Abstract
Brilliant cresyl blue (BCB) is a super vital stain that has been used to select competent oocytes in different species. The objectives of present studies were to determine mRNA abundance for select TGFβ superfamily components, SMAD2/3 and SMAD1/5 phosphorylation levels and transcript abundance for other oocyte (JY1) and cumulus cell (CTSB, CTSK, CTSS and CTSZ) markers of oocyte quality in bovine oocytes and or adjacent cumulus cells classified based on developmental potential using BCB staining. The ability of exogenous FST, JY1, or cathepsin inhibitor treatment to enhance development of embryos derived from poor quality oocytes selected based on BCB staining was also determined. Cumulus oocyte complexes (COCs) from abattoir derived ovaries were subjected to BCB staining and GV stage oocytes and cumulus cells harvested from control, BCB+ and BCB- (poor oocyte quality) groups for real time PCR or Western blot analysis. Remaining COCs underwent in vitro maturation, in vitro fertilization and embryo culture in presence or absence of above described treatments. Levels of FST, JY1, BMP15 and SMAD1, 2, 3 and 5 transcripts were higher in BCB+ oocytes whereas abundance of CTSB, CTSK, CTSS and CTSZ mRNAs was higher in cumulus cells surrounding poor quality BCB- oocytes. Western blot analysis revealed SMAD1/5 and SMAD2/3 phosphorylation were higher in BCB+ than BCB− oocytes. Embryo culture studies demonstrated that follistatin and cathepsin inhibitor treatment but not JY-1 treatment can promote developmental competence of BCB- oocytes. Results provide further understanding of molecular indices of oocyte competence.
Keywords: G6PDH, FST, JY-1, E-64, cumulus cell cathepsins, SMAD
INTRODUCTION
Oocyte developmental competence is generally referred to as the ability of the oocyte to resume meiosis, cleave post fertilization, promote embryonic development, and bring a pregnancy to term in good health (Sirard et al. 2006). Oocyte quality is one of the major limitations to in vitro embryo production efficiency (Boni 2012; Rajput et al. 2013). Hence, identification of molecular markers of oocyte quality with diagnostic and/or therapeutic applications has been of significant interest in livestock species and humans.
Numerous studies have reported potential association of paracrine/autocrine mediators, including members of the transforming growth factor TGFβ superfamily with embryo developmental progression (Bettegowda et al. 2008; Mamo et al. 2011; Patel et al. 2007; Ruvolo et al. 2013; Torner et al. 2008; Wang and Sun 2007). For example, the oocyte specific protein JY-1 is functionally required for meiotic maturation, cumulus expansion and embryo development post fertilization (Bettegowda et al. 2007; Lee et al. 2014b). Our previous studies (Bettegowda et al. 2008; Patel et al. 2007) in the prepubertal calf model of poor oocyte competence have identified intrinsic oocyte (FST) and cumulus derived factors (CTSB, CTSK, CTSS, CTSZ) associated with oocyte competence. The oocyte derived TGFβ superfamily members GDF9 and BMP15 are key regulators of fertility (Abir et al. 2014; Barzegari et al. 2010; Persani et al. 2014). GDF9 signals intracellularly through SMAD2/3, whereas BMP15 utilizes the SMAD1/5/8 signaling pathway (Shimasaki et al. 2004). Expression of mRNA for SMAD signal transduction factors in human granulosa cells is correlated with oocyte quality (Kuo et al. 2011). Furthermore, supplementation with the pro-mature complex form of BMP15 during in vitro maturation improves bovine early embryonic development (Sudiman et al. 2014). Yet, the global significance of above factors to oocyte competence is not completely understood.
Despite recent advances in reproductive and developmental biology, morphological criteria remain the most widely used method for oocyte selection because molecular marker (RNA)-based evaluation of oocytes is invasive, costly and or not readily amenable to rapid classification. Brilliant Cresyl Blue (BCB) staining is a non-invasive method for oocyte evaluation that has been used to select competent oocytes in different species including cattle, goats, sheep, pigs (Opiela and Katska-Ksiazkiewicz 2013), buffalo (Manjunatha et al. 2007), horses (Mohammadi-Sangcheshmeh et al. 2011; Pereira et al. 2014) and mice (Wu et al. 2007). BCB is used to determine the intracellular activity of glucose-6-phosphate dehydrogenase (G6PDH) (Alm et al. 2005) linked to egg quality. The G6PDH is a regulatory enzyme that is synthesized and accumulated during oocyte growth and its activity gradually decreases as oocytes complete their growth phase (Mangia and Epstein 1975). The BCB dye can be reduced by the G6PDH enzyme activity, thus oocytes that have completed their growth phase cannot reduce BCB to a colorless compound and display a blue cytoplasm (BCB+). However, growing oocytes have a high activity of G6PDH and can reduce the blue substrate, resulting in a colorless oocyte cytoplasm (BCB−) (Ericsson et al. 1993; Tian et al. 1998; Wassarman 1988). Using BCB staining as an indicator of oocyte competence, previous studies in cattle and other species demonstrated greater rates of blastocyst development following in vitro fertilization for BCB+ oocytes that have presumably completed the growth phase versus BCB− counterparts (Alm et al. 2005).
The objectives of the current studies were to determine 1) whether transcript abundance for select developmentally important genes in GV oocytes (FST, JY1 CTSB, CTSS, CTSZ, BMP15, GDF9, SMAD1, 2, 3 and 5) and adjacent cumulus cells (FST, CTSB, CTSK, CTSS, and CTSZ) differs in good versus poor quality oocytes selected based on BCB staining, 2) differences in activity of SMAD1/5 and SMAD2/3 signaling pathways in good versus poor quality oocytes selected based on BCB staining and 3) to determine the effect of exogenous follistatin and JY-1 protein treatment during embryo culture, and cysteine-proteinase (cathepsin) inhibitor treatment during in vitro maturation on embryonic development following in vitro fertilization of good versus poor quality oocytes selected based on BCB staining.
