates host defense at barrier surfaces. However, IL-22 has also been linked to several conditions involving inflammatory tissue pathology. In this review, we will assess the current understanding of this cytokine, including its physiologic and pathologic effects on epithelial cell function.
Keywords: Interleukin-22, epithelial cells, tissue regeneration
INTRODUCTION
Interleukin-22 (IL-22), which was originally named IL-10-related T cell-derived inducible factor (IL-TIF), has generated considerable interest in recent years making it one of the best-studied members of the IL-10 family of cytokines, which also includes IL-10, IL-19, IL-20, IL-24 and IL-26, as well as the lambda-interferons IL-28A, IL-28B and IL-29. Initially described as a TH17 cytokine, as its production has been observed within the same cells that produce IL-17, subsequent evidence has indicated it can be generated independently of IL-17. Unlike most cytokines, which target hematopoietic cells, the main impact of IL-22 is on non-hematopoietic epithelial cells and fibroblasts in tissues as diverse as lung, liver, kidney, thymus, pancreas, breast, gut, skin and the synovium. Although it can have a profound effect on the regeneration of epithelial tissues following injury, largely by promoting survival by inducing proliferation and inhibiting apoptosis of epithelial cells, this same function has also been implicated in pathological states such as malignancy and psoriasis. In addition, a role for IL-22 has also been described in host defense within barrier tissues such as the intestine, oral mucosa, skin and lung. Given this widespread action in regeneration, host defense and pathology, IL-22 is an attractive target for clinical development. In this review, we detail the current state of knowledge about the biology of IL-22 in animals and humans, its sources and targets, and its role in tissue pathology and regeneration.
2). STRUCTURE, GENE AND SIGNALING
Gene
The human IL22 gene is located at chromosome 12q15 in the vicinity of the genes encoding IFN-γ and IL-26 (1). The 537bp open reading frame of the IL22 gene encodes a 179 amino acid protein that shares 79% homology between mouse and human (2). The active, secreted form of the cytokine is a 146 amino acid protein (3, 4). Although IL-22 is active as a monomer, and crystal structures have been solved for this form, dimers as well as tetramers of IL-22 have been observed in high concentrations in solution (4, 5). Although early studies found constitutive expression of IL-22 only in the thymus and brain (6), subsequent studies have found induced expression in other tissues including the gut, liver, lung, skin, pancreas and spleen (7).
Structure
The 3D crystal structure of human IL-22 has been solved in both E. coli and D. melanogaster, which revealed virtually identical structures. The IL-22 monomer is composed of six α-helices (labeled A–F) and a small N-terminal helix, forming a compact six-helix bundle cytokine (4, 8). Structural analysis has revealed three potential glycosylation sites located in helices A and C as well as in the AB loop (4), and mass spectroscopy indicated that hexasaccharides (two N-acetyl glucosamines, three mannoses and one fucose) are attached to the IL-22 protein expressed in D. melanogaster (9). Comparison with IL-10 (for which it shares 22% homology in mouse and 25% in human) indicated a putative binding site comprised of helix A, loop AB and helix F (10).
IL-22 receptor
The IL-22 receptor (IL-22R) is a Type 2 cytokine receptor and member of the IL-10 family of receptors along with the receptors for IL-10, IL-19, IL-20, IL-24, IL-26, IL-28 and IL-29 (Figure 1). It is composed of two heterodimeric subunits, IL-22R1 and IL-10R2 (2, 3, 11–13). The human gene encoding IL-22R1 is located at chromosome 1p36.11, whereas the gene encoding IL-10R2 is located on chromosome 21q22.11 (3). Binding studies have revealed that IL-22 has a high affinity for IL-22R1 (KD=20nM), but no affinity for IL-10R2, however, there is a strong binding affinity of the IL-10R2 subunit for the IL-22-IL-22R1 complex (KD=7–45µM) (11, 14–18). These data suggest that initial binding of IL-22 to the IL-22R1 subunit enables secondary binding of the IL-10R2 subunit, thereby enabling downstream signaling (19). This two-stage cytokine receptor binding and signaling has also been reported for the IL-10 system (20). Binding of IL-22 to IL-22R occurs in a 1:1 complex (14, 17) and recent work has shown that phosphorylation by GSK3β of IL-22R1 at the serine residue at positions 410 and 414 stabilizes IL-22R and prevents its degradation by the ubiquitin proteasome (21).
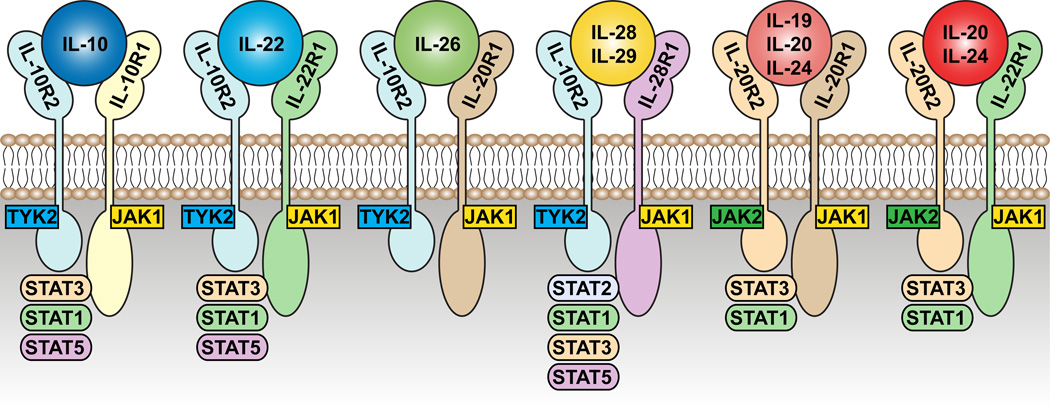
Figure 1. Receptors and JAK-STAT molecules of the IL-10 family of cytokines.
IL-22 is a member of the IL-10 family of cytokines, all of which share common features in their receptors. IL-22R is composed of two subunits: the common IL10R2 subunit, which is shared with the receptors for IL10, IL-26, IL-28 and IL-29; and the IL-22R1 subunit, which itself can also pair an IL-20R2 subunit forming the receptor for IL-20 and IL-24. IL-20 and IL-24 (in addition to IL-19) can also signal through another receptor composed of IL-20R1 and IL-20R2. Each of these receptors signals through components of the JAK-STAT pathway, although there is evidence that IL-22 can also signal through p38 and MAP kinase pathways.
Signaling
Although STAT-3 phosphorylation appears to be the primary mediator of IL-22 signaling, phosphorylation of STAT-1 and STAT-5 has also been observed (22, 23). Phosphorylation of STAT-3 upon ligation of the IL-22-IL-22R1-IL-10R2 complex occurs at the tyrosine residue at position 705, however, it has also been found to phosphorylate a serine residue at position 727, potentially enhancing transactivation (22). This enables recruitment of the Jak signaling molecules Jak1 and Tyk2 to the receptor complex (12, 24). In addition to STAT signaling, IL-22 binding has also been found to activate the MAPK and p38 pathways (25–28).
3). CELLULAR SOURCES OF IL-22
Primarily cells of the lymphoid lineage produce IL-22; encompassing cells of both the innate and adaptive immune systems, including αβ T cells, γδ T cells, NKT cells and innate lymphoid cells (ILCs) (Figure 2). However, some studies have also described some myeloid and non-hematopoietic cells as capable of producing IL-22 in various tissues and states, including macrophages, neutrophils and fibroblasts.
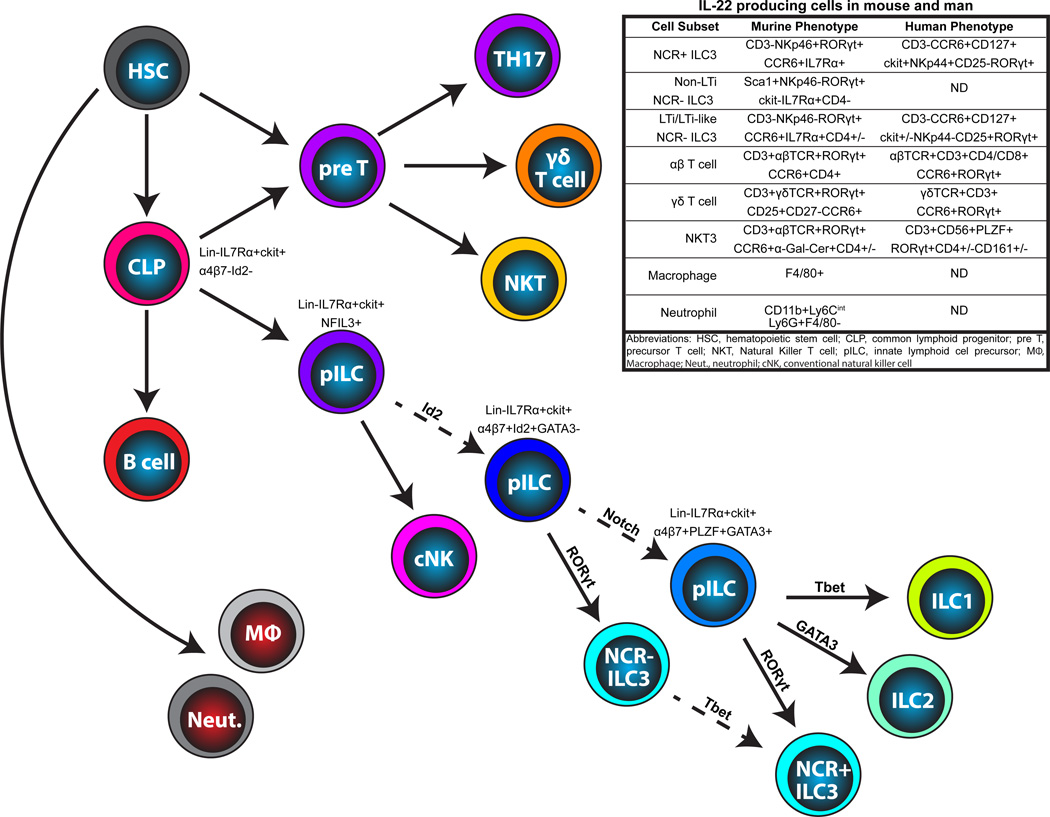
Figure 2. Cellular sources of IL-22 and their lineage relationships.
All IL-22 producing cells derive from a hematopoietic stem cell located in the bone marrow (BM). TH17 cells, γδ T cells and NKT cells develop in the thymus, branching off from conventional T cell development at different stages: NKT cells at the DN3 stage of thymocyte development; γδ T cells upon TCR rearrangement; and TH17 cells at the naïve T cell stage outside the thymus. All ILCs derive from a common ckit+IL7Rα+ common lymphoid progenitor (CLP). Several recent papers have described ILC precursors (pILC) with varying capacity for differentiation. An NFIL3+ pILC has recently been described with the capacity to differentiate into all ILC subsets as well as conventional NK cells. This precursor gives rise to an Id2+ cell giving rise to all ILC populations except for NK cells. A PLZF+ pILC has also been described, differentiating into ILC1, ILC2 and NCR+ ILC3, but not into conventional NK cells or into NCR− ILC3. Evidence also suggests that NCR− group 3 ILC can differentiate into NCR+ ILC3. Solid lines represent known, and dashed lines represent putative, progenitor-progeny relationships.
αβ T cells
IL-22 was first cloned in conventional T cells activated with IL-9 (6, 29). Human production of IL-22 by T cells has been identified in TH1, TH17 and TH22 cells; where it is estimated that approximately 33% of IL-22-producing CD4+ T cells in peripheral blood are TH1 cells (co-expressing IFNγ), 50% produce IL-22 alone (TH22), and 15% are TH17 cells also producing IL-17 (30). In mouse T cells, production of IL-22 has been largely attributed to TH17 cells (31–34). Despite their divergent functions, both TH17 cells and Tregs require TGFβ for their formation (35). Additional signals from IL-6 drive T cell polarization towards the TH17 lineage (36, 37), promoting expression of RORγt and IL-23R (35, 38, 39); although the latter could be indirectly induced by RORγt (40). RORγt is crucial for the formation of TH17 cells (41) and IL-23 signaling is critical for the production of IL-22 by TH17 cells (34, 42–45). TGFβ concentration regulates the balance between Treg and TH17 cells; at low concentrations induction of TH17 cells occurs via upregulation of RORγt and IL-23R, while at higher concentrations, TGFβ represses IL-23R and induces FoxP3 (46). Similarly, high concentration of TGFβ can inhibit the production of IL-22 (47, 48), although this can be overcome by IL-23 (46). In addition to CD4+ T cells, a subset of human CD8+ αβ T cells has also been identified that expresses both IL-22 and IL-17 (49–51), as well as a subset that solely expresses IL-22, partly under the control of IL-21 (52) and particularly in the setting of inflammation (53).
γδ T cells
In addition to conventional αβ T cells, significant IL-22 production has been described in γδ T cells; which like other IL-22 producing cells, express RORγt and are stimulated to produce IL-22 in response to IL-23 (54–57). However, although IL-23 signaling appears to be important for their IL-22 production, γδ T cells can also respond to innate ligands through Toll-like receptors (TLRs) (54, 58). γδ T cells diverge at the DN3 (CD25+CD44−) stage of thymocyte development (59), however, unlike αβ T cells antigen recognition in the thymus is not required for their development. Differential antigen recognition in the thymus during development determines the subset fate of γδ T cell: IFNγ-producing or IL-22 and IL-17-producing (60). These subsets of γδ T cells can be differentiated by their expression of CD25, CD27 and CCR6 (61–63)
Innate lymphoid cells (ILCs)
ILCs are a newly described heterogeneous population of immune cells possessing lymphoid ontogeny, but lacking the rearranged T cell and B cell receptors of lymphoid cells belonging to the adaptive immune system. They have no demonstrated cytotoxic capacity, and they appear to function primarily by producing cytokines. ILCs can be defined by their differential expression of transcription factors (including Tbet, GATA3 or RORγt) and cytokines (such as IFNγ, IL-13, or IL-22) (64), largely mirroring CD4 helper T cell subsets. A working group has recently created a nomenclature guideline to define ILC subsets (65). Group 3 ILCs (ILC3), which express RORγt and are potent producers of IL-22 (65), are themselves a heterogenous population that can be broadly stratified based on their expression of Natural Killer cell cytotoxicity receptors (NCR) such as Nkp44 in humans (66) and Nkp46 in mice (67–69). IL-23 is the most potent inducer of IL-22 production by both NCR+ and NCR− ILC3 cells (66, 68).
One of the first cellular sources of IL-22 to be characterized were a subset of NK cells (29) that were designated NK22 cells (70, 71). Further analysis revealed that these cells were distinct from the NK cell lineage (72, 73) and, due to their expression of RORγt and IL-22, were designated as a subset of ILC3 in human and mouse tissues (65). NCR+ ILC3, as they are now referred, express typical NK cell markers such as CD56 (in human) and NKG2D (in mouse), but also express NKp44 in human and NKp46 in mice and do not express cytokines typical of NK cell function such as IFNγ (66–69, 71, 74). NCR+ ILC3 are dependent on IL-15 (like conventional NK cells) and RORγt for their development (66–69).
Comprising approximately 15–20% of the ILC3 population after birth (75, 76), lymphoid tissue-inducer (LTi) cells were the first of the ILC family to be described. Identified in fetal development due to their unusual CD3−CD4+ phenotype and ability to colonize lymphoid tissue anlagen (77), LTi cells were studied extensively for their important role in the formation of secondary lymphoid tissues such as lymph nodes and Peyer’s patches (78–81), the generation of tertiary lymphoid structures (82–86), as well as the maturation of thymic epithelial cells (87); although these functions are largely attributed to their production of lymphotoxin (LT) α1β2 (83). LTi cells can be phenotypically defined by expression of IL-7Rα and RORγt, lack of expression of CD3 and NKp46 (76), and variable expression of CD4 (approximately half of LTi cells) (88, 89). More recently LTi-like cells were described in the adult mouse and human as potent producers of IL-22 (76, 90) and IL-17 (91). There remains considerable uncertainty in the distinction between LTi cells and other group 3 ILC that do not express NCR, with limited understanding of the identity and function of non-LTi NCR− ILC3 cells (65, 92). The most prominent description of which was reported by the Powrie group, who identified a subpopulation of Sca1+RORγt+IL-23R+IL-7Rα−CD4−ckit− NCR− ILC3 that co-expressed IL-17, IL-22 and, surprisingly, IFNγ; causing considerable pathology in experimental models of colitis (93). For the purposes of this review, unless specifically describing the functional capacity of lymphoid tissue induction, we will refer to all NCR− group 3 ILC, including LTi and LTi-like cells, as NCR− ILC3.
Although in one study no lineage relationship between NCR− and NCR+ RORγt+ ILC3 was found (76), there is increasing evidence that LTi-like NCR− ILC3 cells may give rise to NCR+ ILC3 (74, 91). This was confirmed by fate-mapping studies performed by Diefenbach and colleagues, providing evidence to suggest NCR− ILC3 stably upregulate their expression of NKp46 (72). Interestingly, the Diefenbach study went on to show that these newly converted NCR+ ILC3 subsequently downregulate expression of RORγt, which is associated with a shift in cytokine production towards IFNγ (72). This was consistent with the findings from the Colonna group identifying an NCR+ ILC3 that downregulates expression of IL-22 in favor of upregulation of IFNγ, under the control of IL-2 (70). This latter differentiation step appears to be under the control of the transcription factor T-bet (94, 95), which induces NKp46 expression and IFNγ production by ILCs (96); and is prevented by stimulation of AhR (97).
All ILC, including NCR+ and NCR− ILC3 are thought to derive from common lymphoid progenitors (CLP) (98, 99). Group 1, 2 and 3 ILCs appear to arise downstream of an Id2+ precursor (100, 101), although this precursor does not appear to give rise to NK cells (102). B cell development appears to be the default pathway for CLPs (103) and EBF, a transcription factor critical for B cell development (104), binds and represses Id2 expression, thereby inducing B-lineage over an ILC-lineage fate on CLPs (105). However, in contrast, Id2 suppresses the effects of E proteins such as E2A (106), another factor critical for B cell development (107). It is unclear what diverts CLPs from their B-lineage bias, however Notch signals may play an important role at this early stage of programing, but then needs to be shut off for efficient RORγt+ ILC3 generation (98, 108). Recent work has revealed additional insight into the ILC developmental hierarchy, with identification of an Id2+α4β7+ precursor of all ILC (but not conventional NK cell) (102); and a PLZF-expressing precursor capable of generating ILC1 and ILC2, and NCR+ but not NCR− ILC3 (109). Furthermore, two studies have now established Nfil3 as a crucial transcription factor regulating development of all ILC subsets including conventional NK cells (110, 111) (Figure 2). This could be due to the ability of Nfil3 to bind to the regulatory region of Id2 and promote its transcription (112). In addition, GATA3 which is another factor involved at multiple stages of T cell development (113) and was originally thought to be only involved with ILC2 lineage development (101, 114, 115), also seems to play a crucial role in ILC3 differentiation (116).
NKT cells
A subset of NKT cells has also been described as being capable of producing IL-22 (117–120). Much like γδ T cells, TH17 cells and ILC3, IL-22 producing NKT cells express CCR6, IL-23R and RORγt (121–124), and IL-23 stimulates their production o f IL-22 (119, 120, 125, 126). Unlike αβ TCR IL-22 producing cells, but similar to ILCs and γδT cells, NKT do not require IRF4 signaling for IL-22 production (127). In contrast to other IL-22 producing cells, but consistent with NKT populations as a whole, IL-22-producing NKT cells required TCR-CD1d interaction to stimulate IL-22 production (125).
NKT cell development occurs in the thymus, where at the CD4+CD8+ double positive thymocyte stage, cells that have recombined their αβ TCR to enable binding of the CD1d molecule (expressed by other DP cells) are driven towards the NKT cell lineage (128). At least 4 distinct NKT cell developmental stages have been identified that can be defined by their differential expression of CD24, CD44 and NK1.1 and under the control of transcription factors such as PLZF, c-Myc, Egr2, RelA and T-bet (121, 128). Although the development of IL-22 producing NKT cells is still poorly understood, they seem to branch off early in NKT cell development at what is known as “stage 0” (CD24+CD44loNK1.1−), potentially under the control of RORγt (129). Like ILCs, recent reports suggest that these NKT cells require IL-7 for their maintenance and survival (130).
Non-lymphoid production of IL-22
Although primarily cells of the lymphoid lineage have been described as producers of IL-22, some reports have revealed non-lymphoid sources of IL-22. These include macrophages in the lung of human and mouse, which produce IL-22 in response to lung injury (131); neutrophils, which are induced to express IL-22 in the gastrointestinal tract during experimental models of colitis (132); and even one report that describes fibroblast expression of IL-22 in patients with rheumatoid arthritis (RA) (26).
4). REGULATION OF IL-22 PRODUCTION
IL-23
IL-23, which like IL-22 signals through STAT3 (133), is one of the primary inducers of IL-22 production (34, 42–45), and the phenotype of Il23a−/− mice mirrors that of Il22−/− (35, 44, 93). IL-23 is a heterodimer of a specific p19 subunit and a shared p40 subunit, which can also combine with the specific p35 subunit to form IL-12 (134). TH17 cells, ILC3, γδT cells and NKT cells all express the IL-23R, and in vitro stimulation with IL-23 induces production of IL-22 (44, 54–57, 93, 119, 120, 125, 135–138). Dendritic cells (DCs) are potent source of IL-23, particularly the CD103+ subset (138–141), however, there is also evidence that CX3CR1+ cells may be important producers of IL-23 as well (142–144). Several distinct cytokines and pathways have been described as regulating IL-23 production, including TNFα, IFNα/β, IFNγ, CD40L, CCR4, PGE2, lymphotoxin, microRNAs, toll-like receptor (TLR) signaling, and sensory neurons.
