Abstract
The microenvironment of the cancer cell is pivotal to its phenotypic regulation. One of the central components of the microenvironment is temperature. An elevation in environmental temperature has been shown to increase the cancer cell's susceptibility to chemo- and radiation therapy. The goal of the studies described here was to identify some of the pathways that are modified by a mild increase in temperature in cancer cells. Using prostate cancer cells as a model system we found that in addition to the well described and anticipated up-regulation of the heat shock family of proteins, there is a significant down-regulation of certain members of the “cold shock” family of proteins such as, RNA binding motif protein 3 (RBM3) and cold inducible RNA binding protein (CIRBP). siRNA-mediated down-regulation of the cold shock protein (CSP) encoding mRNAs dramatically attenuates cell survival in the absence of any heat application. Furthermore, we also demonstrate that knocking down the CSPs can enhance the therapeutic response of prostate cancer cells to chemotherapy. Our findings suggest that down-regulating CSPs in cancer cells may “mimic” the stress response the cells experience when exposed to heat treatment rendering them more susceptible to therapy. Thus, the pharmacological modulation of RBM3 and CIRBP may represent novel therapeutic approaches for prostate cancer.
Keywords: Cold Shock Proteins, Chemotherapy, Prostate Cancer
The microenvironment plays a pivotal role in tumor cell activity. While classically studies on the environment have focused on cell–cell and cell–extracellular matrix interactions, other environmental features that appear to influence tumor biology remain relatively unexplored. Heat is an important microenvironmental and epigenetic regulatory factor in many biological processes including growth and differentiation [Ferguson and Joanen, 1982; Guan et al., 2006]. An innate cellular response to microenvironmental temperature fluctuations includes not only the heat shock family of proteins but several other pathways that ultimately minimize the deleterious effects of heat stress, prevent apoptosis and ensure cell survival [Shamovsky and Nudler, 2008].
At the same time, numerous clinical and basic studies have demonstrated that tumor cells are less adaptive to heat stress resulting in decreased survival both in vivo and in vitro [Dewhirst et al., 2005]. Further, with many tumor types a mild increase in heat (41–43°C) synergizes the response with other therapies such as radiation therapy, chemotherapy, and immunotherapy. Perhaps one of the most clinically relevant examples of heat synergism may be seen in testicular cancer. It has been suggested that metastatic testicular cancer cells that spread into the body cavity–and thus experience a relatively higher temperature–may be more sensitive to therapeutically induced cell death by radiation or chemotherapy [Coffey et al., 2006].
The most sensitive cellular target of heat is the nuclear matrix, a dynamic scaffold that organizes many functions within the nucleus. It plays an important role in several cellular activities such as maintaining cell morphology, three-dimensional organization of the nucleus, DNA replication and transcription. As a result, the nuclear matrix is intimately associated with cell proliferation and differentiation, as well as carcinogenesis [Berezney et al., 1995; Pederson, 2000]. In cancer cells, the nuclear matrix is not only abnormal in morphology, but is also different in its composition [Getzenberg et al., 1996]. Studies of the thermal effects on the nucleus by Roti Roti et al. [1998] and Lepock et al. [2001] have demonstrated thermally induced unfolding of the nuclear matrix and subsequent changes in the binding of specific proteins to the matrix. Taken together these data strongly suggest that changes in the structure and composition of the nuclear matrix in response to heat treatment of cancer cells may account for some of the underlying mechanisms. However, despite significant progress in uncovering the effects of heat shock on cellular processes [Roti Roti, 2008], understanding the molecular mechanisms underlying its effects in treating cancer still represent significant challenges.
In this article a comprehensive study aimed at uncovering molecular changes in prostate cancer cells in response to mild heat was performed. During the course of these investigations it was observed that in addition to the expected up-regulation of the heat shock proteins (HSPs), there is a significant down-regulation of the cold shock proteins (CSPs), namely RNA binding motif protein 3 (RBM3) and cold inducible RNA binding protein (CIRBP). The CSPs are a conserved group of proteins that are induced upon hypothermia and other forms of cellular stress such as UV radiation and hypoxia [De Leeuw et al., 2007]. Experimentally down-regulating their expression has a dramatic impact of prostate cancer cell survival even in the absence of heat treatment. Furthermore, when coupled to drug treatment, there is a significant increase in cell death. Taken together, the present findings may have identified a new pathway that can be pharmacologically manipulated to enhance therapeutic approach for prostate cancer.
Materials and Methods
Cell Culture and Treatment
The prostate cancer cell lines PC-3 and LNCaP were obtained from the American Type Culture Collection (Rockville, MD). The cells were cultured at 37°C in routine RPMI (Invitrogen, Carlsbad, CA) media supplemented with 10% fetal bovine serum (FBS) and treated by mild heat for an indicated duration in an incubator at 41°C, or simultaneously treated with chemical drug for 4 h.
RNA Isolation, Microarray Experiments, and Data Analysis
Total RNA isolation, fragmentation and microarray hybridization and scanning procedures were carried out following the suppliers protocol (Agilent Technologies). Lowess normalization was used to normalize the intensity log ratio M of the non-control probes. All computations were performed under R environment (http://www.r-project.org).
Reverse Transcription (RT) and Real-Time PCR
RNA samples were treated with DNase I (Invitrogen), and cDNA was synthesized using the iScript cDNA synthesis kit (BioRad, Hercules, CA). Real-time PCR was done in triplicate on an iCycler iQ Multicolor Real-time PCR Detection system (BioRad). Target gene expression was related to TATA box binding protein (TBP) for normalization. PCR sequences used were shown in Supplemental Table I.
Western Blotting
Twenty-five micrograms of protein were separated on 10–20% SDS–PAGE and transferred onto PVD filters (Millipore, Bedford, MA). Membranes were incubated with primary antibodies overnight at 4°C followed by horseradish peroxidase-conjugated secondary antibodies, and developed with the Super Signal West Dura Extended Duration Substrate kit (Pierce). Rbm3 antibody was generated in a rabbit against a peptide (Sigma-Gneosys, The Woodlands, TX). Hspa1a and cirbp antibodies were obtained from Lifespan Biosciences (ProteinTech Group, Chicago, IL). Phosphohistone H2A.X (γ-H2A.X) antibody was obtained from Millipore. Other antibodies were purchased from Cell Signaling (Danvers, MA) and Santa Cruz (Santa Cruz, CA).