RESULTS
Specific oocyte and cumulus expressed transcripts are associated with higher developmental competence of bovine oocyte
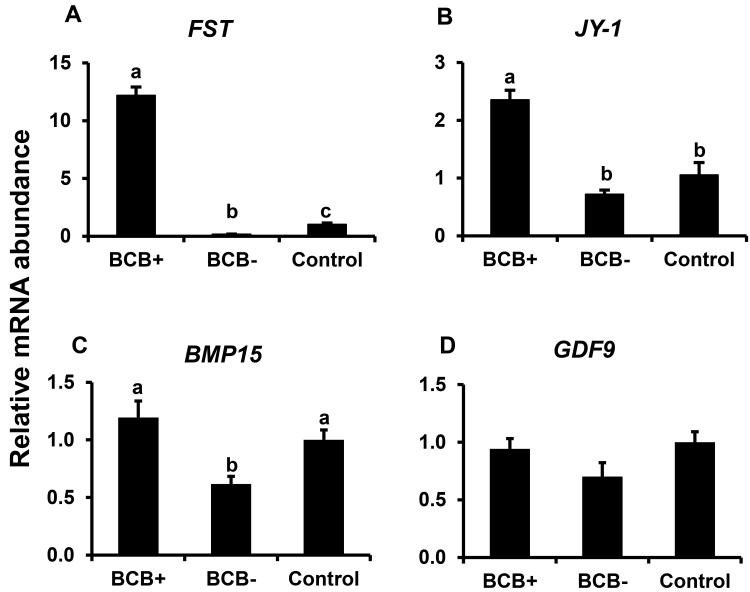
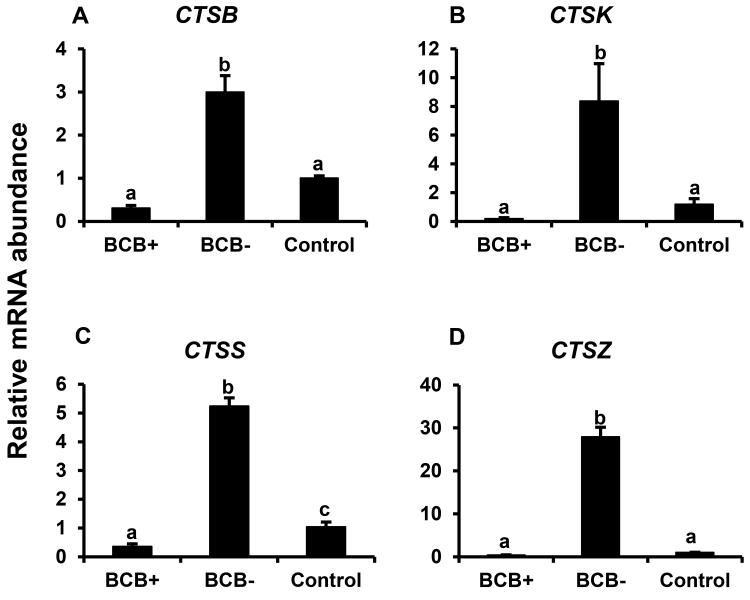
Expression of specific developmentally important genes were analyzed in BCB selected GV stage oocytes (FST, JY-1, CTSB, CTSS, CTSZ, BMP15, and GDF9) and adjacent cumulus cells (CTSB, CTSK, CTSS, and CTSZ) to investigate factors associated with higher oocyte competence. Real time PCR analysis showed > 20 fold higher abundance of mRNA for FST in good quality GV stage (BCB+) oocytes compared to poor quality (BCB−) oocytes (P < 0.05; Fig. 1A). Oocyte FST mRNA abundance was also lower in BCB− oocytes relative to control oocytes selected based on morphological criteria. Likewise, abundance of oocyte JY-1 and BMP15 mRNA was > 2 fold higher (P < 0.05) in BCB+ versus BCB− and control oocytes, however was not different between control versus BCB- oocytes (Fig. 1B-C respectively). In contrast, oocyte mRNA abundance for GDF9 did not differ between control oocytes versus oocytes that were positive or negative for BCB staining (Fig. 1D). Abundance of transcripts for CTSB, CTSK, CTSS, and CTSZ was higher in cumulus cells surrounding poor quality oocytes (based on BCB staining) than those surrounding good quality BCB+ and control oocytes (P< 0.05; Fig 2A-D). Abundance of CTSS, but not CTSB, CTSK or CTSZ mRNA, was also lower in BCB+ relative to control oocytes. Results support a positive relationship of oocyte FST, JY1 and BMP15 mRNA expression with its developmental potential as determined by measurement of G6PDH activity (BCB staining), and a negative relationship between cumulus cell cathepsin B, S and Z mRNA and oocyte quality based on BCB staining criteria.
Figure 1.
Expression of FST (A), JY1 (B) BMP15 (C), and GDF9 mRNAs (D) in BCB-screened GV-stage bovine oocytes. Quantitative reverse transcriptase PCR analysis of mRNA abundance was performed on samples of control, BCB+ and BCB− GV-stage oocytes (n = 4 pools of 10 oocytes for each oocyte group). Data were normalized relative to abundance of endogenous control (RPS18) and are shown as mean ± standard error. Values with different superscripts across treatments indicate significant differences (P < 0.05).
Figure 2.
Cumulus cell cathepsins expression in BCB-screened GV-stage COCs. Quantitative reverse transcriptase PCR analysis of mRNA abundance for CTSB (A) , CTSK (B) , CTSS (C) and CTSZ (D) in bovine cumulus cells harvested from GV stage control, BCB+ and BCB− bovine COCs (n = 4 pools of 10 COCs for each oocyte group). Data were normalized relative to abundance of endogenous control (RPS18) and are shown as mean ± standard error. Values with different superscripts across treatments indicate significant differences (P < 0.05)
Cumulus cell expression of FST and oocyte expression of CTSB, CTSS, and CTSZ: relationship with BCB staining and oocyte quality
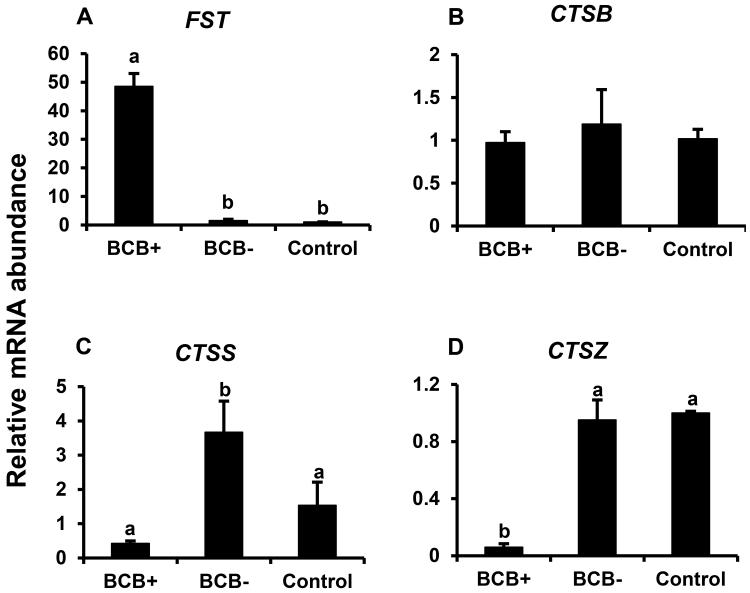
We also examined oocyte expression of above described cumulus cell egg quality markers (CTSB, CTSS, and CTSZ) and cumulus cell expression of FST to determine if observed relationship between transcript abundance and oocyte quality (based on BCB staining) was shared in both the oocyte and cumulus cell compartments. Cumulus cell expression of FST was > 40 fold higher in BCB+ relative to BCB− and control groups, but not different between control and BCB− oocytes (P< 0.05; Fig 3A). Abundance of CTSB mRNA was similar between control oocytes versus oocytes that were positive or negative for BCB staining (Fig 3B). CTSS mRNA abundance was higher in BCB−relative to BCB+ and control oocytes, but did not differ between BCB+ and control oocytes (Fig. 3C). In contrast, oocyte CTSZ mRNA was lowest for the good quality BCB+ oocytes, but did not differ between poor quality BCB− and control oocytes (Fig, 3D). Results indicate similar negative relationship between oocyte and cumulus cell expression of CTSS, but not CTSZ and CTSB mRNAs and oocyte quality based on BCB staining and elevated expression of FST mRNA is shared in the cumulus cells and oocytes that stain positive for BCB relative to poor quality oocytes that are negative for BCB staining.
Figure 3.