Stimulation of multiple TLR pathways, including TLR-2, -4, -5, and -7/8, induces production of IL-23 production by DCs (135, 139, 145–149). Although ligation of TLRs alone is sufficient to drive IL-23 production, additional signals through nucleotide oligomerization domain-containing protein 2 (NOD2), Dectin-1 receptor, and IL-23 itself have been shown to induce maximal induction (135, 145, 146, 150). NOD2 appears to be a key regulator of IL-23 production, as it is also able to inhibit its production (151), although potentially only when NOD2 is stimulated alone with no concomitant stimulation of TLR or Dectin-1 (150). One mechanism by which TLR signaling induces IL-23 production is through directly regulating miR-10a, which inhibits expression of the p40 common subunit (152). CD40L, which is capable of directly inducing IL-23, can also enhance production stimulated by TLR ligation (153, 154). Given that TLR stimulation can trigger both IL-12 and IL-23 production, the crosstalk between IFNγ and TLR pathways appears to be important for influencing differential regulation of the p35 and p19 subunits; inducing p35 and thus IL-12 at the expense of p19 and IL-23 (135, 155, 156). In contrast, there is evidence to suggest that TNFα is global regulator of both IL-12 and IL-23 by directly reducing expression of the common p40 subunit (157). Of note, TLR2 stimulation has been shown to also directly induce IL-22 production by ILC3 in human tonsil (58); and γδ T cells express TLR1 and TLR2 as well as Dectin-1, ligation of which could enhance production of IL-22 (54). However it is unclear how widespread this mechanism of IL-22 induction is.
Dendritic cell localization also seems to be important for regulating IL-23 production. A recent elegant study provided the first evidence to suggest that the peripheral nervous system could control IL-23 production in the skin; with IL-23 producing dermal DCs located in close proximity to sensory neurons (158). In addition, Prostaglandin E2 (PGE2), which is derived from mesenchymal cells and has previously been shown to promote the survival of DCs (159), induces the production of IL-23 by DCs (160–162).
There is increasing evidence to suggest that signaling through LT pathway also has a key role in regulating the production of IL-22. Although LT signaling appears to be dispensable for normal development of ILCs and other producers of IL-22 (163), mice deficient for the LTβ receptor (LTβR) fail to produce IL-22 in response to C. rodentium or those fed a high fat diet; and treatment with an LTβR agonist rescues this defective IL-22 production (164–166). Expression of LTβR has been largely attributed to epithelial cells and stromal cells (167); however, mice with LTβR deleted only in CD11c+ dendritic cells could recapitulate this phenotype (165); and exogenous administration of IL-23 could rescue the phenotype in these mice (164). Together, this strongly suggests that LT signaling is a key regulator of IL-23 expression by DCs. Although the mechanisms by which this occurs is still unclear, intriguingly, LTα1β2 is expressed by many of the same cells that produce IL-22, such as ILC3 (78, 81, 168), thus creating a feedback loop were ILC3 can induce more production of IL-23 by DCs (165).
Interleukin-1β
NKT, ILC3 and TH17 cells can all be induced to produce IL-22 under the control of IL-1β (55, 120, 125, 169–171). IL-1β is produced by multiple cell types, including macrophages, DCs, neutrophils, B and T cells, endothelial cells and epithelial cells (172), and in the gut appears to be dependent on stimulation by the microbiota (173). Interestingly, IL-22 production can be induced by IL-1β, independently of IL-23, although combined they act synergistically (55). However, unlike IL-23, constant IL-1β signaling was required for sustained IL-22 production (174). Moreover, in addition to its role in promoting IL-22 expression, IL-1β also has the capacity to stimulate proliferation and expansion of ILCs and was sufficient to inhibit progression towards IFNγ-producing ILC differentiation (174).
Interleukin-7
Common amongst most lymphoid cells, including those that exhibit the capacity to produce IL-22, is their regulation by IL-7 (175). IL-7 is absolutely critical for the generation and development of all lymphoid IL-22 producing cells such as conventional αβ T cells and γδ T cells (176, 177), however its role in the formation of IL-22-producing NKT cells and ILCs is less clear (175). Although numbers of NKT and ILC3 are reduced in IL-7 or IL-7Rα deficient animals, they are not completely absent suggesting that IL-7 is important for their expansion but not their development (70, 72, 73, 130, 178, 179). IL-7 signaling appears to be semi-redundant with TSLP, which also signals though IL-7Rα and can rescue ILC numbers in Il7−/− mice (180). The importance of IL-7Rα signaling for LTi expansion is highlighted by the fact that secondary lymphoid organogenesis is abnormal in mice lacking components of the IL-7 pathway (such as Il7−/−, Il7ra−/−, γc−/− or Jak3−/−) (178, 179, 181–183), although the effects of IL-7 deficiency are primarily observed in Peyer’s patches. Little decrease in ILCs was observed in the LN of Il7ra−/− mice (81, 178, 179). Reduction in ILC number likely plays an important role in the abnormal lymphoid organogenesis noted above. However, the effect of IL-7 deficiency can also be explained by its regulation of LT signaling, which is crucial for secondary lymphoid organ formation (184). Blockade of IL-7 signaling or IL-7Rα deficiency led to reduction in the expression of LTα1β2 by LTi; and incubation with IL-7 led to upregulation of LTα1β2 (88, 179, 185, 186). Although IL-7 has not been shown to directly regulate IL-22 production, it does appear to stabilize RORγt expression (72), itself critical for the differentiation of all IL-22-producing subsets and for optimal IL-22 gene expression (40, 187). Interestingly, much like other lymphopenic mice such as Rag−/− (138, 188, 189), production of IL-22 by ILC3 is actually increased on a per cell basis in Il7−/− and Il7ra−/− mice (138). Thus, it seems that IL-7 is important for the expansion of IL-22 producing cells (in particular NCR+ ILC3), but may not be required for their functional production of IL-22.
Aryl Hydrocarbon Receptor (AhR)
AhR is critical for ILC3-derived IL-22 production, either as a direct regulator of IL-22 gene expression or as a regulator of ILC3 and TH17 development (187, 190, 191). AhR is located in the cytoplasm in a complex with Hsp90 until ligand binding induces a conformational change that leads to the exchange of Hsp90 with the nuclear translocation component ARNT (192). Potential AhR ligands could be derived endogenously, from the diet, or from the gut microbiota (190), including indol-3-carbinol, 6-formylindolo[3,2-b]carbazole (FICZ) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (193, 194). In addition to these ligands, physical shear stress, cyclic AMP and Ca2+ as well as serum components can activate AhR (195–197). It has been reported that microbial-derived AhR ligands may not be required for ILC development, athough these ligands might be important for IL-22 transcription (187, 190, 191, 198–200).
Notch
Notch signaling, which is required for normal lymphopoiesis (201), is also required for the specific development and differentiation of IL-22 producing subsets such as NCR+ and NCR− ILC3 (95, 98, 202). However, in addition to this role in cellular development, Notch signaling also seems to provide an important signal in regulating IL-22 production (203). RBP-Jκ mice, which are deficient for Notch signals, are extremely susceptible to Con-A mediated hepatitis and experimental autoimmune uveoretinitis, phenotypes that are reversed with the administration of IL-22 (203–205). Mechanisms underlying this regulation of IL-22 include the capacity of Notch signaling to directly regulate RORγt expression as well as other IL-22 regulatory elements such as AhR and Stat3 (203, 206, 207). In fact, Notch signaling, through its target Hes1, is able to extend the phosphorylation of STAT-3 (208), potentially affecting IL-22 targets as well as IL-22 induction.
IL-22 Binding Protein (IL-22BP)
One of the key regulators of IL-22 signaling is a soluble form of the IL-22R1 subunit, known as IL-22 binding protein (IL-22BP). The gene encoding human IL-22BP is located at chromosome 6q23.3 and is closely situated between genes encoding other Type 2 cytokine receptors; 13kb away from the gene encoding IFNγR (Ifngr) and 78kb from the gene encoding for IL20R (Il20r1) (209–211). The overall structure of the mouse gene was found to be very similar to the human gene and encodes an open reading frame of 693 base pairs on chromosome 10. The gene encodes a 210 amino acid protein that shares 34% sequence homology to the extracellular domain of the IL-22R1 subunit (210–214). IL-22BP binds to IL-22 at an overlapping site to IL-22R1, thereby directly interfering with binding to the membrane-bound receptor (215, 216). Moreover, affinity of IL-22 to IL-22BP is up to 1000 fold higher than the membrane bound IL-22R1 form (KD=1pM), primarily as the Koff rate for IL-22BP is up to 4.7 days compared to 7 minutes for IL-22 in complex with IL-22R1, further adding to the inhibitory effect of IL-22BP (14, 217).
IL-22BP has been identified in tissues as diverse as placenta, skin, lung, pancreas, appendix, lung, gastrointestinal tract, breast, lymph nodes, bladder, kidney and thymus (209, 210, 212, 214). Although expression has been primarily identified in dendritic cells (including CD103+ DCs that are also producers of IL-23 (218), IL-22BP has also been found in epithelial cells as well as macrophages (211–214), and there has been one report of IL-22BP expression by B cells (219). Interestingly given its ability to directly inhibit the effects of IL-22, expression of IL-22BP does not increase as levels of IL-22 increase. In models of mouse colitis or LPS administration, when IL-22 levels are significantly increased, amounts of IL-22BP actually decrease (209, 217). However, in later periods after IL-22 has been induced, levels of IL-22BP can increase indicating a regulatory role after the initial early effects of IL-22 (217, 220, 221).
Negative Regulators of IL-22 Production
Although TGFβ is required for TH17 differentiation (37), and can regulate the expression of both RORγt and IL-23R (35, 38, 39), it can also act as an inhibitor of IL-22 production in a dose-dependent fashion (44, 47, 48, 222). IRF4, which is a downstream target of TGFβ, and much like TGFβ is required for TH17 differentiation (223), also inhibits the production of IL-22 possibly through direct binding of the IL-22 promoter (224). However, IL-23 is able to overcome the inhibitory effect of TGFβ and induce the production of IL-22 (46). In addition to TGFβ, IL-27 and ICOS have both also been identified as inhibitors of IL-22 production (225–227). Interestingly, common to the TGFβ, IL-27 and ICOS pathways is the transcription factor c-Maf, which appears to be critical for the inhibition of IL-22 production (48, 227, 228). Furthermore, IL-38, which signals through IL-36Ra, has recently been found to be a regulator of IL-22 production in a dose-dependent fashion; low concentrations of IL-38 inhibit IL-22, while high concentrations modestly induce its production (229, 230).
5). TARGET TISSUES: PATHOPHYSIOLOGY AND REGENERATIVE BIOLOGY
IL-22 is expressed in a broad array of tissues (Figure 3), including liver, lung, skin, thymus, pancreas, kidney, gastrointestinal tract, synovial tissues, heart, adipose tissue, breast and eye (7, 231–240); and its receptor is expressed on the stromal and epithelial cells of those tissues. As IL-22 is produced at sites of inflammation, it may be mediating a physiologic response to repair local tissue damage, or it may be contributing to pathophysiologic inflammation. There is extensive evidence that IL-22 mediates protection and regeneration of epithelial tissues in experimental models, including hepatitis, pancreatitis, colitis, and thymic injury (7, 138, 231, 232). However, this is not true in all models, and IL-22 can induce expression of pro-inflammatory molecules, including IL-1, IL-6, IL-8, IL-11, GCSF, GMCSF, and LPS binding protein (25, 232, 241). This has led to conflicting conclusions as to whether IL-22 is tissue-protective or pro-inflammatory, although these two options are not mutually exclusive. Stimulating epithelial cell survival, proliferation, and production of innate antimicrobials during acute tissue damage can be protective, while chronic overexpression of IL-22 in an otherwise healthy tissue could lead to hyperproliferation, production of chemokmines and other inflammatory signals, and subsequent recruitment of pathologic effector cells to the inflamed tissues. The molecular processes are the same, but the context determines protection vs. pathology.
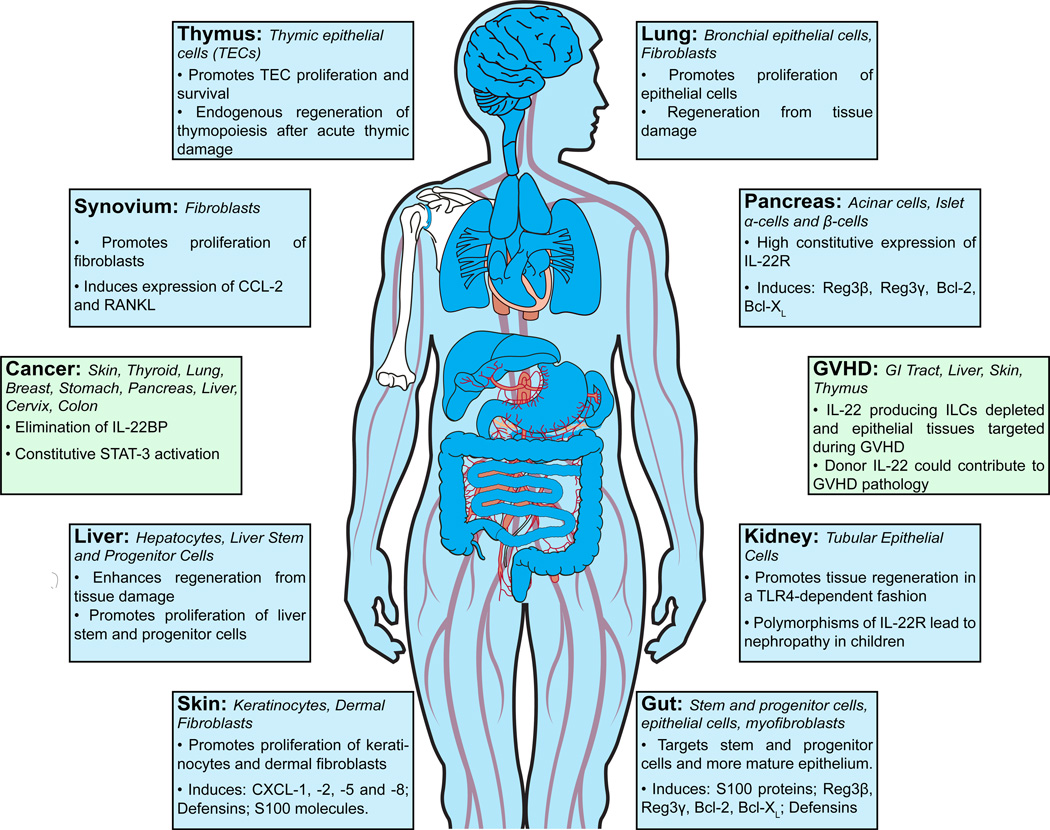
Figure 3. Target tissues and physiological effects of IL-22.
A role for IL-22 has been described in numerous tissues including gut, liver, lung, skin, thymus, kidney, heart, pancreas and synovial tissue. IL-22R is expressed on epithelial cells and some fibroblasts in those tissues. IL-22 can promote their cellular proliferation, resistance to apoptosis, and wound healing. In barrier organs such as gut, lung and skin, IL-22 also promotes the production of antimicrobial molecules such as S100, Reg3β and Reg3γ, as well as defensins, thereby aiding in host defense and barrier function. IL-22 is important for promoting tissue regeneration after injury in several organs. However, studies have also indicated that IL-22 is associated with malignancies of the skin, thyroid, breast, stomach, pancreas, liver, cervix and colon, as well as other inflammatory pathology including psoriasis. In BMT, ILC-derived IL-22 is important for tissue regeneration after injury caused by alloreactive T cells, however, there is also some evidence that IL-22 produced by donor T cells can contribute to inflammation and GVHD pathology.
In addition to the context of IL-22 production, potential pathologic effects of IL-22 could be related to other cytokines expressed with IL-22. It has been proposed that IL-22-related pathology may be due to tissue “priming” by type I interferons that shift IL-22 signaling away from STAT-3 toward a STAT-1 response (242). Furthermore, IL-17 and IL-19 have additive effects with IL-22 in inducing production of other inflammatory molecules (243). Prominent examples of such interplay in inflammation are: in oral mucosa, where IL-1β and IL-22 cooperate in inducing CCL20 expression by human gingival fibroblasts, leading to recruitment of Th17 cells and periodontal disease (244); in a model of psoriasis, where a combination of IL-22, IL-17A, IL-1α, OSM and TNFα synergistically increased chemokine and antimicrobial-peptide expression (245); in the lungs, IL-22 reduced bleomycin-related lung toxicity in IL-17 deficient mice, but promoted airway inflammation if IL-17 was present (246); in chronic hepatitis, where IL-22-related pathology was dependent on the recruitment of Th17 cells (247); and in an anti-CD40 administration model colitis, where IL-22 was reported to exacerbate colon inflammation that was dependent on IL-23 and IFNγ for intestinal pathology (72, 248, 249). Moreover, IL-22 had no effect on anti-CD40-induced colitis in mice lacking ILCs (93, 248), further suggesting that pathologic effects of IL-22 may be dependent on co-expression with other inflammatory cytokines.
Systemic expression in peripheral blood
IL-22 expression has been reported in murine and human peripheral blood under stress conditions, however the baseline serum expression is extremely low (137, 217). Mirroring findings in mice, where TBI conditioning and T cell-depleted bone marrow transplantation (BMT) led to increased serum IL-22 protein in two different murine strains (137), elevated expression of IL-22 has also been reported clinically in patients with inflammatory conditions, including IBD, RA and malignancy (217, 250). Serum IL-22 is also elevated in patients with IL-23 receptor polymorphisms associated with Crohn’s disease, which could indicate a pathologic role for IL-22 in IBD, a compensatory IL-22 response in IBD patients, or a coincidental finding, as Crohn’s disease may actually be dependent on other pathways that are also activated by IL-23 receptor signaling (251). Furthermore, the overall relevance of serum IL-22 expression to disease states is unclear, as this could represent systemic dissemination of an immune response, or leakage into the vascular compartment from a compromised tissue site where IL-22 may be intended to mediate tissue repair. Interestingly, IL-22 can induce production of coagulation factors including fibrinogen (252). While this may simply be indicative of IL-22 function in target tissues such as the liver, regulation of systemically disseminated molecules suggests that serum IL-22 may indeed function in coordination of a general inflammatory response.
Gastrointestinal Tract
IL-22 and its receptor have been described in the human upper GI tract, including the oral cavity, salivary glands, tonsils, stomach, and esophagus, where IL-22 expression has been associated with inflammation and antimicrobial immunity as well as malignancy (66, 91, 253–258). IL-22 levels are increased in colonic tissue from patients with Crohn’s disease and ulcerative colitis (25), and in murine colon during DSS-induced and T cell transfer-mediated colitis (259). Colon IL-22 expression is also increased in experimental colitis due to infection with Clostridium difficile and C rodentium (45, 260). Intestinal IL-22 is critical for orchestrating antibacterial immunity against C. rodentium, which is used in experimental murine modeling of enterohemorrhagic and enteropathogenic E. coli colitis (45, 187). Although the adaptive immune response does contribute to immunity against C. rodentium, ILCs are the dominant source of IL-22 critical for initial resistance against the infection (45, 69, 136). Moreover, ILCs appear to be the essential source of IL-22 for protection against DSS-induced colitis and protection against GI graft versus host disease (GVHD) (137, 261), although other innate sources of IL-22 such as neutrophils may contribute as well (132).
The single common function of IL-22 in the GI tract relates to maintenance of the epithelial barrier. Sonnenberg and colleagues demonstrated that ILC-derived IL-22 was important for routine containment of Alcaligenes species and preventing subsequent systemic inflammation (262). However, the phenotype of IL-22 deficiency in the gut is much more pronounced after challenge with pathologic inflammatory injury, whether infectious, chemical, or alloreactive (7, 231, 232). IL-22-deficient mice exhibit only minimal epithelial perturbations at baseline. However they demonstrate severely exacerbated colitis in response to C. rodentium infection, which can be reversed by exogenous administration of IL-22 (45, 136). Similarly, mice lacking ILC3 are highly susceptible to DSS-induced colitis (263).
IL-22-mediated barrier maintenance involves support of the epithelial lining’s structural integrity as well as its functional activation. IL-22 provides direct support to intestinal epithelial cells by inducing expression of genes regulating proliferation, wound healing, and apoptosis (243, 264). It has also been suggested that IL-22 may regulate tight junctions between intestinal epithelial cells (265). This is consistent with a recent report describing upregulation of mRNA expression for the tight junction protein claudin-1 in human intestinal epithelium after culture with IL-22 in vitro (266). In addition to these examples of IL-22-mediated regulation of the physical barrier, IL-22 provides functional barrier support by inducing the production of innate antimicrobials, including defensins, Reg family molecules, and S100 proteins (23, 28, 34, 45, 262, 264, 267). Bridging the structural and functional barrier, IL-22 also protects goblet cells during colitis and induces production of mucins (Muc1, 3, 10, and 13), which are necessary to create a protective layer between the epithelium and enteric contents (221).
The intestinal barrier regulated by IL-22 is dynamic, involving cross-talk between ILC3 and the intestinal microbiota (69). IL-22 is critical for bacterial containment (262), and AhR stimulation by microbiota-derived ligands may be important for suppression of colitis by mediating bacterial containment and prevention of inflammation induced by gut microbes (199). In addition, the gut microbiota can contribute to retinoic acid production by DCs (268). Retinoic acid has been shown to attenuate colitis due to DSS administration and C. rodentium infection by promoting binding of the retinoic acid receptor to the IL-22 promoter, thereby enhancing IL-22 production by ILCs and γδ T cells (57). Conversely, retinoic acid can also enhance DC production of the negative regulator IL-22BP (218), and gut flora can inhibit IL-22 expression via induction of enterocyte IL-25 production (263). Therefore, there is a fine balance in the GI tract between the microbiota, the immune system, and the epithelial barrier, where flora can induce the IL-22 production that is necessary for its containment. Dysregulation of this system can result in inflammatory epithelial pathology.
