Abstract
Overexpression and mutational activation of the epidermal growth factor receptor (EGFR) plays an important role in the pathogenesis of non–small cell lung cancer (NSCLC). EGFR tyrosine-kinase inhibitors (TKIs) are given as a primary therapy for advanced patients with EGFR-activating mutations; however, the majority of these tumors relapse and patients eventually develop resistance to TKIs. To address a potential role of protein kinase C (PKC) isozymes in the resistance to TKIs, we used the isogenic NSCLC H1650 cell line and its erlotinib-resistant derivative H1650-M3, a cell line that displays a mesenchymal-like morphology driven by transforming growth factor-β signaling. We found that H1650-M3 cells display remarkable PKCα upregulation and PKCδ downregulation. Notably, silencing PKCα from H1650-M3 cells using RNA interference caused a significant reduction in the expression of epithelial-to-mesenchymal transition (EMT) markers vimentin, Zeb2, Snail, and Twist. Moreover, pharmacological inhibition or PKCα RNA interference depletion and PKCδ restoring sensitized H1650-M3 cells to erlotinib. Whereas ectopic overexpression of PKCα in parental H1650 cells was not sufficient to alter the expression of EMT genes or to confer resistance to erlotinib, it caused downregulation of PKCδ expression, suggesting a unidirectional crosstalk. Finally, mechanistic studies revealed that PKCα upregulation in H1650-M3 cells is driven by transforming growth factor-β. Our results identified important roles for specific PKC isozymes in erlotinib resistance and EMT in lung cancer cells, and highlight PKCα as a potential target for lung cancer treatment.
Introduction
Lung cancer remains one of the major causes of mortality worldwide, accounting for more deaths than any other cancer (Kanne, 2014; Ferlay et al., 2015). Diagnosis of lung cancer normally occurs in late stages of the disease, thus limiting the options for treatment. The most common type of lung cancer (approximately 85%) is non–small cell lung cancer (NSCLC), which has three main types: squamous cell carcinoma, adenocarcinoma, and large cell carcinoma (Molina et al., 2008; Shames and Wistuba, 2014). Genetic alterations in NSCLC tumors primarily include oncogenic mutations in the epidermal growth factor receptor (EGFR) and KRAS, as well as inactivation of tumor suppressor genes such as p53, PTEN, Rb, and p16 (Hollstein et al., 1991; Reissmann et al., 1993; Jin et al., 2010). Mutations in the EGFR gene, particularly deletion of exon 19 and L858R mutation in exon 21, occur in 10–50% of NSCLC patients (Gazdar, 2009; Cooper et al., 2013). Small molecule tyrosine-kinase inhibitors (TKIs) that reversibly inhibit EGFR at the ATP pocket domain, such as erlotinib and gefitinib, currently represent the first line of therapy for EGFR-mutated NSCLC patients (Antonicelli et al., 2013; Steins et al., 2014). Although these therapies are initially efficacious, ultimately most patients develop resistance. Whereas resistance has been attributed in some cases to the acquisition of secondary EGFR mutations or MET amplification (Kobayashi et al., 2005; Engelman et al., 2007), the mechanisms behind the resistance to TKIs are only partially understood. Dissecting the signaling mechanisms driving resistance is crucial for designing combinational therapy regimes to overcome this hurdle and extend life expectancy of NSCLC patients.
Protein kinase C (PKC) represents a group of serine-threonine kinases involved in a variety of cellular functions, including mitogenesis, survival, and motility. The PKC family is composed of 10 members classified into three classes: calcium-dependent or conventional PKCs (cPKCα, cPKCβI, cPKCβII, and cPKCγ), calcium-independent or novel PKCs (nPKCδ, nPKCε, nPKCη, and nPKCθ), and phorbol ester/diacylgycerol unresponsive or atypical PKCs (aPKCζ and aPKCι/λ) (Barry and Kazanietz, 2001; Newton, 2001; Griner and Kazanietz, 2007; Garg et al., 2014; Parker et al., 2014). Decades of research have established key roles for different members of the PKC family in the progression of cancer. It became clear that individual PKC isozymes could act either as tumor promoters or tumor suppressors. For example, PKCβ has been proposed to be involved in lung tumorigenesis, and the PKCβ inhibitor enzastaurin has been examined as a potential therapeutic agent for lung cancer patients (Tekle et al., 2008; Willey et al., 2010; Vansteenkiste et al., 2012; El Osta et al., 2014). Our laboratory recently showed that PKCε, a kinase implicated in cell cycle progression and motility, is required for the tumorigenic and metastatic activities of NSCLC cells (Caino et al., 2012a,b). On the other hand, PKCα and PKCδ negatively modulate NSCLC cell cycle progression (Nakagawa et al., 2005; Santiago-Walker et al., 2005; Oliva et al., 2008; Xiao et al., 2008). Most recently, Hill et al. (2014) provided direct evidence for a tumor suppressive role for PKCα in KRAS tumorigenesis. The fact that PKCα promotes NSCLC cell migration (Cheng et al., 2009; O'Neill et al., 2011) suggests divergent roles for this kinase in different stages of lung cancer progression. Likewise, diverse roles for PKCα and other members of the PKC family have been established in survival of NSCLC cells and other cancer cell types (Garg et al., 2014). In addition, the overexpression of some PKC family members has also been associated with low sensitivity to the irreversible TKI afatinib in lung cell line models (Coco et al., 2014).
Toward the goal of determining a potential involvement of PKC isozymes in TKI resistance in lung cancer, here we took advantage of an isogenic NSCLC cell model of erlotinib resistance generated by culturing the parental H1650 cell line in the presence of a high concentration of the inhibitor. Erlotinib-resistant H1650 cells display features of epithelial-to-mesenchymal transition (EMT), a phenotype that is maintained by the transforming growth factor-β (TGF-β) pathway (Yao et al., 2010). Our study identified discrete roles for PKC isozymes, specifically PKCα and PKCδ, in erlotinib resistance and EMT in NSCLC cells.
Materials and Methods
Reagents.
Erlotinib hydrochloride was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The pan-PKC inhibitor GF109203X (2-[1-(3-dimethylaminopropyl)-1H-indol-3-yl]-3-(1H-indol-3-yl)maleimide) was purchased from Enzo Life Sciences (Plymouth Meeting, PA). The cPKC inhibitor Gö6976 (5,6,7,13-tetrahydro-13-methyl-5-oxo-12H-indolo[2,3-a]pyrrolo[3,4-c]carbazole-12-propanenitrile) was obtained from LC Laboratories (Woburn, MA).
Cell Culture.
The H1650-M3 cell line was derived from parental H1650 cells in the laboratory of Dr. Raffaella Sordella (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) (Yao et al., 2010). Both cell lines were kindly provided by Dr. Sordella, and were cultured in RPMI 1640 medium supplemented with 5% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin, and maintained at 37°C in a humidified 5% CO2 atmosphere.
Real-Time Polymerase Chain Reaction.
Total RNA was extracted from subconfluent cell cultures using the RNeasy kit from Qiagen (Valencia, CA). Total RNA (1 μg) was reverse transcribed to cDNA using the TaqMan reverse transcription reagent kit (Applied Biosystems, Branchburg, NJ). Real-time quantitative polymerase chain reaction (qPCR) was performed essentially as described (Wang et al., 2014) using an ABI PRISM 7700 detection system (Applied Biosystems). The reaction was carried out in triplicate samples containing TaqMan Universal PCR MasterMix (Applied Biosystems), target primers (300 nM), fluorescent probe (200 nM), and 4 μl transcribed cDNA (6× dilution). TaqMan primers 5′-end labeled with 6-carboxyfluorescein for PKCα, PKCδ, E-cadherin, Snail, Twist1, vimentin, Zeb2, and 18S rRNA (housekeeping gene) were purchased from Applied Biosystems. PCR product amplification was continuously monitored using the sequence detection system software (version 1.7; Applied Biosystems). Triplicate cycle threshold (Ct) values were averaged and normalized to an average 18S Ct value to calculate the ΔCt. The Δ (ΔCt) was determined by subtracting the control ΔCt value from the experimental ΔCt value. Fold changes were expressed as 2−Δ(ΔCt).
Western Blot Analysis.
