Abstract
The tumor suppressor p53 has long been known to play a central role in maintaining a stable genome in the face of toxic insults through its role in promoting cell-cycle checkpoints, DNA repair, and apoptosis. However, p53 null cells still retain some function of certain checkpoint and repair processes, reducing the genomic changes that otherwise would occur if these mechanisms were absent. Accumulating evidence suggests that mutant forms of p53 proteins may drastically perturb these residual genome-stabilizing mechanisms through gain-of-function interactions with multiple proteins leading to a higher level of genomic instability than in p53 null cells. This review summarizes the current body of evidence that mutp53 plays a role in promoting various forms of genomic instability and provides an overview of current mechanistic proposals.
Keywords: MUTANT P53, GENOMIC INSTABILITY, ANEUPLOIDY, TRANSLOCATIONS, SPINDLE ASSEMBLY CHECKPOINT, DNA DAMAGE CHECKPOINT
The tumor suppressor p53 plays a pivotal role in the response of normal cells to noxious insults. Upon genotoxic stress, stabilization of p53 leads to transcription of genes involved in cell-cycle arrest and DNA repair or apoptosis. In addition, p53 protein can directly promote mitochondrial outer membrane permeabilization (MOMP) to trigger apoptosis by modulating the MOMP governing Bcl-2 family [Vaseva and Moll, 2009].
If the genotoxic damage is repaired, the cell may restart DNA synthesis and proceed through the remainder of the cycle. Failure of DNA repair results in apoptosis, preventing further mutagenesis of unrepaired DNA during replication by eliminating the irreparable cell from the organism. With an intact p53 system in place, accumulation of mutations becomes very difficult for a normal cell. However, despite this strongly protective system, premalignant lesions and early clinical stage cancers still find ways to subvert or deregulate several important hallmark pathways of cancer suppression without mutating the p53 gene or the p53 pathway. Mutations of p53 are typically seen in the later clinical stages of cancer and most likely have a driving role in promoting the aggressive evolution of these life-threatening tumors [Baker et al., 1990; Hruban et al., 2000; Olivier et al., 2006].
For many years, the mechanisms as to why mutations in p53 may lead to rapid tumor progression mainly focused on the role of wild-type (wt) p53 in cell-cycle checkpoint responses and apoptosis. This concept centered on the loss-of-wild-type function (LOF) that occurs when p53 becomes mutated in tumors. However, it has now become clear that many mutant (mut) p53 proteins, beyond the “simple” loss of wt suppressor functions, also exert an active role in promoting tumor progression [Sigal and Rotter, 2000; Brosh and Rotter, 2009]. Several different knock-in mouse models provided convincing evidence that mutp53 proteins play an important gain-of-function (GOF) role in promoting invasion and metastasis of tumors of different tissue origins including bone, lung, skin, muscle, and pancreas, illustrating that both epithelial as well as mesenchymal cell lineages are susceptible to mutp53’s powerful ability to drive tumor progression to a more invasive and metastatic phenotype [Lang et al., 2004; Olive et al., 2004; Hingorani et al., 2005; Caulin et al., 2007; Doyle et al., 2010].
Recent studies have shed some light on the mechanistic basis behind mutp53’s GOF. These studies have focused on signaling pathways known to be involved in promoting invasion and metastasis, including TGF-beta mediated motility and invasion, increased EGFR and integrin signaling by enhanced receptor recycling to the cell surface, and Pin1-mediated activation of a pro-aggressive transcriptional program by mutp53 [Adorno et al., 2009; Muller et al., 2009; Girardini et al., 2011]. Importantly, a common theme behind these mechanistic models is the inactivation of the p53 family member TAp63, a transcription factor with metastasis suppressing activity. A thorough review of these mechanisms has recently been published and will not be discussed here [Muller et al., 2011].
Genomic instability has long been proposed to be a mechanism by which a cell may acquire the necessary properties for invasion and metastasis. According to traditional theory, genetic diversity within the primary tumor, produced by genomic instability, allows for a rich tapestry of genetically distinct cells. Some of these variant cells harbor the necessary properties required for colonization of distant organs. Upon leaving the primary site, these cells will seed distant sites and expand, producing secondary life-threatening tumors. Recently, advanced genotyping of individual human metastatic pancreatic carcinomas was performed [Campbell et al., 2010; Yachida et al., 2010]. Of note, similar signature mutations, called “founder” mutations that included p53 were found throughout the entire primary tumor and within all metastatic clones in a given patient. In addition, de novo genetic evolutionary changes were present within metastatic clones but absent in most of the primary tumor. Therefore they were called “progressor” mutations. However, upon careful geographical dissection and sequencing of individual tissue sections, the “progressor” mutations could also be found in clonal expansions in certain locations within the corresponding primary tumor, consistent with the traditional Darwinian evolutionary view. Thus, genomic instability in a primary tumor may be an important evolutionary driver of metastatic ability. The frequent occurrence of p53 mutations and the presence of genomic instability in metastatic human cancers and the fact that mutp53 knockin mouse models have an increased metastatic load provided further support for the notion that mutp53 might be an important driver of genomic instability.