The intestinal epithelium contains diverse subsets of functionally distinct cell types (269), and the specific cellular targets of IL-22 remain to be fully elucidated. While it is known that goblet cells produce mucins in the intestines, there is little direct evidence indicating that intestinal mucin production downstream of IL-22 is due to a direct effect of IL-22 on goblet cells. Similarly, although IL-22 is thought to regulate Paneth cell production of Reg3 molecules and defensins, and while Paneth cells are reported to produce these molecules (270, 271), there is little direct evidence indicating that IL-22 is instructing this function. Reg3β and Reg3γ can be produced by crypt epithelium and mature enterocytes (272). Furthermore, mice with Paneth cell deficiency due to GVHD demonstrate increased expression of Reg3γ, indicating that enterocytes are potent sources of intestinal antimicrobials (273). Additionally, we have found that IL-22R is expressed on Lgr5+ intestinal stem cells (137), although the functional significance of this is unknown.
Secondary lymphoid tissue
Intestinal IL-22+ ILCs are present in cryptopatches within the lamina propria, and they contribute to the development of secondary lymphoid tissues and mucosa associated lympohoid tissues (MALT), including lymph nodes, cryptopatches, isolated lymphoid follicles (ILFs), and Peyer’s patches (79, 80, 82, 83, 86, 88). It is thought that cryptopatch development is dependent on activation of stromal cells by LT from ILCs and recruitment of DCs and other hematopoietic cells (184, 274–276). Initial cryptopatch development seems intrinsically regulated, but subsequent post-natal recruitment of B cells may induce formation of ILFs in a microbiota-dependent fashion, as ILFs are absent in mice treated with broad antibiotics to clear the microbial flora (86, 277, 278). While the RORγt+ LTi cells critical for lymph node development are robust producers of IL-22, and mice deficient in IL-22+ ILCs due to loss of either RORγt or AhR are lacking both crypotopatches and ILFs, it does not appear that IL-22 itself is critical for the formation of either of these structures (45). Cryptopatches in the small intestine, their large intestine counterpart colonopatches, and ILFs throughout the GI tract were reported by Zheng and colleagues to be present in IL-22-deficient mice. Interestingly though, after C. rodentium infection they found that treatment with an anti-IL-22 neutralizing antibody led to the loss of both colonopatches and ILFs, suggesting that while IL-22 is dispensable for normal development and maintenance of MALT, it may have an important role in maintenance of intestinal lymphoid structures in the setting of inflammatory pathology. It is unknown however if this role of IL-22 is due to direct maintenance of the structures or indirect support of the structures by maintaining the epithelium and/or controlling the infection.
Skin
IL-22 has multiple effects on keratinocytes, such as induction of proliferation, migration, tissue remodeling and secretion of anti-microbial peptides and chemokines, as well as delayed differentiation (reviewed in (7)). IL-22 induces keratinocytes to produce anti-microbial proteins, including β-defensin 2, β-defensin 3, S100A7, S100A8, S100A9 and lipocalin 2 (279–281). IL-22 also promotes the production of neutrophil-attracting chemokines, such as CXC-chemokine ligand 1 (CXCL1), CXCL2, CXCL5 and CXCL8, and inhibits the expression of CCL22, which attracts Th17 and Th2 (282). IL-22 can also induce the expression of extracellular matrix (ECM)-degrading enzymes matrix metalloproteinases (MMP)-1 and -3, which are required for epithelial migratory capacity during epithelial repair (280). In addition to its role in innate immunity by promoting anti-microbial molecules or attracting neutrophils, IL-22 also inhibits differentiation and maturation of keratinocytes through downregulation of molecules such as keratin 1 (KRT1), KRT10, profilaggrin, involucrin, loricin, kallikrein 7, desmocollin 1 and the late cornified envelope protein 1B (280, 282).
IL-22 also increases the expression of IL-20, which shares the IL-22R1 chain in its receptor complex (IL-22R1/IL-20R2) and has similar effects as IL-22 on keratinocytes, resulting in a positive feedback loop to amplify the effects of IL-22 (282). IL-22 has major effects on epidermal architecture, resulting in acanthosis (thickening of stratum spinosum in epidermis), parakeratosis (nuclear remnants in stratum corneum due to dysfunctional keratinocyte cornification) and hypogranularity in both 3-D tissue models, as well as IL-22 overexpressing transgenic mice (282, 283). These architectural changes of the epidermis are similar to those seen in psoriatic plaques. In addition, psoriatic plaques contain neutrophils and increased expression of IL-20 and STAT-3, which could be related to the increased IL-22 levels in the plaques (282, 284–286).
There has been considerable pathology related to IL-22 documented in the skin, where IL-22 is thought to contribute to psoriasis (7, 44). Psoriasis is common chronic skin disease, which affects approximately 2% of the population (287), and shares features with other chronic inflammatory diseases, such as IBD, diabetes mellitus, as well as an associated arthropathy: psoriatic arthritis. The characteristic skin lesions are raised, well-demarcated scaly plaques due to hyperproliferation of the epidermis with premature maturation of keratinocytes and incomplete cornification and retention of nuclei in the stratum corneum (287). The skin lesions contain an inflammatory infiltrate, which consists of dendritic cells, macrophages and T cells in the dermis, as well as neutrophils and T cells in the epidermis. The pathophysiology of psoriasis is multi-factorial with roles for autoimmunity, dysregulation of the innate immune system, proinflammatory cytokines, as well as imbalance of epithelial homeostasis, angiogenesis and genetic and environmental risk factors. However, dendritic and T cells seem to play a predominant role and the primary axis seems to be the production of IL-23 and IL12 by dermal myeloid dendritic cells, which subsequently activates Th1, Th17 and Th22 cells to secrete IL-17, IFN-γ, TNF and IL-22 (288).
Polymorphisms in the IL-22 gene have been associated with psoriasis in a Japanese population (289) and a polymorphism in the promoter region of IL-22 is associated with increased production of IL-22 and increased risk of psoriasis onset in childhood (290). In addition, copy number variation of IL-22 gene exon1 was significantly associated with psoriasis severity (291). Finally, polymorphisms in both IL-23R and IL-12B (coding for common p40 chain) are associated with psoriasis (292).
Initial studies regarding the effects of IL-22 in pathologies of the skin demonstrated that IL-22 levels were increased in T cell dermatoses, such as psoriasis and atopic dermatitis (23, 53). Consistent with a role in its pathophysiology, psoriasis patients have increased levels of IL-22 in their blood and serum, which has been associated with disease severity (293–295). Interestingly, psoriatic plaques also have increased expression of IL-20 and IL-24 (285, 286), which both bind to IL-22R1 and seem to have similar effects as IL-22 on keratinocytes in vitro and in transgenic mice (282, 283, 296, 297).
Notably, effective therapy for psoriasis results in a decrease in IL-22 levels both in the serum and the psoriatic skin lesions (28). A series of recent studies have demonstrated increased numbers of NCR+ ILC3 in the peripheral blood and lesions of psoriasis patients and therapeutic response to anti-TNF resulted in a decrease in the numbers of circulating NCR+ ILC3 (298–300). Interestingly, NCR− ILC3 could be shown to be converted to NCR+ ILC3 upon culture with IL-1beta and IL-23, which are both involved in psoriasis (299). Studies with IL-23 administration in IL-22-deficient mice demonstrated that IL-22 is required for both acanthosis and neutrophil infiltration (44) and injection of IL-22 into human normal skin xenografts in immunodeficient AGR129 mice resulted in changes similar to psoriasis, including epidermal thickening, expression of the psoriasis-associated marker K16 and increased vascularization (301). In addition, administration of neutralizing antibodies to IL-22 prevented the development of disease, acanthosis, inflammatory infiltrates, and expression of Th17 cytokines in experimental models as well as in human xenografts (301, 302).
IL-22 neutralization has been tested in a phase 1 study evaluating a single subcutaneous dose of anti-IL-22 antibody (ILV-094) in individuals with moderate-to-severe chronic plaque psoriasis (ClinicalTrials.gov NCT01010542) as well as in a phase 2 randomized double-blind placebo-controlled study of six infusions of ILV-094 in subjects with atopic dermatitis (ClinicalTrials.gov NCT01941537). However, given that the other IL-22R1 ligands, IL-20 and IL-24, also have a primary role in the pathology of psoriasis (283), inhibition of IL-22R1 could be a more effective clinical approach than neutralization of IL-22.
Lung
Consistent with other tissues, IL-22 production in the lung is primarily regulated by alveolar macrophages and dendritic cells (reviewed in (303). Alveolar macrophages and CD11b+ DCs can be stimulated through their innate pattern recognition receptors (PRRs), such as TLRs, NODs and Dectin, resulting in the secretion of IL-1beta, IL-23 and other pro-inflammatory cytokines (such as IL-18 and IL-6), which can stimulate NF-kappaB and STAT3 in lymphocytes (both CD4+ and ILCs) resulting in the expression of RORγt, Th17 differentiation and the production of IL-17 and IL-22. Conversely, stimulation of plasmacytoid and CD103+ DCs by viral antigens results in the secretion of type I IFNs, which counteract Th17-polarizing cytokines, as well as IFN-gamma and IL-27, which activates STAT1 and suppresses IL-17/22 production in lymphocytes.
The epithelium in the lung has two important functions: barrier to pathogens and facilitation of gas exchange. IL-22 has several effects on the lung epithelium: (a) production of antibacterial proteins, including (in combination with IL-17) lipocalin-2 and β-defensin 2 (42), and chemokines, (b) proliferation and (c) repair after injury. IL-22 can synergize with IL-17 to induce production of G-CSF and IL-6 (42). The combination of IL-17 and IL-22 can induce MMPs, which play a role in the pathophysiology of emphysema (304). However, IL-22 can have multiple and even opposite effects in the lung depending on the stressor, kinetics and environment.
IL-22 levels in the serum are increased in the peripheral blood of patients with asthma and in the lungs of mice in preclinical asthma models (305, 306). IL-22 has a dual role in asthma: it can promote the initiation of asthma in a preclinical model, but ameliorates inflammation during antigen challenges and exacerbation (305). During allergic airway inflammation IL-22 neutralization with an antibody had dual effects: neutralization during allergen sensitization resulted in less lung pathology and lower levels of IL-5 and eosinophils, whereas IL-22 neutralization during antigen challenge increased lung inflammation and pathology (such as goblet cell hyperplasia) (305, 307). Conversely, subcutaneous (but not intraperitoneal) IL-22 administration during sensitization resulted in worse inflammation (305, 307–309), whereas IL-22 administration during antigen challenge could reduce eosinophil infiltration, chemokine expression, production of Th2 cytokines (such as IL-13 and IL-25), goblet cell hyperplasia and airway constriction (305, 307, 309). In this same model IL-17 had pro-inflammatory effects in wild type mice, but was anti-inflammatory in IL-22−/− mice. The mechanisms behind this dual role during allergic airway inflammation are not clear, but could involve inhibition of CCL17 and IL-25 production by airway epithelium (307, 309), epithelial repair (120), and possibly involve IL-10 (308).
IL-22 also plays an important role in host defense against several pulmonary pathogens. During Streptococcus pneumoniae infection, DC activation involving MyD88 results in rapid accumulation of ILC3 in the lung tissue to produce IL-22 (310). Administration of the TLR5 agonist flagellin increases IL-22 production by ILC3 resulting in protection against S. pneumoniae infection. During infection with Klebsiella pneumonia IL-22, produced primarily by NK cells (311), has a protective effect (42) and during infection with the intracellular bacterium Chlamydia muridarum IL-22 levels in the lung rapidly increase (312). IL-22 inhibition with neutralizing anti-IL-22 monoclonal antibodies resulted in impaired Th1 and Th17 responses and exacerbation of disease. Intranasal administration of IL-22 significantly enhanced the Th17 response and resulted in protection following chlamydial lung infection.
Il22−/− mice have an increased fungal burden after infection with Aspergillus fumigatus demonstrating a role in the clearance of this pathogen; through activation of Dectin-1 which results in IL-23 production (313). Studies with human PBMCs pointed at CD4+ T cells as the major IL-22 producers after exposure to A. fumigatus involving IL-1 signaling (314). In contrast, studies also suggest IL-22 may contribute to lung pathology during chronic exposure to A. fumigatus (315). Interestingly, during Candida albicans infection IL-22 produced by ILCs reduces the fungal burden and provides cross-protection against Pseudomonas aeruginosa possibly through stimulation of epithelial repair (316).
During influenza infection NK and NKT cells produce IL-22 in response to IL-23 and IL-1β (secreted by DCs) resulting in epithelial regeneration of airway epithelium, as well as parenchymal epithelium, which has upregulated IL-22R expression after influenza infection (120, 317–319). IL-22−/− mice infected with influenza display defective epithelial regeneration with increased collagen deposition. αβ and γδ T cells can also produce IL-22 during influenza infection and might be involved in survival after superinfection with S. pneumonia through protective effects on lung epithelium (320). IL-22 can also protect against lung fibrosis in a model based on repeated exposure to Bacillus subtilis. Neutralization of IL-22 resulted in enhanced collagen deposition in the lungs, whereas IL-22 administration could decrease lung fibrosis (321). In addition, IL-22 can have a protective effect in a rat model of barotrauma resulting in decreased pulmonary disintegration and edema (322).
The role of IL-22 in Mycobacterium tuberculosis infection is still unclear, although it has been found that IL-22 producing γδ T cells can migrate into the granulomas during M. Tuberculosis infection (323). A recent study using primary epithelial cells demonstrated that mycobacteria might subvert the host defense and establish chronic infection, bypassing NF-kB signaling through GSK3 inhibition and promoting the anti-inflammatory transcription factor CREB resulting in IL-10 and IL-22 production (324). However, IL-22 produced by NK cells seems to be required for effective BCG vaccination for M. tuberculosis and NK cell depletion during vaccination resulted in increased numbers of Tregs and bacilli (325). IL-22 administration could overcome the effects of NK depletion and enhanced memory CD4+ T cell generation in the lungs.
As proposed by McAleer and Kolls (303), IL-22 in the lung has a role in non-redundant epithelial regeneration as well as host defense after pulmonary infection or trauma. Its dual pro- vs anti-inflammatory role in lung inflammation might be particularly dependent on the presence or absence of IL-17, therefore from a therapeutic standpoint, it may be better to inhibit IL-17 without affecting IL-22 levels or administer IL-22 in combination with IL-17 inhibition.
Liver
In the liver hepatocytes are the main targets of IL-22; in which IL-22 is thought to induce the production of acute-phase proteins such as Amyloid A, α1-antichymotrypsin, haptoglobin, and LPS-binding protein (2, 23, 217, 252, 326). Consistent with other tissues, IL-22 also induces proteins involved with protection and regeneration from damage such as the anti-apoptotic proteins Bcl-2 and Bcl-XL, cyclin-dependent kinase 4 (CDK4), cyclin D1, c-myc and p21 (327–330). In fact, IL-22 has been shown to have a considerable protective role in several different experimental models of hepatic injury, including T cell-mediated hepatitis, acute hepatitis, liver ischemia-reperfusion injury, bacterial and parasitic infection, as well as models of acute and chronic alcohol-induced liver damage (327–334). IL-22 promotes the proliferation of liver stem and progenitor cells (335), inducing their expression of cyclin D as well as the anti-apoptotic proteins Bcl-2 and Bcl-XL in a STAT-3 dependent fashion (335). This stimulation of hepatic stem and progenitor cells is consistent with its ability to promote the regenerative response in the liver to partial hepatectomy (330, 336). Further supporting this role of IL-22 in liver regeneration, levels of IL-22 are increased in patients with hepatitis B or C viral infections (330, 337), and receptor expression is upregulated by hepatocytes in periods of injury (330, 331, 333).
Kidney
Low constitutive expression of IL-22R was identified in kidney in the earliest studies looking for its expression (13, 23, 338, 339). Subsequent analysis has revealed that tubular epithelial cells are the main cells expressing IL-22R (340). Consistent with these findings, a human kidney cell line, TK-10, has been shown to respond to IL-22, inducing STAT-3 phosphorylation (3).
Although little is known about its regulation or role in kidney function, IL-22 is induced in the kidney after LPS stimulation (2) as well as in a model of polymicrobial peritonitis (341). However, there is some discrepancy regarding the role of IL-22 in kidney regeneration. In several studies IL-22 has been shown to have a detrimental effect on kidney regeneration where blockade of IL-22 with an IL-22BP-Fc led to reduced kidney injury and enhanced bacterial clearance in polymicrobial peritonitis (341); and in vitro incubation with IL-22 suppresses the growth of the renal carcinoma cell line A498 (342). In contrast, and consistent with other tissues such as gut, thymus, lung and liver, Kulkarni and colleagues have recently found that IL-22 can in fact enhance kidney regeneration in a TLR4-dependent manner (340). In addition, polymorphisms in the IL-22R are associated with development of nephropathy (343). The discrepancies in these studies will clearly need to be reconciled in order to develop IL-22 (or anti-IL-22) as a potential therapy for kidney pathology.
Pancreas
Initial studies of tissues expressing IL-22R1 revealed the pancreas as the tissue with the highest levels of expression (23, 338, 344), likely due to the high relative proportion of cells within the pancreas expressing IL-22R as both acinar cells as well as islet α and β cells express IL-22R1 (338, 345). Within acinar cells, IL-22 is thought to induce production of molecules such as Reg3β, Reg3γ, and osteopontin (338, 346), as well as inducing the anti-apoptotic proteins Bcl-2 and Bcl-XL (335, 338, 345). IL-22 is also thought to induce Reg1 and Reg2 expression within islet cells (347). These findings have led to the idea that IL-22 could be used to treat pancreatitis, which has proven experimentally successful using transgenic overexpression of IL-22 as well as small molecule activators of AhR (346, 348).
Synovial tissue
Although in most tissues epithelial cells are the predominant targets of IL-22, fibroblasts have also been described as targets of IL-22 in the skin, synovial tissues, lung and colon (22, 25, 26, 282); although at least in the skin they appear to express the IL-22R at considerably lower levels than keratinocytes (282). Much like epithelial tissues, but in contrast to colonic or lung fibroblasts (25, 349), IL-22 has been shown to induce proliferation of synovial fibroblasts (26), likely contributing to the pathophysiology of RA. In fact, IL-22 is increased in the synovial tissue and in the serum, and IL-22+ CD4 T cells are increased in the peripheral blood of patients with RA (26, 350–353). In addition to promoting proliferation amongst synovial fibroblasts, IL-22 also induces the production of CCL2 (26), thereby attracting monocytes to the synovium, as well as RANKL which induces their differentiation into osteoclasts (354).
6). SYSTEMIC EFFECTS OF IL-22
Cancer
Perhaps the most dramatic example of pathologic production of IL-22 occurs in the setting of malignancy, where the tissue regenerative effect of IL-22 can be coopted to contribute to tumor growth. IL-22 has been associated with cancers in several sites, including skin, thyroid, lung, breast, stomach, pancreas, liver, cervix, and colon (355–365). Kirchberger and colleagues identified IL-22+ cells in human colon cancer specimens and found that a model of bacteria-induced colon cancer led to accumulation of double positive IL-17+IL-22+ ILCs (366). IL-22 in this experimental cancer model activated STAT-3 in colon epithelium and contributed to colon cancer progression. In an experimental model of colon cancer induced by DSS and azoxymenthane, it was also shown that IL-22 could contribute to tumor development (220). IL-22 protected mice from the initial colitis that was induced by the chemical treatment, however excessive IL-22 expression due to elimination of IL-22BP increased tumor growth. While IL-22 was functionally linked to tumor progression in both of these model systems, IL-22 was not shown to have a causative role in tumor transformation. Rather, malignancy was induced by the experimental model and IL-22 then contributed to progression. Additionally, the IL-22/STAT-3 axis has been shown to activate STAT-3 in human colon cancer cells, inducing Dot1L expression and promoting tumorigenic potential (356), although this still may reflect a property of transformed cells using IL-22, rather than IL-22 playing a causative role in malignant transformation. Therefore, given the connection between inflammation and colon cancer, it is possible that IL-22 could have both protective and pathologic effects in malignant transformation. IL-22 could prevent tumorigenesis by inducing tissue regeneration, promoting barrier function, and reducing chronic inflammation. However, if IL-22 is overexpressed due to chronic inflammation, then malignant cells could utilize this trophic signal to feed their progression.
Graft versus host disease (GVHD)
Contrasting effects of IL-22 have also been reported in GVHD, where recipient-derived IL-22 was shown to reduce mortality and tissue pathology in the liver and GI tract, whereas donor-derived IL-22 increased mortality and target tissue inflammation (137, 367–369). Protective recipient-derived IL-22 was produced by tissue resident ILCs (137). In contrast, donor T cells producing IL-22 led to an overall heightened inflammatory state in recipient mice, with concomitant reduction in regulatory T cells. This dichotomy in experimental GVHD models is supported by clinical investigations indicating that ILC activation and recovery from chemotherapy is associated with reduced susceptibility to mucositis and acute GVHD (370), but acute GVHD skin lesions demonstrate increased infiltration with CD4+ T cells (371). Therefore, the distinct effects of IL-22 in GVHD could be due to 1) the timepoint post-BMT when IL-22 is produced by distinct cellular sources, 2) distinct additional cytokines co-produced by these cellular sources or there neighbors, or 3) distinct localization and cellular targets of donor T cells and recipient ILCs within tissues. Although recipient-derived IL-22 was shown to be protective for gut tissue after allogeneic BMT, the potential benefit of IL-22 was limited during GVHD due to the elimination of recipient-derived IL-22+ ILCs by alloreactive donor T cells (137). This pathophysiology of IL-22 deficiency in GVHD may have a natural parallel in autoimmune polyendocrinopathy - candidiasis - ectodermal dystrophy (APECED) patients who have neutralizing antibodies to cytokines including IL-22, contributing to their susceptibility to candidal infections (372, 373). Furthermore, colon biopsy samples from patients with IBD have also demonstrated a deficiency of IL-22+ cells, and impaired T cell and innate cell IL-22 production has been reported in a primate model of celiac disease, indicating that intestinal pathology due to loss of IL-22-producing cells is not limited to GVHD pathogenesis and the transplant setting (374, 375).