Cumulus cell expression of FST (A) and oocyte expression of CTSB (B), CTSS (C), and CTSZ (D) in oocytes and cumulus cells from BCB-screened, GV-stage bovine COCs. Quantitative reverse-transcriptase PCR analysis of mRNA abundance for FST in cumulus cells, and CTSB, CTSS, and CTSZ in oocytes from control, BCB+ and BCB− groups (n = 4 pools of 10 oocytes/cumulus cells from 10 COCs each per group). Data were normalized relative to abundance of endogenous control (RPS18) and are shown as mean ± standard error. Values with different superscripts across treatments indicate significant differences (P < 0.05)
A possible link between SMAD signaling and oocyte competence
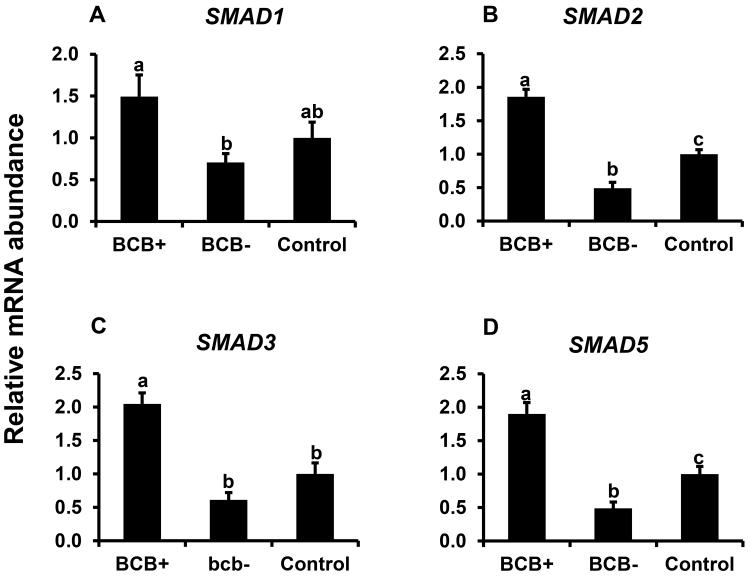
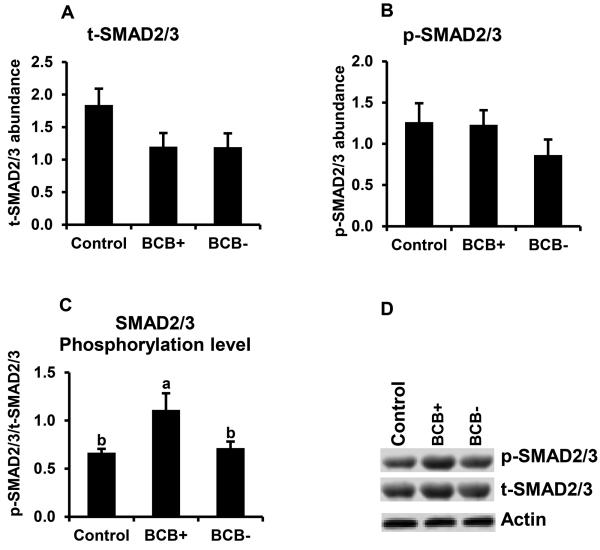
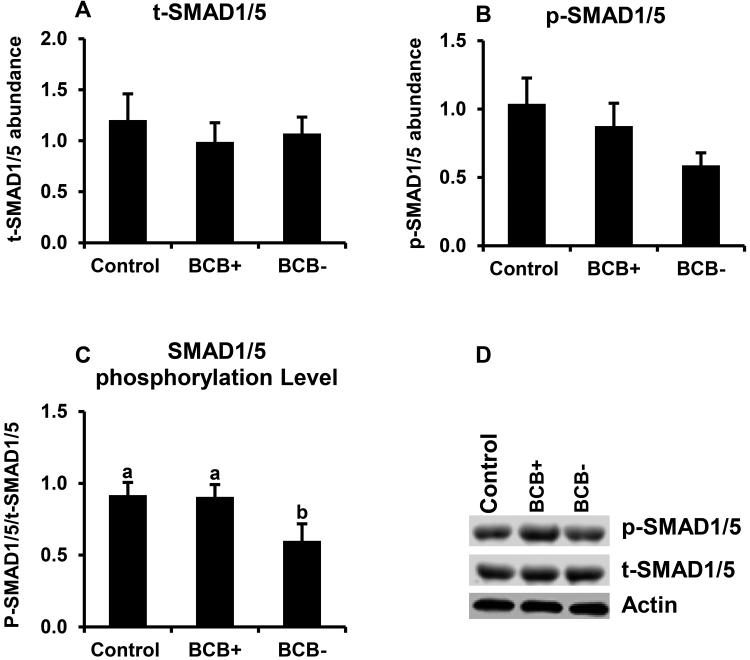
Above studies revealed a positive relationship between BMP15 transcript abundance and oocyte quality based on BCB staining. Hence, the potential relationship between oocyte quality and TGFβ-SMAD signaling was further examined. Results demonstrated higher mRNA abundance for SMAD1, SMAD2, SMAD3, and SMAD5 s in BCB+ relative to BCB− oocytes (P < 0.05; Fig. 4A-D). Oocyte SMAD2, and 5 mRNAs were significantly lower in poor quality (BCB−) oocytes relative to controls, but SMAD1 and SMAD3 mRNAs were not different. The potential relationship between SAMD signaling and bovine oocyte quality was further evaluated by Western blot analysis of SMAD2/3 and SMAD1/5 phosphorylation of GV stage oocytes harvested from control, BCB+, and BCB− groups. We observed > 1.5 fold higher phosphorylation level of SMAD2/3 (Fig. 5) and SMAD1/5 (Fig. 6) in good quality (BCB+) compared to both control and poor quality (BCB−) groups but no significant difference in SMAD phosphorylation was found between BCB− and control oocytes. Results suggesting a positive relationship between both SMAD2/3 and SMAD1/5 phosphorylation and oocyte quality determined by BCB staining.
Figure 4.
Expression of mRNAs for SMAD1 (A), SMAD2 (B), SMAD3 (C) and SMAD5 (D) in BCB-screened, GV-stage oocytes. Quantitative reverse-transcriptase PCR analysis was on oocytes from control, BCB+ and BCB− groups (n = 7 pools of 10 oocytes each per group). Data were normalized relative to abundance of endogenous control (RPS18) and are shown as mean ± standard error. Values with different superscripts across treatments denote significant differences (P < 0.05).
Figure 5.
SMAD 2/3 phosphorylation in BCB-screened, GV-stage oocytes. Samples of control, BCB+ and BCB− GV stage oocytes (n=6 replicates of 20 oocytes/group) were subjected to Western blot analysis of total (t)-SMAD2/3 (A), phosphorylated (p)-SMAD2/3 (B). Expression levels were normalized relative to abundance of endogenous control (actin). Phosphorylation level (C) was expressed as p -SMAD2/3 / t-SMAD2/3 Data are shown as mean ± standard error. Values with different superscripts across treatments indicate significant differences (P < 0.05). Representative Western blot (D).
Figure 6.
SMAD1/5 phosphorylation in BCB-screened, GV-stage oocytes. Samples of control, BCB+ and BCB− GV stage oocytes (n=6 replicates of 20 oocytes/group) were subjected to Western blot analysis of total (t)-SMAD1/5 (A), phosphorylated (p)-SMAD1/5 (B). Expression levels were normalized relative to abundance of endogenous control (actin). Phosphorylation level (C) was expressed as p-SMAD1/5/t-SMAD1/5 Data are shown as mean ± standard error. Values with different superscripts across treatments indicate significant differences (P < 0.05). Representative Western blot (D).
Effects of exogenous follistatin supplementation on indices of embryo developmental progression for embryos derived from good and poor quality oocytes based on BCB staining
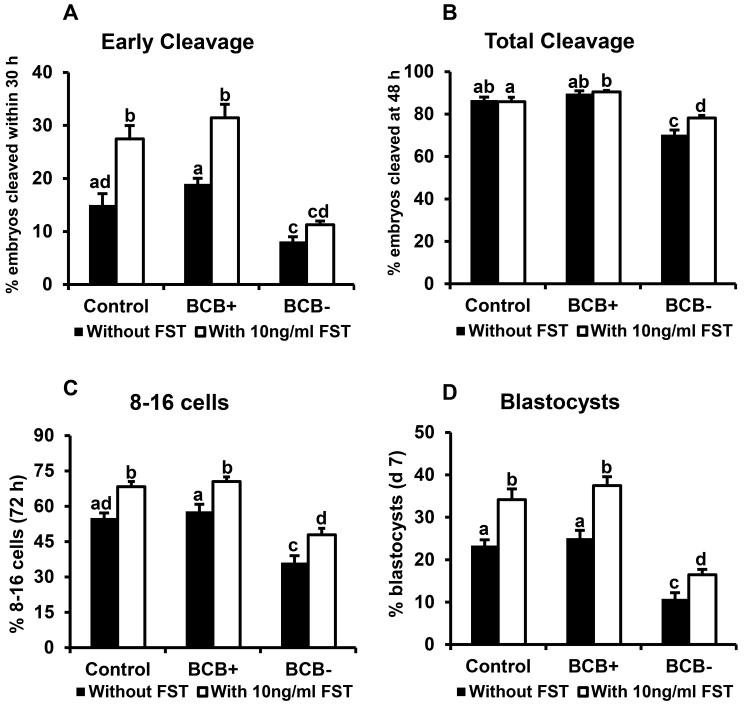
Our results revealed higher FST mRNA abundance in oocytes with high developmental potential (BCB+). Previous studies (Lee et al. 2009) established that exogenous follistatin supplementation enhances the developmental potential of early bovine embryos. Hence, the effects of exogenous follistatin on the developmental capacity of embryos derived from BCB screened oocytes were determined. Presumptive zygotes derived from control, BCB+ and BCB− oocytes were subjected to initial 72 hr of in vitro culture in the presence or absence of maximal stimulatory dose of exogenous follistatin (10 ng/ml) (Lee et al. 2009), then cultured follistatin free until d 7. For embryos cultured in the absence of FST, all developmental endpoints measured (early cleavage, total cleavage, development to 8- to 16-cell and blastocyst stages were lower for embryos derived from poor quality (BCB−) oocytes versus good quality (BCB+) and control oocytes selected based on morphological criteria (Fig.7A-D). Percentage of embryos undergoing early cleavage and rates of development to 8- to 16-cell and blastocyst stages were increased similarly in response to follistatin treatment for BCB+ and control oocytes. Follistatin treatment of embryos derived from poor quality (BCB−) oocytes did not increase early cleavage), but did increase total cleavage rates and rates of development to 8- 16 cell and blastocyst stages (16.4 vs 10.8%), but to levels lower than observed for embryos derived from control and BCB+ oocytes cultured in the absence of follistatin (Fig 7A-D). Results indicate that follistatin treatment during embryo culture can enhance developmental capacity of embryos derived from poor quality oocytes selected based on BCB staining.