Western blot analysis was carried out essentially as previously described (Oliva et al., 2008). Briefly, cells were harvested in lysis buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 0.08% bromophenol blue, and 5% β-mercaptoethanol). Samples were resolved in 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MD). After blocking with 5% milk in 1% Tween 20/phosphate-buffered saline (PBS), membranes were incubated with one of the following primary antibodies: anti-PKCα (EMD Millipore, Billerica, MA), anti-PKCε (Santa Cruz Biotechnology), anti-PKCι (Abcam, Cambridge, MA), anti-vinculin (Sigma-Aldrich, St. Louis, MO), anti-PKCδ, anti-vimentin, anti–E-cadherin, anti-Snail, or anti–phospho-Smad2 (Cell Signaling Technology, Danvers, MA). We used either anti-mouse or anti-rabbit antibodies conjugated with horseradish peroxidase (Bio-Rad, Hercules, CA) as secondary antibodies. Bands were visualized by the enhanced chemiluminescence Western blotting detection system, and images were captured using a Fujifilm LAS-3000 system (Tokyo, Japan).
RNA Interference.
RNA interference (RNAi) duplexes for silencing PKCα were purchased from Dharmacon (Lafayette, CO). The target sequences were as follows: PKCα RNAi 1, CCAUCCGCUCCACACUAAA; and PKCα RNAi 2, GAACAAGGAAUGACUU (Oliva et al., 2008). Control silencer RNAi was purchased from Ambion (Austin, TX). For transfection of RNAi duplexes (25 nM), we used Lipofectamine RNAi/MAX (Invitrogen, Carlsbad, CA).
Adenoviral Infections.
Cells were infected with adenoviruses (AdVs) for PKCα, PKCδ, or LacZ (control) using different multiplicities of infection (MOIs), as previously described (Oliva et al., 2008). Adenoviral infections were carried out in RPMI 1640 medium supplemented with 2% fetal bovine serum. Four hours later, complete medium was added. Experiments were carried at different times after infection, as indicated.
Cell Viability Assay.
Cell viability was determined using the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI), a colorimetric assay that contains MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] and phenazine ethosulfate with enhanced chemical stability. Cells seeded into 96-well plates (1 × 104 cells/well) were treated with different concentrations of erlotinib for different times, as indicated. One hour after addition of the One Solution Reagent, absorbance was recorded at 490 nm using a 96-well plate reader.
Flow Cytometry.
Subconfluent H1650 cells were detached using 0.02% EDTA in PBS, washed, pelleted, and resuspended in FACS buffer (PBS, pH 7.2, 0.2% bovine serum albumin). Then, 5 × 106 cells were costained with phycoerythrin-conjugated anti–human CD24 and allophycocyanin-conjugated anti–human CD44 antibodies (BD Biosciences, San Jose, CA). Labeling was performed for 1 hour at room temperature in the dark. Labeled cells were washed three times with the FACS buffer and sorted using a BD FACS Aria II cell sorter. Gates were set either at high or low expressions for CD24 and CD44, and subpopulations of cells were collected in FACS buffer for RNA extraction.
Statistical Analysis.
All statistical analyses were done using GraphPad Prism software (version 5.03; GraphPad Software, San Diego, CA). Data were analyzed using a two-way analysis of variance. A P value <0.05 was considered statistically significant.
Results
Erlotinib-Resistant Cells Display Altered Expression of PKC Isozymes.
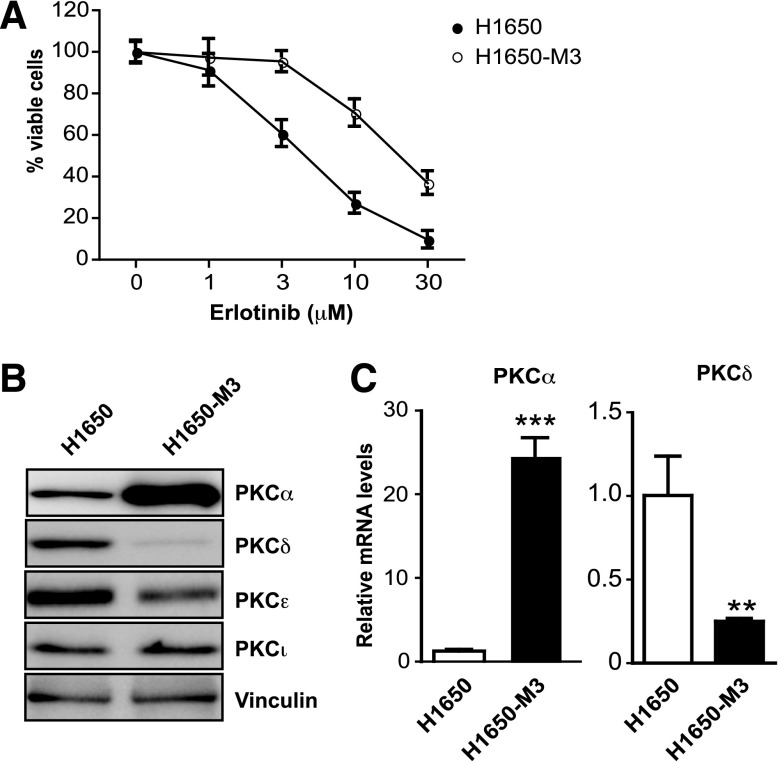
Changes in the expression levels of PKC isozymes have been associated with the progression of many types of cancers, including lung cancer, as well as with resistance to chemotherapeutic agents (Basu et al., 1996; Clark et al., 2003; Bae et al., 2007; Felber et al., 2007; Garg et al., 2014). To determine whether PKC isozymes are implicated in erlotinib resistance, we took advantage of a well characterized isogenic NSCLC cell model: the parental H1650 cell line and its erlotinib-resistant derivative H1650-M3 (Yao et al., 2010) (Fig. 1A). H1650 cells express cPKCα, nPKCδ, nPKCε, and aPKCs. Western blot analysis revealed a remarkable upregulation of PKCα in erlotinib-resistant H1650-M3 cells. H1650-M3 cells also have reduced PKCδ levels relative to parental H1650 cells. A slight reduction in PKCε levels was also observed in H1650-M3 cells (Fig. 1B). Densitometric analysis revealed the following levels relative to parental H1650 cells (n = 3): 10.90 ± 1.61 (PKCα), 0.40 ± 0.07 (PKCδ), 0.85 ± 0.06 (PKCε), and 1.08 ± 0.13 (PKCι). Determination of mRNA levels for PKCα and PKCδ led to similar conclusions. Indeed, H1650-M3 cells have 25-fold higher PKCα mRNA levels than parental H1650 cells, whereas PKCδ mRNA levels are reduced by 5-fold in the erlotinib-resistant cell line (Fig. 1C).
Fig. 1.
Altered expression of PKC isozymes in erlotinib-resistant NSCLC cells. (A) Parental (H1650) and erlotinib-resistant (H1650-M3) cells were treated with erlotinib at indicated concentrations and cell viability was determined 24 hours later using an MTS assay. (B) Expression of PKC isozymes in parental (H1650) and erlotinib-resistant (H1650-M3) cells was analyzed by Western blotting. Similar results were observed in three individual experiments. (C) PKCα and PKCδ mRNA levels in H1650 and H1650-M3 cells were measured by qPCR. Human 18S rRNA was used as an endogenous control for normalization. Results (relative to H1650 cells) are expressed as the mean ± S.D. of triplicate samples. Similar results were observed in three additional experiments. **P < 0.01; ***P < 0.001.
PKCα Is Required But Not Sufficient to Induce Erlotinib Resistance.