Regulation of adaptive immunity
Interestingly, in some of the earliest studies of IL-22, constitutive expression of both IL-22 and IL-22BP were found to be highest in the thymus (6, 209, 210, 212). In fact, the thymus was the tissue chosen to isolate IL-22 cDNA for sequencing (1). Although IL-22 does not seem to be required for normal thymus organogenesis or maintenance, as there is no gross defect in Il22−/− mice (138), constitutive expression of high levels of IL-22 in the steady-state does seem to lead to some thymic involution (252). However, in periods of thymic injury, IL-22 is critical for the endogenous regeneration of thymic tissue and restoration of T cell development (138). Despite its importance for generating a diverse adaptive immune repertoire, the thymus is extremely sensitive to injury, which can be caused by infection, shock, and cytoreductive chemo- or radiation therapy (376, 377). It also has a remarkable capacity for repair, although prolonged T cell deficiency is a major clinical hurdle in patients receiving common cancer cytoreductive therapies and in BMT recipients (377, 378). Post-transplant T cell deficiency is associated with an increased risk of infections, relapse of malignancy, the development of secondary malignancies, and impairment in application of immunotherapeutic strategies, such as vaccination against microbes or tumors (379–383).
Although initial studies did not identify IL-22R expression in the thymus by PCR (279, 338), subsequent analysis of protein expression revealed a population of thymic epithelial cells (138). These discrepancies can likely be explained by the rare thymic epithelial cell (TEC) population not being assayed by common dissociation protocols (384). IL-22R signaling promotes TEC proliferation and survival (138), and one recent report suggested that IL-22 might even regulate FoxN1 (385), a forkhead box transcription factor that is critical for TEC development, maintenance, and regeneration (386–390). However, while Pan and colleagues did observe increased expression of FoxN1 at the same time as there was increased expression of IL-22 (385), there is not yet direct evidence supporting the regulation of FoxN1 by IL-22. Intriguingly, in addition to the role of IL-22 in adaptive immunity by regulating thymopoiesis, ILCs themselves appear regulate CD4+ T cells in the gut (391). Murine intestinal ILC were found to express MHC II, but not co-stimulatory molecules, thus leading to tolerance of mucosal T cells to enteric contents.
7). CONCLUDING REMARKS
In summary, IL-22 has a variety of functions, most notably its trophic effect on non-hematopoietic cells, especially epithelial cells. IL-22 is involved in epithelial regeneration and pathology in several organs depending on the context and/or cytokine milieu. Its involvement in a variety of diseases makes it an attractive target for clinical development. However, therapeutic strategies will have to be individualized based on organ and/or pathology; in some cases the therapeutic goal would be to decrease IL-22 levels (for example: psoriasis or malignancy), whereas in other situations, therapeutic IL-22 administration could be considered (for example: tissue damage such as that caused during GVHD). Successful implementation of either of these strategies will require further investigation into the molecular mechanisms and cellular targets of IL-22 in health and disease.
Table 1.
Role of IL-22 in host defense against infection
| Infection | Type | Tissue | Role | Refs |
|---|---|---|---|---|
| Klebsiella pneumoniae | Bacterial | Lung | Maintenance of barrier function; induction of defence mechanisms; mediates recruitment of leukocytes | (42) |
| Citrobacter rodentium | Bacterial | Gut | Maintenance of barrier function; induction of defence mechanisms; mediates recruitment of leukocytes | (33, 45) |
| M avium | Bacterial | Intestine | No apparent role | (392) |
| Toxoplasma gondii | Protozoa | Intestine | Neutralizing IL-22 leads to less tissue pathology despite equivalent parasite burdens | (392) |
| Listeria monocytogenes | Bacterial | Systemic | No apparent role | (392, 393) |
| Salmonella enterica | Bacterial | Intestine | No apparent role | (332) |
| Commensal Bacteria | Bacterial | Intestine | Regulates composition of microbiota | (262, 391, 394) |
| Aspergillus fumigatus | Yeast | Lung | Controls fungal burden | (313) |
| Candida albicans | Yeast | Mucosa, Skin | Aides in preventing dissemination; mucocutaneous defence | (395, 396) |
| Influenza | Viral | Lung | Limits tissue damage | (120, 317, 319, 320) |
| Hepatitis B | Viral | Liver | Limits tissue damage | (328, 330, 335, 397) |
Table 2.
Known regulators of IL-22 production
| Molecule | Type of regulation |
Mode of action | Refs |
|---|---|---|---|
| IL-23 | Positive | Main stimulator of IL-22 production, mediated by STAT-3 | (34, 42–45, 133) |
| IL-1β | Positive | Directly induces IL-22 production by TH17, NKT and ILCs and inhibits progression of ILCs into IFNγ-producing subsets | (55, 120, 125, 169–171, 174) |
| IL-7 | Positive | Differentiation and expansion of IL-22 producing cells and stabilizing RORγt expression | (70, 72, 175–177) |
| AhR | Positive | Promotes development of ILCs and TH17 cells and directly promotes IL-22 production | (187, 190, 191) |
| Notch | Positive | Differentiation of IL-22 producing lymphoid cells as well as promoting IL-22 production by mediating AhR signals | (95, 98, 202, 203, 205) |
| IL-22BP | Negative | Soluble IL-22R with 1000-fold higher affinity for IL-22 than membrane bound receptor | (14, 215–217) |
| TGFβ | Negative | Inhibits IL-22, potentially binding to IL-22 promoter but also mediated through c-Maf | (224) |
| IL-27 | Negative | (225, 226) | |
| ICOS | Negative | Induction of c-Maf and reduced expression of IL-22 | (227, 228) |
| c-Maf | Negative | Common pathway of TGFβ, IL-27 and ICOS mediated negative regulation of IL-22 | (48, 227, 228) |
| IL-25 | Negative | Produced by epithelial cells, inhibits the production of IL-22 by ILCs | (263) |
ACKNOWLEDGEMENTS
The authors would like to thank Dale Godfrey and Lauren Young for helpful discussions. This work was supported by the following awards from the National Institutes of Health: K99-CA176376 (J.A.D.), K08-HL115355 (A.M.H), R01-HL069929 (M.R.M. vdB.), R01-AI100288 (M.R.M.vdB.), R01-AI080455 (M.R.M.vdB.), R01-AI101406 (M.R.M.vdB), P01-CA023766 (M.R.M.vdB). Support was also received from the U.S National Institute of Allergy and Infectious Diseases (NIAID Contract HHSN272200900059C), the European Union (award GC220918, C. Blackburn), The Experimental Therapeutics Center of MSKCC funded by Mr. William H. Goodwin and Mrs. Alice Goodwin, The Lymphoma Foundation, Alex's Lemonade Stand, The Geoffrey Beene Cancer Research Center at MSKCC, The Susan and Peter Solomon Divisional Genomics Program, and The Lucille Castori Center for Microbes, Inflammation & Cancer. J.A.D. was supported by a CJ Martin fellowship from the Australian National Health and Medical Research Council and a Scholar Award from the American Society of Hematology. A.M.H was supported by a Scholar Award from the American Society of Hematology, a New Investigator Award from the American Society for Blood and Marrow Transplantation, and the Amy Strelzer Manasevit Research Program for the Study of Post-Transplant Complications..
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review. Patent applications have been filed on the therapeutic use of IL-22 (US 61/487,517; US 61/901,151) with J.A.D, A.M.H and M.R.M.vdB listed as inventors.
LITERATURE CITED
- 1.Dumoutier L, Van Roost E, Ameye G, Michaux L, Renauld JC. IL-TIF/IL-22: genomic organization and mapping of the human and mouse genes. Genes Immun. 2000;1:488–494. doi: 10.1038/sj.gene.6363716. [DOI] [PubMed] [Google Scholar]
- 2.Dumoutier L, Van Roost E, Colau D, Renauld JC. Human interleukin-10-related T cell-derived inducible factor: molecular cloning and functional characterization as an hepatocyte-stimulating factor. Proc. Natl Acad. Sci. USA. 2000;97:10144–10149. doi: 10.1073/pnas.170291697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie MH. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2–4 and IL-22R. J. Biol. Chem. 2000;275:31335–31339. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]
- 4.Nagem RA. Crystal structure of recombinant human interleukin-22. Structure. 2002;10:1051–1062. doi: 10.1016/s0969-2126(02)00797-9. [DOI] [PubMed] [Google Scholar]
- 5.de Oliveira Neto M. Interleukin-22 forms dimers that are recognized by two interleukin-22R1 receptor chains. Biophys. J. 2008;94:1754–1765. doi: 10.1529/biophysj.107.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumoutier L, Louahed J, Renauld JC. Cloning and characterization of IL-10-related T cell-derived inducible factor (IL-TIF), a novel cytokine structurally related to IL-10 and inducible by IL-9. J. Immunol. 2000;164:1814–1819. doi: 10.4049/jimmunol.164.4.1814. [DOI] [PubMed] [Google Scholar]
- 7.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov. 2014;13:21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 8.Xu T, Logsdon NJ, Walter MR. Structure of insect-cell-derived IL-22. Acta Crystallogr D Biol Crystallogr. 2005;61:942–950. doi: 10.1107/S0907444905009601. [DOI] [PubMed] [Google Scholar]
- 9.Xu T, Logsdon NJ, Walter MR. Crystallization and X-ray diffraction analysis of insect-cell-derived IL-22. Acta Crystallogr D Biol Crystallogr. 2004;60:1295–1298. doi: 10.1107/S0907444904010492. [DOI] [PubMed] [Google Scholar]
- 10.Nagem RA, Ferreira Junior JR, Dumoutier L, Renauld JC, Polikarpov I. Interleukin-22 and its crystal structure. Vitam Horm. 2006;74:77–103. doi: 10.1016/S0083-6729(06)74004-3. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Tomkinson KN, Tan XY, Wu P, Yan G, Spaulding V, Deng B, Annis-Freeman B, Heveron K, Zollner R, De Zutter G, Wright JF, Crawford TK, Liu W, Jacobs KA, Wolfman NM, Ling V, Pittman DD, Veldman GM, Fouser LA. Temporal associations between interleukin 22 and the extracellular domains of IL-22R and IL-10R2. Int Immunopharmacol. 2004;4:693–708. doi: 10.1016/j.intimp.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J. 1997;16:5894–5903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotenko SV. Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10Rbeta]) is a common chain of both the IL-10 and IL-22 (IL-10-related T cell-derived inducible factor, IL-TIF) receptor complexes. J. Biol. Chem. 2001;276:2725–2732. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- 14.Jones BC, Logsdon NJ, Walter MR. Structure of IL-22 bound to its high-affinity IL-22R1 chain. Structure. 2008;16:1333–1344. doi: 10.1016/j.str.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Logsdon NJ. The IL-10R2 binding hot spot on IL-22 is located on the N-terminal helix and is dependent on N-linked glycosylation. J. Mol. Biol. 2004;342:503–514. doi: 10.1016/j.jmb.2004.07.069. [DOI] [PubMed] [Google Scholar]
- 16.Yoon SI. Structure and mechanism of receptor sharing by the IL-10R2 common chain. Structure. 2010;18:638–648. doi: 10.1016/j.str.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolk K. Is there an interaction between interleukin-10 and interleukin-22? Genes Immun. 2005;6:8–18. doi: 10.1038/sj.gene.6364144. [DOI] [PubMed] [Google Scholar]
- 18.Logsdon NJ, Jones BC, Josephson K, Cook J, Walter MR. Comparison of interleukin-22 and interleukin-10 soluble receptor complexes. J. Interferon Cytokine Res. 2002;22:1099–1112. doi: 10.1089/10799900260442520. [DOI] [PubMed] [Google Scholar]
- 19.Bleicher L. Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism. FEBS Lett. 2008;582:2985–2992. doi: 10.1016/j.febslet.2008.07.046. [DOI] [PubMed] [Google Scholar]
- 20.Reineke U, Schneider-Mergener J, Glaser RW, Stigler RD, Seifert M, Volk HD, Sabat R. Evidence for conformationally different states of interleukin-10: binding of a neutralizing antibody enhances accessibility of a hidden epitope. J Mol Recognit. 1999;12:242–248. doi: 10.1002/(SICI)1099-1352(199907/08)12:4<242::AID-JMR461>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 21.Weathington NM, Snavely CA, Chen BB, Zhao J, Zhao Y, Mallampalli RK. Glycogen Synthase Kinase-3β Stabilizes the Interleukin (IL)-22 Receptor from Proteasomal Degradation in Murine Lung Epithelia. Journal of Biological Chemistry. 2014;289:17610–17619. doi: 10.1074/jbc.M114.551747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lejeune D, Dumoutier L, Constantinescu S, Kruijer W, Schuringa JJ, Renauld JC. Interleukin-22 (IL-22) activates the JAK/STAT, ERK, JNK, and p38 MAP kinase pathways in a rat hepatoma cell line. Pathways that are shared with and distinct from IL-10. J Biol Chem. 2002;277:33676–33682. doi: 10.1074/jbc.M204204200. [DOI] [PubMed] [Google Scholar]
- 23.Wolk K. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Spencer SD, Di Marco F, Hooley J, Pitts-Meek S, Bauer M, Ryan AM, Sordat B, Gibbs VC, Aguet M. The orphan receptor CRF2–4 is an essential subunit of the interleukin 10 receptor. J Exp Med. 1998;187:571–578. doi: 10.1084/jem.187.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andoh A. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 2005;129:969–984. doi: 10.1053/j.gastro.2005.06.071. [DOI] [PubMed] [Google Scholar]
- 26.Ikeuchi H, Kuroiwa T, Hiramatsu N, Kaneko Y, Hiromura K, Ueki K, Nojima Y. Expression of interleukin-22 in rheumatoid arthritis: Potential role as a proinflammatory cytokine. Arthritis & Rheumatism. 2005;52:1037–1046. doi: 10.1002/art.20965. [DOI] [PubMed] [Google Scholar]
- 27.Mitra A, Raychaudhuri SK, Raychaudhuri SP. IL-22 induced cell proliferation is regulated by PI3K/Akt/mTOR signaling cascade. Cytokine. 2012;60:38–42. doi: 10.1016/j.cyto.2012.06.316. [DOI] [PubMed] [Google Scholar]
- 28.Wolk K. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur. J. Immunol. 2006;36:1309–1323. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- 29.Wolk K, Kunz S, Asadullah K, Sabat R. Cutting edge: immune cells as sources and targets of the IL-10 family members? Journal of Immunology. 2002;168:5397–5402. doi: 10.4049/jimmunol.168.11.5397. [DOI] [PubMed] [Google Scholar]
- 30.Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol. 2009;10:857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- 31.Chung Y. Expression and regulation of IL-22 in the IL-17-producing CD4+ T lymphocytes. Cell Res. 2006;16:902–907. doi: 10.1038/sj.cr.7310106. [DOI] [PubMed] [Google Scholar]
- 32.Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, Basham B, Smith K, Chen T, Morel F, Lecron JC, Kastelein RA, Cua DJ, McClanahan TK, Bowman EP, de Waal Malefyt R. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8:950–957. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 33.Basu R, O Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W, Hatton RD, Weaver CT. Th22 Cells Are an Important Source of IL-22 for Host Protection against Enteropathogenic Bacteria. Immunity. 2012;37:1061–1075. doi: 10.1016/j.immuni.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang SC, Tan X-Y, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. The Journal of Experimental Medicine. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 36.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 37.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 39.Morishima N, Mizoguchi I, Takeda K, Mizuguchi J, Yoshimoto T. TGF-beta is necessary for induction of IL-23R and Th17 differentiation by IL-6 and IL-23. Biochem Biophys Res Commun. 2009;386:105–110. doi: 10.1016/j.bbrc.2009.05.140. [DOI] [PubMed] [Google Scholar]
- 40.Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 41.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 42.Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, Husain S, Kreindler JL, Dubin PJ, Pilewski JM, Myerburg MM, Mason CA, Iwakura Y, Kolls JK. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munoz M. Interleukin (IL)-23 mediates Toxoplasma gondii-induced immunopathology in the gut via matrixmetalloproteinase-2 and IL-22 but independent of IL-17. J. Exp. Med. 2009;206:3047–3059. doi: 10.1084/jem.20090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, Ouyang W. Interleukin-22, a TH17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 45.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 46.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, Ziegler SF, Littman DR. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Penel-Sotirakis K, Simonazzi E, Peguet-Navarro J, Rozieres A. Differential capacity of human skin dendritic cells to polarize CD4+ T cells into IL-17, IL-21 and IL-22 producing cells. PLoS One. 2012;7:e45680. doi: 10.1371/journal.pone.0045680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rutz S, Noubade R, Eidenschenk C, Ota N, Zeng W, Zheng Y, Hackney J, Ding J, Singh H, Ouyang W. Transcription factor c-Maf mediates the TGF-beta-dependent suppression of IL-22 production in T(H)17 cells. Nat Immunol. 2011;12:1238–1245. doi: 10.1038/ni.2134. [DOI] [PubMed] [Google Scholar]
- 49.Hamada H, Garcia-Hernandez Mde L, Reome JB, Misra SK, Strutt TM, McKinstry KK, Cooper AM, Swain SL, Dutton RW. Tc17, a unique subset of CD8 T cells that can protect against lethal influenza challenge. J Immunol. 2009;182:3469–3481. doi: 10.4049/jimmunol.0801814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kondo T, Takata H, Matsuki F, Takiguchi M. Cutting edge: Phenotypic characterization and differentiation of human CD8+ T cells producing IL-17. J Immunol. 2009;182:1794–1798. doi: 10.4049/jimmunol.0801347. [DOI] [PubMed] [Google Scholar]
- 51.Ciric B, El-behi M, Cabrera R, Zhang GX, Rostami A. IL-23 drives pathogenic IL-17-producing CD8+ T cells. J Immunol. 2009;182:5296–5305. doi: 10.4049/jimmunol.0900036. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Yang B, Ma J, Wang H, Huang F, Zhang J, Chen H, Wu C. Interleukin-21 induces the differentiation of human Tc22 cells via phosphorylation of signal transducers and activators of transcription. Immunology. 2011;132:540–548. doi: 10.1111/j.1365-2567.2010.03399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nograles KE. IL-22-producing "T22" T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J. Allergy Clin. Immunol. 2009;123:1244–1252. doi: 10.1016/j.jaci.2009.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin B, Hirota K, Cua DJ, Stockinger B, Veldhoen M. Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31:321–330. doi: 10.1016/j.immuni.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 55.Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KH. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–341. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Mabuchi T, Takekoshi T, Hwang ST. Epidermal CCR6+ gammadelta T cells are major producers of IL-22 and IL-17 in a murine model of psoriasiform dermatitis. J Immunol. 2011;187:5026–5031. doi: 10.4049/jimmunol.1101817. [DOI] [PubMed] [Google Scholar]
- 57.Mielke LA, Jones SA, Raverdeau M, Higgs R, Stefanska A, Groom JR, Misiak A, Dungan LS, Sutton CE, Streubel G, Bracken AP, Mills KHG. Retinoic acid expression associates with enhanced IL-22 production by γδ T cells and innate lymphoid cells and attenuation of intestinal inflammation. The Journal of Experimental Medicine. 2013;210:1117–1124. doi: 10.1084/jem.20121588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crellin NK, Trifari S, Kaplan CD, Satoh-Takayama N, Di Santo JP, Spits H. Regulation of Cytokine Secretion in Human CD127+ LTi-like Innate Lymphoid Cells by Toll-like Receptor 2. Immunity. 2010;33:752–764. doi: 10.1016/j.immuni.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 59.Ciofani M, Zuniga-Pflucker JC. Determining gammadelta versus alphass T cell development. Nat Rev Immunol. 2010;10:657–663. doi: 10.1038/nri2820. [DOI] [PubMed] [Google Scholar]
- 60.Jensen KD, Su X, Shin S, Li L, Youssef S, Yamasaki S, Steinman L, Saito T, Locksley RM, Davis MM, Baumgarth N, Chien YH. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity. 2008;29:90–100. doi: 10.1016/j.immuni.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ, Girardi M, Borst J, Hayday AC, Pennington DJ, Silva-Santos B. CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nat Immunol. 2009;10:427–436. doi: 10.1038/ni.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haas JD, Gonzalez FH, Schmitz S, Chennupati V, Fohse L, Kremmer E, Forster R, Prinz I. CCR6 and NK1.1 distinguish between IL-17A and IFN-gamma-producing gammadelta effector T cells. Eur J Immunol. 2009;39:3488–3497. doi: 10.1002/eji.200939922. [DOI] [PubMed] [Google Scholar]
- 63.Shibata K, Yamada H, Nakamura R, Sun X, Itsumi M, Yoshikai Y. Identification of CD25+ gamma delta T cells as fetal thymus-derived naturally occurring IL-17 producers. J Immunol. 2008;181:5940–5947. doi: 10.4049/jimmunol.181.9.5940. [DOI] [PubMed] [Google Scholar]