Genomic instability is defined as an increase in the rate of DNA alterations compared to normal cells. There are four types of genomic instability (IN): subtle sequence instabilities at the nucleotide (NIN) and mismatch (MIN) level, chromosomal instability (CIN), and amplification instability (AIN) [Lengauer et al., 1998]. Wild-type p53 has been implicated either directly or indirectly in suppression of each type of instability. So far, GOF p53 mutants have been implicated in promoting two types of instabilities, chromosomal (CIN) and amplification (AIN) instability.
ANEUPLOIDY
CIN can be divided into aneuploidy (the numerical gain or loss of whole chromosomes) and structural changes (i.e., translocations and deletions). Aneuploidy and translocations can be separate from one another, as certain cells may have aneuploidy without translocations and vice versa. Aneuploidy is an almost universal aspect of human solid tumors and initially may be an unfavorable state for the preneoplastic cell [Williams et al., 2008]. However, over time, aneuploidy is thought to give rise to clonal evolution of tumor cells that harbor advantageous combinations of chromosomes.
Early reports implicating mutp53 in promoting aneuploidy were based on increased rates of aneuploidy present in mutp53-expressing cells compared to cells that contained no p53. Two well known numerical checkpoints exist in the cell cycle, the mitotic spindle assembly checkpoint (SAC) in M-phase and the tetraploidy checkpoint at G1/S. Mounting evidence suggests that p53-null cells have disrupted only the tetraploidy G1/S checkpoint, while mutp53 harboring cells have disrupted both the G1/S and M checkpoints.
For example, fibroblasts cultured from Li-Fraumeni patients harboring p53 missense mutations including R175H were able to undergo S-phase reentry and polyploidization after disruption of their mitotic spindles, while cells expressing a truncated p53 protein (equivalent to no p53) were blocked from re-entry. This suggests that p53 mutants cells can actively disrupt or at least bypass the mitotic checkpoint and exit as tetraploid cells, while p53 null cells get stopped by this checkpoint [Gualberto et al., 1998]. In the absence of a p53-mediated tetraploid checkpoint response at G1, tetraploid cells may undergo asymmetric chromosome divisions in the next M-phase, resulting in aneuploidy [Lanni and Jacks, 1998; Andreassen et al., 2001].
The SAC protein BubR1 plays a key role in proper centrosome maintenance and chromosomal stability. Upon sensing kinetochores with improper microtubule attachment, BubR1 inhibits the anaphase promoting complex (APC) from initiating metaphase, thereby preventing sister chromatid separation. BubR1 can physically associate with and activate wtp53 during SAC checkpoint activation, and wtp53 has been shown to reciprocally activate transcription of BubR1, potentially forming a positive feedback loop [Oikawa et al., 2005; Ha et al., 2007]. Not surprisingly, BubR1 is expressed at low levels in early passage p53−/− MEFs containing extensive aneuploidy. However, BubR1 expression rises in late passage p53−/− MEFs presumably through secondary mutational mechanisms, resulting in subsequent stabilization of an aneuploid karyotype. The p53 family member TAp73 has also been shown to play an important role in SAC by associating with BubR1 and potentiating its ability to phosphorylate downstream checkpoint effectors [Tomasini et al., 2009]. Possibly TAp73 may perform this secondary karyotype stabilization in the absence of p53.
Conversely, in mutp53-harboring cells binding and inactivation of TAp73 by mutp53 protein may lead to inefficient spindle checkpoint function and aneuploidy, instead of secondary karyotype stabilization that is seen in p53-null cells. Additional mechanisms independent of inhibition of p53 family members may also be at play, as suggested by aberrant centrosome amplification and aneuploidy in skin squamous cell carcinomas in the absence of mutp53 binding to p63 and p73 [Caulin et al., 2007]. Although not formally tested, mutant p53 proteins may have a more direct role in SAC inhibition by physically associating with BubR1, similar to wtp53. However, instead of activating BubR1, the high levels of mutp53 protein may sequester and inhibit BubR1’s checkpoint functions, preventing the karyotype stabilization seen in p53−/− cells and resulting in persistent CIN and a high level of genetic variation at the level of aneuploidy.