Figure 7.
Effects of exogenous follistatin supplementation on indices of development for embryos derived from BCB-screened, GV-stage oocytes. Presumptive zygotes derived from in vitro fertilization of oocytes from control, BCB+ and BCB− groups were cultured with or without 10 ng/ml follistatin (n=25-30 presumptive zygotes/group; n = 4 replicates) for 72 hr, then cultured in fresh medium (minus FST) until d 7. Effect of exogenous follistatin on (A) proportion of embryos that reached the 2-cell stage within 30 h post fertilization (early cleaving), (B) total cleavage rate (determined 48 hpi), (C) proportion of embryos developing to the 8-16 cell stage (72 hpi) and (D) proportion of embryos developing to the blastocyst stage (d 7). Data are shown as mean ± standard error. Values with different superscripts across treatments indicate significant differences (P < 0.05).
Effects of exogenous JY-1 treatment during initial 72 hr of in vitro embryo culture on developmental capacity of bovine embryos derived from BCB screened and control oocytes
We also determined the effects of treatment with exogenous recombinant JY-1 protein (rJY-1) during initial 72 hr of in vitro embryo culture on similar developmental endpoints as described above for control oocytes selected based on morphological criteria and oocytes selected based on BCB staining. Reduced developmental capacity as indicated by early cleavage, total cleavage and development to 8-16 cell and blastocyst stages was observed for BCB- oocytes relative to control and BCB+ oocytes as observed above. However, stimulatory effects of exogenous rJY-1 treatment were not observed for any developmental endpoints for embryos derived from control, BCB+ and BCB− oocytes (Supplemental data figure 1). Results indicate exogenous JY-1 supplementation during embryo culture does not enhance developmental capacity of embryos derived from control oocytes or oocytes of good and poor quality selected based on BCB staining.
Effects of cathepsin inhibitor (E-64) treatment during meiotic maturation on developmental capacity of in vitro fertilized embryos derived from control oocytes and oocytes classified based on BCB staining
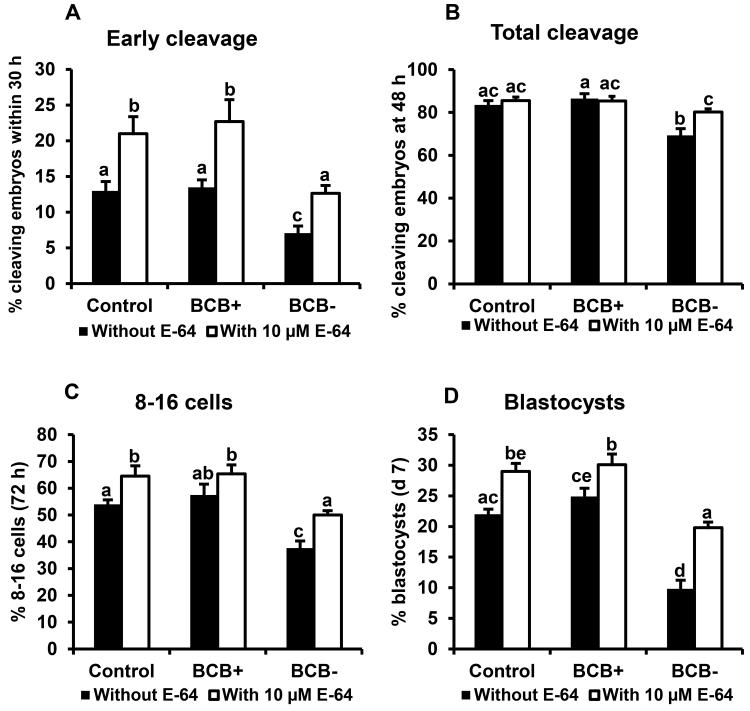
Above studies revealed a negative relationship between cumulus cell CTSB, CTSS, CTSZ mRNA and oocyte competence based on BCB staining and previous studies demonstrated beneficial effects of cathepsin inhibitor treatment (during meiotic maturation) on subsequent early embryonic development. Hence, the effects of cathepsin inhibitor (E-64) treatment during in vitro meiotic maturation on indices of developmental progression for embryos derived from control oocytes selected based on morphological criteria versus oocytes screened for developmental competence based on BCB staining (BCB+ and BCB−) were investigated. Reduced developmental capacity as indicated by early cleavage, total cleavage and development to 8-16 cell and blastocyst stages was observed for BCB− oocytes relative to control and BCB+ oocytes in this experiment as observed above. Treatment with E-64 during meiotic maturation similarly increased early cleavage and rates of development to 8-16 cell and blastocyst stages for embryos derived from BCB+ and control oocytes relative to embryos derived from untreated oocytes in each group (Fig 8A-D). Furthermore, treatment with E-64 improved developmental competence of BCB− oocytes to a level similar to untreated control oocytes in terms of early cleavage and development to 8-16 cell and blastocyst stages (Fig. 8A-D).
Figure 8.
Effects of cathepsin inhibitor (E-64) treatment during meiotic maturation on developmental capacity of in vitro fertilized embryos derived from BCB-screened, GV-stage oocytes. Cumulus oocyte complexes (COCs) from control, BCB+ and BCB− oocytes were cultured with or without 10 µM cathepsin inhibitor E-64 during in vitro maturation (45-50 COCs per group, 4 replicates). After subsequent in vitro fertilization, presumptive zygotes were cultured until d 7. Effect of E-64 supplementation during meiotic maturation on (A) early cleavage rate (determined at 30 hpi), (B) total cleavage rate (determined 48 hpi), (C) proportion of embryos developing to the 8-16 cell stage (72 hpi) and (D) blastocyst formation rate (d 7). Data are shown as mean ± standard error. Values with different superscripts across treatments indicate significant differences (P < 0.05).
Discussion
The results of the current studies demonstrated higher levels of FST, JY1, BMP15 and SMAD1, 2, 3 and 5 transcripts in BCB+ oocytes relative to poorer quality (BCB−) oocytes and greater abundance of CTSB, CTSK, CTSS and CTSZ mRNAs in cumulus cells surrounding BCB− oocytes, supporting a similar relationship of expression of above genes with oocyte competence as reported in our previous studies (Bettegowda et al. 2008; Patel et al. 2007). Furthermore, follistatin treatment during in vitro embryo culture and cathepsin inhibitor treatment during meiotic maturation were able to enhance developmental competence of poor quality oocytes selected based on BCB staining, but exogenous JY-1 supplementation during embryo culture had no effect on developmental progression on embryos derived from control, BCB+ and BCB− oocytes. As our previous studies have shown that exogenous JY-1 added to oocyte/embryo culture medium can rescue oocytes after JY-1 knockdown (Lee et al. 2014b), these results suggest that differences in oocyte FST, but not JY-1 expression, and cumulus cathepsin expression are functionally related to reduced developmental capacity of growing oocytes selected based on BCB staining.