- 64.Spits H, Cupedo T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu Rev Immunol. 2012;30:647–675. doi: 10.1146/annurev-immunol-020711-075053. [DOI] [PubMed] [Google Scholar]
- 65.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, Powrie F, Vivier E. Innate lymphoid cells - a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 66.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, Doherty JM, Mills JC, Colonna M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, Chasson L, Hardwigsen J, Anguiano E, Banchereau J, Chaussabel D, Dalod M, Littman DR, Vivier E, Tomasello E. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 68.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, Diefenbach A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Satoh-Takayama N. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 70.Cella M, Otero K, Colonna M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc Natl Acad Sci U S A. 2010;107:10961–10966. doi: 10.1073/pnas.1005641107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Colonna M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 2009;31:15–23. doi: 10.1016/j.immuni.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 72.Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, Flach M, Bengsch B, Thimme R, Holscher C, Honig M, Pannicke U, Schwarz K, Ware CF, Finke D, Diefenbach A. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Satoh-Takayama N, Lesjean-Pottier S, Vieira P, Sawa S, Eberl G, Vosshenrich CAJ, Di Santo JP. IL-7 and IL-15 independently program the differentiation of intestinal CD3−NKp46+ cell subsets from Id2-dependent precursors. J Exp Med. 2010;207:273–280. doi: 10.1084/jem.20092029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crellin NK, Trifari S, Kaplan CD, Cupedo T, Spits H. Human NKp44+IL-22+ cells and LTi-like cells constitute a stable RORC+ lineage distinct from conventional natural killer cells. J Exp Med. 2010;207:281–290. doi: 10.1084/jem.20091509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rankin L, Groom J, Mielke LA, Seillet C, Belz GT. Diversity, function, and transcriptional regulation of gut innate lymphocytes. Front Immunol. 2013;4:22. doi: 10.3389/fimmu.2013.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, Langa F, Di Santo JP, Eberl G. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 77.Kelly KA, Scollay R. Seeding of neonatal lymph nodes by T cells and identification of a novel population of CD3−CD4+ cells. Eur J Immunol. 1992;22:329–334. doi: 10.1002/eji.1830220207. [DOI] [PubMed] [Google Scholar]
- 78.Mebius RE, Rennert P, Weissman IL. Developing lymph nodes collect CD4+CD3− LTbeta+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 1997;7:493–504. doi: 10.1016/s1074-7613(00)80371-4. [DOI] [PubMed] [Google Scholar]
- 79.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 80.Eberl G, Littman DR. The role of the nuclear hormone receptor RORgamma(t) in the development of lymph nodes and Peyer's patches. Immunological Reviews. 2003;195:81–90. doi: 10.1034/j.1600-065x.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- 81.Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nature Immunology. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 82.Cupedo T, Jansen W, Kraal G, Mebius RE. Induction of secondary and tertiary lymphoid structures in the skin. Immunity. 2004;21:655–667. doi: 10.1016/j.immuni.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 83.Finke D. Fate and function of lymphoid tissue inducer cells. Curr Opin Immunol. 2005;17:144–150. doi: 10.1016/j.coi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 84.Withers DR, Kim MY, Bekiaris V, Rossi SW, Jenkinson WE, Gaspal F, McConnell F, Caamano JH, Anderson G, Lane PJ. The role of lymphoid tissue inducer cells in splenic white pulp development. Eur J Immunol. 2007;37:3240–3245. doi: 10.1002/eji.200737541. [DOI] [PubMed] [Google Scholar]
- 85.Scandella E, Bolinger B, Lattmann E, Miller S, Favre S, Littman DR, Finke D, Luther SA, Junt T, Ludewig B. Restoration of lymphoid organ integrity through the interaction of lymphoid tissue-inducer cells with stroma of the T cell zone. Nature Immunology. 2008;9:667–675. doi: 10.1038/ni.1605. [DOI] [PubMed] [Google Scholar]
- 86.Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, Ivanov II, Itoh K, Littman DR, Fagarasan S. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 2008;29:261–271. doi: 10.1016/j.immuni.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 87.Rossi SW, Kim MY, Leibbrandt A, Parnell SM, Jenkinson WE, Glanville SH, McConnell FM, Scott HS, Penninger JM, Jenkinson EJ, Lane PJ, Anderson G. RANK signals from CD4(+)3(−) inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J Exp Med. 2007;204:1267–1272. doi: 10.1084/jem.20062497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoshida H, Honda K, Shinkura R, Adachi S, Nishikawa S, Maki K, Ikuta K, Nishikawa SI. IL-7 receptor alpha+ CD3(−) cells in the embryonic intestine induces the organizing center of Peyer's patches. Int Immunol. 1999;11:643–655. doi: 10.1093/intimm/11.5.643. [DOI] [PubMed] [Google Scholar]
- 89.van de Pavert SA, Olivier BJ, Goverse G, Vondenhoff MF, Greuter M, Beke P, Kusser K, Hopken UE, Lipp M, Niederreither K, Blomhoff R, Sitnik K, Agace WW, Randall TD, de Jonge WJ, Mebius RE. Chemokine CXCL13 is essential for lymph node initiation and is induced by retinoic acid and neuronal stimulation. Nat Immunol. 2009;10:1193–1199. doi: 10.1038/ni.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takatori H, Kanno Y, Watford WT, Tato CM, Weiss G, Ivanov II, Littman DR, O'Shea JJ. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cupedo T, Crellin NK, Papazian N, Rombouts EJ, Weijer K, Grogan JL, Fibbe WE, Cornelissen JJ, Spits H. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10:66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 92.Hazenberg MD, Spits H. Human innate lymphoid cells. Blood. 2014 doi: 10.1182/blood-2013-11-427781. [DOI] [PubMed] [Google Scholar]
- 93.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, Powrie F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sciume G, Hirahara K, Takahashi H, Laurence A, Villarino AV, Singleton KL, Spencer SP, Wilhelm C, Poholek AC, Vahedi G, Kanno Y, Belkaid Y, O'Shea JJ. Distinct requirements for T-bet in gut innate lymphoid cells. J Exp Med. 2012;209:2331–2338. doi: 10.1084/jem.20122097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, Mielke LA, McKenzie ANJ, Carotta S, Nutt SL, Belz GT. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d'Hargues Y, Goppert N, Croxford AL, Waisman A, Tanriver Y, Diefenbach A. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 97.Hughes T, Briercheck EL, Freud AG, Trotta R, McClory S, Scoville SD, Keller K, Deng Y, Cole J, Harrison N, Mao C, Zhang J, Benson DM, Yu J, Caligiuri MA. The Transcription Factor AHR Prevents the Differentiation of a Stage 3 Innate Lymphoid Cell Subset to Natural Killer Cells. Cell Rep. 2014;8:150–162. doi: 10.1016/j.celrep.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Possot C, Schmutz S, Chea S, Boucontet L, Louise A, Cumano A, Golub R. Notch signaling is necessary for adult, but not fetal, development of RORgammat(+) innate lymphoid cells. Nat Immunol. 2011;12:949–958. doi: 10.1038/ni.2105. [DOI] [PubMed] [Google Scholar]
- 99.Mebius RE, Miyamoto T, Christensen J, Domen J, Cupedo T, Weissman IL, Akashi K. The fetal liver counterpart of adult common lymphoid progenitors gives rise to all lymphoid lineages, CD45+CD4+CD3− cells, as well as macrophages. J Immunol. 2001;166:6593–6601. doi: 10.4049/jimmunol.166.11.6593. [DOI] [PubMed] [Google Scholar]
- 100.Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 101.Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, Diefenbach A. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Klose Christoph SN, Flach M, Möhle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, Domingues Rita G, Veiga-Fernandes H, Arnold Sebastian J, Busslinger M, Dunay Ildiko R, Tanriver Y, Diefenbach A. Differentiation of Type 1 ILCs from a Common Progenitor to All Helper-like Innate Lymphoid Cell Lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 103.Welner RS, Pelayo R, Kincade PW. Evolving views on the genealogy of B cells. Nat Rev Immunol. 2008;8:95–106. doi: 10.1038/nri2234. [DOI] [PubMed] [Google Scholar]
- 104.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 105.Nechanitzky R, Akbas D, Scherer S, Gyory I, Hoyler T, Ramamoorthy S, Diefenbach A, Grosschedl R. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol. 2013;14:867–875. doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- 106.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 108.Cherrier M, Sawa S, Eberl Gr. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. The Journal of Experimental Medicine. 2012;209:729–740. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014 doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, Geary CD, O’Sullivan TE, van den Brink MR, Pamer EG, Hanash AM, Sun JC. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. The Journal of Experimental Medicine. 2014 doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, Huntington ND, Belz GT, Carotta S. Nfil3 is required for the development of all innate lymphoid cell subsets. The Journal of Experimental Medicine. 2014 doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Male V, Nisoli I, Kostrzewski T, Allan DSJ, Carlyle JR, Lord GM, Wack A, Brady HJM. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. The Journal of Experimental Medicine. 2014 doi: 10.1084/jem.20132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klein Wolterink RG, Serafini N, van Nimwegen M, Vosshenrich CA, de Bruijn MJ, Fonseca Pereira D, Veiga Fernandes H, Hendriks RW, Di Santo JP. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc Natl Acad Sci U S A. 2013;110:10240–10245. doi: 10.1073/pnas.1217158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mjosberg J, Bernink J, Golebski K, Karrich JJ, Peters CP, Blom B, te Velde AA, Fokkens WJ, van Drunen CM, Spits H. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 2012;37:649–659. doi: 10.1016/j.immuni.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 116.Serafini N, Klein Wolterink RG, Satoh-Takayama N, Xu W, Vosshenrich CA, Hendriks RW, Di Santo JP. Gata3 drives development of RORgammat+ group 3 innate lymphoid cells. J Exp Med. 2014;211:199–208. doi: 10.1084/jem.20131038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goto M, Murakawa M, Kadoshima-Yamaoka K, Tanaka Y, Nagahira K, Fukuda Y, Nishimura T. Murine NKT cells produce Th17 cytokine interleukin-22. Cell Immunol. 2009;254:81–84. doi: 10.1016/j.cellimm.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 118.Juno JA, Keynan Y, Fowke KR. Invariant NKT cells: regulation and function during viral infection. PLoS Pathog. 2012;8:e1002838. doi: 10.1371/journal.ppat.1002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moreira-Teixeira L, Resende M, Coffre M, Devergne O, Herbeuval JP, Hermine O, Schneider E, Rogge L, Ruemmele FM, Dy M, Cordeiro-da-Silva A, Leite-de-Moraes MC. Proinflammatory environment dictates the IL-17-producing capacity of human invariant NKT cells. J Immunol. 2011;186:5758–5765. doi: 10.4049/jimmunol.1003043. [DOI] [PubMed] [Google Scholar]
- 120.Paget C, Ivanov S, Fontaine J, Renneson J, Blanc F, Pichavant M, Dumoutier L, Ryffel B, Renauld JC, Gosset P, Gosset P, Si-Tahar M, Faveeuw C, Trottein F. Interleukin-22 is produced by invariant natural killer T lymphocytes during influenza A virus infection: potential role in protection against lung epithelial damages. J Biol Chem. 2012;287:8816–8829. doi: 10.1074/jbc.M111.304758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 122.Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, Berzins SP, Smyth MJ, Godfrey DI. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1− NKT cell population. Proc Natl Acad Sci U S A. 2008;105:11287–11292. doi: 10.1073/pnas.0801631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Doisne JM, Becourt C, Amniai L, Duarte N, Le Luduec JB, Eberl G, Benlagha K. Skin and peripheral lymph node invariant NKT cells are mainly retinoic acid receptor-related orphan receptor (gamma)t+ and respond preferentially under inflammatory conditions. J Immunol. 2009;183:2142–2149. doi: 10.4049/jimmunol.0901059. [DOI] [PubMed] [Google Scholar]
- 124.Michel ML, Mendes-da-Cruz D, Keller AC, Lochner M, Schneider E, Dy M, Eberl G, Leite-de-Moraes MC. Critical role of ROR-gammat in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci U S A. 2008;105:19845–19850. doi: 10.1073/pnas.0806472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Doisne JM, Soulard V, Becourt C, Amniai L, Henrot P, Havenar-Daughton C, Blanchet C, Zitvogel L, Ryffel B, Cavaillon JM, Marie JC, Couillin I, Benlagha K. Cutting edge: crucial role of IL-1 and IL-23 in the innate IL-17 response of peripheral lymph node NK1.1− invariant NKT cells to bacteria. J Immunol. 2011;186:662–666. doi: 10.4049/jimmunol.1002725. [DOI] [PubMed] [Google Scholar]
- 126.Rachitskaya AV, Hansen AM, Horai R, Li Z, Villasmil R, Luger D, Nussenblatt RB, Caspi RR. Cutting edge: NKT cells constitutively express IL-23 receptor and RORgammat and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J Immunol. 2008;180:5167–5171. doi: 10.4049/jimmunol.180.8.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Raifer H, Mahiny AJ, Bollig N, Petermann F, Hellhund A, Kellner K, Guralnik A, Reinhard K, Bothur E, Huber M, Bauer S, Lohning M, Kiss EA, Ganal SC, Diefenbach A, Korn T, Lohoff M. Unlike alphabeta T cells, gammadelta T cells, LTi cells and NKT cells do not require IRF4 for the production of IL-17A and IL-22. Eur J Immunol. 2012;42:3189–3201. doi: 10.1002/eji.201142155. [DOI] [PubMed] [Google Scholar]
- 128.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 129.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. J Exp Med. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Webster KE, Kim HO, Kyparissoudis K, Corpuz TM, Pinget GV, Uldrich AP, Brink R, Belz GT, Cho JH, Godfrey DI, Sprent J. IL-17-producing NKT cells depend exclusively on IL-7 for homeostasis and survival. Mucosal Immunol. 2014 doi: 10.1038/mi.2013.122. [DOI] [PubMed] [Google Scholar]
- 131.Hansson M, Silverpil E, Linden A, Glader P. Interleukin-22 produced by alveolar macrophages during activation of the innate immune response. Inflamm Res. 2013;62:561–569. doi: 10.1007/s00011-013-0608-1. [DOI] [PubMed] [Google Scholar]
- 132.Zindl CL, Lai JF, Lee YK, Maynard CL, Harbour SN, Ouyang W, Chaplin DD, Weaver CT. IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc Natl Acad Sci U S A. 2013;110:12768–12773. doi: 10.1073/pnas.1300318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kastelein RA, Hunter CA, Cua DJ. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annu Rev Immunol. 2007;25:221–242. doi: 10.1146/annurev.immunol.22.012703.104758. [DOI] [PubMed] [Google Scholar]
- 134.Hunter CA. New IL-12-family members: IL-23 and IL-27, cytokines with divergent functions. Nat Rev Immunol. 2005;5:521–531. doi: 10.1038/nri1648. [DOI] [PubMed] [Google Scholar]
- 135.Gerosa F, Baldani-Guerra B, Lyakh LA, Batoni G, Esin S, Winkler-Pickett RT, Consolaro MR, De Marchi M, Giachino D, Robbiano A, Astegiano M, Sambataro A, Kastelein RA, Carra G, Trinchieri G. Differential regulation of interleukin 12 and interleukin 23 production in human dendritic cells. J Exp Med. 2008;205:1447–1461. doi: 10.1084/jem.20071450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4+ Lymphoid Tissue-Inducer Cells Promote Innate Immunity in the Gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hanash AM, Dudakov JA, Hua G, OȉConnor MH, Young LF, Singer NV, West ML, Jenq RR, Holland AM, Kappel LW, Ghosh A, Tsai JJ, Rao UK, Yim NL, Smith OM, Velardi E, Hawryluk EB, Murphy GF, Liu C, Fouser LA, Kolesnick R, Blazar BR, van den Brink MRM. Interleukin-22 Protects Intestinal Stem Cells from Immune-Mediated Tissue Damage and Regulates Sensitivity to Graft versus Host Disease. Immunity. 2012;37:339–350. doi: 10.1016/j.immuni.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dudakov JA, Hanash AM, Jenq RR, Young LF, Ghosh A, Singer NV, West ML, Smith OM, Holland AM, Tsai JJ, Boyd RL, van den Brink MRM. Interleukin-22 Drives Endogenous Thymic Regeneration in Mice. Science. 2012;336:91–95. doi: 10.1126/science.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, Flavell RA, Littman DR, Pamer EG. Interleukin 23 Production by Intestinal CD103+CD11b+ Dendritic Cells in Response to Bacterial Flagellin Enhances Mucosal Innate Immune Defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Satpathy AT, Briseno CG, Lee JS, Ng D, Manieri NA, Kc W, Wu X, Thomas SR, Lee W-L, Turkoz M, McDonald KG, Meredith MM, Song C, Guidos CJ, Newberry RD, Ouyang W, Murphy TL, Stappenbeck TS, Gommerman JL, Nussenzweig MC, Colonna M, Kopan R, Murphy KM. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Siddiqui KR, Laffont S, Powrie F. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity. 2010;32:557–567. doi: 10.1016/j.immuni.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Manta C, Heupel E, Radulovic K, Rossini V, Garbi N, Riedel CU, Niess JH. CX(3)CR1(+) macrophages support IL-22 production by innate lymphoid cells during infection with Citrobacter rodentium. Mucosal Immunol. 2013;6:177–188. doi: 10.1038/mi.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Niess JH, Adler G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. J Immunol. 2010;184:2026–2037. doi: 10.4049/jimmunol.0901936. [DOI] [PubMed] [Google Scholar]
- 144.Longman RS, Diehl GE, Victorio DA, Huh JR, Galan C, Miraldi ER, Swaminath A, Bonneau R, Scherl EJ, Littman DR. CX3CR1+ mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J Exp Med. 2014 doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Roses RE, Xu S, Xu M, Koldovsky U, Koski G, Czerniecki BJ. Differential production of IL-23 and IL-12 by myeloid-derived dendritic cells in response to TLR agonists. J Immunol. 2008;181:5120–5127. doi: 10.4049/jimmunol.181.7.5120. [DOI] [PubMed] [Google Scholar]
- 146.Paustian C, Taylor P, Johnson T, Xu M, Ramirez N, Rosenthal KS, Shu S, Cohen PA, Czerniecki BJ, Koski GK. Extracellular ATP and Toll-like receptor 2 agonists trigger in human monocytes an activation program that favors T helper 17. PLoS One. 2013;8:e54804. doi: 10.1371/journal.pone.0054804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Peral de Castro C, Jones SA, Ní Cheallaigh Co, Hearnden CA, Williams L, Winter J, Lavelle EC, Mills KHG, Harris J. Autophagy Regulates IL-23 Secretion and Innate T Cell Responses through Effects on IL-1 Secretion. The Journal of Immunology. 2012;189:4144–4153. doi: 10.4049/jimmunol.1201946. [DOI] [PubMed] [Google Scholar]
- 148.Flutter B, Nestle FO. TLRs to cytokines: mechanistic insights from the imiquimod mouse model of psoriasis. Eur J Immunol. 2013;43:3138–3146. doi: 10.1002/eji.201343801. [DOI] [PubMed] [Google Scholar]
- 149.van der Fits L, Mourits S, Voerman JS, Kant M, Boon L, Laman JD, Cornelissen F, Mus AM, Florencia E, Prens EP, Lubberts E. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J Immunol. 2009;182:5836–5845. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- 150.Lyakh L, Trinchieri G, Provezza L, Carra G, Gerosa F. Regulation of interleukin-12/interleukin-23 production and the T-helper 17 response in humans. Immunological Reviews. 2008;226:112–131. doi: 10.1111/j.1600-065X.2008.00700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Brain O, Owens BMJ, Pichulik T, Allan P, Khatamzas E, Leslie A, Steevels T, Sharma S, Mayer A, Catuneanu AM, Morton V, Sun M-Y, Jewell D, Coccia M, Harrison O, Maloy K, Schönefeldt S, Bornschein S, Liston A, Simmons A. The Intracellular Sensor NOD2 Induces MicroRNA-29 Expression in Human Dendritic Cells to Limit IL-23 Release. Immunity. 2013;39:521–536. doi: 10.1016/j.immuni.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 152.Xue X, Feng T, Yao S, Wolf KJ, Liu CG, Liu X, Elson CO, Cong Y. Microbiota downregulates dendritic cell expression of miR-10a, which targets IL-12/IL-23p40. J Immunol. 2011;187:5879–5886. doi: 10.4049/jimmunol.1100535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sender LY, Gibbert K, Suezer Y, Radeke HH, Kalinke U, Waibler Z. CD40 ligand-triggered human dendritic cells mount interleukin-23 responses that are further enhanced by danger signals. Mol Immunol. 2010;47:1255–1261. doi: 10.1016/j.molimm.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 154.Zheng M, Rapaka RR, Yu AC, Shellito JE, Kolls JK. Role of interleukin-23-dependent antifungal immune responses in dendritic cell-vaccinated mice. Infect Immun. 2011;79:3778–3783. doi: 10.1128/IAI.05163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ma X, Chow JM, Gri G, Carra G, Gerosa F, Wolf SF, Dzialo R, Trinchieri G. The interleukin 12 p40 gene promoter is primed by interferon gamma in monocytic cells. J Exp Med. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Qian X, Ning H, Zhang J, Hoft DF, Stumpo DJ, Blackshear PJ, Liu J. Posttranscriptional regulation of IL-23 expression by IFN-gamma through tristetraprolin. J Immunol. 2011;186:6454–6464. doi: 10.4049/jimmunol.1002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zakharova M, Ziegler HK. Paradoxical anti-inflammatory actions of TNF-alpha: inhibition of IL-12 and IL-23 via TNF receptor 1 in macrophages and dendritic cells. J Immunol. 2005;175:5024–5033. doi: 10.4049/jimmunol.175.8.5024. [DOI] [PubMed] [Google Scholar]