Loss or mutational inactivation of wild-type p53 has been reported to lead to abnormal amplification of centrosomes via deregulation of the centrosome duplication cycle and failure to undergo cytokinesis. p53 is proposed to control centrosome duplication via transactivation-dependent (p21Waf1/CDK2/Cyclin E) and transactivation-independent (direct physical interaction with the centrosome) mechanisms [Tarapore and Fukasawa, 2002]. Ectopic expression of mutp53 R172H in p53-null primary mouse mammary epithelial cells resulted in aberrant centrosome amplification, multipolar mitoses, and consequently increased numbers of chromosomes [Murphy et al., 2000]. Interestingly, the p53 R172H mutant protein was found to directly bind to centrosomes [Tarapore et al., 2001]. Moreover, in vivo mouse models of mammary tumors, pancreatic tumors and squamous cell carcinoma of the skin expressing mutp53 R172H all displayed similar findings [Li et al., 1998; Wang et al., 1998; Hingorani et al., 2005; Caulin et al., 2007]. Importantly, centrosome amplification is a frequent finding in certain human mutp53-harboring cancers [Weber et al., 1998; Carroll et al., 1999]. Further research clarifying the role of mutp53 in the centrosome duplication cycle will lead to a better understanding of mutp53-induced aneuploidy.
TRANSLOCATIONS AND AMPLIFICATIONS
Besides aneuploidy, structural chromosomal changes are a common finding in mutp53 human cancers. Several studies investigating the genomic complexity of a variety of human primary tumors have generally shown an increase in genomic aberrations present in mutp53 samples versus wild-type p53 samples. Breast tumors with p53 mutations have a higher average number of comparative genomic hybridization (CGH) alterations compared to p53 wild-type tumors [Jain et al., 2001; Jong et al., 2004]. p53 mutations are sometimes present in chronic myelogenous leukemia (CML) and chronic lymphocytic leukemia (CLL) and have been suggested to be a predisposing factor for development of the additional cytogenetic aberrations associated with clonal evolution and preterminal blast crisis [Guinn et al., 1994; Zenz et al., 2008]. Soft tissue sarcomas with highly complex, unbalanced karyotypes correlate with mutant p53 status, while the ones with simpler karyotypes typically have a wild-type p53 status [Borden et al., 2003]. Osteosarcomas harboring mutp53 show significantly increased genomic complexity, including increased gains and losses of genetic information, compared to wild-type p53 osteosarcomas [Overholtzer et al., 2003]. Interestingly, Mdm2-amplified mutant p53 osteosarcomas had a lower degree of genomic instability compared to Mdm2-nonamplified osteosarcomas, suggesting that high levels of mutp53 protein are important for maintaining high levels of genomic instability [Overholtzer et al., 2003]. However, the stage of malignancy tends to be a confounding factor in these studies, as many, but not all, malignancies containing mutp53 are associated with an advanced clinical stage. Advanced stage tumors tend to have more genomic aberrations than earlier stage tumors simply by virtue of the higher number of mitotic doublings that late stage tumor cells have undergone.
Although in general mouse tumors often lack complex genomic aberrations, mutp53 harboring mouse tumors have shown a striking degree of karyotype complexity. In a mouse model in which the oncogenic K-ras G12D and mutp53R172H were conditionally expressed in the pancreas, highly aggressive carcinomas developed with extensive metastatic disease. The karyotypes of primary cell lines derived from these tumors displayed extensive chromosomal instability with numerous chromatid breaks, fusions, and clonal nonreciprocal translocations (NRTs) [Hingorani et al., 2005]. Presently, a stringent karyotype comparison between mutp53 versus null p53 tumors from the same tissue of origin in the same mouse model has still not been performed. However, using a sensitive PCR-based approach, Song and Liu assessed the levels of interchromosomal T-cell receptor (TCR) translocations between p53 null versus p53 mutant pretumoral thymocytes [Song et al., 2007]. Importantly, this study showed increased translocations present in mutant compared to null thymocytes and provided evidence that mutp53 can facilitate the development of balanced chromosomal translocations in vivo. In addition, the radiation-induced loss of TCR expression in human T-leukemia (p53-null Jurkat cells)—which was interpreted as mutation frequency—was increased upon ectopic expression of mutp53 [Iwamoto et al., 1996]. However, it is not known whether this was due to translocation, deletions, or point mutations since the mutational mechanism was not investigated in this study.