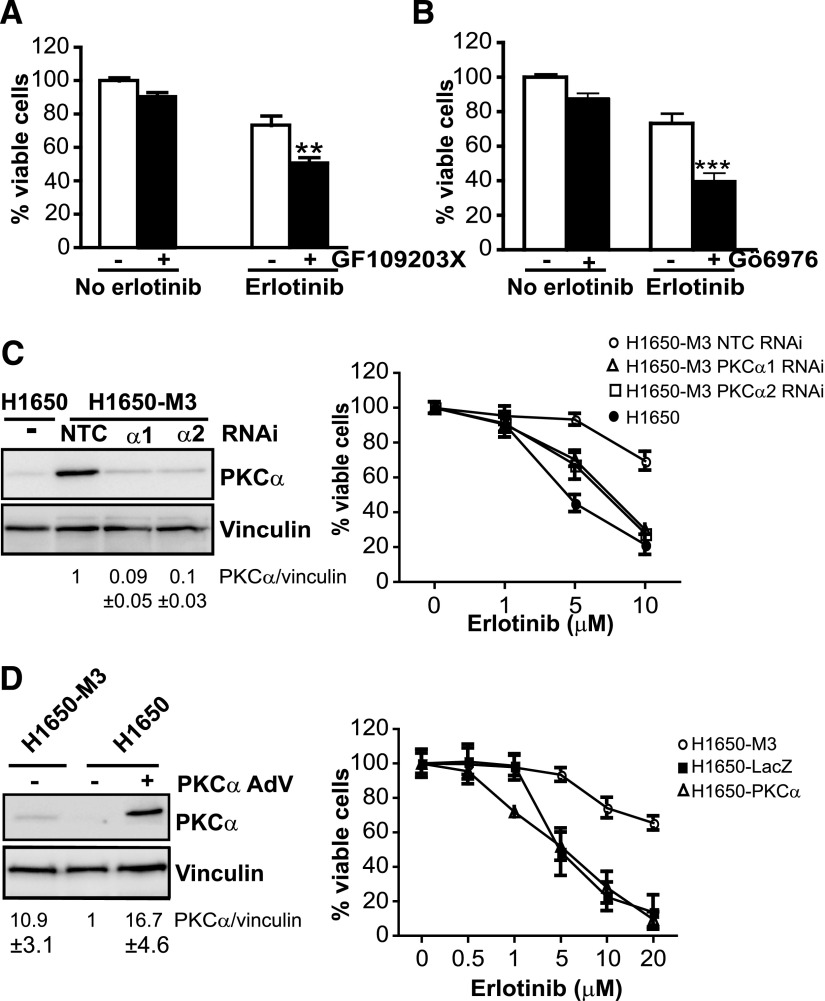
To assess a potential association between altered PKCα expression and erlotinib resistance, we used both pharmacological and RNAi approaches. Because PKCα has been implicated in drug resistance in some cancer types (Chen et al., 2010; Lee et al., 2012; Zhao et al., 2012) and its levels are strikingly high in erlotinib-resistant cells, we speculated that this PKC could be involved in acquired resistance to erlotinib in NSCLC cells. Initial experiments showed that treatment of H1650-M3 cells with the pan-PKC inhibitor GF109203X increases their sensitivity to erlotinib (10 μM) (Fig. 2A). Gö6976, which preferentially inhibits cPKCs (Martiny-Baron et al., 1993), also enhanced the killing effect of erlotinib in H1650-M3 cells (Fig. 2B). PKCα is the most upregulated cPKC in this cell line; thus, it is likely that this PKC mediates erlotinib resistance.
Fig. 2.
PKCα protects H1650-M3 cells from erlotinib-induced cell death. (A) H1650-M3 cells were pretreated for 1 hour with either the pan-PKC inhibitor GF109203X (5 μM) or vehicle. Cells were then treated with erlotinib (10 μM), and cell viability was determined 24 hours later using an MTS assay. **P < 0.01 versus vehicle. (B) H1650-M3 cells were pretreated for 1 hour with either the cPKC inhibitor Gö6976 (5 μM) or vehicle. Cells were then treated with erlotinib (10 μM), and cell viability was determined 24 hours later using an MTS assay. ***P < 0.001 versus vehicle. (C) H1650-M3 cells were transfected with either PKCα (PKCα1 or PKCα2) or nontarget control RNAi duplexes. After 48 hours, cells were treated with erlotinib for 24 hours at the indicated concentrations. The left panel shows PKCα expression by Western blot analysis. The right panel shows cell viability determined using an MTS assay. Parental H1650 cells were included for comparison. (D) Parental H1650 cells were infected with either PKCα AdV or LacZ AdV (MOI = 30 pfu/cell). Five days after infection, cells were treated with erlotinib at the indicated concentrations. The left panel shows PKCα expression by Western blot analysis. The right panel shows cell viability determined 24 hours later. H1650-M3 cells were included for comparison. Data are expressed as the mean ± S.D. of triplicate samples. Similar results were observed in two additional experiments. NTC, nontarget control.
To unambiguously establish a role for PKCα in erlotinib resistance, we used RNAi. Two different PKCα RNAi duplexes were transfected into H1650-M3 cells, which depleted PKCα by 91% (PKCα1 RNAi) and 89% (PKCα2 RNAi) relative to a nontarget control RNAi duplex, as determined by densitometry (Fig. 2C, left panel). A dose-response analysis for inhibition of cell viability by erlotinib revealed an IC50 of approximately 5 μM in nontarget control H1650 cells (which is similar to parental H1650 cells). On the other hand, IC50 in H1650-M3 cells was >20 μM, as also established in a previous study (Yao et al., 2010). Notably, PKCα depletion sensitizes H1650-M3 cells to erlotinib, as judged by the reduction in IC50 (8.7 ± 1.4 μM for PKCα1 RNAi and 9.2 ± 3.0 μM for PKCα2 RNAi) (Fig. 2C, right panel).
To determine whether PKCα upregulation was sufficient to induce erlotinib resistance, PKCα was overexpressed in parental H1650 cells using an AdV. A LacZ AdV was used as control (Fig. 2D, left panel). We found that PKCα overexpression failed to alter the response of H1650 cells to the TKI (IC50 = 4.7 ± 1.3 μM for PKCα AdV and 5.5 ± 2.0 μM for LacZ AdV) (Fig. 2D, right panel). Taken together, these data indicate that although PKCα is required for the resistance of NSCLC cells to erlotinib, overexpression of this kinase is not alone sufficient to induce erlotinib resistance.
PKCδ Alters the Sensitivity of H1650-M3 Cells to Erlotinib.
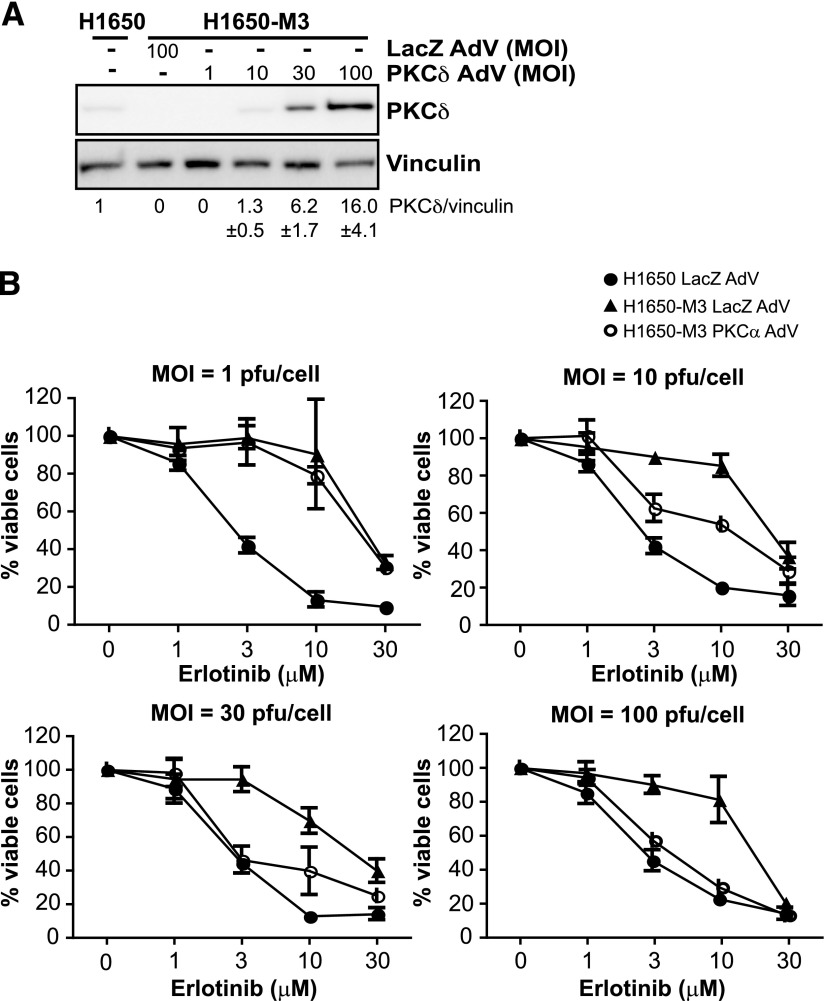
Our results clearly ascribe a role for PKCα in determining the sensitivity of H1650 cells to erlotinib. The fact that H1650-M3 cells display PKCδ downregulation relative to parental H1650 cells prompted us to investigate whether changes in PKCδ levels could also dictate the sensitivity to the TKI. PKCδ was previously shown to mediate the cytotoxic effect of several anticancer drugs (Reyland et al., 1999; Blass et al., 2002). To address this issue, we first overexpressed PKCδ in H1650-M3 cells using a PKCδ AdV (Fig. 3A). As shown in Fig. 3B, overexpression of PKCδ in erlotinib-resistant cells caused a reduction in the IC50 for erlotinib. This effect was proportional to the expression levels of PKCδ achieved by infecting cells with different MOIs of the PKCδ AdV. Infection of H1650-M3 cells with an MOI equal to 1 plaque-forming unit/cell did not cause any significant PKCδ overexpression or sensitization to erlotinib (IC50 = 24.2 ± 0.6 μM for PKCδ AdV and 24.7 ± 2.0 μM for control LacZ AdV). On the other hand, infection with PKCδ AdV at MOI = 10 plaque-forming units/cell caused significant sensitization (IC50 = 8.7 ± 1.9 μM for PKCδ AdV and 26.4 ± 0.4 μM for LacZ AdV). At higher MOIs, the sensitivity of H1650-M3 cells was essentially similar to that observed in parental H1650 cells (MOI = 30: IC50 = 6.3 ± 0.5 μM for PKCδ AdV and 22.2 ± 0.4 μM for LacZ AdV; MOI = 100: IC50 = 4.5 ± 0.4 μM for PKCδ AdV and 19.5 ± 1.0 μM for LacZ AdV). Thus, PKCδ downregulation in H1650-M3 cells contributes to erlotinib resistance.