- 158.Riol-Blanco L, Ordovas-Montanes J, Perro M, Naval E, Thiriot A, Alvarez D, Wood JN, von Andrian UH. Nociceptive sensory neurons drive interleukin-23-mediated psoriasiform skin inflammation. Nature. 2014 doi: 10.1038/nature13199. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vassiliou E, Sharma V, Jing H, Sheibanie F, Ganea D. Prostaglandin E2 promotes the survival of bone marrow-derived dendritic cells. J Immunol. 2004;173:6955–6964. doi: 10.4049/jimmunol.173.11.6955. [DOI] [PubMed] [Google Scholar]
- 160.Schirmer C, Klein C, von Bergen M, Simon JC, Saalbach A. Human fibroblasts support the expansion of IL-17-producing T cells via up-regulation of IL-23 production by dendritic cells. Blood. 2010;116:1715–1725. doi: 10.1182/blood-2010-01-263509. [DOI] [PubMed] [Google Scholar]
- 161.Sheibanie AF, Tadmori I, Jing H, Vassiliou E, Ganea D. Prostaglandin E2 induces IL-23 production in bone marrow-derived dendritic cells. Faseb J. 2004;18:1318–1320. doi: 10.1096/fj.03-1367fje. [DOI] [PubMed] [Google Scholar]
- 162.Schnurr M, Toy T, Shin A, Wagner M, Cebon J, Maraskovsky E. Extracellular nucleotide signaling by P2 receptors inhibits IL-12 and enhances IL-23 expression in human dendritic cells: a novel role for the cAMP pathway. Blood. 2005;105:1582–1589. doi: 10.1182/blood-2004-05-1718. [DOI] [PubMed] [Google Scholar]
- 163.Satoh-Takayama N, Lesjean-Pottier S, Sawa S, Vosshenrich CAJ, Eberl G, Di Santo JP. Lymphotoxin-β receptor-independent development of intestinal IL-22-producing NKp46+ innate lymphoid cells. European Journal of Immunology. 2011;41:780–786. doi: 10.1002/eji.201040851. [DOI] [PubMed] [Google Scholar]
- 164.Ota N, Wong K, Valdez PA, Zheng Y, Crellin NK, Diehl L, Ouyang W. IL-22 bridges the lymphotoxin pathway with the maintenance of colonic lymphoid structures during infection with Citrobacter rodentium. Nat Immunol. 2011;12:941–948. doi: 10.1038/ni.2089. [DOI] [PubMed] [Google Scholar]
- 165.Tumanov AV, Koroleva EP, Guo X, Wang Y, Kruglov A, Nedospasov S, Fu Y-X. Lymphotoxin Controls the IL-22 Protection Pathway in Gut Innate Lymphoid Cells during Mucosal Pathogen Challenge. Cell host & microbe. 2011;10:44–53. doi: 10.1016/j.chom.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Upadhyay V, Poroyko V, Kim TJ, Devkota S, Fu S, Liu D, Tumanov AV, Koroleva EP, Deng L, Nagler C, Chang EB, Tang H, Fu YX. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nat Immunol. 2012;13:947–953. doi: 10.1038/ni.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ware CF. Network communications: lymphotoxins, LIGHT, and TNF. Annu Rev Immunol. 2005;23:787–819. doi: 10.1146/annurev.immunol.23.021704.115719. [DOI] [PubMed] [Google Scholar]
- 168.Tumanov AV, Grivennikov SI, Shakhov AN, Rybtsov SA, Koroleva EP, Takeda J, Nedospasov SA, Kuprash DV. Dissecting the role of lymphotoxin in lymphoid organs by conditional targeting. Immunol Rev. 2003;195:106–116. doi: 10.1034/j.1600-065x.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- 169.Monteiro M, Almeida CF, Agua-Doce A, Graca L. Induced IL-17-producing invariant NKT cells require activation in presence of TGF-beta and IL-1beta. J Immunol. 2013;190:805–811. doi: 10.4049/jimmunol.1201010. [DOI] [PubMed] [Google Scholar]
- 170.Lee Y, Kumagai Y, Jang MS, Kim JH, Yang BG, Lee EJ, Kim YM, Akira S, Jang MH. Intestinal Lin− c-Kit+ NKp46− CD4− population strongly produces IL-22 upon IL-1beta stimulation. J Immunol. 2013;190:5296–5305. doi: 10.4049/jimmunol.1201452. [DOI] [PubMed] [Google Scholar]
- 171.Chen VL, Surana NK, Duan J, Kasper DL. Role of murine intestinal interleukin-1 receptor 1-expressing lymphoid tissue inducer-like cells in salmonella infection. PLoS One. 2013;8:e65405. doi: 10.1371/journal.pone.0065405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nat Rev Immunol. 2010;10:89–102. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- 173.Shaw MH, Kamada N, Kim YG, Nunez G. Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hughes T, Becknell B, Freud AG, McClory S, Briercheck E, Yu J, Mao C, Giovenzana C, Nuovo G, Wei L, Zhang X, Gavrilin MA, Wewers MD, Caligiuri MA. Interleukin-1[beta] Selectively Expands and Sustains Interleukin-22+ Immature Human Natural Killer Cells in Secondary Lymphoid Tissue. Immunity. 2010;32:803–814. doi: 10.1016/j.immuni.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Vonarbourg C, Diefenbach A. Multifaceted roles of interleukin-7 signaling for the development and function of innate lymphoid cells. Semin Immunol. 2012;24:165–174. doi: 10.1016/j.smim.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 176.von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, Meyer JD, Davison BL. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Adachi S, Yoshida H, Honda K, Maki K, Saijo K, Ikuta K, Saito T, Nishikawa SI. Essential role of IL-7 receptor alpha in the formation of Peyer's patch anlage. Int Immunol. 1998;10:1–6. doi: 10.1093/intimm/10.1.1. [DOI] [PubMed] [Google Scholar]
- 179.Luther SA, Ansel KM, Cyster JG. Overlapping roles of CXCL13, interleukin 7 receptor alpha, and CCR7 ligands in lymph node development. J Exp Med. 2003;197:1191–1198. doi: 10.1084/jem.20021294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chappaz S, Finke D. The IL-7 Signaling Pathway Regulates Lymph Node Development Independent of Peripheral Lymphocytes. Journal of Immunology. 2010;184:3562–3569. doi: 10.4049/jimmunol.0901647. [DOI] [PubMed] [Google Scholar]
- 181.Cao X, Shores EW, Hu-Li J, Anver MR, Kelsall BL, Russell SM, Drago J, Noguchi M, Grinberg A, Bloom ET, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 182.Park SY, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, Miyake K, Nakauchi H, Shirasawa T, Saito T. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 183.Meier D, Bornmann C, Chappaz S, Schmutz S, Otten LA, Ceredig R, Acha-Orbea H, Finke D. Ectopic lymphoid-organ development occurs through interleukin 7-mediated enhanced survival of lymphoid-tissue-inducer cells. Immunity. 2007;26:643–654. doi: 10.1016/j.immuni.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 184.De Togni P, Goellner J, Ruddle NH, Streeter PR, Fick A, Mariathasan S, Smith SC, Carlson R, Shornick LP, Strauss-Schoenberger J, et al. Abnormal development of peripheral lymphoid organs in mice deficient in lymphotoxin. Science. 1994;264:703–707. doi: 10.1126/science.8171322. [DOI] [PubMed] [Google Scholar]
- 185.Honda K, Nakano H, Yoshida H, Nishikawa S, Rennert P, Ikuta K, Tamechika M, Yamaguchi K, Fukumoto T, Chiba T, Nishikawa SI. Molecular basis for hematopoietic/mesenchymal interaction during initiation of Peyer's patch organogenesis. J Exp Med. 2001;193:621–630. doi: 10.1084/jem.193.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yoshida H, Naito A, Inoue J, Satoh M, Santee-Cooper SM, Ware CF, Togawa A, Nishikawa S, Nishikawa S. Different cytokines induce surface lymphotoxin-alphabeta on IL-7 receptor-alpha cells that differentially engender lymph nodes and Peyer's patches. Immunity. 2002;17:823–833. doi: 10.1016/s1074-7613(02)00479-x. [DOI] [PubMed] [Google Scholar]
- 187.Qiu J, Heller JJ, Guo X, Chen Z-mE, Fish K, Fu Y-X, Zhou L. The Aryl Hydrocarbon Receptor Regulates Gut Immunity through Modulation of Innate Lymphoid Cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Korn LL, Thomas HL, Hubbeling HG, Spencer SP, Sinha R, Simkins HM, Salzman NH, Bushman FD, Laufer TM. Conventional CD4+ T cells regulate IL-22-producing intestinal innate lymphoid cells. Mucosal Immunol. 2014 doi: 10.1038/mi.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Spencer SP, Wilhelm C, Yang Q, Hall JA, Bouladoux N, Boyd A, Nutman TB, Urban JF, Wang J, Ramalingam TR, Bhandoola A, Wynn TA, Belkaid Y. Adaptation of Innate Lymphoid Cells to a Micronutrient Deficiency Promotes Type 2 Barrier Immunity. Science. 2014;343:432–437. doi: 10.1126/science.1247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, Colonna M. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 192.Esser C, Rannug A, Stockinger B. The aryl hydrocarbon receptor in immunity. Trends Immunol. 2009;30:447–454. doi: 10.1016/j.it.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 193.Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 195.McMillan BJ, Bradfield CA. The aryl hydrocarbon receptor is activated by modified low-density lipoprotein. Proc Natl Acad Sci U S A. 2007;104:1412–1417. doi: 10.1073/pnas.0607296104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Oesch-Bartlomowicz B, Huelster A, Wiss O, Antoniou-Lipfert P, Dietrich C, Arand M, Weiss C, Bockamp E, Oesch F. Aryl hydrocarbon receptor activation by cAMP vs. dioxin: divergent signaling pathways. Proc Natl Acad Sci U S A. 2005;102:9218–9223. doi: 10.1073/pnas.0503488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Puga A, Ma C, Marlowe JL. The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways. Biochem Pharmacol. 2009;77:713–722. doi: 10.1016/j.bcp.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lowe MM, Mold JE, Kanwar B, Huang Y, Louie A, Pollastri MP, Wang C, Patel G, Franks DG, Schlezinger J, Sherr DH, Silverstone AE, Hahn ME, McCune JM. Identification of Cinnabarinic Acid as a Novel Endogenous Aryl Hydrocarbon Receptor Ligand That Drives IL-22 Production. PLoS One. 2014;9:e87877. doi: 10.1371/journal.pone.0087877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Qiu J, Guo X, Chen Z-ming E, He L, Sonnenberg Gregory F, Artis D, Fu Y-X, Zhou L. Group 3 Innate Lymphoid Cells Inhibit T-Cell-Mediated Intestinal Inflammation through Aryl Hydrocarbon Receptor Signaling and Regulation of Microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 201.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 202.Mielke LA, Groom JR, Rankin LC, Seillet C, Masson F, Putoczki T, Belz GT. TCF-1 Controls ILC2 and NKp46+RORγt+ Innate Lymphocyte Differentiation and Protection in Intestinal Inflammation. The Journal of Immunology. 2013;191:4383–4391. doi: 10.4049/jimmunol.1301228. [DOI] [PubMed] [Google Scholar]
- 203.Alam MS, Maekawa Y, Kitamura A, Tanigaki K, Yoshimoto T, Kishihara K, Yasutomo K. Notch signaling drives IL-22 secretion in CD4+ T cells by stimulating the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2010;107:5943–5948. doi: 10.1073/pnas.0911755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Xu M, Morishima N, Mizoguchi I, Chiba Y, Fujita K, Kuroda M, Iwakura Y, Cua DJ, Yasutomo K, Mizuguchi J, Yoshimoto T. Regulation of the development of acute hepatitis by IL-23 through IL-22 and IL-17 production. Eur J Immunol. 2011;41:2828–2839. doi: 10.1002/eji.201141291. [DOI] [PubMed] [Google Scholar]
- 205.Bhuyan ZA, Asanoma M, Iwata A, Ishifune C, Maekawa Y, Shimada M, Yasutomo K. Abrogation of Rbpj attenuates experimental autoimmune uveoretinitis by inhibiting IL-22-producing CD4+ T cells. PLoS One. 2014;9:e89266. doi: 10.1371/journal.pone.0089266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mukherjee S, Schaller MA, Neupane R, Kunkel SL, Lukacs NW. Regulation of T cell activation by Notch ligand, DLL4, promotes IL-17 production and Rorc activation. J Immunol. 2009;182:7381–7388. doi: 10.4049/jimmunol.0804322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat Cell Biol. 2004;6:547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- 208.Murano T, Okamoto R, Ito G, Nakata T, Hibiya S, Shimizu H, Fujii S, Kano Y, Mizutani T, Yui S, Akiyama-Morio J, Nemoto Y, Tsuchiya K, Nakamura T, Watanabe M. Hes1 promotes the IL-22-mediated antimicrobial response by enhancing STAT3-dependent transcription in human intestinal epithelial cells. Biochem Biophys Res Commun. 2014;443:840–846. doi: 10.1016/j.bbrc.2013.12.061. [DOI] [PubMed] [Google Scholar]
- 209.Weiss B. Cloning of murine IL-22 receptor alpha 2 and comparison with its human counterpart. Genes Immun. 2004;5:330–336. doi: 10.1038/sj.gene.6364104. [DOI] [PubMed] [Google Scholar]
- 210.Gruenberg BH. A novel, soluble homologue of the human IL-10 receptor with preferential expression in placenta. Genes Immun. 2001;2:329–334. doi: 10.1038/sj.gene.6363786. [DOI] [PubMed] [Google Scholar]
- 211.Kotenko SV, Izotova LS, Mirochnitchenko OV, Esterova E, Dickensheets H, Donnelly RP, Pestka S. Identification, cloning, and characterization of a novel soluble receptor that binds IL-22 and neutralizes its activity. J Immunol. 2001;166:7096–7103. doi: 10.4049/jimmunol.166.12.7096. [DOI] [PubMed] [Google Scholar]
- 212.Dumoutier L, Lejeune D, Colau D, Renauld JC. Cloning and characterization of IL-22 binding protein, a natural antagonist of IL-10-related T cell-derived inducible factor/IL-22. J. Immunol. 2001;166:7090–7095. doi: 10.4049/jimmunol.166.12.7090. [DOI] [PubMed] [Google Scholar]
- 213.Wei CC, Ho TW, Liang WG, Chen GY, Chang MS. Cloning and characterization of mouse IL-22 binding protein. Genes Immun. 2003;4:204–211. doi: 10.1038/sj.gene.6363947. [DOI] [PubMed] [Google Scholar]
- 214.Xu W, Presnell SR, Parrish-Novak J, Kindsvogel W, Jaspers S, Chen Z, Dillon SR, Gao Z, Gilbert T, Madden K, Schlutsmeyer S, Yao L, Whitmore TE, Chandrasekher Y, Grant FJ, Maurer M, Jelinek L, Storey H, Brender T, Hammond A, Topouzis S, Clegg CH, Foster DC. A soluble class II cytokine receptor, IL-22RA2, is a naturally occurring IL-22 antagonist. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9511–9516. doi: 10.1073/pnas.171303198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wu PW. IL-22R, IL-10R2, and IL-22BP binding sites are topologically juxtaposed on adjacent and overlapping surfaces of IL-22. J. Mol. Biol. 2008;382:1168–1183. doi: 10.1016/j.jmb.2008.07.046. [DOI] [PubMed] [Google Scholar]
- 216.de Moura PR. Crystal structure of a soluble decoy receptor IL-22BP bound to interleukin-22. FEBS Lett. 2009;583:1072–1077. doi: 10.1016/j.febslet.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 217.Wolk K. IL-22 induces lipopolysaccharide-binding protein in hepatocytes: a potential systemic role of IL-22 in Crohn's disease. J. Immunol. 2007;178:5973–5981. doi: 10.4049/jimmunol.178.9.5973. [DOI] [PubMed] [Google Scholar]
- 218.Martin JC, Beriou G, Heslan M, Chauvin C, Utriainen L, Aumeunier A, Scott CL, Mowat A, Cerovic V, Houston SA, Leboeuf M, Hubert FX, Hemont C, Merad M, Milling S, Josien R. Interleukin-22 binding protein (IL-22BP) is constitutively expressed by a subset of conventional dendritic cells and is strongly induced by retinoic acid. Mucosal Immunol. 2014;7:101–113. doi: 10.1038/mi.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lecart S, Morel F, Noraz N, Pene J, Garcia M, Boniface K, Lecron JC, Yssel H. IL-22, in contrast to IL-10, does not induce Ig production, due to absence of a functional IL-22 receptor on activated human B cells. Int Immunol. 2002;14:1351–1356. doi: 10.1093/intimm/dxf096. [DOI] [PubMed] [Google Scholar]
- 220.Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, Hedl M, Zhang W, O'Connor W, Murphy AJ, Valenzuela DM, Yancopoulos GD, Booth CJ, Cho JH, Ouyang WJ, Abraham C, Flavell RA. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 2012;491:259-+. doi: 10.1038/nature11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Volpe E, Touzot M, Servant N, Marloie-Provost MA, Hupe P, Barillot E, Soumelis V. Multiparametric analysis of cytokine-driven human Th17 differentiation reveals a differential regulation of IL-17 and IL-22 production. Blood. 2009;114:3610–3614. doi: 10.1182/blood-2009-05-223768. [DOI] [PubMed] [Google Scholar]
- 223.Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 224.Valdez PA, Vithayathil PJ, Janelsins BM, Shaffer AL, Williamson PR, Datta SK. Prostaglandin E2 suppresses antifungal immunity by inhibiting interferon regulatory factor 4 function and interleukin-17 expression in T cells. Immunity. 2012;36:668–679. doi: 10.1016/j.immuni.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wang H, Li Z, Yang B, Yu S, Wu C. IL-27 suppresses the production of IL-22 in human CD4(+) T cells by inducing the expression of SOCS1. Immunol Lett. 2013;152:96–103. doi: 10.1016/j.imlet.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 226.Liu H, Rohowsky-Kochan C. Interleukin-27-mediated suppression of human Th17 cells is associated with activation of STAT1 and suppressor of cytokine signaling protein 1. J Interferon Cytokine Res. 2011;31:459–469. doi: 10.1089/jir.2010.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Paulos CM, Carpenito C, Plesa G, Suhoski MM, Varela-Rohena A, Golovina TN, Carroll RG, Riley JL, June CH. The inducible costimulator (ICOS) is critical for the development of human T(H)17 cells. Sci Transl Med. 2010;2:55ra78. doi: 10.1126/scitranslmed.3000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, Kuchroo VK. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.van de Veerdonk FL, Stoeckman AK, Wu G, Boeckermann AN, Azam T, Netea MG, Joosten LA, van der Meer JW, Hao R, Kalabokis V, Dinarello CA. IL-38 binds to the IL-36 receptor and has biological effects on immune cells similar to IL-36 receptor antagonist. Proc Natl Acad Sci U S A. 2012;109:3001–3005. doi: 10.1073/pnas.1121534109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Tortola L, Rosenwald E, Abel B, Blumberg H, Schafer M, Coyle AJ, Renauld JC, Werner S, Kisielow J, Kopf M. Psoriasiform dermatitis is driven by IL-36-mediated DC-keratinocyte crosstalk. J Clin Invest. 2012;122:3965–3976. doi: 10.1172/JCI63451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Witte E, Witte K, Warszawska K, Sabat R, Wolk K. Interleukin-22: A cytokine produced by T, NK and NKT cell subsets, with importance in the innate immune defense and tissue protection. Cytokine & Growth Factor Reviews. 2010;21:365–379. doi: 10.1016/j.cytogfr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 232.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 233.Cordero-Coma M, Calleja S, Llorente M, Rodriguez E, Franco M, Ruiz de Morales JG. Serum cytokine profile in adalimumab-treated refractory uveitis patients: decreased IL-22 correlates with clinical responses. Ocul Immunol Inflamm. 2013;21:212–219. doi: 10.3109/09273948.2013.770888. [DOI] [PubMed] [Google Scholar]
- 234.Dalmas E, Venteclef N, Caer C, Poitou C, Cremer I, Aron-Winewsky J, Lacroix-Desmazes S, Bayry J, Kaveri SV, Clement K, Andre S, Guerre-Millo M. T cell-derived IL-22 amplifies IL-1beta-driven inflammation in human adipose tissue: relevance to obesity and type 2 diabetes. Diabetes. 2014 doi: 10.2337/db13-1511. [DOI] [PubMed] [Google Scholar]
- 235.Fabbrini E, Cella M, McCartney SA, Fuchs A, Abumrad NA, Pietka TA, Chen Z, Finck BN, Han DH, Magkos F, Conte C, Bradley D, Fraterrigo G, Eagon JC, Patterson BW, Colonna M, Klein S. Association between specific adipose tissue CD4+ T-cell populations and insulin resistance in obese individuals. Gastroenterology. 2013;145:366–374. e1–e3. doi: 10.1053/j.gastro.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kong Q, Xue Y, Wu W, Yang F, Liu Y, Gao M, Lai W, Pan X. IL-22 exacerbates the severity of CVB3-induced acute viral myocarditis in IL-17A-deficient mice. Mol Med Rep. 2013;7:1329–1335. doi: 10.3892/mmr.2013.1323. [DOI] [PubMed] [Google Scholar]
- 237.Rainard P, Cunha P, Bougarn S, Fromageau A, Rossignol C, Gilbert FB, Berthon P. T helper 17-associated cytokines are produced during antigen-specific inflammation in the mammary gland. PLoS One. 2013;8:e63471. doi: 10.1371/journal.pone.0063471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sugita S, Kawazoe Y, Imai A, Kawaguchi T, Horie S, Keino H, Takahashi M, Mochizuki M. Role of IL-22− and TNF-alpha-producing Th22 cells in uveitis patients with Behcet's disease. J Immunol. 2013;190:5799–5808. doi: 10.4049/jimmunol.1202677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sugita S, Kawazoe Y, Imai A, Usui Y, Takahashi M, Mochizuki M. Suppression of IL-22-producing T helper 22 cells by RPE cells via PD-L1/PD-1 interactions. Invest Ophthalmol Vis Sci. 2013;54:6926–6933. doi: 10.1167/iovs.13-12703. [DOI] [PubMed] [Google Scholar]
