Abstract
Over the past few decades, our understanding of the bacterial protein toxins that modulate G proteins has advanced tremendously through extensive biochemical and structural analyses. This article provides an updated survey of the various toxins that target G proteins, ending with a focus on recent mechanistic insights in our understanding of the deamidating toxin family. The dermonecrotic toxin from Pasteurella multocida (PMT) was recently added to the list of toxins that disrupt G-protein signal transduction through selective deamidation of their targets. The C3 deamidase domain of PMT has no sequence similarity to the deamidase domains of the dermonecrotic toxins from Escherichia coli (cytotoxic necrotizing factor (CNF)1-3), Yersinia (CNFY) and Bordetella (dermonecrotic toxin). The structure of PMT-C3 belongs to a family of transglutaminase-like proteins, with active site Cys–His–Asp catalytic triads distinct from E. coli CNF1.
Keywords: cysteine protease, cytotoxic necrotizing factor, deamidation, dermonecrotic toxin, GTPase, heterotrimeric G proteins, Pasteurella multocida toxin, signal transduction, transglutamination
Bacterial protein toxins comprise a formidable arsenal for modulating host–pathogen interactions. From extensive genetic and biochemical studies over the past few decades, coupled with the numerous crystal structures now available, we have made enormous progress in our understanding of toxin-mediated disease processes. Information gleaned from these studies has also enabled scientists to exploit many of them as selective and efficient tools in research applications to dissect signaling mechanisms within eukaryotic cells, and as therapeutic agents in clinical applications [1–9].
A number of these toxins share the common feature of being large multipartite enzymes (A–B toxins) or effector proteins (sometimes named exoenzymes), which catalyze reactions that can interfere with host cell signal transduction and physiological processes. The functional B parts of A–B toxins bind host cell receptors, and mediate entry into and traffic within the host cell, as well as subsequent delivery of the functional toxic A part into the host cell cytosol. The toxic effector proteins, on the other hand, do not have B parts but, instead, are delivered directly from the bacterial cell into the eukaryotic cell through specialized bacterial secretion systems, the best studied of which are the type III secretion system (T3SS) and type IV secretion system (T4SS). Many of these delivered toxic units are highly specialized enzymes that alter the activity of cellular target proteins, most often through covalent modification. The enzyme activities reported for these toxins include ADP ribosylation [10], glucosylation [11], DNA degradation [12], deadenylation [13], acetylation [14], protein phosphorylation/dephosphorylation [15], proteolysis [16], actin-crosslinking [17] and deamidation/transglutamination [18–20].
G proteins are the molecular targets for a large number of intracellularly acting toxins (Table 1). This article will begin with a brief overview of the different G-protein-targeting toxins, and then will hone in on recent structural and mechanistic advances that have been made regarding the deamidating toxin family, particularly in light of the determination that the structural fold and active site of the Pasteurella multocida toxin (PMT) deamidase is distinct from that of the cytotoxic necrotizing factor (CNF)-1-related family members [21].
Table 1.
G proteins and their modulating toxins.
| G-protein targets | Modulating toxins | Modification | Effect on G protein | Ref. |
|---|---|---|---|---|
| Small GTPases | ||||
| RhoA, Rac1,Cdc42 | TcdA, TcdB, TcsH, TcsL, Tcnα | Monoglucosylation | Inactivation | [59,60] |
| Rap1, Rap2 | TcdA, TcsL | Monoglucosylation | Inactivation | [61,62] |
| RhoA, RhoB, RhoC | clostridial C3 | ADP-ribosylation | Inactivation | [51] |
| Ras | ExoS | ADP-ribosylation | Inactivation | [79–82] |
| RhoA, Rac1,Cdc42 | ExoS, ExoT, SptP, YopE | GAP-like activity | Inactivation | [72–74] |
| Rab1 | LepB | GAP-like activity | Inactivation | [86] |
| RhoA, Rac1 | YpkA, YopO | GDI-like activity | Inactivation | [85] |
| RhoA, Rac1,Cdc42 | YopT, Avr/PhpB | Proteolysis | Inactivation | [61] |
| RhoA, RhoB, RhoC, Rac1, Cdc42 | MARTX | Unknown | Inactivation | [92] |
| RhoA, Rac1, Cdc42 | SopE, SifA, SifB, lpgB1,lpgB2, Map, EspM, EspT | GEF-like activity | Activation | [69] |
| Arf | RalF | GEF-like activity | Activation | [71] |
| Rab1 | DrrA/SidM | GEF-like activity | Activation | [86–88] |
| Rab1 | DrrA/SidM | GDF-like activity | Activation | [86–88] |
| RhoA, Rac1, Cdc42 | CNF1,CNF2, CNF3 | Deamidation† | Activation | [19] |
| RhoA | CNFY | Deamidation† | Activation | [100] |
| RhoA, Rac1, Cdc42 | DNT | Transglutamination‡ | Activation | [19] |
| Heterotrimeric Gα subunits | ||||
| Gi, Go Gt | PT | ADP ribosylation | Inactivation§ | [45,48] |
| Gq | YpkA | Phosphorylation | Inactivation | [84] |
| Gs, Golf, Gt | CT, HLT | ADP ribosylation | Activation§ | [45,48] |
| Gq, G13, Gi | PMT | Deamidation | Activation§ | [9,21,32,112,113,135] |
| Large, multidomain GTPases | ||||
| EF-2 | DT, ExoA | ADP ribosylation | nactivation | [41] |
Deamidation > transglutamination.
Transglutamination > deamidation.
Followed by uncoupling of receptor-effector signaling.
CNF: Cytotoxic necrotizing factor; CT: Cholera toxin; DNT: Dermonecrotic toxin; GDF: GDI-displacement factor; GDI: Guanine nucleotide dissociation inhibitor; GET: Guanine nucleotide exchange factor; HLT: Heat-labile enterotoxin; PMT: Pasteurella multocida toxin; PT: Pertussis toxin.
G proteins as targets of bacterial toxins
G proteins are GTPases that bind and hydrolyze GTP and act as regulatory molecular switches in various signaling processes by cycling between an inactive GDP-bound state and an active GTP-bound state [22–25]. There are three large families of G proteins that serve as targets for toxins: small GTPases of 20–25 kDa, the 40–45-kDa α-subunits of heterotrimeric G proteins and the large (~100-kDa) multidomain elongation factors that regulate protein synthesis through their GTPase activity.
In their inactive GDP-bound state, small G proteins are often complexed with a guanine nucleotide dissociation inhibitor (GDI), which stabilizes the GDP–GTPase complex (Figure 1). Activation of the G protein occurs by exchange of the bound GDP with GTP through the interaction with guanine nucleotide exchange factors (GEFs). In the active GTP-bound state, small GTPases can interact with their target effector proteins and thereby modulate numerous downstream signaling processes involved in cytoskel-etal function, cell polarity, secretion, vesicle trafficking, gene transcription and cell cycle progression [26]. For instance, members of the Rho GTPase family RhoA, Rac and Cdc42, act to promote formation of stress fibers, lamellipodia and filopedia, respectively, whereas RhoD acts in opposition to RhoA to disassemble actin stress fibers.
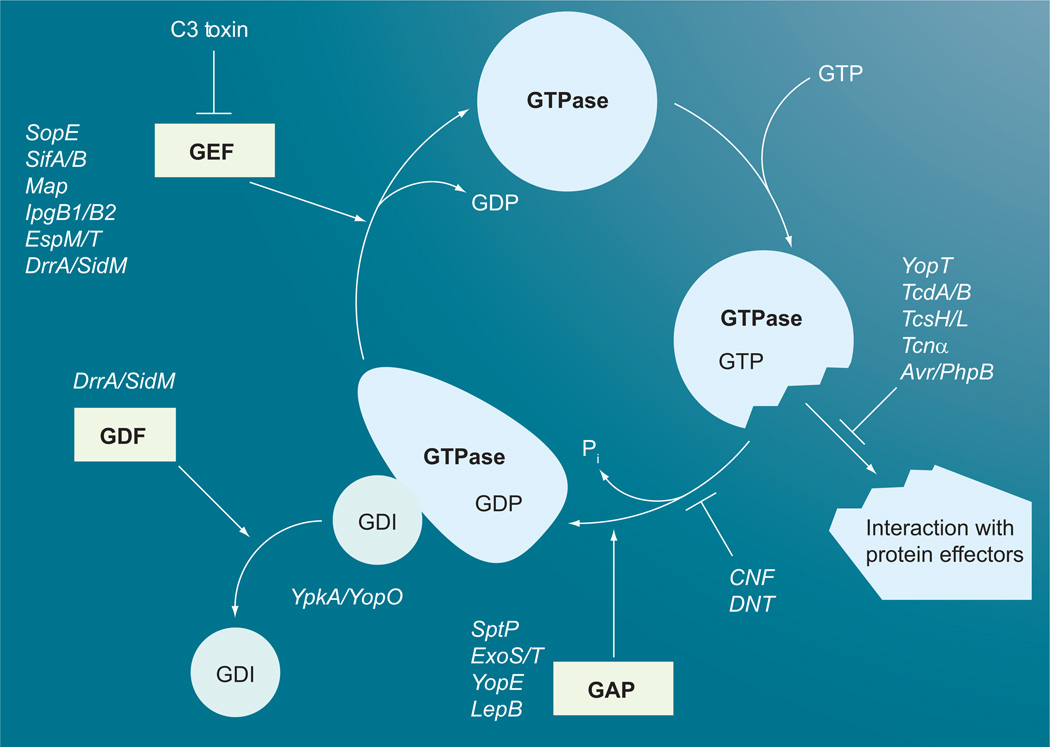
Figure 1. GTPase cycle of small G proteins and points of toxin interactions.
Small GTPase binds GDI in the inactive GDP-bound form. GEF facilitates the release of GDI and GDP and the GTPase then binds GTP. The active GTP-bound form interacts with its downstream effectors. Subsequent interaction with GAP stimulates the hydrolysis of GTP to GDP, which converts the GTPase back into its inactive form. Large clostridial toxins (TcdA, TcdB, TcsH, TcsL and Tcnα), YopT and Avr/PhpB interfere with the GTPase interaction with effectors. CNF and DNT block GTPase activity, while the type III secretion system (T3SS) effectors SptP, ExoS, ExoT and YopE, and the T4SS effector LepB, act as GAPs to stimulate GTP hydrolysis. The T3SS effector YpkA/YopO acts as a GDI to prevent release of GDP. The clostridial C3 toxin blocks GEF interaction with the G protein, while the T3SS effectors SopE, SifA, SifB, Map, EspM, EspT, IpgB1 and lpgB2 act as GEFs. The T4SS effector DrrA/SidM acts as both a GEF and a GDF.
CNF: Cytotoxic necrotizing factor; DNT: Dermonecrotic toxin; GAP: GTPase activating protein; GDI: Guanine nucleotide dissociation inhibitor; GEF: Guanine nucleotide exchange factor.
Heterotrimeric G proteins, comprised of α, β and γ subunits, constitute a large family of GTPases that transduce extracellular hormonal signals from ligand-bound integral membrane receptors to eukaryotic effector proteins involved in various signal transduction pathways and metabolic processes [27]. The α subunits of heterotrimeric G proteins from invertebrates and vertebrates are distinguished into four main classes based on sequence and functional similarities: Gs (Gs, Golf and Ggus), Gi (Gi1/i2/i3, GoA/oB, Gt1/t2 and Gz), G12 (G12 and G13), and Gq (Gq, G11, G14 and G15/16) [28]. As shown in Figure 2, Gα subunits cycle between an inactive receptor-bound, Gβγ-complexed state, with GDP bound, and an active GTP-bound state, where the Gα subunit is dissociated from the activated ligand-bound receptor and the Gβγ subunits [27]. Each of the Gα subunits has its own downstream signaling effector protein(s) that it interacts with when in the GTP-bound state. The Gβγ subunit complex increases the binding affinity of the Gα subunit for the corresponding receptor, and also regulates its own set of effectors when dissociated from the Gα subunit.