Nuclear Matrix Protein (NMP) Isolation, Two-Dimensional Gel Electrophoresis, and Protein Identification
NMP extraction and high resolution, two-dimensional electrophoresis was performed as previously described [Inoue et al., 2008]. Protein identification was done by LC matrix-assisted laser desorption/ionization mass spectrometry (LC/MALDI MS) using an ABI Tempo LC MALDI mass spectrometer in the reflector mode using delayed extraction. Peptides were analyzed by collisioninduced dissociation (CID) using nano-LC tandem mass spectrometry analysis on a LTQ (Thermo Fisher Scientific, Waltham, MA) [Shevchenko et al., 1996]. Peptide sequences were identified by screening the fragmentation data against the NCBI non-redundant database (uniprot_sprot_20070123) using the Mascot sequence query search engine (Matrix Science, Boston, MA). Identified sequences were confirmed by manually inspecting CID spectra.
Cell Viability Inhibition Assay
Five thousand cells per well were seeded in 96-well plates. Seventy-two hours after drug treatment, cell proliferation reagent WST-1 (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate) was added to each well, as specified by the supplier (Roche, Nutley, NJ). After 4-h incubation, WST-1 absorbance at 450 nm was measured.
Sirna Transfections
RBM3 and CIRBP ON-TARGETplus SMARTpool siRNA were obtained from Dharmacon (Lafayette, CO; sequences available on request). siRNA was transfected with 2.5 × 103 PC-3 cells or 5 × 103 LNCaP cells at 100 nM total oligo concentration, using 0.2 μl DharmaFECT-3 transfection reagent, and plated simultaneously in a 96-well plate. Chemical treatment was conducted 24–48 h after transfection.
Clonogenic Assays
PC-3 and LNCaP cells were transfected with 100 nM RBM3 or CIRBP siRNA. Forty-eight hours after transfection, cells were plated at 500 cells (PC-3) or 1,000 cells (LNCaP) in 10-cm dish and grown without being disturbed. Colonies were counted after 2 weeks. Only colonies containing more than 50 cells were included.
Facs Analysis of Apoptosis and Cell Cycle
Apoptosis and cell cycle were detected on a Guva system (Guava Technologies, Hayward, CA) using Terminal Transferase dUTP Nick End Labeling (TUNEL) kit (Guava Technologies) and Guava Cell Cycle Reagent (Guava Technologies), respectively.
Statistical Analysis
Comparisons were made using the Student's t-test. Two-sided P of less than 0.05 was considered significant.
Results
In order to elucidate the changes in cellular pathways in response to mild heat, an experimental design which focuses on a single mild temperature increase was implemented (Fig. 1). Earlier in vitro studies have demonstrated that 1–4 h of heating (depending on the sequence of modalities) at 41°C produces substantial heat-induced radiosensitization with little or no cell killing by heat alone [Myerson et al., 2004]. This temperature is also one which is clinically feasible. The goal of this study was to examine the designed heat response in prostate cancer cells lines. The androgen-responsive LNCaP prostate cancer cells and the aggressive androgen-insensitive PC-3 cancer cells representing relatively diverse forms of the disease were utilized. Cells were grown at the normal temperature of 37°C or at 41°C to discern their response to heat-shock for the designated time periods (Fig. 1).
Fig. 1.

Experimental design for mapping cellular response to mild heat treatment. Prostate cancer cells PC-3 and LNCaP were cultured at 41°C for indicated duration and subjected to RNA or protein analysis.
Changes in NMP Composition in Response to Heat
Since changes in NMP composition have often been accompanied by changes in cellular pathology, proteomic analysis of the nuclear matrix preparations from both cell lines was performed. Several spots appeared to be distinctly over-expressed upon heat shock that were identified by mass-spectrometric peptide sequencing as heat shock 70 kDa protein 1A (hspa1a), heat shock 70 kDa protein 8 (hspa8) and heat shock 70 kDa protein 6 (hspa6) (Supplemental Fig. 1 and Supplemental Table II). However, no protein spots were observed that appeared to be significantly down-regulated upon heat treatment at 41°C.
Mild Heat Treatment Up-Regulates Heat Shock Proteins but Down-Regulates Cold Shock Proteins
In order to correlate the changes in protein expression with concomitant changes in gene expression global gene expression profiles were interrogated using DNA microarrays. Only those genes that had Odds ratios of ≥10 and a fold change of ≥ 1.5 were selected as being significantly different. In LNCaP and PC-3 cells, 216 and 158 genes are dysregulated, respectively (Supplemental Tables III and IV). Of the dysregulated genes, 54 genes are shared by both cell lines (Table I) suggesting that there may be common pathways that cancer cells utilize to counter the stress response. The majority of the differentially regulated genes are up-regulated genes in both cell lines. Of these, as one would expect, the most abundant genes are those encoding the HSPs and proteins related to modulating HSPs such as AHSA1, corroborating the results from the proteomic analysis. Among other classes of genes are those that encode transcription factors such as CAMTA2 and WBP5, transporters such as SLC1A3, and those related to apoptosis such as BAG3 and THAP2. Unlike the proteomic analysis, gene expression profiling also revealed genes that are significantly down-regulated including those encoding the RNA-binding proteins many of which belong to the “cold shock protein” family such as RBM3 and CIRBP (Table I).
Table I.