Fig. 3.
PKCδ alters the sensitivity of H1650-M3 cells to erlotinib. (A) H1650-M3 cells were infected with either PKCδ AdV or LacZ AdV at the indicated MOIs. Expression of PKCδ was determined using Western blot analysis. Densitometric analysis is shown as the mean ± S.D. (n = 3). (B) A viability assay using MTS was carried out 48 hours after infection. Data are expressed as the mean ± S.D. of triplicate samples. Similar results were observed in two additional experiments. pfu, plaque-forming unit.
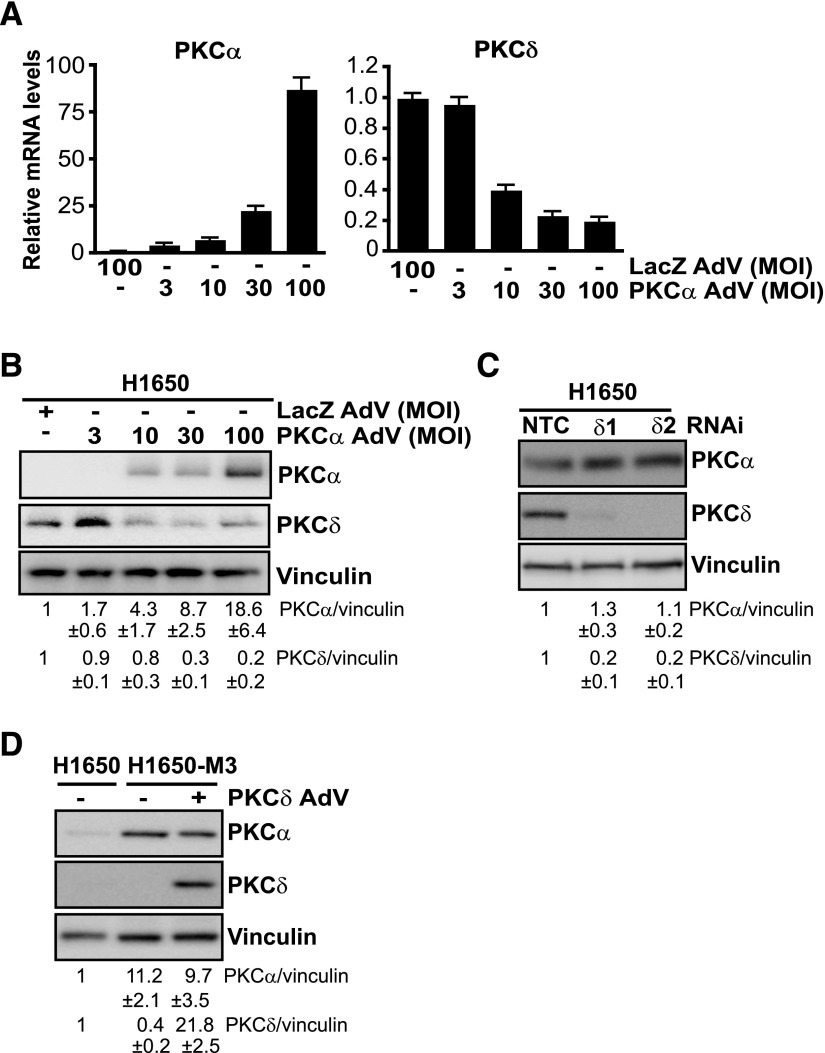
Previous studies have shown that overexpression of one PKC isozyme could alter the expression of other PKC family members. For example, overexpression of PKCα alters the expression of PKCδ and PKCε in various cellular models (Ways et al., 1995; Romanova et al., 1998; Tonetti et al., 2000). Because erlotinib-resistant H1650 cells display PKCα overexpression and PKCδ downregulation relative to the parental cell line, we asked whether there is a mutual regulation between these PKCs. To test our hypothesis, we either overexpressed PKCα or depleted PKCδ in parental H1650 cells. Interestingly, PKCα overexpression by adenoviral means reduced PKCδ expression, both at mRNA and protein levels. These effects were proportional to the PKCα overexpression levels achieved by using increased MOIs of the PKCα AdV (Fig. 4, A and B). Next, to assess whether downregulation of PKCδ alters PKCα expression levels, we silenced PKCδ expression from parental H1650 cells using RNAi. As shown in Fig. 4C, both control and PKCδ-depleted H1650 cells display similar PKCα levels. Furthermore, adenoviral overexpression of PKCδ in erlotinib-resistant H1650-M3 cells failed to induce changes in PKCα expression (Fig. 4D). These results argue for a unidirectional crosstalk whereby overexpression of PKCα in erlotinib-resistant H1650-M3 cells contributes to PKCδ downregulation; however, PKCδ was unable to influence PKCα expression.
Fig. 4.
PKCα modulates the expression of PKCδ in H1650 cells. (A) H1650 cells were infected with either PKCα AdV or LacZ AdV at the indicated MOIs. PKCα and PKCδ mRNA levels were determined by qPCR 72 hours after infection. Data are expressed as the mean ± S.D. of triplicate samples. Results are expressed as the fold change relative to LacZ AdV. (B) Expression of PKCα and PKCδ was determined by Western blot 72 hours after infection with either PKCα AdV or LacZ AdV. (C) Parental H1650 cells were transfected with either PKCδ (PKCδ1 or PKCδ2) or NTC RNAi duplexes. PKCα and PKCδ levels were analyzed 72 hours later by Western blot analysis. (D) H1650-M3 cells were infected with either PKCδ AdV or LacZ AdV (MOI = 100 pfu/cell). PKCδ and PKCα levels were analyzed 96 hours later by Western blotting. Similar results were observed in three independent experiments. Densitometric analysis is shown as the mean ± S.D. (n = 3). NTC, nontarget control.
PKCα Is Required for the Maintenance of Mesenchymal Phenotype of H1650-M3 Cells.
Erlotinib-resistant H1650 cells exhibit mesenchymal properties, driven by the TGF-β pathway (Yao et al., 2010). The mesenchymal phenotype is a hallmark of cancer cells exhibiting an aggressive phenotype (Tam et al., 2013). A recent study in breast cancer showed that PKCα is upregulated in cells that had undergone EMT (Tam et al., 2013). Thus, we speculated that this kinase might contribute to the maintenance of the mesenchymal phenotype of erlotinib-resistant H1650 cells.