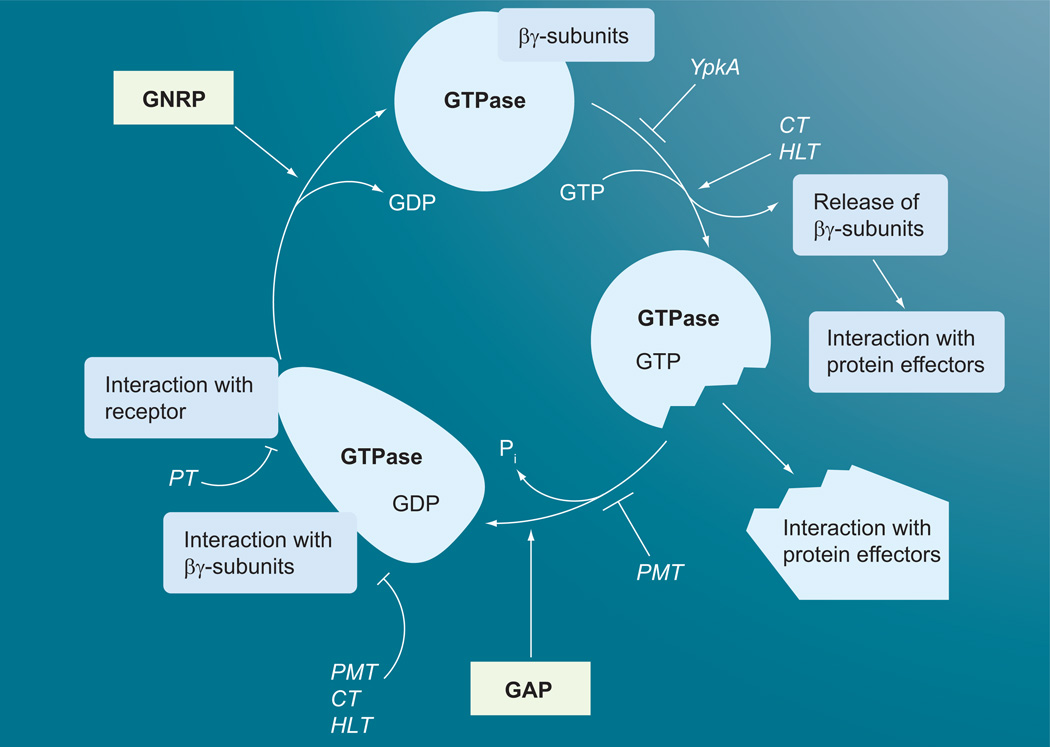
Figure 2. GTPase cycle of heterotrimeric G proteins and points of toxin interactions.
Heterotrimeric GTPase α subunit binds βγ subunits in the inactive GDP-bound form. The G-protein-coupled receptor bound to its ligand acts as GNRP to stimulate the release of GDP and the α subunit then binds GTP and the βγ subunits dissociate to interact with their downstream effectors. The dissociated active GTP-bound α subunit then interacts with its downstream effectors. PMT, CT and HLT lock the α subunit in its active form and prevent interaction with the βγ subunits and the receptor. PT locks the G protein in its heterotrimeric inactive form and prevents its interaction with the receptor. YpkA prevents GDP/GTP binding to the α subunit.
CT: Cholera toxin; GNRP: Guanine nucleotide release protein; HLT: Heat-labile enterotoxins; PMT: Pasteurella multocida toxin; PT: Pertussis toxin.
G proteins have intrinsic GTPase activity that can convert the bound GTP back to GDP and, in doing so, return the G protein back to its inactive state. The intrinsic GTPase activity of most G proteins is low, ranging from 0.02 min−1 for Ras-like small GTPases to 2–5 min−1 for α subunits of heterotrimeric G proteins [23,24,29]. The Gα subunits have two conserved active-site residues, glutamine and arginine, which stabilize the transition state of the GTPase. Small GTPases also have analogous active-site glutamines, but the corresponding active-site arginine is absent, accounting for the relatively low intrinsic GTPase activity for the small GTPases. Other regulatory proteins, known as GTPase-activating proteins (GAPs), can help stimulate this intrinsic GTPase activity by as much as 1000-fold, often by supplying a catalytic arginine group, sometimes referred to as an arginine finger [30–34]. For heterotrimeric Gα proteins, GTP hydrolysis can be stimulated by regulator of G protein signaling (RGS) proteins, which bind the switch regions of Gα subunits and stabilize the GTPase transition state, but do so without providing an arginine finger [35].
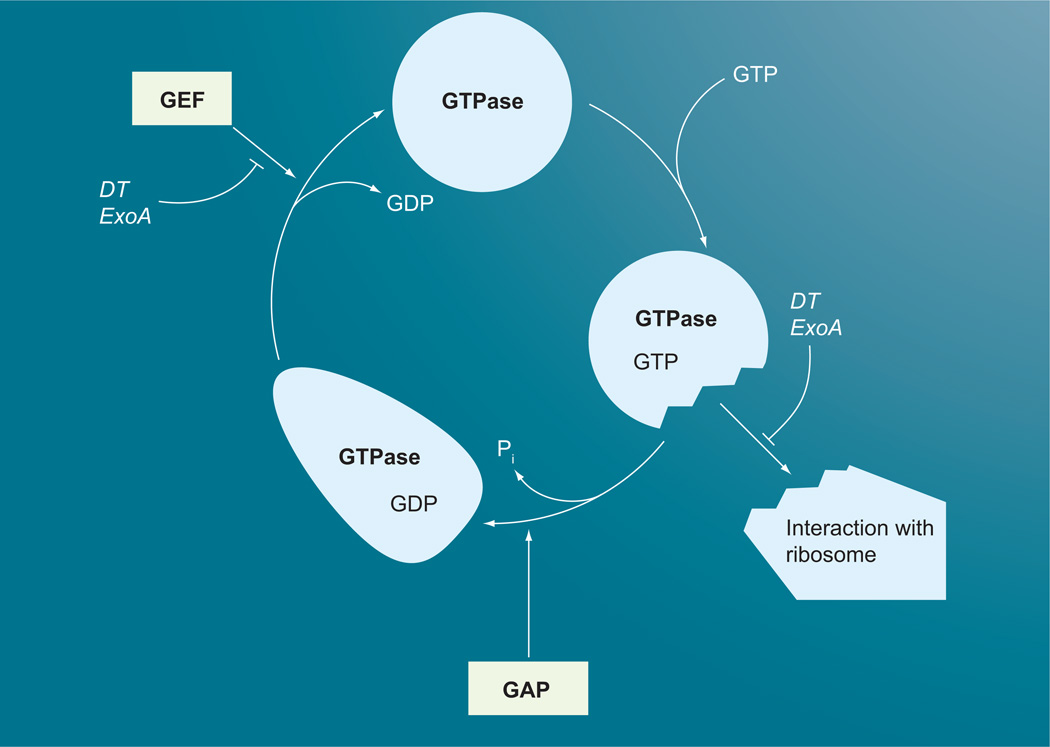
Large, multidomain GTPases, such as the elongation factors involved in protein synthesis, have conserved active-site arginines, but the intrinsic GTPase activity is very low and only enhanced upon binding with the ribosome in the pretranslocation phase of the peptide elongation cycle (Figure 3) [36–40]. The precise mechanism for this ribosome-mediated stimulation is still unclear.
Figure 3. GTPase cycle of large G proteins and points of toxin interactions.
ADP ribosylation of elongation factor (EF)-2 by DT or ExoA blocks interaction of GTP-bound EF-2 with the ribosome in the pretranslocation phase of the peptideelongation cycle, which prevents formation of the high-affinity complex and stimulation of the GAP activity of EF-2 by the ribosome. ADP ribosylation of EF-2 does not interfere with GTP or GDP binding to EF-2, but may inhibit the exchange of GDP with GTP.
DT: Diphtheria toxin; GAP: GTPase activating protein; GEF: Guanine nucleotide exchange factor.
Within this framework, most bacterial toxins that act on G protein targets do so by locking them into either an inactive or an active state (Table 1). Consequently, many signaling pathways modulated by these G proteins have been elucidated through the selective action of bacterial toxins.
Bacterial toxins that modulate G-protein targets through ADP ribosylation
Diphtheria toxin from Corynebacterium diphtheriae and exotoxin A from Pseudomonas aeruginosa catalyze the ADP ribosylation of a unique, highly conserved post-translationally modified His residue at position 715 (diphthamide, 2-[3-carboxyamido-3-(trimethyammonio) propyl] histidine) on the large, multidomain GTPase elongation factor (EF)-2 of eukaryotes [41]. Precisely how this covalent modification alters EF-2 function is still unclear [36]. There is some evidence that ADP ribosylation of EF-2 does not interfere with GTP or GDP binding, but may inhibit the exchange of GDP with GTP [42]. His-715 is near the proposed interaction site of EF-2 with the codon–anticodon duplex, and it is thought to interfere with EF-2 binding to the ribosome in the pretranslocation phase of the peptide elongation cycle [36,42,43], which prevents formation of the high-affinity complex and stimulation of the GAP activity by the ribosome, effectively blocking protein synthesis and resulting in cell death. Archaea have a deamidated form of the diphthamide residue (known as diphthine), which can also serve as a target for diphtheria toxin but at a 1000-fold slower reaction rate [44]. Prokaryotic elongation factors have a lysine at the analogous position instead and, thus, are not substrates for bacterial ADP ribosylating toxins.
Cholera toxin (CT) from Vibrio cholerae and pertussis toxin (PT) from Bordetella pertussis were the first toxins known to act on heterotrimeric G proteins [45,46], and have served as valuable tools for probing G-protein function in ligand–receptor-mediated signal transduction [8]. CT activates Gαs proteins, while PT inhibits Gαi. proteins involved in coupling of hormone receptor-mediated regulation of adenylate cyclase signaling pathways [45]. CT and the closely related heat-labile enterotoxins (HLTs) from Escherichia coli catalyze the ADP ribosylation of an active site arginine (Arg-201) in Gαs subunits. This modification results in dissociation of the Gα subunit from the βγ subunits and locks the Gα subunit in an active state that can stimulate its downstream effectors [47]. PT catalyzes the ADP ribosylation of a Cys residue (Cys-352) four amino acids from the C-terminus of Gαi. proteins. Other Gα proteins do not have a Cys at this position and are often referred to as PT-insensitive G proteins [48]. The preferred substrate for PT is the heterotrimeric form of the Gi protein [49]. ADP ribosylation of Gαi proteins by PT uncouples the Gαi protein interaction with, and activation by, the receptor, which, in turn, impedes GDP/GTP exchange and effectively locks the Gα subunit in its GDP-bound heterotrimeric form [50].