- 240.Zaidi T, Zaidi T, Cywes-Bentley C, Lu R, Priebe GP, Pier GB. Microbiota-Driven Immune Cellular Maturation Is Essential for Antibody-Mediated Adaptive Immunity to Staphylococcus aureus Infection in the Eye. Infect Immun. 2014;82:3483–3491. doi: 10.1128/IAI.01951-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M. Microbiota-Dependent Crosstalk Between Macrophages and ILC3 Promotes Intestinal Homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Muhl H. Pro-Inflammatory Signaling by IL-10 and IL-22: Bad Habit Stirred Up by Interferons? Front Immunol. 2013;4:18. doi: 10.3389/fimmu.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Nagalakshmi ML, Rascle A, Zurawski S, Menon S, de Waal Malefyt R. Interleukin-22 activates STAT3 and induces IL-10 by colon epithelial cells. International Immunopharmacology. 2004;4:679–691. doi: 10.1016/j.intimp.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 244.Hosokawa Y, Hosokawa I, Shindo S, Ozaki K, Matsuo T. IL-22 Enhances CCL20 Production in IL-1beta-Stimulated Human Gingival Fibroblasts. Inflammation. 2014 doi: 10.1007/s10753-014-9939-5. [DOI] [PubMed] [Google Scholar]
- 245.Rabeony H, Petit-Paris I, Garnier J, Barrault C, Pedretti N, Guilloteau K, Jegou JF, Guillet G, Huguier V, Lecron JC, Bernard FX, Morel F. Inhibition of Keratinocyte Differentiation by the Synergistic Effect of IL-17A, IL-22, IL-1alpha, TNFalpha and Oncostatin M. PloS one. 2014;9:e101937. doi: 10.1371/journal.pone.0101937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sonnenberg GF. Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17A. J. Exp. Med. 2010;207:1293–1305. doi: 10.1084/jem.20092054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhao J, Zhang Z, Luan Y, Zou Z, Sun Y, Li Y, Jin L, Zhou C, Fu J, Gao B, Fu Y, Wang FS. Pathological functions of interleukin-22 in chronic liver inflammation and fibrosis with hepatitis B virus infection by promoting T helper 17 cell recruitment. Hepatology. 2014;59:1331–1342. doi: 10.1002/hep.26916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Eken A, Singh AK, Treuting PM, Oukka M. IL-23R+ innate lymphoid cells induce colitis via interleukin-22-dependent mechanism. Mucosal Immunol. 2014;7:143–154. doi: 10.1038/mi.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Uhlig HH, McKenzie BS, Hue S, Thompson C, Joyce-Shaikh B, Stepankova R, Robinson N, Buonocore S, Tlaskalova-Hogenova H, Cua DJ, Powrie F. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–318. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 250.Souza JM, Matias BF, Rodrigues CM, Murta EF, Michelin MA. IL-17 and IL-22 serum cytokine levels in patients with squamous intraepithelial lesion and invasive cervical carcinoma. European journal of gynaecological oncology. 2013;34:466–468. [PubMed] [Google Scholar]
- 251.Schmechel S, Konrad A, Diegelmann J, Glas J, Wetzke M, Paschos E, Lohse P, Goke B, Brand S. Linking genetic susceptibility to Crohn's disease with Th17 cell function: IL-22 serum levels are increased in Crohn's disease and correlate with disease activity and IL23R genotype status. Inflamm Bowel Dis. 2008;14:204–212. doi: 10.1002/ibd.20315. [DOI] [PubMed] [Google Scholar]
- 252.Liang SC, Nickerson-Nutter C, Pittman DD, Carrier Y, Goodwin DG, Shields KM, Lambert A-J, Schelling SH, Medley QG, Ma H-L, Collins M, Dunussi-Joannopoulos K, Fouser LA. IL-22 Induces an Acute-Phase Response. Journal of Immunology. 2010;185:5531–5538. doi: 10.4049/jimmunol.0904091. [DOI] [PubMed] [Google Scholar]
- 253.Ciccia F, Guggino G, Rizzo A, Ferrante A, Raimondo S, Giardina A, Dieli F, Campisi G, Alessandro R, Triolo G. Potential involvement of IL-22 and IL-22-producing cells in the inflamed salivary glands of patients with Sjogren's syndrome. Ann Rheum Dis. 2012;71:295–301. doi: 10.1136/ard.2011.154013. [DOI] [PubMed] [Google Scholar]
- 254.Delsing CE, Bleeker-Rovers CP, van de Veerdonk FL, Tol J, van der Meer JW, Kullberg BJ, Netea MG. Association of esophageal candidiasis and squamous cell carcinoma. Med Mycol Case Rep. 2012;1:5–8. doi: 10.1016/j.mmcr.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hughes T, Becknell B, McClory S, Briercheck E, Freud AG, Zhang X, Mao H, Nuovo G, Yu J, Caligiuri MA. Stage 3 immature human natural killer cells found in secondary lymphoid tissue constitutively and selectively express the TH 17 cytokine interleukin-22. Blood. 2009;113:4008–4010. doi: 10.1182/blood-2008-12-192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kato-Kogoe N, Nishioka T, Kawabe M, Kataoka F, Yamanegi K, Yamada N, Hata M, Yamamoto T, Nakasho K, Urade M, Terada N, Ohyama H. The promotional effect of IL-22 on mineralization activity of periodontal ligament cells. Cytokine. 2012;59:41–48. doi: 10.1016/j.cyto.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 257.Naher L, Kiyoshima T, Kobayashi I, Wada H, Nagata K, Fujiwara H, Ookuma YF, Ozeki S, Nakamura S, Sakai H. STAT3 signal transduction through interleukin-22 in oral squamous cell carcinoma. Int J Oncol. 2012;41:1577–1586. doi: 10.3892/ijo.2012.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zhuang Y. Increased intratumoral IL-22-producing CD4+ T cells and Th22 cells correlate with gastric cancer progression and predict poor patient survival. Cancer Immunol. Immunother. 2012;61:1965–1975. doi: 10.1007/s00262-012-1241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.te Velde AA, de Kort F, Sterrenburg E, Pronk I, ten Kate FJ, Hommes DW, van Deventer SJ. Comparative analysis of colonic gene expression of three experimental colitis models mimicking inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:325–330. doi: 10.1002/ibd.20079. [DOI] [PubMed] [Google Scholar]
- 260.Sadighi Akha AA, Theriot CM, Erb-Downward JR, McDermott AJ, Falkowski NR, Tyra HM, Rutkowski DT, Young VB, Huffnagle GB. Acute infection of mice with Clostridium difficile leads to eIF2alpha phosphorylation and pro-survival signalling as part of the mucosal inflammatory response. Immunology. 2013;140:111–122. doi: 10.1111/imm.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bishop JL, Roberts ME, Beer JL, Huang M, Chehal MK, Fan X, Fouser LA, Ma HL, Bacani JT, Harder KW. Lyn activity protects mice from DSS colitis and regulates the production of IL-22 from innate lymphoid cells. Mucosal Immunol. 2014;7:405–416. doi: 10.1038/mi.2013.60. [DOI] [PubMed] [Google Scholar]
- 262.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, Tardif MIR, Sathaliyawala T, Kubota M, Farber DL, Collman RG, Shaked A, Fouser LA, Weiner DB, Tessier PA, Friedman JR, Kiyono H, Bushman FD, Chang K-M, Artis D. Innate Lymphoid Cells Promote Anatomical Containment of Lymphoid-Resident Commensal Bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Kleinschek M, Cua D, Di Santo JP, Eberl G. RORgamma]t+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 264.Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, Ouyang W, Neurath MF, Becker C. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kim CJ. A role for mucosal IL-22 production and Th22 cells in HIV-associated mucosal immunopathogenesis. Mucosal Immunol. 2012;5:670–680. doi: 10.1038/mi.2012.72. [DOI] [PubMed] [Google Scholar]
- 266.Mizuno S, Mikami Y, Kamada N, Handa T, Hayashi A, Sato T, Matsuoka K, Matano M, Ohta Y, Sugita A, Koganei K, Sahara R, Takazoe M, Hisamatsu T, Kanai T. Cross-talk Between RORgammat+ Innate Lymphoid Cells and Intestinal Macrophages Induces Mucosal IL-22 Production in Crohn's Disease. Inflammatory bowel diseases. 2014 doi: 10.1097/MIB.0000000000000105. [DOI] [PubMed] [Google Scholar]
- 267.Sanos SL, Vonarbourg C, Mortha A, Diefenbach A. Control of epithelial cell function by interleukin-22-producing RORγt+ innate lymphoid cells. Immunology. 2011;132:453–465. doi: 10.1111/j.1365-2567.2011.03410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Konieczna P, Ferstl R, Ziegler M, Frei R, Nehrbass D, Lauener RP, Akdis CA, O'Mahony L. Immunomodulation by Bifidobacterium infantis 35624 in the murine lamina propria requires retinoic acid-dependent and independent mechanisms. PLoS One. 2013;8:e62617. doi: 10.1371/journal.pone.0062617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 270.Salzman NH, Bevins CL. Dysbiosis--a consequence of Paneth cell dysfunction. Semin Immunol. 2013;25:334–341. doi: 10.1016/j.smim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 271.Kolls JK, McCray PB, Jr, Chan YR. Cytokine-mediated regulation of antimicrobial proteins. Nat Rev Immunol. 2008;8:829–835. doi: 10.1038/nri2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Burger-van Paassen N, Loonen LM, Witte-Bouma J, Korteland-van Male AM, de Bruijn AC, van der Sluis M, Lu P, Van Goudoever JB, Wells JM, Dekker J, Van Seuningen I, Renes IB. Mucin Muc2 deficiency and weaning influences the expression of the innate defense genes Reg3beta, Reg3gamma and angiogenin-4. PLoS One. 2012;7:e38798. doi: 10.1371/journal.pone.0038798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Eriguchi Y, Uryu H, Nakamura K, Shimoji S, Takashima S, Iwasaki H, Miyamoto T, Shimono N, Hashimoto D, Akashi K, Ayabe T, Teshima T. Reciprocal expression of enteric antimicrobial proteins in intestinal graft-versus-host disease. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2013;19:1525–1529. doi: 10.1016/j.bbmt.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 274.Reynders A, Yessaad N, Manh TPV, Dalod M, Fenis A, Aubry C, Nikitas G, Escaliere B, Renauld JC, Dussurget O, Cossart P, Lecuit M, Vivier E, Tomasello E. Identity, regulation and in vivo function of gut NKp46(+)ROR gamma t(+) and NKp46(+)ROR gamma t(−) lymphoid cells. Embo Journal. 2011;30:2934–2947. doi: 10.1038/emboj.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Eberl G, Sawa S. Opening the crypt: current facts and hypotheses on the function of cryptopatches. Trends Immunol. 2010;31:50–55. doi: 10.1016/j.it.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 276.Kanamori Y, Ishimaru K, Nanno M, Maki K, Ikuta K, Nariuchi H, Ishikawa H. Identification of novel lymphoid tissues in murine intestinal mucosa where clusters of c-kit+ IL-7R+ Thy1+ lympho-hemopoietic progenitors develop. J Exp Med. 1996;184:1449–1459. doi: 10.1084/jem.184.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- 278.Lorenz RG, Newberry RD. Isolated lymphoid follicles can function as sites for induction of mucosal immune responses. Ann N Y Acad Sci. 2004;1029:44–57. doi: 10.1196/annals.1309.006. [DOI] [PubMed] [Google Scholar]
- 279.Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 Increases the Innate Immunity of Tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 280.Wolk K, Witte E, Wallace E, Docke WD, Kunz S, Asadullah K, Volk HD, Sterry W, Sabat R. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur J Immunol. 2006;36:1309–1323. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- 281.Boniface K. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J. Immunol. 2005;174:3695–3702. doi: 10.4049/jimmunol.174.6.3695. [DOI] [PubMed] [Google Scholar]
- 282.Wolk K. IL-22 and IL-20 are key mediators of the epidermal alterations in psoriasis while IL-17 and IFN-[gamma] are not. J. Mol. Med. 2009;87:523–536. doi: 10.1007/s00109-009-0457-0. [DOI] [PubMed] [Google Scholar]
- 283.Sa SM. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J. Immunol. 2007;178:2229–2240. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- 284.Wolk K. The Th17 cytokine IL-22 induces IL-20 production in keratinocytes: a novel immunological cascade with potential relevance in psoriasis. Eur. J. Immunol. 2009;39:3570–3581. doi: 10.1002/eji.200939687. [DOI] [PubMed] [Google Scholar]
- 285.Romer J. Epidermal overexpression of interleukin-19 and-20 mRNA in psoriatic skin disappears after short-term treatment with cyclosporine A or calcipotriol. J. Invest. Dermatol. 2003;121:1306–1311. doi: 10.1111/j.1523-1747.2003.12626.x. [DOI] [PubMed] [Google Scholar]
- 286.Kunz S. Interleukin (IL)-19, IL-20 and IL-24 are produced by and act on keratinocytes and are distinct from classical ILs. Exp. Dermatol. 2006;15:991–1004. doi: 10.1111/j.1600-0625.2006.00516.x. [DOI] [PubMed] [Google Scholar]
- 287.Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 288.Lowes MA, Suarez-Farinas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32:227–255. doi: 10.1146/annurev-immunol-032713-120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Saeki H, Hirota T, Nakagawa H, Tsunemi Y, Kato T, Shibata S, Sugaya M, Sato S, Doi S, Miyatake A, Ebe K, Noguchi E, Ebihara T, Amagai M, Esaki H, Takeuchi S, Furue M, Nakamura Y, Tamari M. Genetic polymorphisms in the IL22 gene are associated with psoriasis vulgaris in a Japanese population. J Dermatol Sci. 2013;71:148–150. doi: 10.1016/j.jdermsci.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 290.Nikamo P, Cheuk S, Lysell J, Enerback C, Bergh K, Xu Landen N, Eidsmo L, Stahle M. Genetic Variants of the IL22 Promoter Associate to Onset of Psoriasis before Puberty and Increased IL-22 Production in T Cells. J Invest Dermatol. 2014;134:1535–1541. doi: 10.1038/jid.2014.5. [DOI] [PubMed] [Google Scholar]
- 291.Prans E, Kingo K, Traks T, Silm H, Vasar E, Koks S. Copy number variations in IL22 gene are associated with Psoriasis vulgaris. Hum Immunol. 2013;74:792–795. doi: 10.1016/j.humimm.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 292.Johnston A, Gudjonsson JE. 22 Again: IL-22 as a Risk Gene and Important Mediator in Psoriasis. J Invest Dermatol. 2014;134:1501–1503. doi: 10.1038/jid.2014.81. [DOI] [PubMed] [Google Scholar]
- 293.Boniface K, Guignouard E, Pedretti N, Garcia M, Delwail A, Bernard FX, Nau F, Guillet G, Dagregorio G, Yssel H, Lecron JC, Morel F. A role for T cell-derived interleukin 22 in psoriatic skin inflammation. Clin Exp Immunol. 2007;150:407–415. doi: 10.1111/j.1365-2249.2007.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lo YH, Torii K, Saito C, Furuhashi T, Maeda A, Morita A. Serum IL-22 correlates with psoriatic severity and serum IL-6 correlates with susceptibility to phototherapy. J Dermatol Sci. 2010;58:225–227. doi: 10.1016/j.jdermsci.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 295.Shimauchi T, Hirakawa S, Suzuki T, Yasuma A, Majima Y, Tatsuno K, Yagi H, Ito T, Tokura Y. Serum interleukin-22 and vascular endothelial growth factor serve as sensitive biomarkers but not as predictors of therapeutic response to biologics in patients with psoriasis. The Journal of Dermatology. 2013;40:805–812. doi: 10.1111/1346-8138.12248. [DOI] [PubMed] [Google Scholar]
- 296.Blumberg H. Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell. 2001;104:9–19. doi: 10.1016/s0092-8674(01)00187-8. [DOI] [PubMed] [Google Scholar]
- 297.Stenderup K. Interleukin-20 plays a critical role in maintenance and development of psoriasis in the human xenograft transplantation model. Br. J. Dermatol. 2009;160:284–296. doi: 10.1111/j.1365-2133.2008.08890.x. [DOI] [PubMed] [Google Scholar]
- 298.Teunissen MB, Munneke JM, Bernink JH, Spuls PI, Res PC, Te Velde A, Cheuk S, Brouwer MW, Menting SP, Eidsmo L, Spits H, Hazenberg MD, Mjosberg J. Composition of Innate Lymphoid Cell Subsets in the Human Skin: Enrichment of NCR ILC3 in Lesional Skin and Blood of Psoriasis Patients. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.146. [DOI] [PubMed] [Google Scholar]
- 299.Villanova F, Flutter B, Tosi I, Grys K, Sreeneebus H, Perera GK, Chapman A, Smith CH, Di Meglio P, Nestle FO. Characterization of Innate Lymphoid Cells in Human Skin and Blood Demonstrates Increase of NKp44+ ILC3 in Psoriasis. J Invest Dermatol. 2014;134:984–991. doi: 10.1038/jid.2013.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Dyring-Andersen B, Geisler C, Agerbeck C, Lauritsen JP, Gudjonsdottir SD, Skov L, Bonefeld CM. Increased number and frequency of group 3 innate lymphoid cells in nonlesional psoriatic skin. Br J Dermatol. 2014;170:609–616. doi: 10.1111/bjd.12658. [DOI] [PubMed] [Google Scholar]
- 301.Perera GK, Ainali C, Semenova E, Hundhausen C, Barinaga G, Kassen D, Williams AE, Mirza MM, Balazs M, Wang X, Rodriguez RS, Alendar A, Barker J, Tsoka S, Ouyang W, Nestle FO. Integrative biology approach identifies cytokine targeting strategies for psoriasis. Sci Transl Med. 2014;6:223ra22. doi: 10.1126/scitranslmed.3007217. [DOI] [PubMed] [Google Scholar]
- 302.Ma HL. IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation. J. Clin. Invest. 2008;118:597–607. doi: 10.1172/JCI33263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.McAleer JP, Kolls JK. Directing traffic: IL-17 and IL-22 coordinate pulmonary immune defense. Immunol Rev. 2014;260:129–144. doi: 10.1111/imr.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Chen K, Pociask DA, McAleer JP, Chan YR, Alcorn JF, Kreindler JL, Keyser MR, Shapiro SD, Houghton AM, Kolls JK, Zheng M. IL-17RA is required for CCL2 expression, macrophage recruitment, and emphysema in response to cigarette smoke. PLoS One. 2011;6:e20333. doi: 10.1371/journal.pone.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Besnard AG. Dual Role of IL-22 in allergic airway inflammation and its cross-talk with IL-17A. Am. J. Respir. Crit. Care Med. 2011;183:1153–1163. doi: 10.1164/rccm.201008-1383OC. [DOI] [PubMed] [Google Scholar]
- 306.Farfariello V. IL-22 mRNA in peripheral blood mononuclear cells from allergic rhinitic and asthmatic pediatric patients. Pediatr. Allergy Immunol. 2011;22:419–423. doi: 10.1111/j.1399-3038.2010.01116.x. [DOI] [PubMed] [Google Scholar]
- 307.Takahashi K. IL-22 attenuates IL-25 production by lung epithelial cells and inhibits antigen-induced eosinophilic airway inflammation. J. Allergy Clin. Immunol. 2011;128:1067–1076. doi: 10.1016/j.jaci.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 308.Nakagome K. High expression of IL-22 suppresses antigen-induced immune responses and eosinophilic airway inflammation via an IL-10-associated mechanism. J. Immunol. 2011;187:5077–5089. doi: 10.4049/jimmunol.1001560. [DOI] [PubMed] [Google Scholar]
- 309.Taube C. IL-22 is produced by innate lymphoid cells and limits inflammation in allergic airway disease. PLoS ONE. 2011;6:e21799. doi: 10.1371/journal.pone.0021799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Van Maele L, Carnoy C, Cayet D, Ivanov S, Porte R, Deruy E, Chabalgoity JA, Renauld JC, Eberl G, Benecke AG, Trottein F, Faveeuw C, Sirard JC. Activation of Type 3 innate lymphoid cells and interleukin 22 secretion in the lungs during Streptococcus pneumoniae infection. J Infect Dis. 2014;210:493–503. doi: 10.1093/infdis/jiu106. [DOI] [PubMed] [Google Scholar]
- 311.Xu X, Weiss ID, H HZ, Singh SP, Wynn TA, Wilson MS, Farber JM. Conventional NK Cells Can Produce IL-22 and Promote Host Defense in Klebsiella pneumoniae Pneumonia. J Immunol. 2014;192:1778–1786. doi: 10.4049/jimmunol.1300039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Peng Y, Gao X, Yang J, Shekhar S, Wang S, Fan Y, Zhao W, Yang X. IL-22 Promotes Th1/Th17 Immunity in Chlamydial Lung Infection. Mol Med. 2014 doi: 10.2119/molmed.2013.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Gessner MA. Dectin-1-dependent interleukin-22 contributes to early innate lung defense against Aspergillus fumigatus. Infect. Immun. 2012;80:410–417. doi: 10.1128/IAI.05939-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Gresnigt MS, Becker KL, Smeekens SP, Jacobs CW, Joosten LA, van der Meer JW, Netea MG, van de Veerdonk FL. Aspergillus fumigatus-induced IL-22 is not restricted to a specific Th cell subset and is dependent on complement receptor 3. J Immunol. 2013;190:5629–5639. doi: 10.4049/jimmunol.1202601. [DOI] [PubMed] [Google Scholar]
- 315.Lilly LM, Gessner MA, Dunaway CW, Metz AE, Schwiebert L, Weaver CT, Brown GD, Steele C. The beta-glucan receptor dectin-1 promotes lung immunopathology during fungal allergy via IL-22. J Immunol. 2012;189:3653–3660. doi: 10.4049/jimmunol.1201797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Mear JB, Gosset P, Kipnis E, Faure E, Dessein R, Jawhara S, Fradin C, Faure K, Poulain D, Sendid B, Guery B. Candida albicans airway exposure primes the lung innate immune response against Pseudomonas aeruginosa infection through innate lymphoid cell recruitment and interleukin-22-associated mucosal response. Infect Immun. 2014;82:306–315. doi: 10.1128/IAI.01085-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Kumar P, Thakar MS, Ouyang W, Malarkannan S. IL-22 from conventional NK cells is epithelial regenerative and inflammation protective during influenza infection. Mucosal Immunol. 2013;6:69–82. doi: 10.1038/mi.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Guo H, Topham DJ. Interleukin-22 (IL-22) production by pulmonary Natural Killer cells and the potential role of IL-22 during primary influenza virus infection. J Virol. 2010;84:7750–7759. doi: 10.1128/JVI.00187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Pociask DA. IL-22 is essential for lung epithelial repair following influenza infection. Am. J. Pathol. 2013;182:1286–1296. doi: 10.1016/j.ajpath.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Ivanov S. Interleukin-22 reduces lung inflammation during influenza A virus infection and protects against secondary bacterial infection. J. Virol. 2013;87:6911–6924. doi: 10.1128/JVI.02943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Simonian PL. [gamma][delta] T cells protect against lung fibrosis via IL-22. J. Exp. Med. 2010;207:2239–2253. doi: 10.1084/jem.20100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Hoegl S. Protective properties of inhaled IL-22 in a model of ventilator-induced lung injury. Am. J. Respir. Cell. Mol. Biol. 2011;44:369–376. doi: 10.1165/rcmb.2009-0440OC. [DOI] [PubMed] [Google Scholar]