Results of present studies further support a positive relationship of follistatin expression with oocyte quality, as evidenced by greater FST mRNA in BCB+ versus BCB- oocytes. We have previously demonstrated higher abundance of mRNA for the TGFβ growth factor superfamily binding protein follistatin in good quality oocytes collected from adult animals, versus oocytes from prepubertal animals which are known to be of reduced developmental competence (Patel et al. 2007). We also demonstrated a positive relationship between maternal FST transcript abundance in bovine 2-cell embryos and time to first cleavage (a second indicator of developmental competence) and demonstrated embryotrophic effects of exogenous follistatin on early cleavage, blastocyst development, and cell lineage determination in bovine embryos (Lee et al. 2009). Embryotrophic actions of follistatin were also observed on cultured rhesus monkey embryos (VandeVoort et al. 2009). However, follistatin expression and effects of exogenous follistatin treatment during embryo culture were similar for embryos derived from good quality oocytes based on BCB staining versus morphological criteria. Hence, while BCB selection provides a useful tool for selection based on oocyte quality it does not yield greater developmental potential than selection based on rigid morphological criteria in our described system.
We previously established that the novel oocyte specific protein JY-1 is an important regulator of cumulus/granulosa cell function and early embryogenesis (Bettegowda et al. 2007; Lee et al. 2014b). JY-1 gene polymorphisms are associated with reproductive traits, such as the occurrence of early pregnancy in cattle (De Camargo et al. 2014; De Camargo et al. 2013). Furthermore, knockdown of endogenous oocyte JY-1 using siRNA demonstrated that JY-1 is required for meiotic maturation, cumulus expansion and for cleavage divisions post fertilization. Effects of JY-1 knockdown can be rescued by supplementation of culture media with recombinant JY-1 protein (Lee et al. 2014b). While BCB+ oocytes had slightly higher relative levels of JY-1 mRNA than BCB− oocytes or oocytes selected based on morphological criteria, supplementation with exogenous JY-1 protein during initial stages of embryo culture following IVF did not impact development of embryos derived from control, BCB+ or BCB− oocytes. Hence, while functional studies support a requirement of JY-1 for meiotic maturation, cumulus expansion and early embryogenesis, results of present studies suggest that levels of endogenous JY-1 in oocytes selected morphologically and or via BCB staining are not limiting to early embryonic development.
Our previous studies of cumulus cell determinants of oocyte competence revealed cumulus cell cathepsin (CTSB, CTSK, CTSS, and CTSZ) mRNA transcripts are elevated in cumulus cells of prepubertal oocytes which have lower developmental competence than oocytes harvested from adult animals (Bettegowda et al. 2008). We also demonstrated that reduced developmental competence of oocytes from prepubertal animals is linked to increased cumulus cell apoptosis (Bettegowda et al. 2008). Inhibition of cathepsin activity during bovine in vitro maturation (Balboula et al. 2013; Bettegowda et al. 2008) using a cathepsin inhibitor (E-64) reduces cumulus cell apoptosis and improves oocyte developmental competence and blastocyst development. In the current studies, treatment with E-64 during in vitro oocyte maturation increases development of embryos derived from control, BCB+ and BCB− oocytes, with E-64 treatment rescuing development of embryos derived from BCB− oocytes to similar rates as embryos derived from untreated control oocytes. While we cannot discount potential beneficial effects of cathepsin inhibitor treatment during in vitro maturation directly on the oocyte, as oocyte CTSS and CTSZ mRNA was also higher in poor quality BCB− oocytes, our previous studies suggest effects of E-64 treatment are mediated directly on cumulus cells as E-64 treatment of denuded oocytes during in vitro maturation has no effect on developmental potential of resulting embryos. Hence, results suggest that reduced developmental potential of oocytes derived from BCB− embryos is also linked to altered cumulus cell phenotype/function.
Bone morphogenetic protein 15 (BMP15) and growth differentiation factor 9 (GDF9) are TGFβ superfamily members that play an important role in follicular and oocyte development (McNatty et al. 2005; Yan et al. 2001). Our previous studies demonstrated dynamic temporal regulation of mRNA for numerous TGFβ superfamily ligands and receptors during oocyte maturation and early embryonic development (Lee et al. 2014a). Based on results to date, the potential mechanism of action of follistatin in regulation of bovine early embryogenesis is unclear. The mechanisms responsible for observed effects are not clear. Follistatin was initially classified as a high affinity activin binding protein (Nakamura et al. 1990). However, follistatin can also bind and regulate activity of multiple TGFβ superfamily members including inhibins and select BMPs (Balemans and Van Hul 2002; Lin et al. 2003; Otsuka et al. 2001) and similar effects of activin versus follistatin treatment of early embryos were observed in previous studies (Lee et al. 2009). In the current studies, higher levels of FST, BMP15, SMAD1, SMAD5, SMAD2 and SMAD3 mRNAs, and SMAD1/5 and SMAD2/3 phosphorylation were observed in BCB+ versus BCB− oocytes. This may suggest that effects of follistatin on early embryonic development are potentially not mediated by inhibition of ligand induced activity of TGFβ superfamily members and associated decrease in SMAD2/3 and or SMAD1/5 activation. However, divergent effects of follistatin supplementation during in vitro maturation versus embryo culture have been reported previously, with inhibitory effect of exogenous follistatin treatment during in vitro maturation on subsequent embryonic development observed for rhesus monkey (VandeVoort et al. 2009) and bovine oocytes (Silva and Knight 1998) versus stimulatory effects of exogenous follistatin supplementations (during embryo culture) on indices of developmental capacity (Lee et al. 2009). Given stimulatory effects of follistatin on early embryonic development are dose dependent and reversed at higher follistatin concentrations, the inhibitory effects of follistatin observed during in vitro maturation versus embryo culture may be explained in part by contribution of follistatin coming from the cumulus cell layer. Indeed follistatin mRNA was higher in the cumulus layer surrounding BCB+ versus control and BCB− oocytes in the current studies.
In summary, results of present studies provide novel information supporting a positive relationship between oocyte follistatin expression and oocyte competence based on BCB staining and a negative relationship between cumulus cell cathepsin expression and oocyte quality based on BCB staining. Results also conclusively demonstrate that treatment with exogenous follistatin during embryo culture or cathepsin inhibitor (E-64) treatment during meiotic maturation can enhance development post fertilization of poorer quality oocytes selected based on BCB staining. Results also demonstrate a positive relationship of SMAD1/5 and SMAD2/3 activation with oocyte competence and provide further understanding of previously reported indices with oocyte competence.
MATERIALS AND METHODS
Materials
All chemicals and reagents used were obtained from Sigma-Aldrich unless stated otherwise.
Oocyte recovery and Brilliant Cresyl Blue staining
Oocytes were recovered as previously described (Lee et al. 2009). Isolated cumulus oocyte complexes (COCs) were exposed to 26 µM of BCB diluted in Dulbecco’s Phosphate Buffer Saline containing 0.4 % BSA (mDPBS) for 90 min at 38.5°C in a 5 % CO2 humidified air atmosphere (Torner et al. 2008). Control COCs were selected based on morphology as previously described (Lee et al. 2009) and incubated in mDPBS without BCB for 90 min (Su et al. 2012). Following BCB exposure, the COCs were washed twice in a warm solution of mDPBS, and examined under a stereomicroscope. COCs exposed to BCB were divided into two groups according to their cytoplasm coloration: oocytes with any degree of blue coloration to the cytoplasm (BCB+) and oocytes without blue cytoplasm (BCB−) and processed for RNA isolation and quantitative real time RT-PCR or Western blot analysis and remaining COCs utilized for in vitro maturation and embryo culture experiments as described below.
RNA isolation and real time PCR analysis
For RNA analyses, cumulus cells and oocytes were harvested separately from control, BCB+ [+] and BCB− [-] germinal vesicle (GV) stage oocytes (n = 4 pools of 10 oocytes for each oocyte group). Total RNA was extracted using the RNeasy® Micro Kit (Qiagen) according to manufacturer’s instructions. Before RNA extraction, each sample was spiked with 250fg green fluorescent protein (GFP) synthetic RNA as an exogenous control for RNA recovery and efficiency of cDNA synthesis (Bettegowda et al. 2006). Total RNA from each sample was utilized for reverse transcription using iScript cDNA synthesis kit (BioRad) according to manufacturer’s instructions. After termination of cDNA synthesis, each RT reaction was then diluted with nuclease-free water to a final volume of 40 μl.