A number of ADP ribosylating toxins act on small GTPases of the Rho and Ras families to inactivate them [51]. These small GTPases modulate cellular processes, such as actin cytoskeletal organization and dynamics, membrane trafficking, cell proliferation and apoptosis [26,52]. The exoenzyme C3 from Clostridium botulinum specifically catalyzes the ADP ribosylation of an Asn residue (Asn-41) of Rho GTPases (RhoA, RhoB and RhoC, but not Rac, Cdc42 or Ras), inactivating the Rho proteins and causing depolymerization of actin filaments [53]. C3-like ADP ribosyltransferases are also produced by Clostridium limosum [54], Staphylococcus aureus [55,56] and Bacillus cereus [57]. ADP ribosylation of Rho at Asn-41 has little or no effect on GDP/GTP binding, or on the intrinsic GTPase activity, but appears to interfere with the activation of the G protein by its GEF, Lbc [58].
Bacterial toxins that modulate G-protein targets through glucosylation
RhoA and other related GTPases, such as Rac and Cdc42, are targets of the Clostridium difficile toxins TcdA and TcdB, Clostridium sordellii TcsH and TcsL, and Clostridium novyi Tcnα. These toxins, whose catalytic domains all belong structurally to the type A family of glycosyltransferases, inactivate their G-protein targets by monoglucosylation at a specific threonine residue (Thr-37 in Rho, Thr-35 in Rac and Cdc42), thereby inducing reorganization of actin and cell rounding [59,60]. In addition, TcdA and TcsL monoglucosylate the Ras-like proteins Rap1 and Rap2 [61,62]. With the exception of Tcnα, which also utilizes UDP-N-acetylglucosamine, all the other clostridial glucosyltransferases use UDP-glucose as a cosubstrate. Inactivation of the small GTPases by glucosylation prevents the G protein from interacting with its downstream effector proteins [58].
Bacterial toxins that modulate G-protein targets through proteolysis
The Yersinia pseudotuberculosis T3SS effector protein (YopT) and the related Pseudomonas syringae T3SS effector protein (Avr/PhpB) also inactivate the small GTPases RhoA, Rac and Cdc42, but they do so by proteolytic cleavage of a C-terminal peptide that contains a post-translationally modified Cys residue (with an isoprene moiety) [63]. Loss of this peptide prevents membrane localization of the G protein and uncouples downstream signaling.
Bacterial toxins that modulate G-protein targets through noncovalent interactions
Several T3SS effector proteins from Salmonella (SopE, SifA and SifB) [64,65], Shigella (IpgB1 and IpgB2) [66], E. coli (Map, EspM and EspT) [67–69], and Citrobacter rodentium (EspM and EspT) [67,68,70] belong to a family of proteins containing a WxxxE motif, which activate Rho GTPases through GEF-like mechanisms [71]. Although originally proposed to be Rho GTPase mimics [72], structural and biochemical evidence have shown that these WxxxE-motif-containing T3SS effector proteins are, indeed, Rho GEFs, with structurally similar GEF-like domains that selectively regulate RhoA, Rac1 or Cdc42 signaling pathways by interacting with the switch I and switch II regions of the GTPases [64,65,69,71]. By contrast, the GEF-like domain of the T4SS effector RalF from Legionella pneumophila, which subverts Arf GTPase signaling has no structural similarity with the WxxE-motif family of bacterial GEFs, but does have structural similarity to the GEF domain of the mammalian Dbl/Sec7 protein family of Rho GEFs [73].
Bacterial toxins that modulate multiple G-protein targets through multiple activity domains
Pseudomonas aeruginosa ExoS and ExoT are bifunctional cytotoxic T3SS effector proteins that share 76% protein sequence homology [74]. The N-terminal domain of both cytotoxins acts as a GAP for the Rho GTPases, Rho, Rac and Cdc42, by supplying a catalytic arginine residue (Arg-146 of ExoS) to stabilize the GTPase transition state [75,76], which results in actin depolymerization and interference with phagocytosis. Other T3SS effector proteins with similar GAP-like domains include the protein tyrosine phosphatase from Salmonella enterica serovar Typhimurium (SptP) [77,78] and the YopE protein from Y. pseudotuberculosis [79]. Although the primary amino acid sequences of the ExoS, SptP and YopE GAP domains have limited similarity, their crystal structures are strikingly similar [80]. Interestingly, while the RhoGAP domain of ExoS is a functional mimic of eukaryotic GAPs, their structures do not share any similarity [76]. The C-terminal domains of ExoS and ExoT both have ADP ribosyltransferase activity but, in contrast to their RhoGAP domains, the ADP ribosyltransferase domains do not have the same substrate specificity [74]. The C-terminal domain of ExoS catalyzes the ADP ribosylation of a number of host signaling proteins [81,82], including two arginine residues (Arg-41, Arg-128) of Ras GTPases [83], which uncouples mitogenic signal transduction by preventing activation of Ras by its GEF Cdc25 [84]. By contrast, the C-terminal domain of ExoT specifically ADP ribosylates Crk-I and Crk-II, which are Src homology 2–3 domain-containing adaptor proteins involved in regulation of focal adhesion and phagocytosis [85].
Another bifunctional T3SS effector protein has been identified from Yersinia – YpkA in Y. pseudotuberculosis and YopO in Y. enterocolitica [86]. YpkA has two domains that act synergistically to disrupt host actin organization and prevent phagocytosis. The C-terminal domain is a Rho GDI that inhibits GDP/GTP exchange of small GTPases of the Rho family [87]. The N-terminal domain of YpkA is a serine/threonine protein kinase that phosphorylates Gαq at Ser-47, a key residue in the diphosphate-binding site of the GTPase domain and, thereby, blocks GDP/GTP binding and inhibits Gαq signaling [86].
Legionella pneumophila uses the bifunctional DrrA/SidM T4SS effector protein to activate and recruit the Golgi–endoplasmic reticulum (ER) vesicle-trafficking regulator Rab1 to the specialized Legionella-containing vacuole through both Rab1-specific GEF-like activity and GDI-displacement factor (GDF)-like activity [88,89]. DrrA/SidM has extensive interactions with the switch I and II regions of Rab1, which result in displacement of the switch I region [90]. In addition, L. pneumophila has another T4SS effector protein, LepB, which has Rab1 GAP-like activity that, in conjunction with DrrA/ SidM, functions in membrane cycling of Rab1 by promoting hydrolysis of GTP and release of Rab1 from the membrane [88].
Another family of large bacterial toxins with multiple activity domains is the multifunctional autoprocessing repeats of toxins (MARTX) of the Vibrio, Aeromonas, Photorhabdus and Yersinia genera [17]. The MARTX from Vibrio cholerae (Mr ~460 kDa), and the related one from V. vulnificus, cause the ‘rounding up’ of cells. One of the functional domains that these proteins possess is a Rho-inactivation domain (RID), which causes inactivation of Rho GTPases through a currently unknown mechanism [91], but these large proteins are comprised of several additional domains that contribute to cytotoxicity. The flanking N- and C-termini contain extensive repeat regions, which are involved in membrane pore formation and translocation of the multiple effector domains, including the RID, an actin-crosslinking domain (ACD) with sequence similarity to glutamine synthetases [92–94], an α-β hydrolase-like domain of unknown function, and an autocatalytic, inositol hexakisphosphate-dependent caspase-like cysteine protease domain (CPD) that processes the MARTX to release the effector domains into eukaryotic cells [95–97]. MARTX proteins illustrate how toxins can have multiple effects on multiple aspects of host cell processes, including G-protein function.
Bacterial toxins that modulate small G-protein targets through deamidation and/or transglutamination
In addition to inactivation of the G-protein targets, toxin-catalyzed modifications can also lead to activation. Modifications by the CNFs from E. coli and Yersinia (CNF1, CNF2, CNF3 and CNFY) and the dermonecrotic toxin (DNT) from Bordetella spp. activate the small GTPases, RhoA, Rac and Cdc42, but not RhoD [19]. The switch I and switch II regions of Rho proteins are involved in protein–protein interactions between GTPases and their effectors. The CNFs modify Rho proteins by deamidation of a specific Gln residue, Gln-63 of RhoA [98–100] and Gln-61 of Rac and Cdc42 [101], located in the switch II region, while CNFY appears to have more stringent substrate specificity for RhoA and does not modify Rac or Cdc42 [102]. CNF1 has also been shown to transglutaminate RhoA at the same residue (Gln-63), although to a lesser extent [103]. DNT, on the other hand, activates Rho proteins primarily through transglutamination of the corresponding residues (Gln-63/61) [104–106], with putrescine, spermidine and spermine serving as the in vivo cosubstrates for the transglutamination reaction [107,108]. Modeling of the DNT active site based on the structure for CNF1 suggests that this preference may stem from DNT having a substantial negative charge in the active-site pocket, which may be able to accommodate positively charged primary amines [109]. Both GDP-bound RhoA and GTP-bound RhoA can serve as substrates for CNF1, but DNT prefers GDP-bound RhoA [106]. The resulting modifications inhibit the intrinsic and GAP-stimulated GTPase activity of the targets, resulting in constitutive activation. Interestingly, a Glu residue (Glu-64 in RhoA) adjacent to the target Gln (Gln-63) appears to be critical for substrate recognition by the toxins; indeed, exchange of the equivalent residues in RhoD (Gln-75, Asp-76) to Gln–Glu converted RhoD into a substrate for CNF1 and DNT [110].
Pasteurella multocida toxin: a bacterial toxin that modulates heterotrimeric G proteins through deamidation
The Gαq family of Gα subunits were first distinguished as the class of heterotrimeric G proteins that mediated activation of phospholipase Cβ (PLCβ) signaling pathways that were resistant to PT treatment [48,111–113], and for years there were no biochemical tools for studying the role of these Gq proteins in mitogenic and calcium signal-transduction pathways. This changed dramatically with the discovery that the Pasteurella multocida dermonecrotic toxin (PMT) strongly stimulates, but subsequently uncouples, PLCβ signal transduction through its action on the Gαq subunit, but not the closely related Gα11 subunit [9,114,115]. PMT-mediated activation of Gαq leads to stimulation of calcium signaling through PLCβ hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) to release inositol trisphosphate (IP3) and diacylglycerol (DAG) [114–121], activation of mitogenic signaling through MAPK and STAT protein phosphorylation [122–126], and Rho-dependent actin cytoskeletal rearrangements [126–132].
Exposure to PMT directly facilitated Gαq -mediated activation of PLCβ1 and, to a lesser extent, PLCβ3, but not PLCβ2 [114], in-keeping with known cellular responses elicited by Gαq -coupled receptors [111,112]. This strong initial response was potentiated by release of the Gαq subunit from the heterotrimeric complex through either PT-mediated sequestration of the Gβγ subunits or through dissociation of the Gα subunit from Gβγ by using antibodies against the Gβ subunit [114]. PMT action on Gαq is irreversible and persistent [114,133] and independent of interaction with G-protein-coupled receptors [133]. Furthermore, overexpression of Gαq enhanced the PMT-induced response, while decreased expression of Gαq or treatment with GDPβS, a known inhibitor of Gα signaling, blocked the PMT-induced response [114], suggesting that the monomeric form of Gαq is the preferred substrate of PMT. Recently, the biochemical activity of PMT was determined to be through deamidation of Gαq at Gln-209 [21]. Interestingly, Gln-209 is functionally equivalent to Gln-63 of the small GTPase RhoA, which serves as the target of the CNFs and DNT.