In vitro data also suggest that mutp53 can facilitate structural chromosomal abnormalities by interacting with and inhibiting the genome caretaker proteins of DNA repair. The fidelity of DNA double strand break repair plays a central role in preventing translocations. In response to DNA double strand breaks, cells invoke two distinct repair pathways: homologous recombination (HR) and nonhomologous end joining (NHEJ) [Jackson, 2002]. The choice between these pathways involves multiple factors, including the cell-cycle stage, tissue type (somatic vs. germ cells), and the integrity of the two pathways [Takata et al., 1998; Essers et al., 2000]. Mre11 is a DNA binding protein with roles in both HR and NHEJ [Zha et al., 2009]. Several mutp53 proteins were proposed to bind and sequester Mre11 away from double strand DNA breaks. Loss of functional Mre11 would be predicted to increase the amount of spontaneous chromosome and chromatid breaks and translocations, which occurs in Mre11 null cells [Buis et al., 2008]. Abrogation of Mre11 function limits the phosphorylation and activation of ATM, the major double strand break sensor in cells, resulting in bypassing of the G2/M DNA damage checkpoint. Loss of this checkpoint severely reduces the ability for efficient HR, resulting in less conservation of genetic information [Song et al., 2007].
Topoisomerase I has an important enzymatic function in DNA homeostasis. By creating single strand breaks followed by unwinding, this protein prevents excessive torsion due to overwinding of the DNA double helix created by such processes as replication, transcription, and DNA repair. Wild-type p53 was shown to associate with and increase topoisomerase I activity during times of DNA stress in a regulated fashion, facilitating DNA repair [Gobert et al., 1999]. This association is retained in the face of p53 mutations since the amino acids responsible for interaction, aa 302–320, are not commonly perturbed by p53 mutations. In addition, p53 mutants are capable of inducing topoisomerase I activity just like wtp53. Thus, the high levels of nuclear mutp53 in cancer cells is speculated to lead to inappropriate topoisomerase I activity, resulting in an increase in recombinogenic events [Gobert et al., 1999].
Telomere maintenance may also play a role in mutp53 GOF. Intriguingly, in the mutp53 expressing pancreatic cancer model discussed above [Hingorani et al., 2005], significant stretches of telomeric sequence were observed at the fusion points of translocations, while the telomere sequences at the ends of chromatids were conserved, a phenotype reminiscent of cells which lost the function of telomere capping proteins [de Lange, 2002; Smogorzewska et al., 2002]. This finding suggests that maintenance of telomere protection, rather than shortening of telomere sequence, may play an additional role in the generation of chromosome breaks and translocations by mutp53 in vivo. Further research comparing telomere maintenance between p53 mutant versus null cells may elucidate a novel GOF in this context.
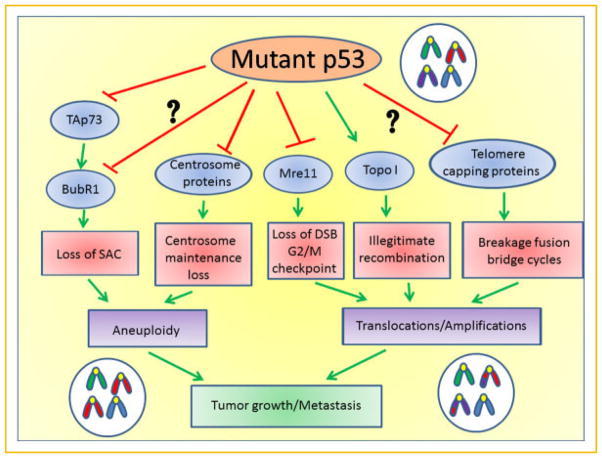
Gene amplification is commonly a late stage event in tumorigenesis and often occurs in mutp53 tumors [Lengauer et al., 1998]. This is probably facilitated in part by loss of the DNA damage response. Wild-type p53 plays an important role in suppression of gene amplification [Livingstone et al., 1992; Yin et al., 1992]. Conversely, gene amplification can be promoted in p53 null cells expressing ectopic mutp53, and can be further enhanced by the topoisomerase 1 specific agent camptothecin, reflecting mutp53’s enhancement of topoisom-erase 1 activity [El-Hizawi et al., 2002]. Moreover, enhanced double strand breaks followed by improper repair can also result in gene amplification, particularly in NHEJ deficient cells [Zhu et al., 2002]. Thus, amplification can also be an indirect consequence of the mutp53-mediated defective DNA double stranded break repair described above. A summary of the proposed mutp53-mediated mechanisms of CIN and AIN is shown in Figure 1.
Fig. 1.