The quantification of all gene transcripts was done by real-time quantitative RT-PCR using The CFX96™ Real-time PCR System (BioRad). All PCR primers used in this study were designed using PerlPrimer® Software v1.1.21 (Marshall 2004). Primer sequences and GenBank accession numbers for transcripts analyzed are shown in Table 1. Transcript abundance for genes of interest was normalized relative to abundance of endogenous control RPS18.
Table (1).
Sequence of primers used for real-time PCR
| Gene | GenBank Accession number | Primer Sequence* |
|---|---|---|
| BMP15 | AY572412 | F: 5'-TTGGACAGAGATGGATATCATGGA-3' R: 5'-CGGCGCCCCTTGTGA-3' |
| CTSB | BF868324 | F: 5'-CGATGCCCGGGAACAGT-3' R: 5'-GAGCCAGGATCCCTGATC-3' |
| CTSK | BF230198 | F: 5'-CATATGAACTGGCCATGAACCA-3' R:5'-TGAGTCCAGTCATCTTCTGAACCA-3' |
| CTSS | BE482678 | F: 5'-TCGTGGTTGGCTATGGTAACC-3' R: 5'-TGCAGGCCCCAGCTGTT-3' |
| CTSZ | BE752253 | F: 5'-GGGAGAAGATGATGGCAGAAAT-3' R: 5'-TCTTTTCGGTTGCCATTATGC-3' |
| FST | BF774514 F | F: 5l-CAGAGCTGCAAGTCCAGTACCA-3' R: 5'-CATGTAGAGCTGCCTGGACAGA-3' |
| GDF9 | AF307092 | F: 5'-TGACCAGAAGAGAGGGCTGTCT-3' R: 5'-CGGTGACGGGACAATCTTACA-3' |
| JY-1 | EF642497 | F: 5'-TTGGAACTTCCATGGACGACC-3' R: 5'- TCATTTTGTGGCTTCCATTCTG-3' |
| RPS18 | BC102293 | F: 5 '-GTGGTGTTGAGGAAAGCAGACA-3' R: 5'-TGATCACACGTTCCACCTCATC-3' |
| SMAD1 | BC116117 | F: 5'-CACCATGAACTGAAACCATTGG-3' R: 5'-GATGCACACCTCCTTCTGCTT-3' |
| SMAD2 | BC123801 | F: 5'-TGCCGAGTGCCTAAGTGACA-3' R: 5'-GGTGCCAGCCATATCTCTGATT-3' |
| SMAD3 | XM_593090 | F: 5'-GCTGCGGGCCATGGA-3' R: 5'-ATTCACGCAGACCTCGTCCTT-3' |
| SMAD5 | DV821574 | F: 5'-GCAACGTTTCCTGATTCTTTCC-3' R: 5'-GGCGGGTAGGGACTATTTGG-3' |
F, forward; R, reverse.
Western blot analysis
Samples were collected in RIPA buffer (150 mM NaCl, 1.0% IGEPAL®, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0, Sigma) supplemented with 1X protease, phosphatase inhibitor cocktail (Roche Applied Science) and stored at −80 °C until used for western blotting. The samples were mixed with 5X sample buffer and denatured at 95 °C for 10 min. Equal amount of protein (20 oocytes/group) were separated by SDS-PAGE electrophoresis using 4-20% per-cast gels (Bio-Rad) and transferred on to a polyvinylidene difluoride (PVDF, Millipore, Bedford, MA, USA) membrane with tank electrophoretic transfer apparatus (Bio-Rad). After the transfer, membranes were blocked for 1 hr in 5% BSA in Tris Buffered Saline with Tween® 20 (TBST, 137 mM Sodium Chloride, 20 mM Tris, 0.5% Tween-20, pH 7.5) at room temperature before overnight incubation with the appropriate primary antibody (listed below) diluted in TBST with 5% BSA at 4°C. Membranes were probed sequentially with primary rabbit anti-phosphorylated (p) SMAD2/3 polyclonal antibody [1:1000 (vol/vol), Santa Cruz, sc-11769] or primary rabbit anti p-SMAD1/5/8 polyclonal antibody [1:1000 (vol/vol), Santa Cruz, sc-12353]. After detection of p-SMAD2/3 and p-sMAD1/5, membranes were striped with restore Western blot stripping buffer (Thermo Scientific) for 30 min at room temperature and re-probed with Rabbit anti total (t)-SMAD2/3 polyclonal antibody [1:1000 (vol/vol), Santa Cruz, sc-8332] or rabbit anti t-SMAD1/5/8 polyclonal antibody [1:1000 (vol/vol), Santa Cruz, sc-6031-R]. After detection of t-SMAD2/3 or t-SMAD1/5, membranes were striped and re-probed with mouse anti-actin monoclonal antibody (1:5000 (vol/vol) Millipore; MAB1501). HRP-conjugated Anti-rabbit-IgG (Cell Signaling Technology] and Anti-mouse-IgG (Thermo Scientific) were used as secondary antibodies at a 1:5000 (vol/vol) dilution. Protein signals were detected with SuperSignal West Dura Chemiluminescent Substrate (Thermo scientific, Waltham, MA, USA). Images were scanned using myECL Imager (Thermo Scientific). Then, intensities of protein bands were quantified by ImageJ software (Schneider et al. 2012) and normalized relative to abundance of actin level in each lane. Phosphorylation level was then expressed as corrected signal intensity for p-SMAD/t-SMAD.
In vitro embryo production and FST and JY-1 supplementation
After completion of BCB staining protocol, BCB+, BCB− and control oocytes were matured in TCM-199 media [supplemented with 0.2 mM sodium pyruvate, 5 mg/ml Gentamycin sulfate, 156 nM bovine LH (Sioux Biochemical, Sioux Center, IA), 15.6 nM bovine FSH (Sioux Biochemical), 3.67 nM 17β-estradiol, and 10% v/v defined FBS (Hyclone, Logan, UT)] at 38.5 °C in a 5 % CO2 humidified air atmosphere for 24 hrs. Then, the matured COCs were co-incubated with frozen thawed bovine spermatozoa separated by Percoll gradient techniques for 20 hr in fertilization media FIV (114 mM NaCl, 25 mM NaHCO3, 3.2 mM KCl, 0.34 mM NaH2PO4, 0.183 mM penicillin-G, 16.6 mM sodium lactate, 0.5 mM MgCl2.6H2O, 2.7 mM CaCl2.2H2O, 0.2 mM sodium pyruvate, 6 mg/ml BSA, and 1.5 U of heparin) in 38.5 °C in 5 % CO2 humidified air. After IVM and IVF, the cumulus cells were stripped using 0.1% hyaluronidase and vortexed for 5 min. Then, presumptive zygotes were cultured in KSOM medium supplemented with 0.3 % BSA and 1) with or without 10 ng/ml follistatin (25-30 presumptive zygotes per group, 4 replicates) and 2) with or without 1 ng/ml JY-1 (25-30 presumptive zygotes per group, 4 replicates). The 8-16 cell stage embryos were then separated 72 h post fertilization and cultured in fresh KSOM medium supplemented with 0.3 % BSA and 10 % FBS until d 7.
Cysteine-Proteinase (Cathepsin) Inhibitor Treatment
For determination of effects of cathepsin inhibitor (E-64) treatment on embryonic development following IVF for BCB+, BCB− and control oocytes, in vitro maturation, in vitro fertilization and embryo culture were performed as described above except that treatments consisted of 0 or 10 µM E-64 in in vitro maturation medium (45-50 COCs per group, 4 replicates).
Statistical analysis
For gene expression studies, differences in mRNA expression were analyzed by ANOVA using the general linear models procedure of SAS (SAS Institute Inc., NC, USA). For Western blot data, similar models were used to analyze the differences in protein expression and phosphorylation level. For experiments studying the effects of Follistatin, JY1 and cysteine proteinase (cathepsin) inhibitor (E-64) supplementation on early embryonic development, data were arcsin transformed prior to analysis by mixed linear models analysis procedures. In all cases data are presented as untransformed mean ± SEM.