Pasteurella multocida dermonecrotic toxin is also a potent activator and subsequent uncoupler of Gαi. signaling, converting the G protein into a form that is no longer sensitive to PT treatment [134]. Treatment of intact wild-type, Gαq/11-deficient or Gα12/13-deficient mouse embryonic fibroblasts with PMT leads to inhibition of isoproterenol and forskolin-mediated stimulation of adenylate cyclase activity, as well as cAMP accumulation through Gs -coupled receptors, while enhancing the inhibition of cAMP accumulation by lysophosphatidic acid (LPA) through Gi-coupled receptors. PT treatment blocked LPA-mediated inhibition of cAMP accumulation, yet was unable to block PMT-mediated activation of Gαi. or inhibition of cAMP accumulation. Moreover, pretreatment of cells with PMT prevented PT-induced ADP ribosylation of Gαi2, in keeping with the proposed model where PMT acts on the monomeric Gα subunit to irreversibly convert it into an active state [114]. This effectively shifts the equilibrium to dissociate the heterotrimeric complex and release the Gβγ subunits [135], which can then interact with their downstream effector proteins. Since the preferred substrate for PT is the heterotrimeric G protein, and not the monomeric Gα subunit [136], PMT deamidation of the Gαi2 subunit at Gln-205 converts it into a form that is no longer a substrate for PT [21].
Identification of Gαi2 as a substrate for PMT also enabled further study of the effect of PMT on the GTPase activity of the Gαi. subunit. PMT treatment of cells reduced both basal and LPA-induced hydrolysis of GTP by the Gαi. protein in membrane preparations [134]. A similar effect was observed with the use of the wasp venom peptide mastoparan, a widely used receptor-independent activator of Gαi, suggesting that PMT-mediated activation of Gαi and subsequent inhibition of adenylate cyclase may be caused by PMT-mediated inhibition of the intrinsic GTPase activity of Gαi. However, PMT also inhibited LPA-receptor-stimulated binding of GTPγS to Gαi. [134]. This finding supports the model for PMT action, where PMT first locks the monomeric Gαi subunit in its active form through deamidation, which inhibits its GTPase activity [21]. This action prevents reassociation of the Gαi. subunit with the Gβγ subunits, as evidenced by the failure of PMT-exposed Gαi2 to bind to Gβγ and serve as a substrate for PT [21], and essentially leads to the functional uncoupling of the G protein from its receptor, similar to what was observed for PMT action on Gαq [114].
In addition to the activation of Gαq and Gαi proteins, PMT activates Gα12/13 signaling pathways [137], resulting in formation of actin stress fibers and assembly of focal adhesions through indirect activation of RhoA mediated by the regulator of G-protein signalling (RGS) domains of Rho GEFs, such as LARG, p115-RhoGEF, PDZ-RhoGEF or Dbl [138–141]. Gα12 and Gα13 share 67% sequence identity, except for their N-terminal 30 residues, which only share 16% identity and confer receptor specificity [142]. In GGαq/11-deficient fibroblasts, RhoA activation by PMT was inhibited by dominant-negative Gα13, whereas in Gα12/13 -deficient cells, RhoA activation by PMT could be reconstituted by infection with retrovirus encoding Gα13 [137]. Although PMT-mediated activation of Gα12 signaling was not tested in this study and direct deamidation of Gα12 and Gα13 by PMT has not yet been demonstrated, both Gα12 and Gα13 have analogous switch II Gln residues (Gln-229) that could serve as PMT targets for deamidation. However, it should be noted that Gαq and Gα11 share even greater homology (88% sequence identity) with each other, including the switch II Gln-209 residue [113], but only Gαq is a substrate for PMT [21,115]. The reason for this difference in substrate specificity between Gαq and Gα11 is not known; however, exchange of two residues (Glu-105 and Asn-109) in the helical domain of Gα11 with the corresponding His residues of Gαq rendered the mutant Gα11 now capable of mediating PMT-induced activation of PLCβ in Gαq/11-deficient fibroblasts [143]. It is not yet known whether this differential interaction is due to differences in PMT substrate recognition of Gαq versus Gα11 or due to differential interaction of the Gαq and Gα11 proteins with the PLCβ1 effector protein.
Structural comparisons
PMT & related dermonecrotic toxins
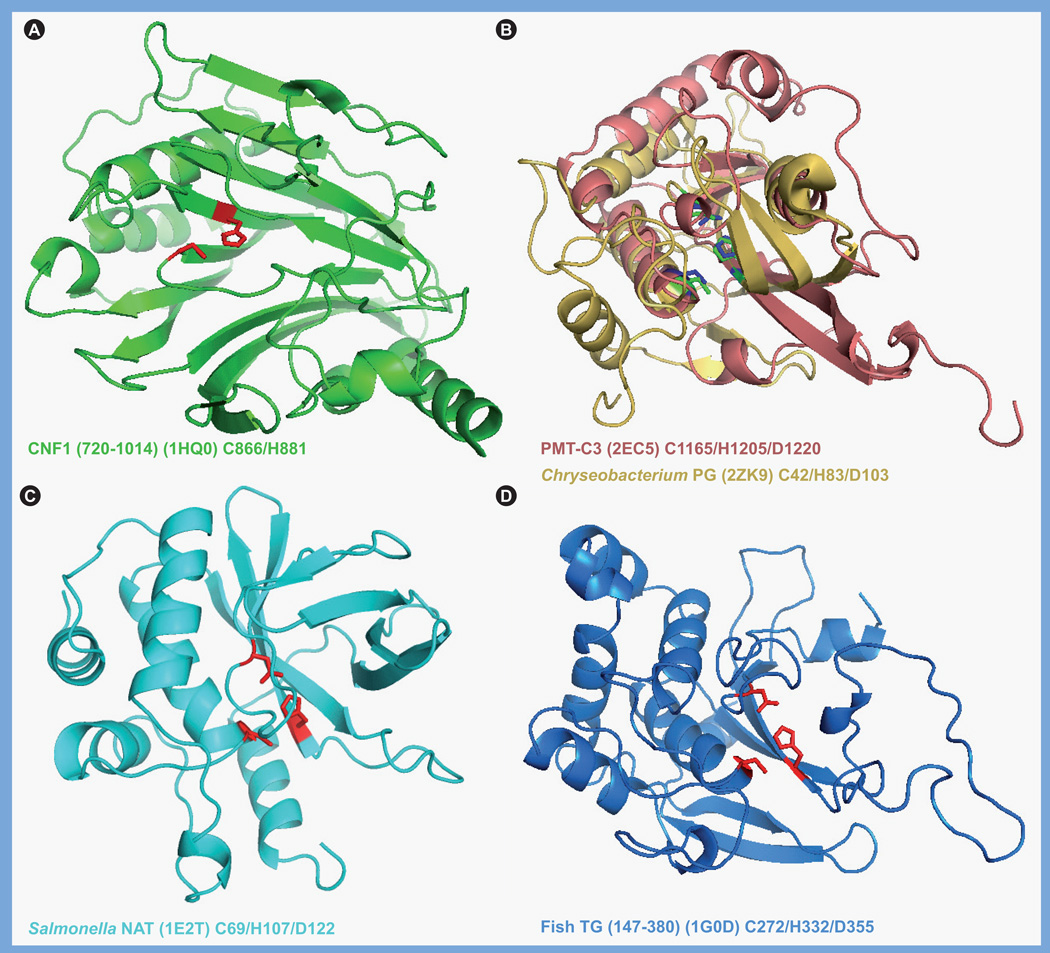
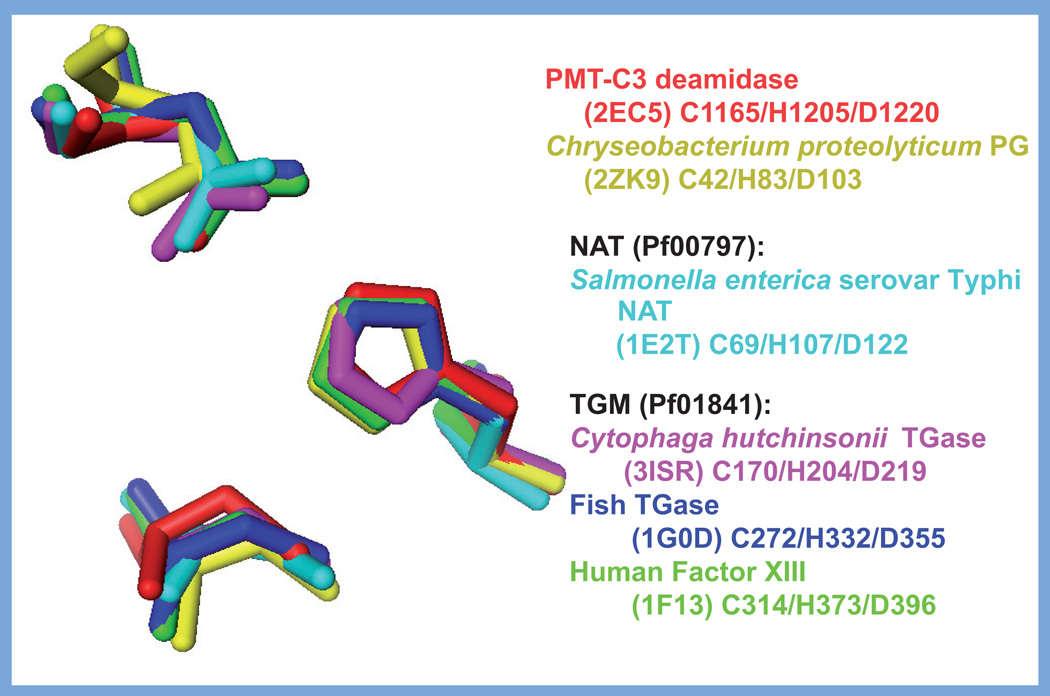
When the sequences of PMT and the related CNFs and DNT were first examined, there were no matches found in the databases other than with each other and, thus, only biochemical and structural analyses provided insights into their functional organization. The receptor-binding domain is located in the N-terminus of each of these toxins [144–146], whereas the intracellular activity domain resides in the C-terminus [109,145–149]. The crystal structure of the catalytic domain of CNF1 (PDB 1HQ0) has been solved [109], and since the CNFs and DNT share significant sequence similarities (27–32%) in their C-terminal domains (residues 720–1014 in the CNFs, 1176–1464 in DNT), as well as similar target substrates and catalytic activities, it is presumed that their activity domains also have similar overall structures. Indeed, the Cys and His residues located in a putative active-site pocket (Figure 4) are not only conserved in all members of the CNF/DNT family (Cys-866 and His-881 in CNF1, and Cys-1305 and His-1320 in DNT), but are also essential for catalytic activity [103].
Figure 4. Structures of toxin-like deamidase/transglutaminase domains.
Shown are the structural folds of the catalytic domains of representative members of the cytotoxic necrotizing factor/dermonecrotic toxin-like family and the PMT-like family of deamidases/ TGases, with the respective active site His–Cys dyad or His–Cys–Asp triad indicated. (A) Catalytic domain of CNF1 (PDB 1HQ0), residues 720–1014 shown in green, with the Cys-866 and His-881 shown in red. (B) Superimposition of the catalytic domains of PMT (PDB 2EC5), residues 1105–1285 shown in pink, with Cys-1165, His-1205 and Asp-1220 shown in blue, and the protein glutaminase from Chryseobacterium proteolyticum (PDB 2ZK9), shown in yellow, with Cys-42, His-83, and Asp-103 shown in green. (C) Catalytic domain of the arylamine N-acetyltransferase from Salmonella enterica serovar Typhimurium (PDB 1E2T), residues 1–197 shown in cyan, with the Cys-69, His-107, and Asp-122 shown in red. (D) Catalytic domain of the fish-derived TGase from red sea bream (PDB 1G0D), residues 147–380 shown in blue, with Cys-272, His-332 and Asp-355 shown in red.