- 323.Yao S, Huang D, Chen CY, Halliday L, Zeng G, Wang RC, Chen ZW. Differentiation, distribution and gammadelta T cell-driven regulation of IL-22-producing T cells in tuberculosis. PLoS Pathog. 2010;6:e1000789. doi: 10.1371/journal.ppat.1000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Lutay N, Hakansson G, Alaridah N, Hallgren O, Westergren-Thorsson G, Godaly G. Mycobacteria Bypass Mucosal NF-kB Signalling to Induce an Epithelial Anti-Inflammatory IL-22 and IL-10 Response. PLoS One. 2014;9:e86466. doi: 10.1371/journal.pone.0086466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Dhiman R, Periasamy S, Barnes PF, Jaiswal AG, Paidipally P, Barnes AB, Tvinnereim A, Vankayalapati R. NK1.1+ cells and IL-22 regulate vaccine-induced protective immunity against challenge with Mycobacterium tuberculosis. J Immunol. 2012;189:897–905. doi: 10.4049/jimmunol.1102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Zhang Y. A proinflammatory role for interleukin-22 in the immune response to hepatitis B virus. Gastroenterology. 2011;141:1897–1906. doi: 10.1053/j.gastro.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Pan H, Hong F, Radaeva S, Gao B. Hydrodynamic gene delivery of interleukin-22 protects the mouse liver from concanavalin A-, carbon tetrachloride-, and Fas ligand-induced injury via activation of STAT3. Cell. Mol. Immunol. 2004;1:43–49. [PubMed] [Google Scholar]
- 328.Radaeva S, Sun R, Pan HN, Hong F, Gao B. Interleukin 22 (IL-22) plays a protective role in T cell-mediated murine hepatitis: IL-22 is a survival factor for hepatocytes via STAT3 activation. Hepatology. 2004;39:1332–1342. doi: 10.1002/hep.20184. [DOI] [PubMed] [Google Scholar]
- 329.Zenewicz LA. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity. 2007;27:647–659. doi: 10.1016/j.immuni.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Park O. In vivo consequences of liver-specific interleukin-22 expression in mice: implications for human liver disease progression. Hepatology. 2011;54:252–261. doi: 10.1002/hep.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Ki SH. Interleukin-22 treatment ameliorates alcoholic liver injury in a murine model of chronic-binge ethanol feeding: role of signal transducer and activator of transcription 3. Hepatology. 2010;52:1291–1300. doi: 10.1002/hep.23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Schulz SM. Protective immunity to systemic infection with attenuated Salmonella enterica serovar enteritidis in the absence of IL-12 is associated with IL-23-dependent IL-22, but not IL-17. J. Immunol. 2008;181:7891–7901. doi: 10.4049/jimmunol.181.11.7891. [DOI] [PubMed] [Google Scholar]
- 333.Chestovich PJ. Interleukin-22: implications for liver ischemia-reperfusion injury. Transplantation. 2012;93:485–492. doi: 10.1097/TP.0b013e3182449136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Mastelic B. IL-22 protects against liver pathology and lethality of an experimental blood-stage malaria infection. Front. Immunol. 2012;3:85. doi: 10.3389/fimmu.2012.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Feng D. Interleukin-22 promotes proliferation of liver stem/progenitor cells in mice and patients with chronic hepatitis B virus infection. Gastroenterology. 2012;143:188–198. doi: 10.1053/j.gastro.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Ren X, Hu B, Colletti LM. IL-22 is involved in liver regeneration after hepatectomy. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;298:G74–G80. doi: 10.1152/ajpgi.00075.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Dambacher J. The role of interleukin-22 in hepatitis C virus infection. Cytokine. 2008;41:209–216. doi: 10.1016/j.cyto.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 338.Aggarwal S, Xie MH, Maruoka M, Foster J, Gurney AL. Acinar cells of the pancreas are a target of interleukin-22. J. Interferon Cytokine Res. 2001;21:1047–1053. doi: 10.1089/107999001317205178. [DOI] [PubMed] [Google Scholar]
- 339.Tachiiri A, Imamura R, Wang Y, Fukui M, Umemura M, Suda T. Genomic structure and inducible expression of the IL-22 receptor alpha chain in mice. Genes and Immunity. 2003;4:153–159. doi: 10.1038/sj.gene.6363934. [DOI] [PubMed] [Google Scholar]
- 340.Kulkarni OP, Hartter I, Mulay SR, Hagemann J, Darisipudi MN, Kumar VrS, Romoli S, Thomasova D, Ryu M, Kobold S, Anders HJ. Toll-Like Receptor 4-Induced IL-22 Accelerates Kidney Regeneration. J Am Soc Nephrol. 2014 doi: 10.1681/ASN.2013050528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Weber GF, Schlautkotter S, Kaiser-Moore S, Altmayr F, Holzmann B, Weighardt H. Inhibition of interleukin-22 attenuates bacterial load and organ failure during acute polymicrobial sepsis. Infect Immun. 2007;75:1690–1697. doi: 10.1128/IAI.01564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Zhang F, Shang D, Zhang Y, Tian Y. Interleukin-22 suppresses the growth of A498 renal cell carcinoma cells via regulation of STAT1 pathway. PLoS One. 2011;6:e20382. doi: 10.1371/journal.pone.0020382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Suh JS, Cho SH, Chung JH, Moon A, Park YK, Cho BS. A polymorphism of interleukin-22 receptor alpha-1 is associated with the development of childhood IgA nephropathy. J Interferon Cytokine Res. 2013;33:571–577. doi: 10.1089/jir.2012.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Parrish-Novak J, Xu W, Brender T, Yao L, Jones C, West J, Brandt C, Jelinek L, Madden K, McKernan PA, Foster DC, Jaspers S, Chandrasekher YA. Interleukins 19, 20, and 24 signal through two distinct receptor complexes. Differences in receptor-ligand interactions mediate unique biological functions. J Biol Chem. 2002;277:47517–47523. doi: 10.1074/jbc.M205114200. [DOI] [PubMed] [Google Scholar]
- 345.Shioya M, Andoh A, Kakinoki S, Nishida A, Fujiyama Y. Interleukin 22 receptor 1 expression in pancreas islets. Pancreas. 2008;36:197–199. doi: 10.1097/MPA.0b013e3181594258. [DOI] [PubMed] [Google Scholar]
- 346.Xue J, Nguyen DT, Habtezion A. Aryl hydrocarbon receptor regulates pancreatic IL-22 production and protects mice from acute pancreatitis. Gastroenterology. 2012;143:1670–1680. doi: 10.1053/j.gastro.2012.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Hill T. The involvement of interleukin-22 in the expression of pancreatic beta cell regenerative Reg genes. Cell Regener. 2013;2:2. doi: 10.1186/2045-9769-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Feng D. Interleukin-22 ameliorates cerulein-induced pancreatitis in mice by inhibiting the autophagic pathway. Int. J. Biol. Sci. 2012;8:249–257. doi: 10.7150/ijbs.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Lo Re S. IL-17A-producing [gamma][delta] T and Th17 lymphocytes mediate lung inflammation but not fibrosis in experimental silicosis. J. Immunol. 2010;184:6367–6377. doi: 10.4049/jimmunol.0900459. [DOI] [PubMed] [Google Scholar]
- 350.Shen H, Goodall JC, Hill Gaston JS. Frequency and phenotype of peripheral blood Th17 cells in ankylosing spondylitis and rheumatoid arthritis. Arthritis Rheum. 2009;60:1647–1656. doi: 10.1002/art.24568. [DOI] [PubMed] [Google Scholar]
- 351.Leipe J. Interleukin 22 serum levels are associated with radiographic progression in rheumatoid arthritis. Ann. Rheum. Dis. 2011;70:1453–1457. doi: 10.1136/ard.2011.152074. [DOI] [PubMed] [Google Scholar]
- 352.Zhang L. Elevated Th22 cells correlated with Th17 cells in patients with rheumatoid arthritis. J. Clin. Immunol. 2011;31:606–614. doi: 10.1007/s10875-011-9540-8. [DOI] [PubMed] [Google Scholar]
- 353.da Rocha LF., Jr Increased serum interleukin 22 in patients with rheumatoid arthritis and correlation with disease activity. J. Rheumatol. 2012;39:1320–1325. doi: 10.3899/jrheum.111027. [DOI] [PubMed] [Google Scholar]
- 354.Kim KW. Interleukin-22 promotes osteoclastogenesis in rheumatoid arthritis through induction of RANKL in human synovial fibroblasts. Arthritis Rheum. 2012;64:1015–1023. doi: 10.1002/art.33446. [DOI] [PubMed] [Google Scholar]
- 355.Kim K, Kim G, Kim JY, Yun HJ, Lim SC, Choi HS. Interleukin-22 promotes epithelial cell transformation and breast tumorigenesis via MAP3K8 activation. Carcinogenesis. 2014 doi: 10.1093/carcin/bgu044. [DOI] [PubMed] [Google Scholar]
- 356.Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, Vatan L, Szeliga W, Dou Y, Owens S, Zgodzinski W, Majewski M, Wallner G, Fang J, Huang E, Zou W. IL-22+CD4+ T Cells Promote Colorectal Cancer Stemness via STAT3 Transcription Factor Activation and Induction of the Methyltransferase DOT1L. Immunity. 2014 doi: 10.1016/j.immuni.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Fukui H, Zhang X, Sun C, Hara K, Kikuchi S, Yamasaki T, Kondo T, Tomita T, Oshima T, Watari J, Imura J, Fujimori T, Sasako M, Miwa H. IL-22 produced by cancer-associated fibroblasts promotes gastric cancer cell invasion via STAT3 and ERK signaling. Br J Cancer. 2014 doi: 10.1038/bjc.2014.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Liu F, Pan X, Zhou L, Zhou J, Chen B, Shi J, Gao W, Lu L. Genetic Polymorphisms and Plasma Levels of Interleukin-22 Contribute to the Development of Nonsmall Cell Lung Cancer. DNA Cell Biol. 2014 doi: 10.1089/dna.2014.2432. [DOI] [PubMed] [Google Scholar]
- 359.Qin S, Ma S, Huang X, Lu D, Zhou Y, Jiang H. Th22 cells are associated with hepatocellular carcinoma development and progression. Chin J Cancer Res. 2014;26:135–141. doi: 10.3978/j.issn.1000-9604.2014.02.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Wen Z, Liao Q, Zhao J, Hu Y, You L, Lu Z, Jia C, Wei Y, Zhao Y. High expression of interleukin-22 and its receptor predicts poor prognosis in pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2014;21:125–132. doi: 10.1245/s10434-013-3322-x. [DOI] [PubMed] [Google Scholar]
- 361.Jiang R, Zhang C, Xia Y, Qian X, Wang X, Sun B. Reply: To PMID 21674558. Hepatology. 2014;59:1208. doi: 10.1002/hep.26613. [DOI] [PubMed] [Google Scholar]
- 362.Waidmann O, Kronenberger B, Scheiermann P, Koberle V, Muhl H, Piiper A. Interleukin-22 serum levels are a negative prognostic indicator in patients with hepatocellular carcinoma. Hepatology. 2014;59:1207. doi: 10.1002/hep.26528. [DOI] [PubMed] [Google Scholar]
- 363.Kobold S. Interleukin-22 Is frequently expressed in small- and large-cell lung cancer and promotes growth in chemotherapy-resistant cancer cells. J. Thorac. Oncol. 2013;8:1032–1042. doi: 10.1097/JTO.0b013e31829923c8. [DOI] [PubMed] [Google Scholar]
- 364.Zhang S, Fujita H, Mitsui H, Yanofsky VR, Fuentes-Duculan J, Pettersen JS, Suarez-Farinas M, Gonzalez J, Wang CQ, Krueger JG, Felsen D, Carucci JA. Increased Tc22 and Treg/CD8 ratio contribute to aggressive growth of transplant associated squamous cell carcinoma. PLoS One. 2013;8:e62154. doi: 10.1371/journal.pone.0062154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Eun YG, Shin IH, Lee YC, Shin SY, Kim SK, Chung JH, Kwon KH. Interleukin 22 polymorphisms and papillary thyroid cancer. J Endocrinol Invest. 2013;36:584–587. doi: 10.3275/8879. [DOI] [PubMed] [Google Scholar]
- 366.Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, Harrison O, Powrie F. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med. 2013;210:917–931. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Zhao K, Zhao D, Huang D, Song X, Chen C, Pan B, Wu Q, Cao J, Yao Y, Zeng L, Xu K. The identification and characteristics of IL-22-producing T cells in acute graft-versus-host disease following allogeneic bone marrow transplantation. Immunobiology. 2013;218:1505–1513. doi: 10.1016/j.imbio.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 368.Couturier M, Lamarthee B, Arbez J, Renauld JC, Bossard C, Malard F, Bonnefoy F, Mohty M, Perruche S, Tiberghien P, Saas P, Gaugler B. IL-22 deficiency in donor T cells attenuates murine acute graft-versus-host disease mortality while sparing the graft-versus-leukemia effect. Leukemia. 2013;27:1527–1537. doi: 10.1038/leu.2013.39. [DOI] [PubMed] [Google Scholar]
- 369.Zhao K, Zhao D, Huang D, Yin L, Chen C, Pan B, Wu Q, Li Z, Yao Y, Shen E, Zeng L, Xu K. Interleukin-22 Aggravates Murine Acute Graft-Versus-Host Disease by Expanding Effector T Cell and Reducing Regulatory T Cell. J Interferon Cytokine Res. 2014 doi: 10.1089/jir.2013.0099. [DOI] [PubMed] [Google Scholar]
- 370.Munneke JM, Bjorklund AT, Mjosberg JM, Garming-Legert K, Bernink JH, Blom B, Huisman C, van Oers MH, Spits H, Malmberg KJ, Hazenberg MD. Activated innate lymphoid cells are associated with a reduced susceptibility to graft-versus-host disease. Blood. 2014;124:812–821. doi: 10.1182/blood-2013-11-536888. [DOI] [PubMed] [Google Scholar]
- 371.Bruggen MC, Klein I, Greinix H, Bauer W, Kuzmina Z, Rabitsch W, Kalhs P, Petzelbauer P, Knobler R, Stingl G, Stary G. Diverse T-cell responses characterize the different manifestations of cutaneous graft-versus-host disease. Blood. 2014;123:290–299. doi: 10.1182/blood-2013-07-514372. [DOI] [PubMed] [Google Scholar]
- 372.Laakso SM, Kekalainen E, Heikkila N, Mannerstrom H, Kisand K, Peterson P, Ranki A, Arstila TP. In vivo analysis of helper T cell responses in patients with autoimmune polyendocrinopathy - candidiasis - ectodermal dystrophy provides evidence in support of an IL-22 defect. Autoimmunity. 2014:1–7. doi: 10.3109/08916934.2014.929666. [DOI] [PubMed] [Google Scholar]
- 373.Karner J, Meager A, Laan M, Maslovskaja J, Pihlap M, Remm A, Juronen E, Wolff AS, Husebye ES, Podkrajsek KT, Bratanic N, Battelino T, Willcox N, Peterson P, Kisand K. Anti-cytokine autoantibodies suggest pathogenetic links with autoimmune regulator deficiency in humans and mice. Clin Exp Immunol. 2013;171:263–272. doi: 10.1111/cei.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Leung JM, Davenport M, Wolff MJ, Wiens KE, Abidi WM, Poles MA, Cho I, Ullman T, Mayer L, Loke P. IL-22-producing CD4+ cells are depleted in actively inflamed colitis tissue. Mucosal Immunol. 2014;7:124–133. doi: 10.1038/mi.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Xu H, Feely SL, Wang X, Liu DX, Borda JT, Dufour J, Li W, Aye PP, Doxiadis GG, Khosla C, Veazey RS, Sestak K. Gluten-sensitive enteropathy coincides with decreased capability of intestinal T cells to secrete IL-17 and IL-22 in a macaque model for celiac disease. Clin Immunol. 2013;147:40–49. doi: 10.1016/j.clim.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Hollander GA, Krenger W, Blazar BR. Emerging strategies to boost thymic function. Curr Opin Pharmacol. 2010;10:443–453. doi: 10.1016/j.coph.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Boehm T, Swann JB. Thymus involution and regeneration: two sides of the same coin? Nat Rev Immunol. 2013;13:831–838. doi: 10.1038/nri3534. [DOI] [PubMed] [Google Scholar]
- 378.Legrand N, Dontje W, van Lent AU, Spits H, Blom B. Human thymus regeneration and T cell reconstitution. Semin Immunol. 2007;19:280–288. doi: 10.1016/j.smim.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 379.Maury S, Mary JY, Rabian C, Schwarzinger M, Toubert A, Scieux C, Carmagnat M, Esperou H, Ribaud P, Devergie A, Guardiola P, Vexiau P, Charron D, Gluckman E, Socie G. Prolonged immune deficiency following allogeneic stem cell transplantation: risk factors and complications in adult patients. Br J Haematol. 2001;115:630–641. doi: 10.1046/j.1365-2141.2001.03135.x. [DOI] [PubMed] [Google Scholar]
- 380.Storek J, Gooley T, Witherspoon RP, Sullivan KM, Storb R. Infectious morbidity in long-term survivors of allogeneic marrow transplantation is associated with low CD4 T cell counts. Am J Hematol. 1997;54:131–138. doi: 10.1002/(sici)1096-8652(199702)54:2<131::aid-ajh6>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 381.Maraninchi D, Gluckman E, Blaise D, Guyotat D, Rio B, Pico JL, Leblond V, Michallet M, Dreyfus F, Ifrah N, et al. Impact of T-cell depletion on outcome of allogeneic bone-marrow transplantation for standard-risk leukaemias. Lancet. 1987;2:175–178. doi: 10.1016/s0140-6736(87)90763-x. [DOI] [PubMed] [Google Scholar]
- 382.Curtis RE, Rowlings PA, Deeg HJ, Shriner DA, Socie G, Travis LB, Horowitz MM, Witherspoon RP, Hoover RN, Sobocinski KA, Fraumeni JF, Jr, Boice JD., Jr Solid cancers after bone marrow transplantation. N Engl J Med. 1997;336:897–904. doi: 10.1056/NEJM199703273361301. [DOI] [PubMed] [Google Scholar]
- 383.Small TN, Papadopoulos EB, Boulad F, Black P, Castro-Malaspina H, Childs BH, Collins N, Gillio A, George D, Jakubowski A, Heller G, Fazzari M, Kernan N, MacKinnon S, Szabolcs P, Young JW, O'Reilly RJ. Comparison of immune reconstitution after unrelated and related T-cell-depleted bone marrow transplantation: effect of patient age and donor leukocyte infusions. Blood. 1999;93:467–480. [PubMed] [Google Scholar]
- 384.Gray DH, Fletcher AL, Hammett M, Seach N, Ueno T, Young LF, Barbuto J, Boyd RL, Chidgey AP. Unbiased analysis, enrichment and purification of thymic stromal cells. J Immunol Methods. 2008;329:56–66. doi: 10.1016/j.jim.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 385.Pan B, Liu J, Zhang Y, Sun Y, Wu Q, Zhao K, Zeng L, Xu K. Acute ablation of DP thymocytes induces up-regulation of IL-22 and Foxn1 in TECs. Clinical Immunology. 2014;150:101–108. doi: 10.1016/j.clim.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 386.Nehls M, Kyewski B, Messerle M, Waldschutz R, Schuddekopf K, Smith AJ, Boehm T. Two genetically separable steps in the differentiation of thymic epithelium. Science. 1996;272:886–889. doi: 10.1126/science.272.5263.886. [DOI] [PubMed] [Google Scholar]
- 387.Chen L, Xiao S, Manley NR. Foxn1 is required to maintain the postnatal thymic microenvironment in a dosage-sensitive manner. Blood. 2009;113:567–574. doi: 10.1182/blood-2008-05-156265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Bleul CC, Corbeaux T, Reuter A, Fisch P, Monting JS, Boehm T. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature. 2006;441:992–996. doi: 10.1038/nature04850. [DOI] [PubMed] [Google Scholar]
- 389.Bredenkamp N, Nowell CS, Blackburn CC. Regeneration of the aged thymus by a single transcription factor. Development. 2014;141:1627–1637. doi: 10.1242/dev.103614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Zook EC, Krishack PA, Zhang S, Zeleznik-Le NJ, Firulli AB, Witte PL, Le PT. Overexpression of Foxn1 attenuates age-associated thymic involution and prevents the expansion of peripheral CD4 memory T cells. Blood. 2011;118:5723–5731. doi: 10.1182/blood-2011-03-342097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Hepworth MR, Monticelli LA, Fung TC, Ziegler CGK, Grunberg S, Sinha R, Mantegazza AR, Ma H-L, Crawford A, Angelosanto JM, Wherry EJ, Koni PA, Bushman FD, Elson CO, Eberl G, Artis D, Sonnenberg GF. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Wilson MS. Redundant and pathogenic roles for IL-22 in mycobacterial, protozoan, and helminth infections. J. Immunol. 2010;184:4378–4390. doi: 10.4049/jimmunol.0903416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Graham AC. IL-22 production is regulated by IL-23 during Listeria monocytogenes infection but is not required for bacterial clearance or tissue protection. PLoS ONE. 2011;6:e17171. doi: 10.1371/journal.pone.0017171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Zenewicz LA. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J. Immunol. 2013;190:5306–5312. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.De Luca A. IL-22 defines a novel immune pathway of antifungal resistance. Mucosal Immunol. 2010;3:361–373. doi: 10.1038/mi.2010.22. [DOI] [PubMed] [Google Scholar]
- 396.Kisand K. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J. Exp. Med. 2010;207:299–308. doi: 10.1084/jem.20091669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Karow M, Flavell RA. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity. 2007;27:647–659. doi: 10.1016/j.immuni.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]