Representative Common Dysregulated Gene in PC-3 and LNCaP Cells*
| Systematic name | Description | Gene name | PC-3 fold change | LNCaP fold change |
|---|---|---|---|---|
| NM_005345 | Heat shock 70 kDa protein 1A | HSPA1A | 7.7 | 5.8 |
| NM_005527 | Heat shock 70 kDa protein 1-like | HSPA1L | 6.0 | 4.7 |
| NM_006644 | Heat shock 105 kDa/110 kDa protein 1 | HSPH1 | 2.8 | 3.6 |
| NM_004281 | BCL2-associated athanogene 3 | BAG3 | 2.5 | 3.8 |
| NM_014278 | Heat shock 70 kDa protein 4-like | HSPA4L | 2.4 | 2.4 |
| NM_130469 | Jun dimerization protein 2 | JDP2 | 2.1 | 2.2 |
| NM_007034 | Dnaj (Hsp40) homolog, subfamily B, member 4 | DNAJB4 | 2.1 | 3.6 |
| NM_001114 | Adenylate cyclase 7 | ADCY7 | 2.1 | 1.9 |
| NM_001539 | Dnaj (Hsp40) homolog, subfamily A, member 1 | DNAJA1 | 2.1 | 2.1 |
| NM_005347 | Heat shock 70 kDa protein 5 | HSPA5 | 2.0 | 2.7 |
| NM_001540 | Heat shock 27 kDa protein 1 | HSPB1 | 1.9 | 1.7 |
| NM_001040141 | Heat shock protein 90 kDa alpha, class A member 2 | HSP90AA2 | 1.9 | 1.7 |
| NM_004172 | Solute carrier family 1, member 3 | SLC1A3 | 1.8 | 1.7 |
| NM_006145 | Dnaj (Hsp40) homolog, subfamily B, member 1 | DNAJB1 | 1.8 | 2.6 |
| NM_006010 | Arginine-rich, mutated in early stage tumors | ARMET | 1.8 | 2.2 |
| NM_012111 | AHA1, activator of heat shock 90 kDa protein atpase homolog 1 | AHSA1 | 1.8 | 1.6 |
| NM_015099 | Calmodulin binding transcription activator 2 | CAMTA2 | 1.8 | 1.6 |
| NM_001005199 | Olfactory receptor, family 8, subfamily H, member 1 | OR8H1 | 1.7 | 1.7 |
| NM_052957 | Acidic repeat containing | ACRC | 1.7 | 1.8 |
| NM_182972 | Interferon regulatory factor 2 binding protein 2 | IRF2BP2 | 1.7 | 1.7 |
| NM_031435 | THAP domain containing, apoptosis associated protein 2 | THAP2 | 1.7 | 1.6 |
| NM_016303 | WW domain binding protein 5 | WBP5 | 1.6 | 2.1 |
| NM_003124 | Sepiapterin reductase | SPR | 1.6 | 1.6 |
| NM_015513 | Cysteine-rich with EGF-like domains 1 | CRELD1 | 1.6 | 1.9 |
| NM_000164 | Gastric inhibitory polypeptide receptor | GIPR | 1.6 | 2.0 |
| NM_032356 | LSM domain containing 1 | LSMD1 | 1.6 | 1.7 |
| NM_002155 | Heat shock 70 kDa protein 6 | HSPA6 | 1.6 | 2.0 |
| NM_014660 | PHD finger protein 14 | PHF14 | −1.6 | −1.6 |
| NM_001280 | Cold inducible RNA binding protein | CIRBP | −1.6 | −1.7 |
| NM_001356 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | DDX3X | −1.6 | −1.5 |
| NM_005033 | Exosome component 9 | EXOSC9 | −1.6 | −1.5 |
| NM_001017430 | RNA binding motif protein 3 | RBM3 | −1.7 | −1.5 |
| NM_194460 | Ring finger protein 126 | RNF126 | −1.7 | −2.0 |
| NM_031372 | Heterogeneous nuclear ribonucleoprotein D-like | HNRPDL | −1.9 | −1.7 |
All dysregulated gene were selected by Odd ratio ≥10, fold change ≥1.5.
Real-Time RT-PCR Validation of Microarray Data
Gene expression changes identified using microarrays were independently verified by real-time RT-PCR. Representative members from both up- and down-regulated genes were selected. As shown in Figure 2A, both HSPA1A and HSPA6 are also found to be up-regulated upon heat treatment at 41 °C while both the CSPs RBM3 and CIRBP are down-regulated at the same temperature in both the prostate cell lines. Notably, the decrease of CSP expression was enhanced when the exposure time at mild heat was extend from 4 to 24 h, suggesting a dose-dependent modulation of heat on expression of these genes.
Fig. 2.
Validation of representative gene expression changes in microarray analysis. A: mRNA levels were determined by real-time RT-PCR. Values are expressed relative to the expression of TATA box binding protein (TBP) mRNA. Data are represented as mean ±SD. B: Cold shock protein levels are decreased upon heat treatment. PC-3 and LNCaP cells were subjected to 41°C heat treatment for indicated duration or maintained at37°C, and Western blotting was performed to detect hspa1a, rbm3 and cirbp proteins. Actin was used as a control for protein loading.
Immunoblotting Confirms Differential Expression at Protein Level
Having confirmed the gene expression changes at the RNA level, the protein levels were evaluated. Immunoblotting analysis of total cell lysates from both cell lines demonstrated that Hspa1a increased in expression within 4 h of heat administration (Fig. 2B). However, unlike the RNA levels, a sustained increased expression of the Hspa1a protein was observed over the 24 h study period. Alternatively, both the rbm3 and cirbp proteins are significantly down-regulated during the same period (Fig. 2B). Considered together, the data demonstrate that mild heat treatment results in the up-regulation of a family of HSPs and down-regulation of members of the cold shock proteins both at the mRNA as well as the protein level.
Sirna-Mediated Knockdown of Cold Shock Protein Genes Attenuates Prostate Cancer Cell Growth
As a class the CSPs include RNA-binding proteins and have been reported to be up-regulated in response to cold and other forms of stress analogous to the HSPs [Sutherland et al., 2005]. Despite this, the functional significance of this down-regulation in response to heat-induced stress is not readily apparent. In order to gain additional insight, the expression of the cold shock transcripts was reduced to determine if this could, at least in part, “mimic” the heat-shock effect and thereby, enhance the susceptibility of these cells to therapeutic treatment in the absence of any heat. As shown in Figure 3A, siRNAs specific to either RBM3 or CIRBP decreased the levels of their respective mRNAs by 85–95% compared to a nonspecific (scrambled) siRNA used at the same concentration. The rbm3 and cirbp proteins were also similarly effected by the knockdown and as evident from Figure 3B the proteins were reduced to almost undetectable levels.