Images were generated with PyMOL using the indicated PDB data files.
CNF: Cytotoxic necrotizing factor; NAT: N-acetyltransferase; PDB: Protein Data Bank; PMT: Pasteurella multocida toxin; TGase: Transglutaminase.
Although the CNFs and DNT share limited sequence similarity in their N-terminal receptor-binding and translocation domains with PMT, there is no discernable sequence similarity of their C-terminal catalytic domains with the C-terminal intracellular activity domain of PMT. This was confirmed when the crystal structure of a biologically active C-terminal fragment of PMT consisting of residues 569–1285 (PDB 2EBF) became available [148]. The structure of this fragment revealed three distinct domains: a C1 domain (residues 575–719) with sequence, structural and functional similarity to the membrane-targeting domain of the clostridial toxin TcdB [150]; a C2 domain (residues 720–1104) of currently unknown function, and a C3 domain (residues 1105–1285), with a papain-like cysteine protease structural fold. The PMT-C3 domain was subsequently demonstrated to harbor the minimal domain responsible for toxin-mediated activation of calcium and mitogenic signaling [147]. Disruption of the disulfide bond between Cys-1159 and Cys-1165 in the C3 domain through mutation of Cys-1159 to Ser exposed an active-site Cys–His–Asp triad (Cys-1165, His-1205, Asp-1220) [148]. This finding agreed with earlier studies that demonstrated the importance of Cys-1165 [151] and His-1205 [152] in the biological activity of PMT. However, other than the presence of active-site Cys and His residues, there was no indication that the catalytic activity of PMT was like that of the CNF/DNT family, since the protein folds were quite different (compare Figure 4A & B), and even the positioning of the active-site Cys and His groups were different.
Toxin G-protein deamidase/transglutaminase domains with other deamidases/transglutaminases
The Cys–His–Asp triad is commonly involved in catalysis of two types of reactions: acyl hydrolysis (e.g., protease activity) or acyl transfer (e.g., transglutaminase activity). Based on the similarity of the Cys–His–Asp triad in PMT with that of papain, it was proposed that PMT might act as a cysteine protease [148]. However, transglutaminases (TGases), which exchange the amine group of the side-chain γ-carboxyamide of glutamine with the primary amine group of another molecule, have structural folds with active-site Cys–His–Asp triads similar to that of papain-like cysteine proteases [153]. In the absence of a primary amine-containing substrate, TGases can catalyze the hydrolysis of the γ-carboxyamide group of Gln residues, resulting in deamidation. Deamidases, which convert the γ-carboxyamide group to a carboxylate, are closely related to TGases. Thus, the finding that the C-3 domain of PMT has G-protein Gln-deamidase activity [21] is consistent with its structure [148].
The structural fold of the PMT-C3 deamidase domain belongs to a family of structural folds that are closely related to mammalian and some bacterial TGases (Figure 4), typified by the human blood clotting factor XIII (PDB 1FIE) [154], fish-derived TGase from red sea bream (PDB 1G0D) [155], putative TGase-like cysteine protease from Cytophaga hutchinsonnii (PDB 3ISR), and the protein glutaminase from Chryseobacterium proteolyticum (PDB 2ZK9) [156]. The PMT-C3 core structural fold, similar to that of the other TGases, also bears similarity to the arylamine N-acetyltransferase (NAT) from Salmonella enterica serovar Typhimurium (PDB 1E2T) [157]. The active-site Cys–His–Asp triad is nearly superimposable for all five of these structures (Figure 5). The overall PMT-C3 structure most closely resembles that of the protein glutaminase from Chryseobacterium, with both the catalytic triad and the active-site cores clearly overlapping (Figure 4B). Interestingly, the structural fold of the microbial TGase from Streptomyces mobaraensis (PDB 1IU4) does not resemble the TGase-like PMT fold [158,159]. While still possessing a Cys–Asp–His triad at the active site, these catalytic residues in the Streptomyces TGase (Cys-64, Asp-255 and His-274) do not have the same geometry at the active site found for the PMT-C3 family.
Figure 5. Comparison of the active-site catalytic triads of the Pasteurella multocida toxin-like deamidases/transglutaminases.
Superimposed images of the active site Cys, His and Asp side chains of the catalytic triads from: PMT-C3 (PDB 2EC5) in red, Chryseobacterium protein glutatminase (PDB 2ZK9) in yellow, Salmonella NAT (PDB 1E2T) in light blue, Cytophaga hutchlnsonll TGase (PDB 3ISR) in magenta, fish-derived TGase (PDB 1G0D) in purple and human factor XIII (PDB 1F13) in green.
Images were generated with PyMOL using the indicated PDB data files.
NAT: N-acetyltransferase; PDB: Protein Data Bank; PMT: Pasteurella multocida toxin; TGase: Transglutaminases;
A striking finding about the fish TGase and human factor XIII, in comparison with PMT-C3, is that the active sites of these proteins possess an additional Cys residue (Cys-333 in fish TGase and Cys-374 in factor XIII), which is separated from the catalytic Cys of the triad by an active site Tyr residue (Tyr-515 in fish TGase and Tyr-560 in factor XIII) [155]. It was proposed that the Tyr side chain sterically prevents disulfide bond formation, and subsequent inactivation in these proteins. Similar to these proteins, PMT also has another Cys residue (Cys-1159) near the catalytic Cys-1165, with which it does form a disulfide bond. It is interesting to speculate that reduction of the disulfide bond in PMT might occur prior to or during interaction with its substrate proteins.
In contrast to the TGase-like family of structural folds to which PMT belongs, the protein fold of the CNF1 catalytic domain appears to be unique to the CNF/DNT family of deamidating/transglutaminating toxins [109], as well as a few other bacterial proteins. It has been suggested that the CNF1 fold resembles that of the chemoreceptor-modifying deamidases, such as CheD (PDB 2F9Z) from Thermotoga maritima [160]. A similar structural topology has also been found in several proteins of unknown function, including YfiH from Shigella flexneri (PDB 1XAF and 1U05) [161], YlmD from Bacillus stearothermophilus (PDB 1T8H), protein CC_0490 from Caulobacter crescentus (PDB 1XFJ), and the YfiH-like protein from Salmonella enterica serovar Typhi (PDB 1RW0). Unlike CheD and the CNF/DNT toxins, these YfiH-like proteins have a second active-site His residue (His-71), in addition to the Cys–His dyad (Cys-107 and His-124 in YfiH), which acts to coordinate zinc in some of the structures. The role that this zinc plays in the function of these proteins is currently unknown.
Future perspective
Increasing structural data, backed by extensive biochemical studies, are providing new insights into the range of biological functions that can be manipulated by bacterial protein toxins. The consequent depth and scope of our knowledge of their structure and function has enabled us to gain a better picture of the role that these toxins play in host–microbe interactions and bacterial pathogenesis. Indeed, we are now poised to begin contemplating ways to counteract the deleterious effects of these toxins on the host such that we can develop postexposure antitoxin therapeutics. We are even starting to get glimpses of potential roles for these toxins in long-term sequelae to bacterial infection, such as possible involvement of some of the mitogenic toxins, such as PMT in cancer onset or progression [162].
Importantly, our emerging understanding of their biochemical activities and mechanisms of action at the molecular level has been invaluable in making these toxins available as powerful tools to study and manipulate the myriad cellular signaling pathways modulated by G proteins. With the addition of PMT to CT and PT as selective molecular tools for studying heterotrimeric G-protein signaling, we can now begin to discern the functions of the different heterotrimeric G proteins in signal transduction and physiological processes. It remains to be determined which of the other G-protein α-subunits might also be targets for deamidation by PMT, and what the substrate recognition determinants are that discriminate one target protein from another. The consequences to the toxin-modified G-protein molecule also remain unclear.
Although often deleterious to the infected host, a number of these toxins are already beginning to be exploited for beneficial medical applications, owing to the unique selectivity and potency of their activities [2]. Clinical trials are already underway for incorporation of DT, CT, PT, CNF and anthrax toxin into vaccines as adjuvants or as antigen-delivery vehicles. DT, ExoA and several other toxins are being tested as immunotoxins in cancer treatments. A number of toxins, such as CNF, PT, anthrax toxin and botulinum neurotoxin, are being developed as alternative biomedical therapeutics and cosmetics. Further advances will continue to be made as we determine cellular uptake and intracellular targeting mechanisms, decipher the determinants of toxin–substrate specificity, and differentiate the various consequences of toxin action on downstream signaling and cellular function.
Executive summary.
G proteins as targets of bacterial toxins
-
▪
Many bacterial toxins target regulatory G proteins, and act at different points in the GTPase cycle to disrupt G-protein signal transduction.
Bacterial toxins that modulate G-protein targets through ADP ribosylation
-
▪
Diphtheria toxin (DT) and Pseudomonas ExoA ADP ribosylate the large GTPase elongation factor (EF)-2 and block eukaryotic protein synthesis.
-
▪
Cholera toxin (CT) and pertussis toxin (PT) ADP ribosylate heterotrimeric G proteins involved in hormone receptor-mediated signal transduction.
-
▪
Clostridial C3 toxin ADP ribosylates and inactivates small Rho GTPases.
Bacterial toxins that modulate G-protein targets through glucosylation
-
▪
The large clostridial toxins inactivate small Rho GTPases through monoglucosylation.
Bacterial toxins that modulate G-protein targets through proteolysis
-
▪
Some type III secretion system (T3SS) effector proteins (YopT from Yersinia and Avr/PhpB from Pseudomonas) cleave and inactivate small Rho GTPases to prevent membrane localization.
Bacterial toxins that modulate G-protein targets through noncovalent interactions
-
▪
Some T3SS (Salmonella SopE, SifA and SifB; Shigella IpgB1 and lpgB2; Escherichia coli Map, EspM and EspT; and Citrobacter EspM and EspT) and T4SS (Legionella RalF) effector proteins are guanine nucleotide exchange factors (GEFs) that activate Rho GTPases.
Bacterial toxins that modulate multiple G-protein targets through multiple activity domains
-
▪
Some toxins (Pseudomonas ExoS and ExoT, Salmonella SptP, Yersinia YopE, YkpA/YopO, and Vibrio MARTX) have multiple domains with different intracellular activities that modulate small GTPases.
Bacterial toxins that modulate G-protein targets through deamidation &/or transglutamination
-
▪
Deamidation (E. coli and Yersinia cytotoxic necrotizing factor [CNF]s) or transglutamination (Bordetella dermonecrotic toxin [DNT]) of a specific Gln residue (Gln-63 in RhoA) inhibits intrinsic and GAP-stimulated Rho GTPase activity, resulting in constitutive activation.
-
▪
Pasteurella multocida toxin (PMT) deamidates heterotrimeric G proteins, Gαq Gαi2 and Gα13, and possibly other members of the Gq family at a specific Gln residue (Gln-209 in Gαq, Gln-205 in Gαi2), which results in initial stimulation of G-protein-mediated signaling, followed by uncoupling of the signal transduction.
Structural comparison of PMT & related dermonecrotic toxins
-
▪
PMT shares limited sequence similarity with the CNFs or DNT in their N-terminal receptor-binding and translocation domains.
-
▪
PMT shares no sequence or structural similarity with the CNFs or DNT in their C-terminal activity domains (residues 1105–1285 in PMT-C3, 720–1014 in the CNFs, 1176–1464 in DNT).