Proposed pathways how mutant p53 promotes aneuploidy, translocations, and amplifications by multiple gain-of-function mechanisms leading to genomic instability that ultimately promotes tumor growth and metastasis. Expression of mutant p53 in a normal or malignant cell inhibits TAp73, leading to a loss of BubR1 function, a spindle assembly checkpoint defect and aneuploidy. A direct interaction of mutant p53 with BubR1 is also possible. Certain p53 mutants may directly interact with centrosomes, possibly perturbing centrosome maintenance. Alternatively, binding and sequestration of Mre11 by mutant p53 prevents a double strand break checkpoint from occurring, which leads to inefficient DNA repair before cells enter mitosis. Furthermore, topoisomerase I activity is stimulated by p53 mutants, leading to an increase in recombinogenic events between chromosomes. Finally, telomere capping proteins may be inhibited by mutant p53 proteins, leading to loss of telomere protection and breakage—fusion bridge cycles generating chromosomal translocations.
ESCAPE FROM CELL DEATH IN THE FACE OF DNA DAMAGE
Deficient repair in the face of increased levels of DNA damage has devastating consequences for the viability of cells, even in the absence of p53 [Fedier et al., 2003; Tutt et al., 2003]. However, mutp53 cells, instead of being more sensitive, frequently have been found to be as viable and sometimes even more resistant to DNA damage than p53 null cells [Blandino et al., 1999; Bossi et al., 2006; Di Agostino et al., 2006]. This apparent paradox is explained by numerous studies detailing the inhibition of many cell death and checkpoint functions by mutp53. Mutant p53 protein associates with the trimeric transcription factor complex NF-Y at promoters of genes involved in cell-cycle regulation [Di Agostino et al., 2006]. Upon DNA damage, mutp53 recruits the acetyltransferase p300 to NF-Y target genes such as cyclin A, cyclin B2, CDK1, and cdc25 and upregulates their transcription. This abnormal upregulation of cell-cycle genes leads to increased DNA synthesis and proliferation instead of the normal cell-cycle checkpoint response that is mediated by wild-type p53. Besides escape from arrest and death, this perturbation of cell-cycle checkpoints would result in inefficient repair with persistence of double strand breaks, compounding the problems already present from abnormalities of caretaker proteins.
As discussed above, binding and inactivation of p63 and p73 has surfaced as another GOF activity of mutant p53 [Li and Prives, 2007; Rufini et al., 2011]. p63 and p73 can transcribe similar DNA stress response and apoptotic genes as wild-type p53 [Kaghad et al., 1997; Wu et al., 2003; Lokshin et al., 2005]. Primary cells and carcinomas from mice lacking TAp63 or TAp73 show high amounts of chromosome aberrations and instability [Tomasini et al., 2008; Su et al., 2010]. With the p53-independent apoptotic pathways of TAp63 and TAp73 inactivated, the threshold for apoptosis by DNA damage would increase. Accordingly, chemoresistance mediated by mutp53 is correlated with its ability to bind p73 [Bergamaschi et al., 2003; Irwin et al., 2003; Schilling et al., 2010]. Besides inhibition by direct binding, mutp53 may indirectly inactivate p73 through the reduced Chk kinase activity from sequestration of Mre11, since both Chk1 and Chk2 can directly activate p73 transcriptional activity upon DNA damage [Urist et al., 2004]. In addition, the cyclin and CDK deregulation via NF-Y may further augment p73 inhibition, since phosphorylation of p73 by cyclin/CDK complexes can repress its function, further contributing to resistance to DNA damage [Gaiddon et al., 2003]. A summary of the proposed mutp53-mediated mechanisms of escaping cell death is shown in Figure 2.
Fig. 2.
Proposed mechanisms of escape from DNA damage-induced cell death by mutant p53. In response to DNA damage, inhibition of p63 and p73 by mutant p53 leads to loss of p53 independent transcription of apoptosis genes, while damage induced NF-Y/p300/mutp53 transcription complexes enhances cell proliferation by upregulating cell-cycle promoting target genes. Aside from direct inhibition of p73 mediated by mutantp53/p73 complex formation, loss of Chk kinase activity and enhanced transcription of cell-cycle promoting genes can also lead to indirect inhibition of p73.
FUTURE PROSPECTS AND THERAPEUTIC IMPLICATIONS
Much progress has been made in establishing the role of mutp53 in enhancing genomic instability. However, many questions still remain. Does mutp53 have any GOF in promoting nucleotide (NIN) and mismatch (MIN) genetic instabilities? How exactly does mutp53 promote enhanced aneuploidy? Is centrosome amplification with subsequent multipolar mitoses the main mechanism of aneuploidy or is checkpoint inactivation its primary cause? Mutp53 produces structural chromosomal changes by altering DNA repair proteins as well as checkpoint inactivation: what are their relative contributions? Does telomere maintenance play a role? In order to answer these questions, more studies must be conducted on the molecular biology of mutp53 interactions with the factors involved in checkpoint activation and repair, such as Mre11, NF-Y, p63, p73, and topoisomerase. Characterizing the instability phenotype of cells after perturbing these interactions will lead to a better understanding of the main causes of mutant p53-mediated genomic instabilities, which might also be point mutant-specific.