Fig. 3.
Knocking down RBM3 and CIRBP by siRNA in PC-3 and LNCaP cells. PC-3 and LNCaP cells were transfected with 100 nM RBM3 or CIRBP SMART-pool siRNA. Forty-eight hours after transfection, total RNA was isolated and mRNA levels of RBM3 and CIRBP were detected by real-time RT-PCR (A), or cells were lysed and subjected to Western blot to detect Rbm3 and Cirbp levels (B).
Down-Regulating RBM3 and CIRBP Impairs Cell Survival
Having confirmed that the siRNAs abolished the expression of the rbm3 and cirbp proteins, the CIRBP or RBM3 knockdown were evaluated to determine if they could “mimic” the stress effects of heat shock in impairing cancer cell survival. Down-regulation of RBM3 or CIRBP significantly inhibited cell growth in both PC-3 and LNCaP cells (Fig. 4A). Furthermore, colony formation that relies on the intactness of cell survival ability was inhibited by the RBM3 or CIRBP knockdowns by 36% or 62%, respectively, in PC-3 cells and 50% or 46% in LNCaP cells (Fig. 4B). However, unlike in the case of CIRBP where knockdown had a similar effect on cell survival in both cell lines, RBM3 knockdown was more effective in LNCaP cells than in PC-3 cells suggesting that at least a component of RBM3 function may be cell type-dependent.
Fig. 4.
Reducing the expression of RBM3 and CIRBP inhibits cell survival and colony formation in prostate cancer cells. A: PC-3 (1,000 cells) and LNCaP cells (2,500 cells) were transfected with 100 nM RBM3 or CIRBP SMARTpool siRNA. Cell viability was evaluated by WST-1 assay on 3–6 days after transfection. Magnifications 100× on left panels. B: Forty-eight hours after RBM3 or CIRBP transfection, cells were re-plated in 10-cm dish with 500 cells (PC-3) or 1,000 cells (LNCaP) per dish, and colonies were counted after 2-week growing. All experiments were repeated at a minimum of two times. *P<0.01 compared to scrambled siRNA-treated control.
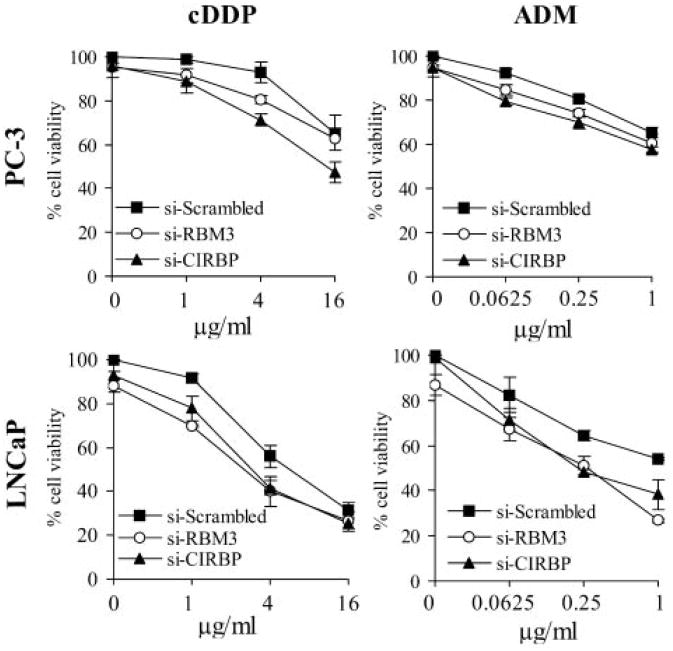
Down-Regulating RBM3 and CIRBP Enhances Chemosensitivity in Prostate Cancer Cells
To investigate whether CSP down-regulation could also “mimic” the synergistic effect of heat on chemotherapy, RBM3 or CIRBP expression was knocked down and the cells subsequently treated with adriamycin (ADM) or cisplatin (cDDP). As shown in Figure 5, reducing the expression of these genes resulted in a marked reduction of cell survival by ∼25–50% in PC-3 and LNCaP cells, respectively compared to the drug treatment alone. Again, RBM3 knockdown showed a larger enhancement on chemosensitivity in LNCaP cells than in PC-3 cells, which is consistent with the above cell survival results. The enhancement of RBM3 and CIRBP knockdown on chemosensitivity was also confirmed by detecting apoptosis using the TUNEL assay (Supplemental Fig. 2). Heating the cells at 41°C combined with drug treatment also resulted in a similar decrease in cell survival (Supplemental Fig. 3) highlighting the heat mimicking effects of the siRNAs on chemotherapy.
Fig. 5.
Knocking down RBM3 and CIRBP by siRNA enhances chemosensitivity in PC-3 cells and LNCaP cells. PC-3 and LNCaP cells were transfected with 100 nM RBM3 or CIRBP SMARTpool siRNA. Twenty-four hours after siRNA transfection, PC-3 cells and LNCaP cells were treated with increasing doses of cisplatin (cDDP) or adriamycin (ADM) for 4 h and then moved back to normal medium for another 72 h. Cell viability inhibition was determined using the WST-1 assay. Data are represented as mean±SD.
RBM3 and CIRBP Act Via Different Mechanisms
To address the mechanism by which RBM3 and CIRBP might modulate the heat mimic, potential cellular pathways were explored. A TUNEL assay revealed that reducing the expression of RBM3 or CIRBP alone (in the absence of any drug) failed to induce apoptosis in either prostate cancer cell line (data not shown). In contrast, RBM3 down-regulation induced a cell cycle arrest prior to the S phase and G2-M phase in LNCaP but not PC-3 cells (Supplemental Fig. 4A and B). Consistent with the cell cycle analysis, knocking down RBM3 reduced cyclin B1 protein levels in LNCaP but not in PC-3 cells (Fig. 6). More importantly, cyclin B1 levels were not increased in response to drug treatment in RBM3 knocked-down LNCaP cells as it was in the scrambled siRNA-treated control cells (Fig. 6B) corroborating the fact that RBM3 down-regulation is more effective in LNCaP cells than in PC-3 cells (Fig. 5).