-
▪
The structure of PMT (575–1285) revealed three domains: C1 membrane-localization domain, C2 domain of unknown function, and C3 domain with cysteine protease-like Cys–His–Asp catalytic triad.
Structural comparison of toxin G-protein deamidase/transglutaminase domains with other deamidases/transglutaminases
-
▪
The structure of the PMT-C3 domain belongs to a family of transglutaminases (TGases; human factor XIII, fish TGase, Chryseobacterium protein glutaminase, Salmonella arylamine N-acetyltransferase and a putative Cytophaga cysteine protease).
-
▪
The PMT-C3 structural fold has no similarity to the CNF/DNT family of deamidases (Thermotoga CheD deamidase; Shigella, Bacillus, Caulobacter, and Salmonella YfiH-like proteins and Streptomyces TGase).
Acknowledgments
Some of this work was supported in part by NIHI/MAID grant AI038396 (to BA Wilson).
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of consideRab1e interest
- 1.Dautin N, Karimova G, Ladant D. Bordetella pertussis adenylate cyclase toxin: a versatile screening tool. Toxicon. 2002;40(10):1383–1387. doi: 10.1016/s0041-0101(02)00158-7. [DOI] [PubMed] [Google Scholar]
- 2.Fabbri A, Travaglione S, Falzano L, Fiorentini C. Bacterial protein toxins: current and potential clinical use. Curr. Med. Chem. 2008;15(11):1116–1125. doi: 10.2174/092986708784221430. [DOI] [PubMed] [Google Scholar]
- 3. Kostrzewa RM, Segura-Aguilar J. Botulinum neurotoxin: evolution from poison, to research tool - onto medicinal therapeutic and future pharmaceutical panacea. Neurotox Res. 2007;12(4):275–290. doi: 10.1007/BF03033911. ▪ Reviews the potential biomedical applications of bacterial toxins.
- 4.Lemonnier M, Landraud L, Lemichez E. Rho GTPase-activating bacterial toxins: from bacterial virulence regulation to eukaryotic cell biology. FEMS Microbiol. Rev. 2007;31(5):515–534. doi: 10.1111/j.1574-6976.2007.00078.x. [DOI] [PubMed] [Google Scholar]
- 5.Mathew M, Verma RS. Humanized immunotoxins: a new generation of immunotoxins for targeted cancer therapy. Cancer Sci. 2009;100(8):1359–1365. doi: 10.1111/j.1349-7006.2009.01192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandvig K, Van Deurs B. Delivery into cells: lessons learned from plant and bacterial toxins. Gene Ther. 2005;12(11):865–872. doi: 10.1038/sj.gt.3302525. [DOI] [PubMed] [Google Scholar]
- 7.Schiavo G, Van Der Goot FG. The bacterial toxin toolkit. Nat. Rev. Mol. Cell Biol. 2001;2(7):530–537. doi: 10.1038/35080089. [DOI] [PubMed] [Google Scholar]
- 8.Ui M, Katada T. Bacterial toxins as probes for receptor-Gi coupling. Adv. Second Messenger Phosphoprotein Res. 1990;24:63–69. [PubMed] [Google Scholar]
- 9. Wilson BA, Ho M. Pasteurella multocida toxin as a tool for studying Gq signal transduction. Rev. Physiol. Biochem. Pharmacol. 2004;152:93–109. doi: 10.1007/s10254-004-0032-6. ▪ Comprehensive review of Pasteurella multocida toxin (PMT) structure–function and interaction with and effects on cellular signaling pathways.
- 10.Deng Q, Barbieri JT. Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu. Rev. Microbiol. 2008;62:271–288. doi: 10.1146/annurev.micro.62.081307.162848. [DOI] [PubMed] [Google Scholar]
- 11.Jank T, Aktories K. Structure and mode of action of clostridial glucosylating toxins: the ABCD model. Trends Microbiol. 2008;16(5):222–229. doi: 10.1016/j.tim.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Thelestam M, Frisan T. Cytolethal distending toxins. Rev. Physiol. Biochem. Pharmacol. 2004;152:111–133. doi: 10.1007/s10254-004-0030-8. [DOI] [PubMed] [Google Scholar]
- 13.Jackson MP. Structure–function analyses of Shiga toxin and the Shiga-like toxins. Microh. Pathog. 1990;8(4):235–242. doi: 10.1016/0882-4010(90)90050-z. [DOI] [PubMed] [Google Scholar]
- 14.Mukherjee S, Hao YH, Orth K. A newly discovered post-translational modification -the acetylation of serine and threonine residues. Trends Biochem. Sci. 2007;32(5):210–216. doi: 10.1016/j.tibs.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Cornells GR. Molecular and cell biology aspects of plague. Proc. Natl Acad. Sci. USA. 2000;97(16):8778–8783. doi: 10.1073/pnas.97.16.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebrun I, Marques-Porto R, Pereira AS, Pereira A, Perpetuo EA. Bacterial toxins: an overview on bacterial proteases and their action as virulence factors. Mini Rev. Med. Chem. 2009;9(7):820–828. doi: 10.2174/138955709788452603. [DOI] [PubMed] [Google Scholar]
- 17.Satchell KJ. MARTX, multifunctional autoprocessing repeats-in-toxin toxins. Infect. Immun. 2007;75(11):5079–5084. doi: 10.1128/IAI.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukui A, Horiguchi Y. Bordetella dermonecrotic toxin exerting toxicity through activation of the small GTPase Rho. J. Biochem. 2004;136(4):415–419. doi: 10.1093/jb/mvh155. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann C, Schmidt G. CNF and DNT. Rev. Physiol. Biochem. Pharmacol. 2004;152:49–63. doi: 10.1007/s10254-004-0026-4. [DOI] [PubMed] [Google Scholar]
- 20.Munro P, Lemichez E. Bacterial toxins activating Rho GTPases. Curr. Top. Microbiol. Immunol. 2005;291:177–190. doi: 10.1007/3-540-27511-8_10. [DOI] [PubMed] [Google Scholar]
- 21. Orth JH, Preuss I, Fester I, Schlosser A, Wilson BA, Aktories K. Pasteurella multocida toxin activation of heterotrimeric G proteins by deamidation. Proc. Natl Acad. Sci. USA. 2009;106(17):7179–7184. doi: 10.1073/pnas.0900160106. ▪▪ Determination that the PMT-C3 intracellular activity domain is a deamidase for Gα subunits.
- 22.Neer EJ, Clapham DE. Roles of G protein subunits in transmembrane signalling. Nature. 1988;333(6169):129–134. doi: 10.1038/333129a0. [DOI] [PubMed] [Google Scholar]
- 23.Sprang SR. G proteins, effectors and GAPS: structure and mechanism. Curr. Opin. Struct. Biol. 1997;7(6):849–856. doi: 10.1016/s0959-440x(97)80157-1. [DOI] [PubMed] [Google Scholar]
- 24.Sprang SR. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 25.Stryer L, Bourne HR. G proteins: a family of signal transducers. Annu. Rev. Cell Biol. 1986;2:391–419. doi: 10.1146/annurev.cb.02.110186.002135. [DOI] [PubMed] [Google Scholar]
- 26.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279(5350):509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 27.Rens-Domiano S, Hamm HE. Structural and functional relationships of heterotrimeric G-proteins. FASEB J. 1995;9(11):1059–1066. doi: 10.1096/fasebj.9.11.7649405. [DOI] [PubMed] [Google Scholar]
- 28.Wilkie TM, Gilbert DJ, Olsen AS, et al. Evolution of the mammalian G protein α subunit multigene family. Nat. Genet. 1992;1(2):85–91. doi: 10.1038/ng0592-85. [DOI] [PubMed] [Google Scholar]
- 29.Graziano MP, Freissmuth M, Gilman AG. Expression of Gs α in Escherichia coli. Purification and properties of two forms of the protein. J. Biol. Chem. 1989;264(1):409–418. [PubMed] [Google Scholar]
- 30.Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 1997;4(9):686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- 31.Mittal R, Ahmadian MR, Goody RS, Wittinghofer A. Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science. 1996;273(5271):115–117. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- 32.Rittinger K, Walker PA, Eccleston JF, et al. Crystal structure of a small G protein in complex with the GTPase-activating protein RhoGAP. Nature. 1997;388(6643):693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- 33.Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ. Structure at 1.65 Å of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389(6652):758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 34.Scheffzek K, Ahmadian MR, Kabsch W, et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277(5324):333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 35.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4-activated G(iα1): stabilization of the transition state for GTP hydrolysis. Cell. 1997;89(2):251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen R, Merrill AR, Andersen GR. The life and death of translation elongation factor 2. Biochem. Soc. Trans. 2006;34(Pt 1):1–6. doi: 10.1042/BST20060001. [DOI] [PubMed] [Google Scholar]
- 37.Parmeggiani A, Sander G. Properties and regulation of the GTPase activities of elongation factors Tu and G, and of initiation factor 2. Mol. Cell Biochem. 1981;35(3):129–158. doi: 10.1007/BF02357085. [DOI] [PubMed] [Google Scholar]
- 38.Rodnina MV, Fricke R, Kuhn L, Wintermeyer W. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J. 1995;14(11):2613–2619. doi: 10.1002/j.1460-2075.1995.tb07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodnina MV, Pape T, Fricke R, Wintermeyer W. Elongation factor Tu, a GTPase triggered by codon recognition on the ribosome: mechanism and GTP consumption. Biochem. Cell Biol. 1995;73(11–12):1221–1227. doi: 10.1139/o95-132. [DOI] [PubMed] [Google Scholar]
- 40.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385(6611):37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 41.Wilson BA, Collier RJ. Diphtheria toxin and Pseudomonas aeruginosa exotoxin A: active-site structure and enzymic mechanism. Curr. Top. Microbiol. Immunol. 1992;175:27–41. doi: 10.1007/978-3-642-76966-5_2. [DOI] [PubMed] [Google Scholar]
- 42.Jorgensen R, Yates SP, Teal DJ, et al. Crystal structure of ADP-ribosylated ribosomal translocase from Saccharomyces cerevisiae . J. Biol. Chem. 2004;279(44):45919–45925. doi: 10.1074/jbc.M406218200. [DOI] [PubMed] [Google Scholar]
- 43.Nygard O, Nilsson L. Kinetic determination of the effects of ADP-ribosylation on the interaction of eukaryotic elongation factor 2 with ribosomes. J. Biol. Chem. 1990;265(11):6030–6034. [PubMed] [Google Scholar]
- 44.Pappenheimer AM, Jr, Dunlop PC, Adolph KW, Bodley JW. Occurrence of diphthamide in archaebacteria. J. Bacteriol. 1983;153(3):1342–1347. doi: 10.1128/jb.153.3.1342-1347.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casey PJ, Gilman AG. G protein involvement in receptor-effector coupling. J. Biol. Chem. 1988;263(6):2577–2580. [PubMed] [Google Scholar]
- 46.Gierschik P. ADP-ribosylation of signal-transducing guanine nucleotide-binding proteins by pertussis toxin. Curr. Top. Microbiol. Immunol. 1992;175:69–96. doi: 10.1007/978-3-642-76966-5_4. [DOI] [PubMed] [Google Scholar]
- 47.Kahn RA, Gilman AG. ADP-ribosylation of Gs promotes the dissociation of its α and β subunits. J. Biol. Chem. 1984;259(10):6235–6240. [PubMed] [Google Scholar]