What is the ultimate phenotypic result of this genomic instability? Is it truly contributing to the increased proliferation, invasion and metastasis seen in tumors of mutp53 mice, and can these results be extended to human tumors? Genomic instability has been proposed to be a common GOF for all p53 mutants, regardless of structural class [Song et al., 2007]. However, a systematic analysis of many p53 mutants in this respect has not yet been conducted.
Finally and most importantly, what is the most appropriate clinical course in the face of this genomic instability? One novel therapeutic strategy involves deliberate abrogation of remaining G2/M checkpoint functions of tumor cells, leading to enhanced genomic instability and mitotic catastrophe when combined with DNA damaging agents [Ashwell and Zabludoff, 2008]. As discussed, mutant p53 cells already have decreased checkpoint function but maintain high viability, raising concerns that this strategy might be inappropriate for mutp53 harboring tumors. There is a pressing need to discover the molecular basis of how mutant p53 cells resist cell death in the face of DNA damage. p63, p73, and NF-Y are potential candidates for this resistance. Thus, effective targeting of these proteins may result in dramatically increased tumor killing in response to conventional DNA damaging chemotherapeutics.
References
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, Parenti AR, Rosato A, Bicciato S, Balmain A, Piccolo S. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwell S, Zabludoff S. DNA damage detection and repair pathways—Recent advances with inhibitors of checkpoint kinases in cancer therapy. Clin Cancer Res. 2008;14:4032–4037. doi: 10.1158/1078-0432.CCR-07-5138. [DOI] [PubMed] [Google Scholar]
- Baker SJ, Preisinger AC, Jessup JM, Paraskeva C, Markowitz S, Willson JK, Hamilton S, Vogelstein B. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990;50:7717–7722. [PubMed] [Google Scholar]
- Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, Yulug I, Merlano M, Numico G, Comino A, Attard M, Reelfs O, Gusterson B, Bell AK, Heath V, Tavassoli M, Farrell PJ, Smith P, Lu X, Crook T. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Blandino G, Levine AJ, Oren M. Mutant p53 gain of function: Differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene. 1999;18:477–485. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- Borden EC, Baker LH, Bell RS, Bramwell V, Demetri GD, Eisenberg BL, Fletcher CD, Fletcher JA, Ladanyi M, Meltzer P, O’Sullivan B, Parkinson DR, Pisters PW, Saxman S, Singer S, Sundaram M, van Oosterom AT, Verweij J, Waalen J, Weiss SW, Brennan MF. Soft tissue sarcomas of adults: State of the translational science. Clin Cancer Res. 2003;9:1941–1956. [PubMed] [Google Scholar]
- Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. Mutant p53 gain of function: Reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene. 2006;25:304–309. doi: 10.1038/sj.onc.1209026. [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: News from the mutant p53 field. Nat Rev. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal SA, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Griffin CA, Burton J, Swerdlow H, Quail MA, Stratton MR, Iacobuzio-Donahue C, Futreal PA. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll PE, Okuda M, Horn HF, Biddinger P, Stambrook PJ, Gleich LL, Li YQ, Tarapore P, Fukasawa K. Centrosome hyperamplification in human cancer: Chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene. 1999;18:1935–1944. doi: 10.1038/sj.onc.1202515. [DOI] [PubMed] [Google Scholar]
- Caulin C, Nguyen T, Lang GA, Goepfert TM, Brinkley BR, Cai WW, Lozano G, Roop DR. An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J Clin Invest. 2007;117:1893–1901. doi: 10.1172/JCI31721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, Piaggio G. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Doyle B, Morton JP, Delaney DW, Ridgway RA, Wilkins JA, Sansom OJ. p53 mutation and loss have different effects on tumourigenesis in a novel mouse model of pleomorphic rhabdomyosarcoma. J Pathol. 2010;222:129–137. doi: 10.1002/path.2748. [DOI] [PubMed] [Google Scholar]
- El-Hizawi S, Lagowski JP, Kulesz-Martin M, Albor A. Induction of gene amplification as a gain-of-function phenotype of mutant p53 proteins. Cancer Res. 2002;62:3264–3270. [PubMed] [Google Scholar]