Fig. 6.
RBM3 and CIRBP knockdown enhances DNA damage induced by chemotherapy. Prostate cancer cells PC-3 (A) and LNCaP (B) were transfected with 100 nM RBM3 or CIRBP SMARTpool siRNA. Forty-eight hours after transfection, cells were treated with 4 μg/ml cDDP or 1 μg/ml ADM for 4 h, and then moved back to normal medium for another 16 h. Cell lysates were subjected to Western blotting to detect cyclin B1, cyclin D1, phospho-histone H2A.X (Ser139) (γ-H2A.X), phospho-p53 (Ser15), and p21. Actin was used as a control for protein loading.
Alternatively, reducing expression of CIRBP in either PC-3 or LNCaP cells did not have a significant impact on either cell cycle or cyclin B1 and D1 levels (Supplemental Fig. 4A and B). However, the levels of cyclin B1 and/or cyclin D1 in CIRBP knocked-down in either cell line were significantly lower than scrambled siRNA-treated control after treating cells with drug (Fig. 6). Together with the fact that CIRBP knockdown similarly enhanced chemosensitivity in both PC-3 and LNCaP cells, these results suggest that CIRBP acts via a mechanism distinct from that of RBM3.
RBM3 and CIRBP May Be Involved In DNA Damage Repair
It is generally accepted that DNA damage and subsequent cell death may be the primary cytotoxic mechanism of ADM and other DNA-binding antitumor drugs. However, since reducing the expression of RBM3 or CIRBP did not induce apoptosis or only partly impeded the cell cycle, it was determined if the effect of knocking down their expression impaired DNA damage repair that results in decreasing cell survival during proliferation and rendering the cells more susceptible to the effects of chemotherapy. Since phosphorylation of H2A.X at serine 139 correlates well with DNA damage [Plesca et al., 2008], we determined the γ-H2A.X content to evaluate the impact of the knockdown on DNA damage and repair response. As shown in Figure 6B, both RBM3 and CIRBP down-regulation significantly enhanced DNA damage triggered by drugs in LNCaP cells as judged by the increase in accumulation of γ-H2A.X. Again, only decreased expression of CIRBP but not RBM3 increased cDDP and ADM induced DNA damage in PC-3 cells, further suggesting that RBM3 function is different between two cell lines (Fig. 6A). Notably, even without any chemical stress, reducing the expression of CIRBP still can induce a slight but identifiable increase of DNA damage marker.
Finally, since p53-p21 proteins play a key role in protecting the cell from DNA damage-induced cell death [Garner and Raj, 2008], their expression in LNCaP cells that expresses wild-type p53 was determined. As expected, knocking down both RBM3 and CIRBP significantly inhibited the activation of p53. In contrast, p53 was activated by cDDP and ADM treatment in scrambled siRNA controls. The up-regulation of p21 induced by ADM was also impeded in LNCaP cells. Furthermore, the significant inhibition in colony formation after knocking-down these two genes in both cell lines in the early experiment also supports the plausible impairment of DNA damage response pathway (Fig. 6). These results suggest that RBM3 and CIRBP may be involved in the DNA damage response process. Consequently, down-regulating them, impairs cell survival and enhances chemotherapy.
Discussion
The benefit of combining heat treatment along with other therapeutic approaches is an area of significant promise in the treatment of cancer. However, the mechanisms leading to favorable clinical results of heat therapy are not fully understood. By interrogating global gene expression profiles and the nuclear matrix-associated proteome, we have identified some of the key heat-induced alterations in the tumor microenvironment using cell models of cancer.
As expected, an overwhelming up-regulation of HSPs and genes related to the cancer cell's innate heat shock response were observed. However, of the dozens of stress-induced HSPs in a cancer cell, only a few have been found to have critical cytoprotective roles in cancer such as HSP27 and HSP70 [Garrido et al., 2006]. Both proteins are powerful chaperones, inhibit key effectors of the apoptotic machinery, and participate in the proteasome-mediated degradation of proteins under stress conditions, thereby contributing to the so called “protein triage.” In cancer cells, both HSP27 and HSP70 appear to participate in oncogenesis and in resistance to chemotherapy [Zhao et al., 2004; Rocchi et al., 2006; Didelot et al., 2007; Kamada et al., 2007]. HSP27 and HSP70 were up-regulated in both cell lines studied here. However, while HSP70 was detected both in gene expression and nuclear matrix proteomic analyses, HSP27 over-expression was only observed in the gene expression profiling studies. Consistent with this result, it has been reported by other investigators that hsp70 binds to the nuclear matrix [Lepock et al., 2001], hsp27, however, interacts indirectly by associating with Safb a constituent of the nuclear matrix [Oesterreich et al., 1997] and thus, may not withstand the harsh extraction procedures underlying nuclear matrix preparations. Interestingly, after a quick increase in response to heat, hsp expression was still maintained at the high level even when exposure time to heat was extended as long as 24 h, at which point the cells become more fragile to survival than those heated for 1 or 4 h. Through exploring the globe gene expression under heat stress, it is of particular interest to identify a decreased expression of two CSPs, which was further confirmed as slow but persistent reduction over heating.
Notably, all organisms from prokaryotes to plants and higher eukaryotes respond to cold shock in a comparatively similar manner. Cold shock invokes the rapid over-expression of a small group of proteins called cold shock proteins, and a coordinated cellular response involving modulation of transcription, translation, metabolism, the cell cycle and the cell cytoskeleton. In mammals however, to date, only two cold shock proteins have been described in detail, cirbp and rbm3 [Al-Fageeh and Smales, 2006]. There is now growing evidence that in addition to their role in cold stress response, the CSPs also play critical roles in cancer cell survival and growth. For example, CIRBP is induced by stresses such as UV irradiation and hypoxia [De Leeuw et al., 2007]. We also found that ADM administration in vitro increased CIRBP expression in LNCaP cells (unpublished observation). Indeed, using the RKO colorectal carcinoma cells, Yang and Carrier [2001] observed that cells expressing reduced levels of CIRBP are more sensitive to UV radiation than those over-expressing CIRBP.