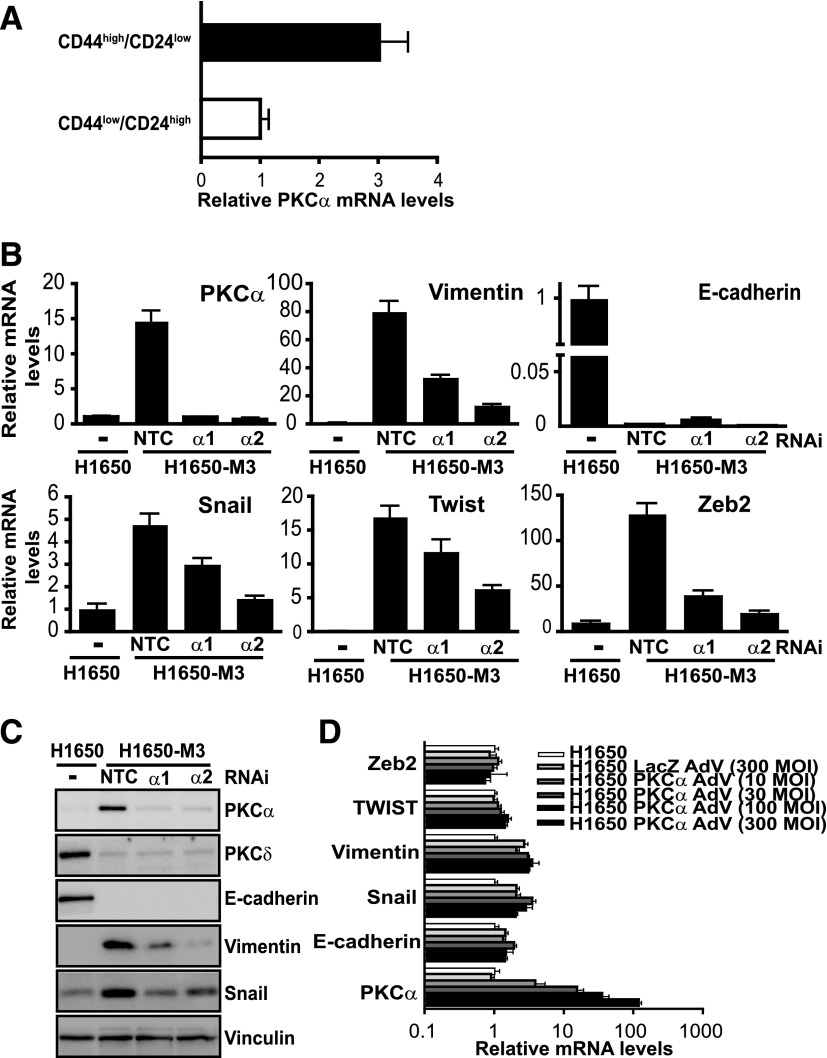
First, we investigated whether PKCα levels were elevated in a subpopulation of H1650 cells that display stem cell–like properties. Parental H1650 cells were sorted into CD44high/CD24low and CD44low/CD24high enriched populations, and PKCα mRNA levels were determined by qPCR. These experiments revealed PKCα upregulation in CD44high/CD24low cells (Fig. 5A). As shown in a previous study (Yao et al., 2010), H1650-M3 cells display elevated levels of genes associated with EMT, including vimentin, Snail, Twist, and Zeb2, as well as reduced levels of E-cadherin. To establish a potential link between PKCα upregulation and the mesenchymal phenotype of H1650-M3 cells, we examined the expression of EMT markers by qPCR after silencing PKCα. Notably, PKCα RNAi depletion caused a significant reduction in vimentin, Snail, Twist, and Zeb expression, suggesting that PKCα mediates the induction of these EMT genes. Expression of the epithelial marker E-cadherin, however, remained unaffected (Fig. 5B). Changes were also validated at the protein level for those markers that could be readily detected by Western blot analysis (64 and 69% reduction for vimentin; 42 and 60% reduction for Snail, using PKCα1 and PKCα2 RNAi, respectively) (Fig. 5C). Despite the PKCα requirement for the expression of EMT markers in H1650-M3 cells, it became apparent that overexpression of this kinase in parental H1650 cells was not sufficient to induce these EMT genes, as determined by qPCR 72 hours after infection with increasing MOIs of the PKCα AdV (Fig. 5D). No changes were observed even 1 week after PKCα AdV infection (data not shown). Altogether, these results indicate that PKCα is required for the expression of genes involved in the maintenance of the mesenchymal phenotype of erlotinib-resistant cells; however, its overexpression is not sufficient to induce this phenotypical change.
Fig. 5.
PKCα is required for the expression of markers of the mesenchymal phenotype. (A) Parental H1650 cells were sorted into CD44high/CD24low and CD44low/CD24high subpopulations by flow cytometry. PKCα mRNA levels were determined by qPCR. Data are expressed as the mean ± S.D. of triplicate samples. (B) H1650-M3 cells were transfected with either PKCα (PKCα1 or PKCα2) or NTC RNAi duplexes. After 72 hours, RNA was extracted for qPCR analysis of selected genes associated with epithelial (E-cadherin) or mesenchymal (vimentin, Snail, Twist, and Zeb2) phenotypes. Results are shown as the fold change relative to parental H1650 cells. Data were expressed as the mean ± S.D. of triplicate samples. (C) Expression of epithelial and mesenchymal markers was determined by Western blot analysis. (D) H1650 cells were infected with either PKCα AdV or LacZ AdV at the indicated MOIs. After 7 days, expression of E-cadherin, vimentin, Snail, Twist, and Zeb2 were determined by qPCR. Similar results were observed in three independent experiments. NTC, nontarget control.
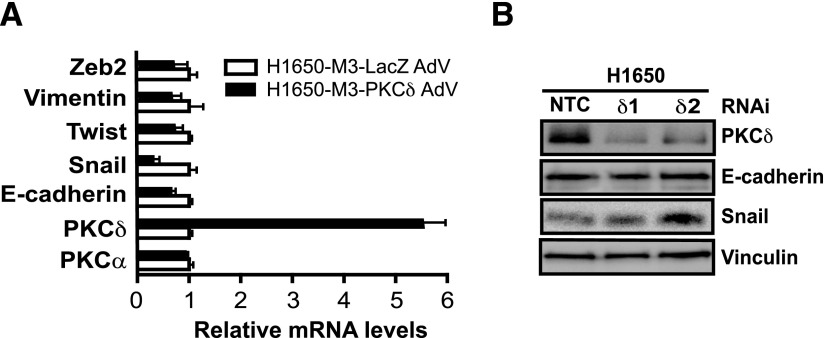
Next, we set to explore whether PKCδ has a role in the expression of genes associated with EMT transition. Because PKCδ is downregulated in H1650-M3 cells, we adenovirally overexpressed PKCδ in these cells and assessed the expression of EMT markers by qPCR. Unlike PKCα silencing, ectopic overexpression of PKCδ in H1650-M3 cells did not change the expression of vimentin, Twist, or Zeb2, although a reduction in Snail levels could be observed. Likewise, PKCδ overexpression did not affect E-cadherin mRNA levels (Fig. 6A). In addition, we also found that PKCδ RNAi depletion from parental H1650 cells failed to change the expression of Snail and E-cadherin (Fig. 6B). Therefore, the involvement of PKCδ is only confined to erlotinib resistance but not to EMT.
Fig. 6.
Genes involved in the mesenchymal phenotype are not regulated by PKCδ. (A) H1650-M3 cells were infected with either PKCδ AdV or LacZ AdV (MOI = 100 pfu/cell). After 96 hours, mRNA levels for various mesenchymal (vimentin, Snail, Twist, and Zeb2) or epithelial (E-cadherin) associated genes were measured by qPCR. Results are shown as the fold change relative to control (LacZ AdV-infected) H1650-M3 cells. Data were expressed as the mean ± S.D. of triplicate samples. (B) Parental H1650 cells were transfected with either PKCδ (PKCδ1 or PKCδ2) or NTC RNAi duplexes. Expression of PKCδ, E-cadherin, and Snail was analyzed by Western blotting 72 hours later. Similar results were observed in three independent experiments. NTC, nontarget control; pfu, plaque-forming unit.
PKCα Upregulation in Erlotinib-Resistant Cells Is Mediated by TGF-β.