- Essers J, van Steeg H, de Wit J, Swagemakers SM, Vermeij M, Hoeijmakers JH, Kanaar R. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedier A, Schlamminger M, Schwarz VA, Haller U, Howell SB, Fink D. Loss of atm sensitises p53-deficient cells to topoisomerase poisons and antimetabolites. Ann Oncol. 2003;14:938–945. doi: 10.1093/annonc/mdg240. [DOI] [PubMed] [Google Scholar]
- Gaiddon C, Lokshin M, Gross I, Levasseur D, Taya Y, Loeffler JP, Prives C. Cyclin-dependent kinases phosphorylate p73 at threonine 86 in a cell cycle-dependent manner and negatively regulate p73. J Biol Chem. 2003;278:27421–27431. doi: 10.1074/jbc.M300251200. [DOI] [PubMed] [Google Scholar]
- Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, Capaci V, Jordan L, Quinlan P, Thompson A, Mano M, Rosato A, Crook T, Scanziani E, Means AR, Lozano G, Schneider C, Del Sal G. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 2011;20:79–91. doi: 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Gobert C, Skladanowski A, Larsen AK. The interaction between p53 and DNA topoisomerase I is regulated differently in cells with wild-type and mutant p53. Proc Natl Acad Sci USA. 1999;96:10355–10360. doi: 10.1073/pnas.96.18.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto A, Aldape K, Kozakiewicz K, Tlsty TD. An oncogenic form of p53 confers a dominant, gain-of-function phenotype that disrupts spindle checkpoint control. Proc Natl Acad Sci USA. 1998;95:5166–5171. doi: 10.1073/pnas.95.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinn BA, Padua RA, Burnett A, Mills K. p53 mutations indicate a changing clonal evolution in a portion of chronic myelocytic leukemia patients. Blood. 1994;84:3591. [PubMed] [Google Scholar]
- Ha GH, Baek KH, Kim HS, Jeong SJ, Kim CM, McKeon F, Lee CW. p53 activation in response to mitotic spindle damage requires signaling via BubR1-mediated phosphorylation. Cancer Res. 2007;67:7155–7164. doi: 10.1158/0008-5472.CAN-06-3392. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- Iwamoto KS, Mizuno T, Ito T, Tsuyama N, Kyoizumi S, Seyama T. Gain-of-function p53 mutations enhance alteration of the T-cell receptor following X-irradiation, independently of the cell cycle and cell survival. Cancer Res. 1996;56:3862–3865. [PubMed] [Google Scholar]
- Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- Jain AN, Chin K, Borresen-Dale AL, Erikstein BK, Eynstein Lonning P, Kaaresen R, Gray JW. Quantitative analysis of chromosomal CGH in human breast tumors associates copy number abnormalities with p53 status and patient survival. Proc Natl Acad Sci USA. 2001;98:7952–7957. doi: 10.1073/pnas.151241198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong YJ, Li LH, Tsou MH, Chen YJ, Cheng SH, Wang-Wuu S, Tsai SF, Chen CM, Huang AT, Hsu MT, Lin CH. Chromosomal comparative genomic hybridization abnormalities in early- and late-onset human breast cancers: Correlation with disease progression and TP53 mutations. Cancer Genet Cytogenet. 2004;148:55–65. doi: 10.1016/s0165-4608(03)00205-x. [DOI] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, El-Naggar AK, Lozano G. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Li Y, Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- Li B, Murphy KL, Laucirica R, Kittrell F, Medina D, Rosen JM. A transgenic mouse model for mammary carcinogenesis. Oncogene. 1998;16:997–1007. doi: 10.1038/sj.onc.1201621. [DOI] [PubMed] [Google Scholar]
- Livingstone LR, White A, Sprouse J, Livanos E, Jacks T, Tlsty TD. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- Lokshin M, Tanaka T, Prives C. Transcriptional regulation by p53 and p73. Cold Spring Harb Symp Quant Biol. 2005;70:121–128. doi: 10.1101/sqb.2005.70.046. [DOI] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, Cromer A, Brugge JS, Sansom OJ, Norman JC, Vousden KH. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KL, Dennis AP, Rosen JM. A gain of function p53 mutant promotes both genomic instability and cell survival in a novel p53-null mammary epithelial cell model. FASEB J. 2000;14:2291–2302. doi: 10.1096/fj.00-0128com. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Okuda M, Ma Z, Goorha R, Tsujimoto H, Inokuma H, Fukasawa K. Transcriptional control of BubR1 by p53 and suppression of centrosome amplification by BubR1. Mol Cell Biol. 2005;25:4046–4061. doi: 10.1128/MCB.25.10.4046-4061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, Varley J, Bignon Y, Uhrhammer N, Winqvist R, Jukkola-Vuorinen A, Niederacher D, Kato S, Ishioka C, Hainaut P, Borresen-Dale AL. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Rao PH, Favis R, Lu XY, Elowitz MB, Barany F, Ladanyi M, Gorlick R, Levine AJ. The presence of p53 mutations in human osteosarcomas correlates with high levels of genomic instability. Proc Natl Acad Sci USA. 2003;100:11547–11552. doi: 10.1073/pnas.1934852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufini A, Agostini M, Grespi F, Tomasini R, Sayan BS, Niklison-Chirou MV, Conforti F, Velletri T, Mastino A, Mak TW, Melino G, Knight RA. p73 in cancer. Genes Cancer. 2011;2:491–502. doi: 10.1177/1947601911408890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling T, Kairat A, Melino G, Krammer PH, Stremmel W, Oren M, Muller M. Interference with the p53 family network contributes to the gain of oncogenic function of mutant p53 in hepatocellular carcinoma. Biochem Biophys Res Commun. 2010;394:817–823. doi: 10.1016/j.bbrc.2010.03.082. [DOI] [PubMed] [Google Scholar]
- Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: The demons of the guardian of the genome. Cancer Res. 2000;60:6788–6793. [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, El-Naggar A, Creighton CJ, Suraokar MB, Wistuba I, Flores ER. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarapore P, Fukasawa K. Loss of p53 and centrosome hyperamplification. Oncogene. 2002;21:6234–6240. doi: 10.1038/sj.onc.1205707. [DOI] [PubMed] [Google Scholar]
- Tarapore P, Tokuyama Y, Horn HF, Fukasawa K. Difference in the centrosome duplication regulatory activity among p53 ’hot spot’ mutants: Potential role of Ser 315 phosphorylation-dependent centrosome binding of p53. Oncogene. 2001;20:6851–6863. doi: 10.1038/sj.onc.1204848. [DOI] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, Iovanna JL, Squire J, Jurisica I, Kaplan D, Melino G, Jurisicova A, Mak TW. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Tsuda C, Lau SK, Wilhelm M, Ruffini A, Tsao MS, Iovanna JL, Jurisicova A, Melino G, Mak TW. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc Natl Acad Sci USA. 2009;106:797–802. doi: 10.1073/pnas.0812096106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt A, Connor F, Bertwistle D, Kerr P, Peacock J, Ross G, Ashworth A. Cell cycle and genetic background dependence of the effect of loss of BRCA2 on ionizing radiation sensitivity. Oncogene. 2003;22:2926–2931. doi: 10.1038/sj.onc.1206522. [DOI] [PubMed] [Google Scholar]
- Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787:414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Greenhalgh DA, Jiang A, He D, Zhong L, Brinkley BR, Roop DR. Analysis of centrosome abnormalities and angiogenesis in epidermal-targeted p53172H mutant and p53-knockout mice after chemical carcinogenesis: Evidence for a gain of function. Mol Carcinog. 1998;23:185–192. doi: 10.1002/(sici)1098-2744(199811)23:3<185::aid-mc7>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Weber RG, Bridger JM, Benner A, Weisenberger D, Ehemann V, Reifenberger G, Lichter P. Centrosome amplification as a possible mechanism for numerical chromosome aberrations in cerebral primitive neuroectodermal tumors with TP53 mutations. Cytogenet Cell Genet. 1998;83:266–269. doi: 10.1159/000015168. [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Nomoto S, Hoque MO, Dracheva T, Osada M, Lee CC, Dong SM, Guo Z, Benoit N, Cohen Y, Rechthand P, Califano J, Moon CS, Ratovitski E, Jen J, Sidransky D, Trink B. DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 2003;63:2351–2357. [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, Velculescu VE, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Tainsky MA, Bischoff FZ, Strong LC, Wahl GM. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- Zenz T, Krober A, Scherer K, Habe S, Buhler A, Benner A, Denzel T, Winkler D, Edelmann J, Schwanen C, Dohner H, Stilgenbauer S. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: Results from a detailed genetic characterization with long-term follow-up. Blood. 2008;112:3322–3329. doi: 10.1182/blood-2008-04-154070. [DOI] [PubMed] [Google Scholar]
- Zha S, Boboila C, Alt FW. Mre11: Roles in DNA repair beyond homologous recombination. Nat Struct Mol Biol. 2009;16:798–800. doi: 10.1038/nsmb0809-798. [DOI] [PubMed] [Google Scholar]
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]