The extraordinary success in treating distant metastasis of testicular cancer is speculated to be due, at least in part, to its susceptibility to the increased body temperature experienced by the metastatic cells [Coffey et al., 2006]. Interestingly, when the mouse testis was exposed to heat stress by experimental cryptorchidism or immersion of the lower abdomen in warm (42°C) water, the expression of cirbp was decreased in the testis within 6 h after either treatment [Nishiyama et al., 1998]. The present results verify that CIRBP expression in cancer cells is also modulated by microenvironmental temperature. Importantly, experimentally lowering CIRBP expression by siRNA to reflect its down-regulation under heat stress decreased cell survival and increased cytotoxic killing, which indeed is an expected therapeutic benefit of heat treatment. Whether testicular cancer cells lose this CSP expression in metastatic sites due to the increased body temperature and therefore result in their high susceptibility to cytotoxic reagents needs to be further clarified. However, targeting CIRBP may represent novel therapeutic approach for prostate cancer.
RBM3 is one of the first proteins synthesized in response to cold shock [Danno et al., 1997]. Although the exact biological function of RBM3 is not fully understood, members of the RBM protein family contain the primary structural motif most commonly referred to as the RNA-recognition motif (RRM) and are thought to function as RNA binding apoptosis regulators [Sutherland et al., 2005]. RBM3 in particular, appears to be a novel proto-oncogene that induces transformation when over-expressed and is essential for cells to progress through mitosis [Sureban et al., 2008]. While overexpression increases cell proliferation and development of compact multicellular spheroids in soft agar, down-regulating RBM3 in HCT116 colon cancer cells with specific siRNA decreases cell growth in culture and tumor xenografts [Sureban et al., 2008]. Down-regulation also increases caspase-mediated apoptosis coupled with nuclear cyclin B1, and phosphorylated cdc25c, chk1 and chk2 kinases, implying that under conditions of RBM3 downregulation, cells undergo mitotic catastrophe [Sureban et al., 2008]. Like in colon cancer, RBM3 is also up-regulated in prostate cancer. However, unlike in colon cancer wherein there is a stage-dependent increase [Sureban et al., 2008] RBM3 is apparently down-regulated in late-stage prostate cancer (www.oncomine.org, unpublished observations). Further, in contrast to colon cancer wherein its down-regulation leads to mitotic catastrophe, the present data indicate that in androgen-sensitive but not in androgen-independent prostate cancer cells, down-regulation of RBM3 leads to a cell cycle arrest and an enhancement of DNA damage induced by drug suggesting tumor-specific differences and warrants further investigation.
Our results on the differential effects of knocking down RBM3 on the cytotoxic drugs in two prostate cancer cell lines may be predicated on the p53 status of the cell models utilized. In response to DNA damage, induction of p53 protein either causes the cells to arrest in different phases of the cell cycle, or if DNA damage is excessive, p53 leads the cells through apoptosis by regulating the bcl-2 family of genes [Weinberg, 1991; Lane, 1992]. Of particular note is the fact that LNCaP cells possess wild-type p53 while PC-3 has mutant p53 that cannot be functionally activated following DNA damage [O'Connor et al., 1997]. It has been shown that the susceptibility of these prostate cancer cells to cDDP and its analogs appears to be linked to the p53 status [Mujoo et al., 2005]. Indeed, in the present study it was observed that knocking down RBM3 impedes p53 activation and the subsequent p21 expression, both of which previously have been shown to render DNA damage repair [Garner and Raj, 2008], when LNCaP cells was subjected to a cytotoxic stress. Considered together, it is tempting to conjecture that RBM3 may be involved in p53-linked DNA damage repair. In contrast, while CIRBP knocked-down similarly altered p53-p21 proteins and increased γ-H2A.X expression under the chemical stress in LNCaP cells, it is also able to enhance DNA damage and cytotoxic killing in PC-3 cells in which p53 regulation is deficient. This p53-independent pathway appears to impair cyclin B1 increase and enhance cyclin D1 decrease after drug treatment. Furthermore, in contrast to the up-regulation of CIRBP by adding ADM to LNCaP cells, RBM3 was significantly down-regulated in response to ADM treatment (unpublished observation). Together, the data suggest that RBM3 and CIRBP appear to involve different cell death resistant mechanisms in different types of cancer cells.
In summary, the down-regulation of the CSPs in response to heat treatment suggests that the CSPs may account for, at least in part, the impairment in cell survival and enhanced sensitivity of heattreated prostate cancer cells to subsequent therapeutic modalities albeit, by different mechanisms. Perhaps, a combination of modalities such as a double knockdown of the HSPs and CSPs for example, coupled with other forms of chemo- or radiation therapy, could further enhance the synergism. Although additional studies are needed, the pharmacological attenuation of their activities in the absence of any heat treatment appears a promising strategy in enhancing the cancer cell's sensitivity to chemo- and/or radiation therapy while minimizing their undesirable effects.
Supplementary Material
Fig. 1. Mapping the nuclear matrix proteins (NMPs) changes in response to heat. Prostate cancer cells PC-3 and LNCaP were cultured at 41°C for 4 h or maintained at 37°C as a control. NMPs were isolated and then subjected to high-resolution, two-dimensional gel electrophoresis followed by a silver staining. The differential expression of the NMPs upon heat treatment was cut out and subjected to mass spectrometry (LC/MALDI MS) analysis. Differential proteins are labeled and marked by circles. All experiments were repeated at a minimum of two times.
Fig. 2. Apoptosis induced by chemotherapy is enhanced by RBM3 or CIRBP knock-down. PC-3 cells (A) and LNCaP cells (B) were transfected with 100 nM RBM3 or CIRBP SMARTpool siRNA. Forty-eight hours after transfection, cells were treated with 4 μg/ml cDDP or 1 μg/ml ADM for 4 h, and then moved back to normal medium for another 16 h. Cells were collected by centrifugation and subjected to Terminal Transferase dUTP Nick End Labeling (TUNEL) assay. Apoptotic and non-apoptotic cells population was divided using flow cytometry. *, P < 0.01 compared to scrambled siRNA-treated control.