- 48.Fields TA, Casey PJ. Signalling functions and biochemical properties of pertussis toxin-resistant G-proteins. Biochem. J. 1997;321(Pt 3):561–571. doi: 10.1042/bj3210561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conklin BR, Bourne HR. Structural elements of G α subunits that interact with G β γ, receptors, and effectors. Cell. 1993;73(4):631–641. doi: 10.1016/0092-8674(93)90245-l. [DOI] [PubMed] [Google Scholar]
- 50.Sunyer T, Monastirsky B, Codina J, Birnbaumer L. Studies on nucleotide and receptor regulation of Gi proteins: effects of pertussis toxin. Mol. Endocrinol. 1989;3(7):1115–1124. doi: 10.1210/mend-3-7-1115. [DOI] [PubMed] [Google Scholar]
- 51.Aktories K, Schmidt G, Just I. Rho GTPases as targets of bacterial protein toxins. Biol. Chem. 2000;381(5–6):421–426. doi: 10.1515/BC.2000.054. [DOI] [PubMed] [Google Scholar]
- 52.Van Aelst L, D’souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11(18):2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 53.Aktories K, Braun U, Rosener S, Just I, Hall A. The Rho gene product expressed in E. coli is a substrate of botulinum ADP-ribosyltransferase C3. Biochem. Biophys. Res. Commun. 1989;158(1):209–213. doi: 10.1016/s0006-291x(89)80199-8. [DOI] [PubMed] [Google Scholar]
- 54.Just I, Mohr C, Schallehn G, et al. Purification and characterization of an ADP-ribosyltransferase produced by Clostridium limosum . J. Biol. Chem. 1992;267(15):10274–10280. [PubMed] [Google Scholar]
- 55.Sugai M, Hashimoto K, Kikuchi A, et al. Epidermal cell differentiation inhibitor ADP-ribosylates small GTP-binding proteins and induces hyperplasia of epidermis. J. Biol. Chem. 1992;267(4):2600–2604. [PubMed] [Google Scholar]
- 56.Wilde C, Chhatwal GS, Schmalzing G, Aktories K, Just I. A novel C3-like ADP-ribosyltransferase from Staphylococcus aureus modifying RhoE and Rnd3. J. Biol. Chem. 2001;276(12):9537–9542. doi: 10.1074/jbc.M011035200. [DOI] [PubMed] [Google Scholar]
- 57.Just I, Selzer J, Jung M, Van Damme J, Vandekerckhove J, Aktories K. Rho-ADP-ribosylating exoenzyme from Bacillus cereus. Purification, characterization, and identification of the NAD-binding site. Biochemistry. 1995;34(1):334–340. doi: 10.1021/bi00001a041. [DOI] [PubMed] [Google Scholar]
- 58.Sehr P, Joseph G, Genth H, Just I, Pick E, Aktories K. Glucosylation and ADP ribosylation of rho Proteins: effects on nucleotide binding, GTPase activity, and effector coupling. Biochemistry. 1998;37(15):5296–5304. doi: 10.1021/bi972592c. [DOI] [PubMed] [Google Scholar]
- 59.Busch C, Aktories K. Microbial toxins and the glycosylation of Rho family GTPases. Curr. Opin. Struct. Biol. 2000;10(5):528–535. doi: 10.1016/s0959-440x(00)00126-3. [DOI] [PubMed] [Google Scholar]
- 60.Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 2005;18(2):247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaves-Olarte E, Weidmann M, Eichel-Streiber C, Thelestam M. Toxins A and B from Clostridium difficile differ with respect to enzymatic potencies, cellular substrate specificities, and surface binding to cultured cells. J. Clin. Invest. 1997;100(7):1734–1741. doi: 10.1172/JCI119698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Popoff MR, Chaves-Olarte E, Lemichez E, et al. Ras, Rap, and Rac small GTP-binding proteins are targets for Clostridium sordellii lethal toxin glucosylation. J. Biol. Chem. 1996;271(17):10217–10224. doi: 10.1074/jbc.271.17.10217. [DOI] [PubMed] [Google Scholar]
- 63.Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109(5):575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 64.Buchwald G, Friebel A, Galan JE, Hardt WD, Wittinghofer A, Scheffzek K. Structural basis for the reversible activation of a rho protein by the bacterial toxin SopE. EMBO J. 2002;21(13):3286–3295. doi: 10.1093/emboj/cdf329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohlson MB, Huang Z, Alto NM, et al. Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe. 2008;4(5):434–446. doi: 10.1016/j.chom.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohya K, Handa Y, Ogawa M, Suzuki M, Sasakawa C. IpgB1 is a novel Shigella effector protein involved in bacterial invasion of host cells. Its activity to promote membrane ruffling via Rac1 and Cdc42 activation. J. Biol. Chem. 2005;280(25):24022–24034. doi: 10.1074/jbc.M502509200. [DOI] [PubMed] [Google Scholar]
- 67.Arbeloa A, Bulgin RR, Mackenzie G, et al. Subversion of actin dynamics by EspM effectors of attaching and effacing bacterial pathogens. Cell Microbiol. 2008;10(7):1429–1441. doi: 10.1111/j.1462-5822.2008.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arbeloa A, Garnett J, Lillington J, et al. EspM2 is a RhoA guanine nucleotide exchange factor. Cell Microbiol. 2010;12(5):654–664. doi: 10.1111/j.1462-5822.2009.01423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang Z, Sutton SE, Wallenfang AJ, et al. Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat. Struct. Mol. Biol. 2009;16(8):853–860. doi: 10.1038/nsmb.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bulgin RR, Arbeloa A, Chung JC, Frankel G. EspT triggers formation of lamellipodia and membrane ruffles through activation of Rac-1 and Cdc42. Cell Microbiol. 2009;11(2):217–229. doi: 10.1111/j.1462-5822.2008.01248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bulgin R, Raymond B, Garnett JA, et al. Bacterial guanine nucleotide exchange factors SopE-like and WxxxE effectors. Infect. Immun. 2010;78(4):1417–1425. doi: 10.1128/IAI.01250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alto NM, Shao F, Lazar CS, et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124(1):133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 73.Amor JC, Swails J, Zhu X, et al. The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J. Biol. Chem. 2005;280(2):1392–1400. doi: 10.1074/jbc.M410820200. [DOI] [PubMed] [Google Scholar]
- 74.Barbieri JT, Sun J. Pseudomonas aeruginosa ExoS and ExoT. Rev. Physiol. Biochem. Pharmacol. 2004;152:79–92. doi: 10.1007/s10254-004-0031-7. [DOI] [PubMed] [Google Scholar]
- 75.Goehring UM, Schmidt G, Pederson KJ, Aktories K, Barbieri JT. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J. Biol. Chem. 1999;274(51):36369–36372. doi: 10.1074/jbc.274.51.36369. [DOI] [PubMed] [Google Scholar]
- 76.Wurtele M, Renault L, Barbieri JT, Wittinghofer A, Wolf E. Structure of the ExoS GTPase activating domain. FEBS Lett. 2001;491(1–2):26–29. doi: 10.1016/s0014-5793(01)02105-6. [DOI] [PubMed] [Google Scholar]
- 77.Fu Y, Galan JE. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature. 1999;401(6750):293–297. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- 78.Kaniga K, Uralil J, Bliska JB, Galan JE. A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium . Mol. Microbiol. 1996;21(3):633–641. doi: 10.1111/j.1365-2958.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 79.Black DS, Bliska JB. The RhoGap activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol. Microbiol. 2000;37(3):515–527. doi: 10.1046/j.1365-2958.2000.02021.x. [DOI] [PubMed] [Google Scholar]
- 80.Evdokimov AG, Tropea JE, Routzahn KM, Waugh DS. Crystal structure of the Yersinia pestis GTPase activator YopE. Protein Sci. 2002;11(2):401–408. doi: 10.1110/ps.34102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coburn J, Dillon ST, Iglewski BH, Gill DM. Exoenzyme S of Pseudomonas aeruginosa ADP-ribosylates the intermediate filament protein vimentin. Infect. Immun. 1989;57(3):996–998. doi: 10.1128/iai.57.3.996-998.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coburn J, Wyatt RT, Iglewski BH, Gill DM. Several GTP-binding proteins, including p21c–h-Ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J. Biol. Chem. 1989;264(15):9004–9008. [PubMed] [Google Scholar]
- 83.Ganesan AK, Frank DW, Misra RP, Schmidt G, Barbieri JT. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J. Biol. Chem. 1998;273(13):7332–7337. doi: 10.1074/jbc.273.13.7332. [DOI] [PubMed] [Google Scholar]
- 84.Ganesan AK, Vincent TS, Olson JC, Barbieri JT. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor-catalyzed nucleotide exchange. J. Biol. Chem. 1999;274(31):21823–21829. doi: 10.1074/jbc.274.31.21823. [DOI] [PubMed] [Google Scholar]
- 85.Sun J, Barbieri JT. Pseudomonas aeruginosa ExoT ADP-ribosylates ct10 regulator of kinase (Crk) proteins. J. Biol. Chem. 2003;278(35):32794–32800. doi: 10.1074/jbc.M304290200. [DOI] [PubMed] [Google Scholar]
- 86.Navarro L, Koller A, Nordfelth R, Wolf-Watz H, Taylor S, Dixon JE. Identification of a molecular target for the Yersinia protein kinase A. Mol. Cell. 2007;26(4):465–477. doi: 10.1016/j.molcel.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 87.Prehna G, Ivanov MI, Bliska JB, Stebbins CE. Yersinia virulence depends on mimicry of host Rho-family nucleotide dissociation inhibitors. Cell. 2006;126(5):869–880. doi: 10.1016/j.cell.2006.06.056. [DOI] [PubMed] [Google Scholar]
- 88.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450(7168):365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 89.Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318(5852):974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- 90.Zhu Y, Hu L, Zhou Y, Yao Q, Liu L, Shao F. Structural mechanism of host Rab1 activatior by the bifunctional Legionella type IV effecto SidM/DrrA. Proc. Natl Acad. Set. USA. 2010;107(10):4699–4704. doi: 10.1073/pnas.0914231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sheahan Kl, Satchell KJ. Inactivation of small Rho GTPases by the multifunctional RTX toxin from Vibrio cholerae . Cell Microbiol. 2007;9(5):1324–1335. doi: 10.1111/j.1462-5822.2006.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Geissler B, Bonebrake A, Sheahan KL, Walker ME, Satchell KJ. Genetic determination of essential residues of the Vibrio cholerae actin cross-linking domain reveals functional similarity with glutamine synthetases. Mol. Microbiol. 2009;73(5):858–868. doi: 10.1111/j.1365-2958.2009.06810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kudryashov DS, Cordero CL, Reisler E, Satchell KJ. Characterization of the enzymatic activity of the actin cross-linking domain from the Vibrio cholerae MARTX Vc toxin. J. Biol. Chem. 2008;283(1):445–452. doi: 10.1074/jbc.M703910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kudryashov DS, Durer ZA, Ytterberg AJ, et al. Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin. Proc. Natl Acad. Sci. USA. 2008;105(47):18537–18542. doi: 10.1073/pnas.0808082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prochazkova K, Satchell KJ. Structure–function analysis of inositol hexakisphosphate-induced autoprocessing of the Vibrio cholerae multifunctional autoprocessing RTx toxin. J. Biol. Chem. 2008;283(35):23656–23664. doi: 10.1074/jbc.M803334200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Prochazkova K, Shuvalova LA, Minasov G, Voburka Z, Anderson WF, Satchell KJ. Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites. J. Biol. Chem. 2009;284(39):26557–26568. doi: 10.1074/jbc.M109.025510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shen A, Lupardus PJ, Albrow VE, et al. Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin. Nat. Chem. Biol. 2009;5(7):469–478. doi: 10.1038/nchembio.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Flatau G, Landraud L, Boquet P, Bruzzone M, Munro P. Deamidation of RhoA glutamine 63 by the Escherichia coli CNF1 toxin requires a short sequence of the GTPase switch 2 domain. Biochem. Biophys. Res. Commun. 2000;267(2):588–592. doi: 10.1006/bbrc.1999.1904. [DOI] [PubMed] [Google Scholar]