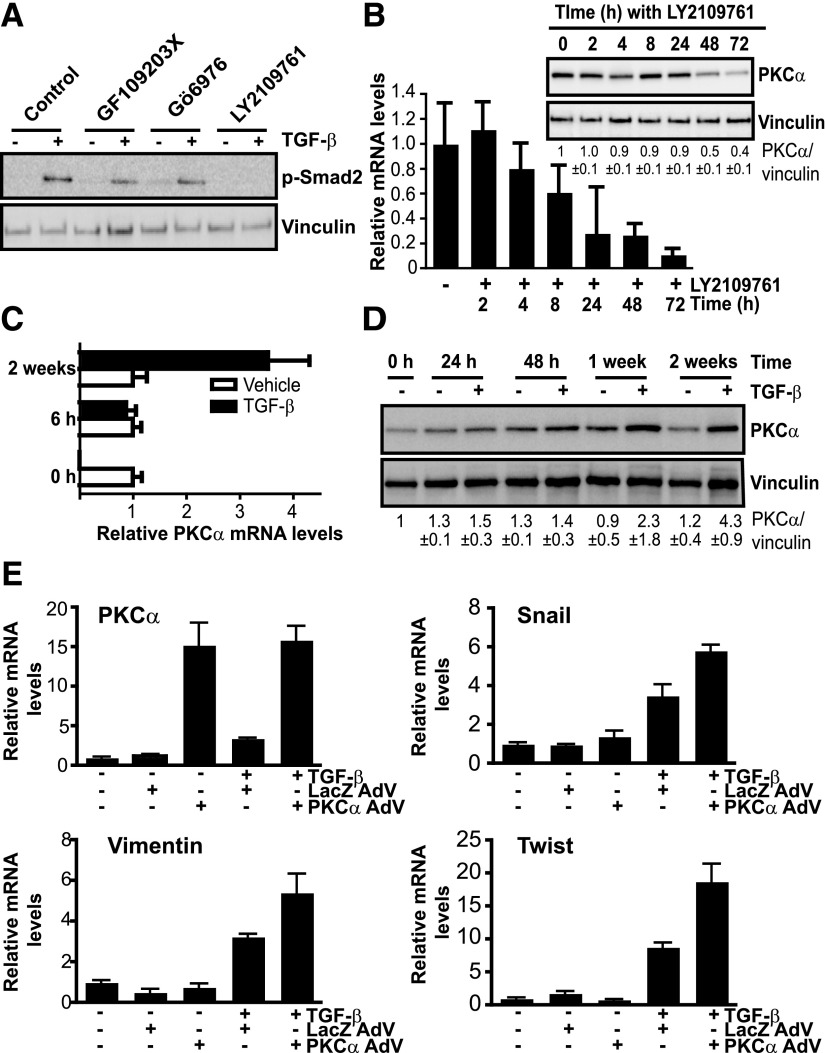
TGF-β has been widely implicated in EMT in multiple cancer types (Massagué, 2012; Moustakas and Heldin, 2012). It was previously established that activation of the TGF-β signaling pathway mediates EMT and erlotinib resistance in H1650 cells (Yao et al., 2010). On the basis of this premise, we sought to establish whether a causal relationship exists between TGF-β signaling and PKCα expression. H1650-M3 cells were treated with the TGF-β receptor inhibitor LY2109761 (4-[5,6-dihydro-2-(2-pyridinyl)-4H-pyrrolo[1,2-b]pyrazol-3-yl]-7-[2-(4-morpholinyl)ethoxy]-quinoline), and its efficacy to inhibit TGF-β signaling was confirmed by its ability to reduce Smad2 phosphorylation (Fig. 7A). PKC inhibitors GF109203X and Gö6976 did not affect Smad2 phosphorylation, suggesting that PKC does not affect the activation of this pathway. Notably, the TGF-β receptor inhibitor caused a time-dependent reduction in PKCα mRNA level. This effect became noticeable at the protein level 48 and 72 hours after LY2109761 treatment (Fig. 7B). Furthermore, when parental H1650 cells were treated with TGF-β for different times, significant PKCα upregulation both at mRNA and protein levels could be observed. This effect was quite remarkable after long-term treatment with TGF-β (Fig. 7, C and D). Therefore, TGF-β signaling is implicated in the overexpression of PKCα observed in erlotinib-resistant cells.
Fig. 7.
TGF-β signaling controls PKCα expression in erlotinib-resistant cells. (A) H1650-M3 cells were pretreated for 1 hour with either the pan-PKC inhibitor GF109203X (5 μM), the cPKC inhibitor Gö6976 (5 μM), the TGF-β receptor inhibitor LY2109761 (5 μM), or vehicle. Cells were then treated with TGF-β (20 ng/ml, 30 minutes) and phospho-Smad2 levels were determined by Western blot analysis. (B) H1650-M3 cells were treated with the TGF-β receptor inhibitor LY2109761 (5 μM) for the indicated times. PKCα mRNA and protein levels were determined by qPCR and Western blot analysis, respectively. Densitometric analysis is shown as the mean ± S.D. (n = 3). (C) PKCα mRNA levels in H1650 cells were measured 6 hours or 2 weeks after TGF-β treatment. (D) H1650 cells were treated with TGF-β (5 ng/ml) for 24 hours, 48 hours, 1 week, or 2 weeks. PKCα levels were determined by Western blot analysis. Densitometric analysis is shown as the mean ± S.D. (n = 3). (E) H1650 cells were infected with either PKCα AdV or LacZ AdV (MOI = 30 pfu/cell). Twenty-four hours after infection, cells were treated with TGF-β (5 ng/ml) for 1 week. mRNA levels for PKCα, Snail, vimentin, and Twist were measured using qPCR. In all cases, data were expressed as the mean ± S.D. of triplicate samples and experiments were reproduced at least three times. pfu, plaque-forming unit.
Finally, we sought to establish an association between PKCα upregulation and TGF-β signaling in the induction of the mesenchymal phenotype. H1650 cells were infected with PKCα AdV (or LacZ AdV as a control) and then subjected to TGF-β treatment. mRNA was extracted 1 week after treatment and EMT markers were determined by qPCR. As shown in Fig. 7E, overexpression of PKCα potentiated TGF-β induction of vimentin, Snail, and Twist, thus establishing the relevance of the TGF-β/PKCα pathway in the induction of the mesenchymal phenotype.
Discussion
Tumor cells harboring activating mutations of EGFR are addicted to this oncogenic stimulus to maintain their proliferative and survival advantages. TKIs such as erlotinib are effective for treatment of advanced NSCLC tumors harboring EGFR-activating mutations. However, many patients treated with erlotinib develop resistance to the targeted molecular therapy (Tang et al., 2013; Steins et al., 2014). PKC isozymes have been recognized as key effectors of known oncogenes implicated in drug resistance such as c-MET, KRAS, and TGF-β (Kermorgant et al., 2004; Sakaguchi et al., 2004; Symonds et al., 2011). Moreover, phorbol esters, which are known activators of PKCs, induce multidrug resistance (Fine et al., 1988; Kalalinia et al., 2012).
Here, we present evidence for the involvement of specific PKC isozymes in erlotinib resistance and EMT in NSCLC cells. Using an isogenic cell model, we found considerable changes in the expression of PKC isozymes that are causally associated with resistance to erlotinib. Erlotinib-resistant H1650-M3 cells exhibit elevated PKCα levels, whereas PKCδ expression in these cells is markedly downregulated. Although this is the first evidence for the involvement of these two PKC isozymes in resistance to this targeted molecular therapy, altered expression of PKCα and PKCδ has been detected in several cancer cell types. For example, elevation of PKCα expression or activity has been reported in pancreatic, colon, prostate, glioma, and gastric cancer cells resistant to chemotherapeutic drugs, including cisplatin, doxorubicin, and vincristine (Matsumoto et al., 1995; Wu et al., 2009; Chen et al., 2010; Zhao et al., 2012). Interestingly, comparable to what we observed in erlotinib-resistant cells, continuous exposure of MCF-7 breast cancer cells to tamoxifen rendered high levels of PKCα and downregulation of PKCδ (Li et al., 2012).
Studies have indicated the importance of PKCα overexpression in protecting cancer cells against drug-induced cell death. For example, PKCα overexpression in colon cancer cells attenuates doxorubicin-induced apoptosis by elevating phosphorylation of Bcl-2, Bad, and decreasing PARP cleavage. More importantly, in several cancer models, PKCα overexpression has been associated with increased drug resistance by elevating expression and phosphorylation of the drug efflux pump P-glycoprotein encoded by the multidrug resistant gene MDR1 (Lee et al., 2012). The functional importance of PKCα overexpression has been further demonstrated by using pharmacological inhibitors and RNAi. For example, inhibition of PKCα using Gö6976 restores the sensitivity of pancreatic cancer cells to chemotherapeutic drugs (Chen et al., 2010), and silencing PKCα by RNAi reverses drug resistance in ovarian cancer cells (Zhao et al., 2012). In our study, we found that RNAi depletion or inhibition of PKCα using Gö6976 sensitizes erlotinib-resistant NSCLC cells to the TKI.
As previously characterized, H1650-M3 cells have elevated expression of genes associated with EMT and display morphologic changes that are reminiscent of the mesenchymal phenotype. Interestingly, parental erlotinib-naive cells possess a small subpopulation of cells that are mesenchymal, erlotinib resistant, and similar to H1650-M3 cells (Yao et al., 2010), indicating that H1650-M3 cells were potentially generated through a selection process that favors the survival of cells that use alternate mechanisms to overcome drug-induced death. A recent study by the Weinberg laboratory established that PKCα preferentially supports the maintenance of the mesenchymal cell state through the regulation of the Fos-related antigen 1 transcription factor. In addition, elevated PKCα expression was found in a subpopulation of normal mammary epithelial cells enriched in the mesenchymal surface marker CD44 (Tam et al., 2013). Similarly, our results indicate a correlation between enrichment of the mesenchymal phenotype and PKCα expression in NSCLC cells. Inhibition of PKCα in H1650-M3 cells also led to a reduction in the expression of genes associated with the mesenchymal phenotype. Interestingly, although exposure to erlotinib resulted in a differential expression of EMT markers, including upregulation of vimentin, Snail, Twist, and Zeb2, as well as downregulation of E-cadherin, the effect of inhibiting PKCα was limited to the genes associated with the mesenchymal phenotype, thus underscoring its role in the maintenance of this phenotype.