Fig. 3. Mild heat treatment enhances chemotherapy in prostate cancer cells. PC-3 cells and LNCaP cells were treated at 41°C for 4h, 24 h, or maintained at 37°C as a control. cDDP or ADM were added during the heat treatment and washed out 4 h later. Following heat and drug treatment, cells were maintained at 37°C under routine condition for another 72 h. Cell viability was determined using the WST-1 assay. All experiments were repeated at a minimum of two times. Data are represented as mean ± SD. *, P < 0.05 compared to 37°C control.
Fig. 4. Knocking down RBM3 but not CIRBP partly impedes cell cycle in LNCaP cells. PC-3 cells (A) and LNCaP cells (B) were transfected with 100 nM RBM3 or CIRBP SMARTpool siRNA. Seventy-two hours after transfection, cells were collected by centrifugation, stained with propidium iodide (PI) and subjected to DNA content analysis using the flow cytometry. *, P < 0.05 compared to scrambled siRNA-treated control.
Primer sequences used in real-time PCR
MALDI-TOF and peptide mass fingerprinting analysis of heat shock response nuclear matrix protein
Dysregulated gene list in PC-3 cells upon heat treatment
Dysregulated gene list in LNCaP cells upon heat treatment
Acknowledgments
This work was supported by an award from the Safeway/Prostate Cancer Foundation S.T.A.R. Program as well as from a generous gift from Mr. Christian Evensen.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Al-Fageeh MB, Smales CM. Control and regulation of the cellular responses to cold shock: The responses in yeast and mammalian systems. Biochem J. 2006;397:247–259. doi: 10.1042/BJ20060166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Mortillaro MJ, Ma H, Wei X, Samarabandu J. The nuclear matrix: A structural milieu for genomic function. Int Rev Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- Coffey DS, Getzenberg RH, DeWeese TL. Hyperthermic biology and cancer therapies: A hypothesis for the “Lance Armstrong effect”. JAMA. 2006;296:445–448. doi: 10.1001/jama.296.4.445. [DOI] [PubMed] [Google Scholar]
- Danno S, Nishiyama H, Higashitsuji H, Yokoi H, Xue JH, Itoh K, Matsuda T, Fujita J. Increased transcript level of RBM3, a member of the glycinerich RNA-binding protein family, in human cells in response to cold stress. Biochem Biophys Res Commun. 1997;236:804–807. doi: 10.1006/bbrc.1997.7059. [DOI] [PubMed] [Google Scholar]
- De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp Cell Res. 2007;313:4130–4144. doi: 10.1016/j.yexcr.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Dewhirst MW, Vujaskovic Z, Jones E, Thrall D. Re-setting the biologic rationale for thermal therapy. Int J Hyperthermia. 2005;21:779–790. doi: 10.1080/02656730500271668. [DOI] [PubMed] [Google Scholar]
- Didelot C, Lanneau D, Brunet M, Joly AL, De Thonel A, Chiosis G, Garrido C. Anti-cancer therapeutic approaches based on intracellular and extracellular heat shock proteins. Curr Med Chem. 2007;14:2839–2847. doi: 10.2174/092986707782360079. [DOI] [PubMed] [Google Scholar]
- Ferguson MW, Joanen T. Temperature of egg incubation determines sex in Alligator mississippiensis. Nature. 1982;296:850–853. doi: 10.1038/296850a0. [DOI] [PubMed] [Google Scholar]
- Garner E, Raj K. Protective mechanisms of p53-p21-pRb proteins against DNA damage-induced cell death. Cell Cycle. 2008;7:277–282. doi: 10.4161/cc.7.3.5328. [DOI] [PubMed] [Google Scholar]
- Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G. Heat shock proteins 27 and 70: Anti-apoptotic proteins with tumorigenic properties. Cell Cycle. 2006;5:2592–2601. doi: 10.4161/cc.5.22.3448. [DOI] [PubMed] [Google Scholar]
- Getzenberg RH, Konety BR, Oeler TA, Quigley MM, Hakam A, Becich MJ, Bahnson RR. Bladder cancer-associated nuclear matrix proteins. Cancer Res. 1996;56:1690–1694. [PubMed] [Google Scholar]
- Guan K, Nayernia K, Maier LS, Wagner S, Dressel R, Lee JH, Nolte J, Wolf F, Li M, Engel W, Hasenfuss G. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199–1203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- Inoue T, Leman ES, Yeater DB, Getzenberg RH. The potential role of purine-rich element binding protein (PUR) alpha as a novel treatment target for hormone-refractory prostate cancer. Prostate. 2008;68:1048–1056. doi: 10.1002/pros.20764. [DOI] [PubMed] [Google Scholar]
- Kamada M, So A, Muramaki M, Rocchi P, Beraldi E, Gleave M. Hsp27 knockdown using nucleotide-based therapies inhibit tumor growth and enhance chemotherapy in human bladder cancer cells. Mol Cancer Ther. 2007;6:299–308. doi: 10.1158/1535-7163.MCT-06-0417. [DOI] [PubMed] [Google Scholar]
- Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Lepock JR, Frey HE, Heynen ML, Senisterra GA, Warters RL. The nuclear matrix is a thermolabile cellular structure. Cell Stress Chaperones. 2001;6:136–147. doi: 10.1379/1466-1268(2001)006<0136:tnmiat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujoo K, Watanabe M, Khokhar AR, Siddik ZH. Increased sensitivity of a metastatic model of prostate cancer to a novel tetravalent platinum analog. Prostate. 2005;62:91–100. doi: 10.1002/pros.20114. [DOI] [PubMed] [Google Scholar]
- Myerson RJ, Roti Roti JL, Moros EG, Straube WL, Xu M. Modelling heat-induced radiosensitization: Clinical implications. Int J Hyperthermia. 2004;20:201–212. doi: 10.1080/02656730310001609353. [DOI] [PubMed] [Google Scholar]