Supplementary Material
Supplemental data Figure 1: Effects of exogenous JY-1 treatment during initial 72 hr of in vitro embryo culture on developmental capacity of bovine embryos derived from BCB-screened, GV-stage oocytes. After in vitro fertilization, presumptive zygotes from control, BCB+ and BCB− groups were cultured with or without 1ng/ml JY-1 (n=25-30 presumptive zygotes/group; n = 4 replicates) for 72 hr, then, cultured in fresh medium (minus JY-1) until d 7. Effect of exogenous JY-1 on (A) proportion of embryos that reached the 2-cell stage within 30 h post fertilization (early cleaving), (B) total cleavage rate (determined 48 hpi), (C) proportion of embryos developing to the 8-16 cell stage (72 hpi) and (D) proportion of embryos developing to the blastocyst stage (d 7). Data are shown as mean ± standard error. Values with different superscripts across treatments indicate significant differences (P < 0.05).
Acknowledgments
Grants: This project was supported by the National Institute of Child Health and Human Development of the National Institutes of Health under award number R01HD072972 and by Michigan State University AgBioResearch. Mohamed Ashry was supported by the Egyptian Government Joint Supervision Grant JS2687.
Abbreviations
- BCB−
Oocytes that reduce the blue color of BCB stain
- BCB
Brilliant Cresyl Blue
- BCB+
Oocytes that retain the blue color of BCB stain
- BMP15
Bone Morphogenetic Protein 15
- CTSB
Cathepsin B
- CTSK
Cathepsin K
- CTSS
Cathepsin S
- CTSZ
Cathepsin Z
- RPS18
Ribosomal protein S18
- FST
Follistatin
- GDF9
Growth Differentiation Factor 9
- GV
Germinal vesicle
- RT-PCR
Real Time – Polymerase Chain Reaction
- TGF-β
Transforming Growth Factor β
- G6PDH
Glucose-6-phosphate dehydrogenase
- COCs
Cumulus Oocyte Complexes
References
- Abir R, Fisch B, Johnson MH. BMP15, fertility and the ovary. Reprod BioMed Online. 2014;29(5):525–526. doi: 10.1016/j.rbmo.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Alm H, Torner H, Lohrke B, Viergutz T, Ghoneim IM, Kanitz W. Bovine blastocyst development rate in vitro is influenced by selection of oocytes by brillant cresyl blue staining before IVM as indicator for glucose-6-phosphate dehydrogenase activity. Theriogenology. 2005;63(8):2194–2205. doi: 10.1016/j.theriogenology.2004.09.050. [DOI] [PubMed] [Google Scholar]
- Balboula AZ, Yamanaka K, Sakatani M, Kawahara M, Hegab AO, Zaabel SM, Takahashi M. Cathepsin B activity has a crucial role in the developmental competence of bovine cumulus-oocyte complexes exposed to heat shock during in vitro maturation. Reproduction. 2013;146(4):407–417. doi: 10.1530/REP-13-0179. [DOI] [PubMed] [Google Scholar]
- Balemans W, Van Hul W. Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev Biol. 2002;250(2):231–250. [PubMed] [Google Scholar]
- Barzegari A, Atashpaz S, Ghabili K, Nemati Z, Rustaei M, Azarbaijani R. Polymorphisms in GDF9 and BMP15 associated with fertility and ovulation rate in Moghani and Ghezel sheep in Iran. Reprod Domest Anim. 2010;45(4):666–669. doi: 10.1111/j.1439-0531.2008.01327.x. [DOI] [PubMed] [Google Scholar]
- Bettegowda A, Patel OV, Ireland JJ, Smith GW. Quantitative analysis of messenger RNA abundance for ribosomal protein L-15, cyclophilin-A, phosphoglycerokinase, beta-glucuronidase, glyceraldehyde 3-phosphate dehydrogenase, beta-actin, and histone H2A during bovine oocyte maturation and early embryogenesis in vitro. Mol Reprod Dev. 2006;73(3):267–278. doi: 10.1002/mrd.20333. [DOI] [PubMed] [Google Scholar]
- Bettegowda A, Patel OV, Lee KB, Park KE, Salem M, Yao J, Ireland JJ, Smith GW. Identification of novel bovine cumulus cell molecular markers predictive of oocyte competence: functional and diagnostic implications. Biol Reprod. 2008;79(2):301–309. doi: 10.1095/biolreprod.107.067223. [DOI] [PubMed] [Google Scholar]
- Bettegowda A, Yao J, Sen A, Li Q, Lee KB, Kobayashi Y, Patel OV, Coussens PM, Ireland JJ, Smith GW. JY-1, an oocyte-specific gene, regulates granulosa cell function and early embryonic development in cattle. Proc Natl Acad Sci U S A. 2007;104(45):17602–17607. doi: 10.1073/pnas.0706383104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni R. Origins and effects of oocyte quality in cattle. Anim Reprod. 2012;9(3):333–340. [Google Scholar]
- De Camargo GM, Costa RB, de Albuquerque LG, Regitano LC, Baldi F, Tonhati H. Association between JY-1 gene polymorphisms and reproductive traits in beef cattle. Gene. 2014;533(2):477–480. doi: 10.1016/j.gene.2013.09.126. [DOI] [PubMed] [Google Scholar]
- De Camargo GMF, Baldi F, Regitano LCA, Tonhati H. Characterization of the Exonic Regions of the JY-1 Gene in Zebu Cattle and Buffaloes. Reprod Domest Anim. 2013;48(6):918–922. doi: 10.1111/rda.12186. [DOI] [PubMed] [Google Scholar]
- Ericsson S, Boice M, Funahashi H, Day B. Assessment of porcine oocytes using brilliant cresyl blue. Theriogenology. 1993;39(1):214. [Google Scholar]
- Kuo FT, Fan K, Ambartsumyan G, Menon P, Ketefian A, Bentsi-Barnes IK, Pisarska MD. Relative expression of genes encoding SMAD signal transduction factors in human granulosa cells is correlated with oocyte quality. J Assist Reprod Genet. 2011;28(10):931–938. doi: 10.1007/s10815-011-9609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KB, Bettegowda A, Wee G, Ireland JJ, Smith GW. Molecular determinants of oocyte competence: potential functional role for maternal (oocyte-derived) follistatin in promoting bovine early embryogenesis. Endocrinology. 2009;150(5):2463–2471. doi: 10.1210/en.2008-1574. [DOI] [PubMed] [Google Scholar]
- Lee KB, Folger JK, Rajput SK, Smith GW. Temporal regulation of mRNAs for select bone morphogenetic proteins (BMP), BMP receptors and their associated SMAD proteins during bovine early embryonic development: effects of exogenous BMP2 on embryo developmental progression. Reprod Biol Endocrinol. 2014a;12:67–76. doi: 10.1186/1477-7827-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KB, Wee G, Zhang K, Folger JK, Knott JG, Smith GW. Functional role of the bovine oocyte-specific protein JY-1 in meiotic maturation, cumulus expansion, and subsequent embryonic development. Biol Reprod. 2014b;90(3):69–1. doi: 10.1095/biolreprod.113.115071. 7. [DOI] [PubMed] [Google Scholar]
- Lin SY, Morrison JR, Phillips DJ, de Kretser DM. Regulation of ovarian function by the TGF-beta superfamily and follistatin. Reproduction. 2003;126(2):133–148. doi: 10.1530/rep.0.1260133. [DOI] [PubMed] [Google Scholar]
- Mamo S, Carter F, Lonergan P, Leal C, Al Naib A, McGettigan P, Mehta J, Evans A, Fair T. Sequential analysis of global gene expression profiles in immature and in vitro matured bovine oocytes: potential molecular markers of oocyte maturation. BMC Genomics. 2011;12(1):14. doi: 10.1186/1471-2164-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangia F, Epstein CJ. Biochemical studies of growing mouse oocytes: preparation of oocytes and analysis of glucose-6-phosphate dehydrogenase and lactate dehydrogenase activities. Dev Biol. 1975;45(2):211–220. doi: 10.1016/0012-1606(75)90061-5. [DOI] [PubMed] [Google Scholar]