In our study, we also identified a functional link between TGF-β and PKCα. TGF-β signaling was shown to be sufficient and required for the induction of erlotinib resistance and EMT in H1650-M3 cells (Yao et al., 2010). We found that inhibition of TGF-β signaling reduced the expression of PKCα in H1650-M3 cells. On the other hand, TGF-β increased the expression of PKCα in parental H1650 cells, indicating that in the process of acquiring an aggressive phenotype, TGF-β upregulates the expression of PKCα. TGF-β is known to control gene expression by activating the Smad transcription factors (Massagué, 2012). The promoter region of PKCα does not display any obvious Smad binding site (data not shown), arguing for the involvement of alternative or indirect mechanisms. It is worth noting that gene profiling analysis in A549 lung adenocarcinoma cells identified PKCα as a TGF-β target gene (Ranganathan et al., 2007).
In summary, our results provide evidence for a role of PKCs in acquired drug resistance to erlotinib and EMT. Elevation of PKCα expression as well as PKCα-dependent downregulation of PKCδ are required for erlotinib resistance, whereas mesenchymal genes are regulated only by PKCα. Our results argue for a potential therapeutic use of PKCα inhibitors to overcome drug resistance and EMT in lung cancer.
Abbreviations
- AdV
adenovirus
- aPKC
atypical protein kinase
- cPKC
conventional protein kinase C
- Ct
cycle threshold
- EGFR
epidermal growth factor receptor
- EMT
epithelial-to-mesenchymal transition
- GF109203X
2-[1-(3-dimethylaminopropyl)-1H-indol-3-yl]-3-(1H-indol-3-yl)maleimide
- Gö6976
5,6,7,13-tetrahydro-13-methyl-5-oxo-12H-indolo[2,3-a]pyrrolo[3,4-c]carbazole-12-propanenitrile
- LY2109761
4-[5,6-dihydro-2-(2-pyridinyl)-4H-pyrrolo[1,2-b]pyrazol-3-yl]-7-[2-(4-morpholinyl)ethoxy]-quinoline
- MOI
multiplicity of infection
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- nPKC
novel protein kinase C
- NSCLC
non–small cell lung cancer
- PBS
phosphate-buffered saline
- PKC
protein kinase C
- qPCR
quantitative polymerase chain reaction
- RNAi
RNA interference
- TKI
tyrosine-kinase inhibitor
Authorship Contributions
Participated in research design: Abera, Kazanietz.
Conducted experiments: Abera.
Performed data analysis: Abera, Kazanietz.
Wrote or contributed to the writing of the manuscript: Abera, Kazanietz.
Footnotes
This research was supported by the National Institutes of Health National Cancer Institute [Grants R01-CA139120 and R01-CA089202].
References
- Antonicelli A, Cafarotti S, Indini A, Galli A, Russo A, Cesario A, Lococo FM, Russo P, Mainini AF, Bonifati LG, et al. (2013) EGFR-targeted therapy for non-small cell lung cancer: focus on EGFR oncogenic mutation. Int J Med Sci 10:320–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae KM, Wang H, Jiang G, Chen MG, Lu L, Xiao L. (2007) Protein kinase C epsilon is overexpressed in primary human non-small cell lung cancers and functionally required for proliferation of non-small cell lung cancer cells in a p21/Cip1-dependent manner. Cancer Res 67:6053–6063. [DOI] [PubMed] [Google Scholar]
- Barry OP, Kazanietz MG. (2001) Protein kinase C isozymes, novel phorbol ester receptors and cancer chemotherapy. Curr Pharm Des 7:1725–1744. [DOI] [PubMed] [Google Scholar]
- Basu A, Weixel K, Saijo N. (1996) Characterization of the protein kinase C signal transduction pathway in cisplatin-sensitive and -resistant human small cell lung carcinoma cells. Cell Growth Differ 7:1507–1512. [PubMed] [Google Scholar]
- Blass M, Kronfeld I, Kazimirsky G, Blumberg PM, Brodie C. (2002) Tyrosine phosphorylation of protein kinase Cdelta is essential for its apoptotic effect in response to etoposide. Mol Cell Biol 22:182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caino MC, Lopez-Haber C, Kim J, Mochly-Rosen D, Kazanietz MG. (2012a) Proteins kinase Cε is required for non-small cell lung carcinoma growth and regulates the expression of apoptotic genes. Oncogene 31:2593–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caino MC, Lopez-Haber C, Kissil JL, Kazanietz MG. (2012b) Non-small cell lung carcinoma cell motility, rac activation and metastatic dissemination are mediated by protein kinase C epsilon. PLoS ONE 7:e31714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yu G, Yu D, Zhu M. (2010) PKCalpha-induced drug resistance in pancreatic cancer cells is associated with transforming growth factor-beta1. J Exp Clin Cancer Res 29:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Hsieh HL, Sun CC, Lin CC, Luo SF, Yang CM. (2009) IL-1 beta induces urokinase-plasminogen activator expression and cell migration through PKC alpha, JNK1/2, and NF-kappaB in A549 cells. J Cell Physiol 219:183–193. [DOI] [PubMed] [Google Scholar]
- Clark AS, West KA, Blumberg PM, Dennis PA. (2003) Altered protein kinase C (PKC) isoforms in non-small cell lung cancer cells: PKCdelta promotes cellular survival and chemotherapeutic resistance. Cancer Res 63:780–786. [PubMed] [Google Scholar]
- Coco S, Truini A, Alama A, Dal Bello MG, Vene R, Garuti A, Carminati E, Rijavec E, Genova C, Barletta G, et al. (2014) Afatinib resistance in non-small cell lung cancer involves the PI3K/AKT and MAPK/ERK signalling pathways and epithelial-to-mesenchymal transition. Target Oncol DOI: 10.1007/s11523-014-0344-7 [published ahead of print]. [DOI] [PubMed] [Google Scholar]
- Cooper WA, Lam DC, O’Toole SA, Minna JD. (2013) Molecular biology of lung cancer. J Thorac Dis 5 (Suppl 5):S479–S490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Osta M, Liu M, Adada M, Senkal CE, Idkowiak-Baldys J, Obeid LM, Clarke CJ, Hannun YA. (2014) Sustained PKCβII activity confers oncogenic properties in a phospholipase D- and mTOR-dependent manner. FASEB J 28:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. (2007) MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316:1039–1043. [DOI] [PubMed] [Google Scholar]
- Felber M, Sonnemann J, Beck JF. (2007) Inhibition of novel protein kinase C-epsilon augments TRAIL-induced cell death in A549 lung cancer cells. Pathol Oncol Res 13:295–301. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. (2015). Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136:E359–E386. [DOI] [PubMed] [Google Scholar]
- Fine RL, Patel J, Chabner BA. (1988) Phorbol esters induce multidrug resistance in human breast cancer cells. Proc Natl Acad Sci USA 85:582–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Benedetti LG, Abera MB, Wang H, Abba M, Kazanietz MG. (2014) Protein kinase C and cancer: what we know and what we do not. Oncogene 33:5225–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF. (2009) Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 28 (Suppl 1):S24–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griner EM, Kazanietz MG. (2007) Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer 7:281–294. [DOI] [PubMed] [Google Scholar]
- Hill KS, Erdogan E, Khoor A, Walsh MP, Leitges M, Murray NR, Fields AP. (2014) Protein kinase Calpha suppresses Kras-mediated lung tumor formation through activation of a p38 MAPK-TGFbeta signaling axis. Oncogene 33:2134–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. (1991) p53 mutations in human cancers. Science 253:49–53. [DOI] [PubMed] [Google Scholar]
- Jin G, Kim MJ, Jeon HS, Choi JE, Kim DS, Lee EB, Cha SI, Yoon GS, Kim CH, Jung TH, et al. (2010) PTEN mutations and relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small cell lung cancers. Lung Cancer 69:279–283. [DOI] [PubMed] [Google Scholar]
- Kalalinia F, Elahian F, Hassani M, Kasaeeian J, Behravan J. (2012) Phorbol ester TPA modulates chemoresistance in the drug sensitive breast cancer cell line MCF-7 by inducing expression of drug efflux transporter ABCG2. Asian Pac J Cancer Prev 13:2979–2984. [DOI] [PubMed] [Google Scholar]
- Kanne JP. (2014) Screening for lung cancer: what have we learned? AJR Am J Roentgenol 202:530–535. [DOI] [PubMed] [Google Scholar]