- Nishiyama H, Danno S, Kaneko Y, Itoh K, Yokoi H, Fukumoto M, Okuno H, Millan JL, Matsuda T, Yoshida O, Fujita J. Decreased expression of cold-inducible RNA-binding protein (CIRP) in male germ cells at elevated temperature. Am J Pathol. 1998;152:289–296. [PMC free article] [PubMed] [Google Scholar]
- O'Connor PM, Jackman J, Bae I, Myers TG, Fan S, Mutoh M, Scudiero DA, Monks A, Sausville EA, Weinstein JN, Friend S, Fornace AJ, Jr, Kohn KW. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–4300. [PubMed] [Google Scholar]
- Oesterreich S, Lee AV, Sullivan TM, Samuel SK, Davie JR, Fuqua SA. Novel nuclear matrix protein HET binds to and influences activity of the HSP27 promoter in human breast cancer cells. J Cell Biochem. 1997;67:275–286. [PubMed] [Google Scholar]
- Pederson T. Half a century of “the nuclear matrix”. Mol Biol Cell. 2000;11:799–805. doi: 10.1091/mbc.11.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesca D, Mazumder S, Almasan A. DNA damage response and apoptosis. Methods Enzymol. 2008;446:107–122. doi: 10.1016/S0076-6879(08)01606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi P, Jugpal P, So A, Sinneman S, Ettinger S, Fazli L, Nelson C, Gleave M. Small interference RNA targeting heat-shock protein 27 inhibits the growth of prostatic cell lines and induces apoptosis via caspase-3 activation in vitro. BJU Int. 2006;98:1082–1089. doi: 10.1111/j.1464-410X.2006.06425.x. [DOI] [PubMed] [Google Scholar]
- Roti Roti JL. Cellular responses to hyperthermia (40–46 degrees C): Cell killing and molecular events. Int J Hyperthermia. 2008;24:3–15. doi: 10.1080/02656730701769841. [DOI] [PubMed] [Google Scholar]
- Roti Roti JL, Kampinga HH, Malyapa RS, Wright WD, vanderWaal RP, Xu M. Nuclear matrix as a target for hyperthermic killing of cancer cells. Cell Stress Chaperones. 1998;3:245–255. doi: 10.1379/1466-1268(1998)003<0245:nmaatf>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky I, Nudler E. New insights into the mechanism of heat shock response activation. Cell Mol Life Sci. 2008;65:855–861. doi: 10.1007/s00018-008-7458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Sureban SM, Ramalingam S, Natarajan G, May R, Subramaniam D, Bishnupuri KS, Morrison AR, Dieckgraefe BK, Brackett DJ, Postier RG, Houchen CW, Anant S. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene. 2008;27:4544–4556. doi: 10.1038/onc.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland LC, Rintala-Maki ND, White RD, Morin CD. RNA binding motif (RBM) proteins: A novel family of apoptosis modulators? J Cell Biochem. 2005;94:5–24. doi: 10.1002/jcb.20204. [DOI] [PubMed] [Google Scholar]
- Weinberg RA. Tumor suppressor genes. Science. 1991;254:1138–1146. doi: 10.1126/science.1659741. [DOI] [PubMed] [Google Scholar]
- Yang C, Carrier F. The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J Biol Chem. 2001;276:47277–47284. doi: 10.1074/jbc.M105396200. [DOI] [PubMed] [Google Scholar]
- Zhao ZG, Ma QZ, Xu CX. Abrogation of heat-shock protein (HSP)70 expression induced cell growth inhibition and apoptosis in human androgen-independent prostate cancer cell line PC-3m. Asian J Androl. 2004;6:319–324. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. 1. Mapping the nuclear matrix proteins (NMPs) changes in response to heat. Prostate cancer cells PC-3 and LNCaP were cultured at 41°C for 4 h or maintained at 37°C as a control. NMPs were isolated and then subjected to high-resolution, two-dimensional gel electrophoresis followed by a silver staining. The differential expression of the NMPs upon heat treatment was cut out and subjected to mass spectrometry (LC/MALDI MS) analysis. Differential proteins are labeled and marked by circles. All experiments were repeated at a minimum of two times.
Fig. 2. Apoptosis induced by chemotherapy is enhanced by RBM3 or CIRBP knock-down. PC-3 cells (A) and LNCaP cells (B) were transfected with 100 nM RBM3 or CIRBP SMARTpool siRNA. Forty-eight hours after transfection, cells were treated with 4 μg/ml cDDP or 1 μg/ml ADM for 4 h, and then moved back to normal medium for another 16 h. Cells were collected by centrifugation and subjected to Terminal Transferase dUTP Nick End Labeling (TUNEL) assay. Apoptotic and non-apoptotic cells population was divided using flow cytometry. *, P < 0.01 compared to scrambled siRNA-treated control.
Fig. 3. Mild heat treatment enhances chemotherapy in prostate cancer cells. PC-3 cells and LNCaP cells were treated at 41°C for 4h, 24 h, or maintained at 37°C as a control. cDDP or ADM were added during the heat treatment and washed out 4 h later. Following heat and drug treatment, cells were maintained at 37°C under routine condition for another 72 h. Cell viability was determined using the WST-1 assay. All experiments were repeated at a minimum of two times. Data are represented as mean ± SD. *, P < 0.05 compared to 37°C control.
Fig. 4. Knocking down RBM3 but not CIRBP partly impedes cell cycle in LNCaP cells. PC-3 cells (A) and LNCaP cells (B) were transfected with 100 nM RBM3 or CIRBP SMARTpool siRNA. Seventy-two hours after transfection, cells were collected by centrifugation, stained with propidium iodide (PI) and subjected to DNA content analysis using the flow cytometry. *, P < 0.05 compared to scrambled siRNA-treated control.
Primer sequences used in real-time PCR
MALDI-TOF and peptide mass fingerprinting analysis of heat shock response nuclear matrix protein
Dysregulated gene list in PC-3 cells upon heat treatment
Dysregulated gene list in LNCaP cells upon heat treatment