- Manjunatha BM, Gupta PS, Devaraj M, Ravindra JP, Nandi S. Selection of developmentally competent buffalo oocytes by brilliant cresyl blue staining before IVM. Theriogenology. 2007;68(9):1299–1304. doi: 10.1016/j.theriogenology.2007.08.031. [DOI] [PubMed] [Google Scholar]
- Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20(15):2471–2472. doi: 10.1093/bioinformatics/bth254. [DOI] [PubMed] [Google Scholar]
- McNatty KP, Juengel JL, Reader KL, Lun S, Myllymaa S, Lawrence SB, Western A, Meerasahib MF, Mottershead DG, Groome NP, Ritvos O, Laitinen MP. Bone morphogenetic protein 15 and growth differentiation factor 9 co-operate to regulate granulosa cell function in ruminants. Reproduction. 2005;129(4):481–487. doi: 10.1530/rep.1.00517. [DOI] [PubMed] [Google Scholar]
- Mohammadi-Sangcheshmeh A, Held E, Ghanem N, Rings F, Salilew-Wondim D, Tesfaye D, Sieme H, Schellander K, Hoelker M. G6PDH-activity in equine oocytes correlates with morphology, expression of candidate genes for viability, and preimplantative in vitro development. Theriogenology. 2011;76(7):1215–1226. doi: 10.1016/j.theriogenology.2011.05.025. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Takio K, Eto Y, Shibai H, Titani K, Sugino H. Activin-binding protein from rat ovary is follistatin. Science. 1990;247(4944):836–838. doi: 10.1126/science.2106159. [DOI] [PubMed] [Google Scholar]
- Opiela J, Katska-Ksiazkiewicz L. The utility of Brilliant Cresyl Blue (BCB) staining of mammalian oocytes used for in vitro embryo production (IVP) Reprod Biol. 2013;13(3):177–183. doi: 10.1016/j.repbio.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Otsuka F, Moore RK, Iemura S, Ueno N, Shimasaki S. Follistatin inhibits the function of the oocyte-derived factor BMP-15. Biochem Biophys Res Commun. 2001;289(5):961–966. doi: 10.1006/bbrc.2001.6103. [DOI] [PubMed] [Google Scholar]
- Patel OV, Bettegowda A, Ireland JJ, Coussens PM, Lonergan P, Smith GW. Functional genomics studies of oocyte competence: evidence that reduced transcript abundance for follistatin is associated with poor developmental competence of bovine oocytes. Reproduction. 2007;133(1):95–106. doi: 10.1530/rep.1.01123. [DOI] [PubMed] [Google Scholar]
- Pereira GR, Lorenzo PL, Carneiro GF, Bilodeau-Goeseels S, Kastelic JP, Esteller-Vico A, Lopez-Bejar M, Liu IK. Selection of developmentally competent immature equine oocytes with brilliant cresyl blue stain prior to in vitro maturation with equine growth hormone. Zygote. 2014;22(4):500–504. doi: 10.1017/S096719941200072X. [DOI] [PubMed] [Google Scholar]
- Persani L, Rossetti R, Di Pasquale E, Cacciatore C, Fabre S. The fundamental role of bone morphogenetic protein 15 in ovarian function and its involvement in female fertility disorders. Hum Reprod Update. 2014;20(6):869–883. doi: 10.1093/humupd/dmu036. [DOI] [PubMed] [Google Scholar]
- Rajput SK, Lee K, Zhenhua G, Di L, Folger JK, Smith GW. Embryotropic actions of follistatin: paracrine and autocrine mediators of oocyte competence and embryo developmental progression. Reprod Fertil Dev. 2013;26(1):37–47. doi: 10.1071/RD13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo G, Fattouh RR, Bosco L, Brucculeri AM, Cittadini E. New molecular markers for the evaluation of gamete quality. J Assist Reprod Genet. 2013;30(2):207–212. doi: 10.1007/s10815-013-9943-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Meth. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimasaki S, Moore RK, Otsuka F, Erickson GF. The bone morphogenetic protein system in mammalian reproduction. Endocr Rev. 2004;25(1):72–101. doi: 10.1210/er.2003-0007. [DOI] [PubMed] [Google Scholar]
- Silva CC, Knight PG. Modulatory actions of activin-A and follistatin on the developmental competence of in vitro-matured bovine oocytes. Biol Reprod. 1998;58(2):558–565. doi: 10.1095/biolreprod58.2.558. [DOI] [PubMed] [Google Scholar]
- Sirard MA, Richard F, Blondin P, Robert C. Contribution of the oocyte to embryo quality. Theriogenology. 2006;65(1):126–136. doi: 10.1016/j.theriogenology.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Su J, Wang Y, Li R, Peng H, Hua S, Li Q, Quan F, Guo Z, Zhang Y. Oocytes selected using BCB staining enhance nuclear reprogramming and the in vivo development of SCNT embryos in cattle. PLoS One. 2012;7(4):e36181. doi: 10.1371/journal.pone.0036181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudiman J, Sutton-McDowall ML, Ritter LJ, White MA, Mottershead DG, Thompson JG, Gilchrist RB. Bone morphogenetic protein 15 in the pro-mature complex form enhances bovine oocyte developmental competence. PLoS One. 2014;9(7):e103563. doi: 10.1371/journal.pone.0103563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian WN, Braunstein LD, Pang J, Stuhlmeier KM, Xi QC, Tian X, Stanton RC. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J Biol Chem. 1998;273(17):10609–10617. doi: 10.1074/jbc.273.17.10609. [DOI] [PubMed] [Google Scholar]
- Torner H, Ghanem N, Ambros C, Holker M, Tomek W, Phatsara C, Alm H, Sirard MA, Kanitz W, Schellander K, Tesfaye D. Molecular and subcellular characterisation of oocytes screened for their developmental competence based on glucose-6-phosphate dehydrogenase activity. Reproduction. 2008;135(2):197–212. doi: 10.1530/REP-07-0348. [DOI] [PubMed] [Google Scholar]
- VandeVoort CA, Mtango NR, Lee YS, Smith GW, Latham KE. Differential effects of follistatin on nonhuman primate oocyte maturation and pre-implantation embryo development in vitro. Biol Reprod. 2009;81(6):1139–1146. doi: 10.1095/biolreprod.109.077198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sun QY. Evaluation of oocyte quality: morphological, cellular and molecular predictors. Reprod Fertil Dev. 2007;19(1):1–12. doi: 10.1071/rd06103. [DOI] [PubMed] [Google Scholar]
- Wassarman M. The mammalian ovum. The Physiology of Reproduction. 1988:62–102. [Google Scholar]
- Wu Y-G, Liu Y, Zhou P, Lan G-C, Han D, Miao D-Q, Tan J-H. Selection of oocytes for in vitro maturation by brilliant cresyl blue staining: a study using the mouse model. Cell Res. 2007;17(8):722–731. doi: 10.1038/cr.2007.66. [DOI] [PubMed] [Google Scholar]
- Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, Prasad SV, Skinner SS, Dunbar BS, Dube JL, Celeste AJ, Matzuk MM. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15(6):854–866. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data Figure 1: Effects of exogenous JY-1 treatment during initial 72 hr of in vitro embryo culture on developmental capacity of bovine embryos derived from BCB-screened, GV-stage oocytes. After in vitro fertilization, presumptive zygotes from control, BCB+ and BCB− groups were cultured with or without 1ng/ml JY-1 (n=25-30 presumptive zygotes/group; n = 4 replicates) for 72 hr, then, cultured in fresh medium (minus JY-1) until d 7. Effect of exogenous JY-1 on (A) proportion of embryos that reached the 2-cell stage within 30 h post fertilization (early cleaving), (B) total cleavage rate (determined 48 hpi), (C) proportion of embryos developing to the 8-16 cell stage (72 hpi) and (D) proportion of embryos developing to the blastocyst stage (d 7). Data are shown as mean ± standard error. Values with different superscripts across treatments indicate significant differences (P < 0.05).