- Kermorgant S, Zicha D, Parker PJ. (2004) PKC controls HGF-dependent c-Met traffic, signalling and cell migration. EMBO J 23:3721–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. (2005) EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 352:786–792. [DOI] [PubMed] [Google Scholar]
- Lee SK, Shehzad A, Jung JC, Sonn JK, Lee JT, Park JW, Lee YS. (2012) Protein kinase Cα protects against multidrug resistance in human colon cancer cells. Mol Cells 34:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang N, Fang J, Huang J, Tian F, Li C, Xie F. (2012) Role of PKC-ERK signaling in tamoxifen-induced apoptosis and tamoxifen resistance in human breast cancer cells. Oncol Rep 27:1879–1886. [DOI] [PubMed] [Google Scholar]
- Martiny-Baron G, Kazanietz MG, Mischak H, Blumberg PM, Kochs G, Hug H, Marmé D, Schächtele C. (1993) Selective inhibition of protein kinase C isozymes by the indolocarbazole Gö 6976. J Biol Chem 268:9194–9197. [PubMed] [Google Scholar]
- Massagué J. (2012) TGFβ signalling in context. Nat Rev Mol Cell Biol 13:616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Tani E, Yamaura I, Miyaji K, Kaba K. (1995) Effects of protein kinase C modulators on multidrug resistance in human glioma cells. Neurosurgery 36:565–571, discussion 572. [DOI] [PubMed] [Google Scholar]
- Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. (2008) Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 83:584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. (2012) Induction of epithelial-mesenchymal transition by transforming growth factor β. Semin Cancer Biol 22:446–454. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Oliva JL, Kothapalli D, Fournier A, Assoian RK, Kazanietz MG. (2005) Phorbol ester-induced G1 phase arrest selectively mediated by protein kinase Cdelta-dependent induction of p21. J Biol Chem 280:33926–33934. [DOI] [PubMed] [Google Scholar]
- Newton AC. (2001) Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev 101:2353–2364. [DOI] [PubMed] [Google Scholar]
- O’Neill AK, Gallegos LL, Justilien V, Garcia EL, Leitges M, Fields AP, Hall RA, Newton AC. (2011) Protein kinase Cα promotes cell migration through a PDZ-dependent interaction with its novel substrate discs large homolog 1 (DLG1). J Biol Chem 286:43559–43568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva JL, Caino MC, Senderowicz AM, Kazanietz MG. (2008) S-Phase-specific activation of PKC alpha induces senescence in non-small cell lung cancer cells. J Biol Chem 283:5466–5476. [DOI] [PubMed] [Google Scholar]
- Parker PJ, Justilien V, Riou P, Linch M, Fields AP. (2014) Atypical protein kinase Cι as a human oncogene and therapeutic target. Biochem Pharmacol 88:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan P, Agrawal A, Bhushan R, Chavalmane AK, Kalathur RK, Takahashi T, Kondaiah P. (2007) Expression profiling of genes regulated by TGF-beta: differential regulation in normal and tumour cells. BMC Genomics 8:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann PT, Koga H, Takahashi R, Figlin RA, Holmes EC, Piantadosi S, Cordon-Cardo C, Slamon DJ, The Lung Cancer Study Group (1993) Inactivation of the retinoblastoma susceptibility gene in non-small-cell lung cancer. Oncogene 8:1913–1919. [PubMed] [Google Scholar]
- Reyland ME, Anderson SM, Matassa AA, Barzen KA, Quissell DO. (1999) Protein kinase C delta is essential for etoposide-induced apoptosis in salivary gland acinar cells. J Biol Chem 274:19115–19123. [DOI] [PubMed] [Google Scholar]
- Romanova LY, Alexandrov IA, Nordan RP, Blagosklonny MV, Mushinski JF. (1998) Cross-talk between protein kinase C-alpha (PKC-alpha) and -delta (PKC-delta): PKC-alpha elevates the PKC-delta protein level, altering its mRNA transcription and degradation. Biochemistry 37:5558–5565. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M, Miyazaki M, Sonegawa H, Kashiwagi M, Ohba M, Kuroki T, Namba M, Huh NH. (2004) PKCalpha mediates TGFbeta-induced growth inhibition of human keratinocytes via phosphorylation of S100C/A11. J Cell Biol 164:979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Walker AE, Fikaris AJ, Kao GD, Brown EJ, Kazanietz MG, Meinkoth JL. (2005) Protein kinase C delta stimulates apoptosis by initiating G1 phase cell cycle progression and S phase arrest. J Biol Chem 280:32107–32114. [DOI] [PubMed] [Google Scholar]
- Shames DS, Wistuba II. (2014) The evolving genomic classification of lung cancer. J Pathol 232:121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steins M, Thomas M, Geißler M. (2014) Erlotinib. Recent Results Cancer Res 201:109–123. [DOI] [PubMed] [Google Scholar]
- Symonds JM, Ohm AM, Carter CJ, Heasley LE, Boyle TA, Franklin WA, Reyland ME. (2011) Protein kinase C δ is a downstream effector of oncogenic K-ras in lung tumors. Cancer Res 71:2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, Wu ZJ, Krall JA, Bierie B, Guo W, et al. (2013) Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell 24:347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Salama R, Gadgeel SM, Sarkar FH, Ahmad A. (2013) Erlotinib resistance in lung cancer: current progress and future perspectives. Front Pharmacol 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekle C, Giovannetti E, Sigmond J, Graff JR, Smid K, Peters GJ. (2008) Molecular pathways involved in the synergistic interaction of the PKC beta inhibitor enzastaurin with the antifolate pemetrexed in non-small cell lung cancer cells. Br J Cancer 99:750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonetti DA, Chisamore MJ, Grdina W, Schurz H, Jordan VC. (2000) Stable transfection of protein kinase C alpha cDNA in hormone-dependent breast cancer cell lines. Br J Cancer 83:782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vansteenkiste J, Ramlau R, von Pawel J, San Antonio B, Eschbach C, Szczesna A, Kennedy L, Visseren-Grul C, Chouaki N, Reck M. (2012) A phase II randomized study of cisplatin-pemetrexed plus either enzastaurin or placebo in chemonaive patients with advanced non-small cell lung cancer. Oncology 82:25–29. [DOI] [PubMed] [Google Scholar]
- Wang H, Gutierrez-Uzquiza A, Garg R, Barrio-Real L, Abera MB, Lopez-Haber C, Rosemblit C, Lu H, Abba M, Kazanietz MG. (2014) Transcriptional regulation of oncogenic protein kinase Cε (PKCε) by STAT1 and Sp1 proteins. J Biol Chem 289:19823–19838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ways DK, Kukoly CA, deVente J, Hooker JL, Bryant WO, Posekany KJ, Fletcher DJ, Cook PP, Parker PJ. (1995) MCF-7 breast cancer cells transfected with protein kinase C-alpha exhibit altered expression of other protein kinase C isoforms and display a more aggressive neoplastic phenotype. J Clin Invest 95:1906–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey CD, Xiao D, Tu T, Kim KW, Moretti L, Niermann KJ, Tawtawy MN, Quarles CC, Lu B. (2010) Enzastaurin (LY317615), a protein kinase C beta selective inhibitor, enhances antiangiogenic effect of radiation. Int J Radiat Oncol Biol Phys 77:1518–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DL, Sui FY, Du C, Zhang CW, Hui B, Xu SL, Lu HZ, Song GJ. (2009) Antisense expression of PKCalpha improved sensitivity of SGC7901/VCR cells to doxorubicin. World J Gastroenterol 15:1259–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Caino MC, von Burstin VA, Oliva JL, Kazanietz MG. (2008) Phorbol ester-induced apoptosis and senescence in cancer cell models. Methods Enzymol 446:123–139. [DOI] [PubMed] [Google Scholar]
- Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, Schlederer M, Johns C, Altorki N, Mittal V, et al. (2010) TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci USA 107:15535–15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LJ, Xu H, Qu JW, Zhao WZ, Zhao YB, Wang JH. (2012) Modulation of drug resistance in ovarian cancer cells by inhibition of protein kinase C-alpha (PKC-α) with small interference RNA (siRNA) agents. Asian Pac J Cancer Prev 13:3631–3636. [DOI] [PubMed] [Google Scholar]