- 99.Flatau G, Lemichez E, Gauthier M, et al. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387(6634):729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 100.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gin 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387(6634):725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 101.Lerm M, Schmidt G, Goehring UM, Schirmer J, Aktories K. Identification of the region of Rho involved in substrate recognition by Escherichia coli cytotoxic necrotizing factor 1 (CNF1) J. Biol. Chem. 1999;274(41):28999–29004. doi: 10.1074/jbc.274.41.28999. [DOI] [PubMed] [Google Scholar]
- 102.Hoffmann C, Pop M, Leemhuis J, Schirmer JA, ktories K, Schmidt G. The Yersinia pseudotuberculosis cytotoxic necrotizing factor (CNFy) selectively activates RhoA. J. Biol. Chem. 2004;279(16):16026–16032. doi: 10.1074/jbc.M313556200. [DOI] [PubMed] [Google Scholar]
- 103.Schmidt G, Selzer J, Lerm M, Aktories K. The Rho-deamidating cytotoxic necrotizing factor 1 from Escherichia coli possesses transglutaminase activity. Cysteine 866 and histidine 881 are essential for enzyme activity. J. Biol. Chem. 1998;273(22):13669–13674. doi: 10.1074/jbc.273.22.13669. [DOI] [PubMed] [Google Scholar]
- 104.Horiguchi Y. Escherichia coli cytotoxic necrotizing factors and Bordetella dermonecrotic toxin: the dermonecrosis-inducing toxins activating Rho small GTPases. Toxicon. 2001;39(11):1619–1627. doi: 10.1016/s0041-0101(01)00149-0. [DOI] [PubMed] [Google Scholar]
- 105.Masuda M, Minami M, Shime H, Matsuzawa T, Horiguchi Y. In vivo modifications of small GTPase Rac and Cdc42 by Bordetella dermonecrotic toxin. Infect. Immun. 2002;70(2):998–1001. doi: 10.1128/iai.70.2.998-1001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schmidt G, Goehring UM, Schirmer J, Lerm M, Aktories K. Identification of the C-terminal part of Bordetella dermonecrotic toxin as a transglutaminase for Rho GTPases. J. Biol. Chem. 1999;274(45):31875–31881. doi: 10.1074/jbc.274.45.31875. [DOI] [PubMed] [Google Scholar]
- 107.Masuda M, Betancourt L, Matsuzawa T, et al. Activation of Rho through a cross-link with polyamines catalyzed by Bordetella dermonecrotizing toxin. EMBOJ. 2000;19(4):521–530. doi: 10.1093/emboj/19.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schmidt G, Goehring UM, Schirmer J, et al. Lysine and polyamines are substrates for transglutamination of Rho by the Bordetella dermonecrotic toxin. Infect. Immun. 2001;69(12):7663–7670. doi: 10.1128/IAI.69.12.7663-7670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Buetow L, Flatau G, Chiu K, Boquet P, Ghosh P. Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1. Nat. Struct. Biol. 2001;8(7):584–588. doi: 10.1038/89610. ▪▪ Crystal structure of cytotoxic necrotizing factor deamidase domain.
- 110.Jank T, Pack U, Giesemann T, Schmidt G, Aktories K. Exchange of a single amino acid switches the substrate properties of RhoA and RhoD toward glucosylating and transglutaminating toxins. J. Biol. Chem. 2006;281(28):19527–19535. doi: 10.1074/jbc.M600863200. [DOI] [PubMed] [Google Scholar]
- 111.Rhee SG, Choi KD. Regulation of inositol phospholipid-specific phospholipase C isozymes. J. Biol. Chem. 1992;267(18):12393–12396. [PubMed] [Google Scholar]
- 112.Sternweis PC, Smrcka AV. Regulation of phospholipase C by G proteins. Trends Biochem. Sci. 1992;17(12):502–506. doi: 10.1016/0968-0004(92)90340-f. [DOI] [PubMed] [Google Scholar]
- 113.Strathmann M, Simon MI. G protein diversity: a distinct class of α subunits is present in vertebrates and invertebrates. Proc. Natl Acad. Sci. USA. 1990;87(23):9113–9117. doi: 10.1073/pnas.87.23.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wilson BA, Zhu X, Ho M, Lu L. Pasteurella multocida toxin activates the inositol triphosphate signaling pathway in Xenopus oocytes via G(q)α-coupled phospholipase C-β1. J. Biol. Chem. 1997;272(2):1268–1275. doi: 10.1074/jbc.272.2.1268. ▪ First identification of Gαq protein as the intracellular target of PMT, providing a new molecular tool for studying heterotrimeric G proteins.
- 115.Zywietz A, Gohla A, Schmelz M, Schultz G, Offermanns S. Pleiotropic effects of Pasteurella multocida toxin are mediated by Gq-dependent and -independent mechanisms. Involvement of Gq but not G11. J. Biol. Chem. 2001;276(6):3840–3845. doi: 10.1074/jbc.M007819200. [DOI] [PubMed] [Google Scholar]
- 116.Aminova LR, Wilson BA. Calcineurin-independent inhibition of 3T3-L1 adipogenesis by Pasteurella multocida toxin: suppression of notch1, stabilization of β-catenin and pre-adipocyte factor 1. Cell Microbiol. 2007;9(10):2485–2496. doi: 10.1111/j.1462-5822.2007.00975.x. [DOI] [PubMed] [Google Scholar]
- 117.Luo S, Ho M, Wilson BA. Application of intact cell-based NFAT-β-lactamase reporter assay for Pasteurella multocida toxin-mediated activation of calcium signaling pathway. Toxicon. 2008;51(4):597–605. doi: 10.1016/j.toxicon.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mullan PB, Lax AJ. Pasteurella multocida toxin stimulates bone resorption by osteoclasts via interaction with osteoblasts. Calcif. Tissue Int. 1998;63(4):340–345. doi: 10.1007/s002239900537. [DOI] [PubMed] [Google Scholar]
- 119.Murphy AC, Rozengurt E. Pasteurella multocida toxin selectively facilitates phosphatidylinositol 4,5-bisphosphate hydrolysis by bombesin, vasopressin, and endothelin. Requirement for a functional G protein. J. Biol. Chem. 1992;267(35):25296–25303. [PubMed] [Google Scholar]
- 120.Staddon JM, Barker CJ, Murphy AC, et al. Pasteurella multocida toxin, a potent mitogen, increases inositol 1,4,5-trisphosphate and mobilizes Ca2+ in Swiss 3T3 cells. J. Biol. Chem. 1991;266(8):4840–4847. [PubMed] [Google Scholar]
- 121.Staddon JM, Chanter N, Lax AJ, Higgins TE, Rozengurt E. Pasteurella multocida toxin, a potent mitogen, stimulates protein kinase C-dependent and -independent protein phosphorylation in Swiss 3T3 cells. J. Biol. Chem. 1990;265(20):11841–11848. [PubMed] [Google Scholar]
- 122.Orth JH, Aktories K, Kubatzky KF. Modulation of host cell gene expression through activation of STAT transcription factors by Pasteurella multocida toxin. J. Biol. Chem. 2007;282(5):3050–3057. doi: 10.1074/jbc.M609018200. [DOI] [PubMed] [Google Scholar]
- 123.Rozengurt E, Higgins T, Chanter N, Lax AJ, Staddon JM. Pasteurella multocida toxin: potent mitogen for cultured fibroblasts. Proc. Natl Acad. Sci. USA. 1990;87(1):123–127. doi: 10.1073/pnas.87.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sabri A, Wilson BA, Steinberg SF. Dual actions of the Gα(q) agonist Pasteurella multocida toxin to promote cardiomyocyte hypertrophy and enhance apoptosis susceptibility. Circ. Res. 2002;90(8):850–857. doi: 10.1161/01.RES.0000016165.23795.1F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Seo B, Choy EW, Maudsley S, Miller WE, Wilson BA, Luttrell LM. Pasteurella multocida toxin stimulates mitogen-activated protein kinase via G(q/11)-dependent transactivation of the epidermal growth factor receptor. J. Biol. Chem. 2000;275(3):2239–2245. doi: 10.1074/jbc.275.3.2239. [DOI] [PubMed] [Google Scholar]
- 126.Wilson BA, Aminova LR, Ponferrada VG, Ho M. Differential modulation and subsequent blockade of mitogenic signaling and cell cycle progression by Pasteurella multocida toxin. Infect. Immun. 2000;68(8):4531–4538. doi: 10.1128/iai.68.8.4531-4538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aepfelbacher M, Essler M. Disturbance of endothelial barrier function by bacterial toxins and atherogenic mediators: a role for Rho/Rho kinase. Cell Microbiol. 2001;3(10):649–658. doi: 10.1046/j.1462-5822.2001.00145.x. [DOI] [PubMed] [Google Scholar]
- 128.Essler M, Hermann K, Amano M, et al. Pasteurella multocida toxin increases endothelial permeability via Rho kinase and myosin light chain phosphatase. J. Immunol. 1998;161(10):5640–5646. [PubMed] [Google Scholar]
- 129.Lacerda HM, Lax AJ, Rozengurt E. Pasteurella multocida toxin, a potent intracellularly acting mitogen, induces p125fak and paxillin tyrosine phosphorylation, actin stress fiber formation, and focal contact assembly in swiss 3T3 cells. J. Biol. Chem. 1996;271(1):439–445. doi: 10.1074/jbc.271.1.439. [DOI] [PubMed] [Google Scholar]
- 130.Ohnishi T, Horiguchi Y, Masuda M, Sugimoto N, Matsuda M. Pasteurella multocida toxin and Bordetella hronchiseptica dermonecrotizing toxin elicit similar effects on cultured cells by different mechanisms. J. Vet. Med. Sci. 1998;60(3):301–305. doi: 10.1292/jvms.60.301. [DOI] [PubMed] [Google Scholar]
- 131.Sagi SA, Seasholtz TM, Kobiashvili M, Wilson BA, Toksoz D, Brown JH. Physical and functional interactions of Gαq with Rho and its exchange factors. J. Biol. Chem. 2001;276(18):15445–15452. doi: 10.1074/jbc.M008961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Thomas W, Pullinger GD, Lax AJ, Rozengurt E. Escherichia coli cytotoxic necrotizing factor and Pasteurella multocida toxin induce focal adhesion kinase autophosphorylation and Src association. Infect. Immun. 2001;69(9):5931–5935. doi: 10.1128/IAI.69.9.5931-5935.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Orth JH, Lang S, Preuss I, Milligan G, Aktories K. Action of Pasteurella multocida toxin on Gα(q) is persistent and independent of interaction with G-protein-coupled receptors. Cell Signal. 2007;19(10):2174–2182. doi: 10.1016/j.cellsig.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 134.Orth JH, Fester I, Preuss I, Agnoletto L, Wilson BA, Aktories K. Activation of Gα (i) and subsequent uncoupling of receptor-Gα(i) signaling by Pasteurella multocida toxin. J. Biol. Chem. 2008;283(34):23288–23294. doi: 10.1074/jbc.M803435200. [DOI] [PubMed] [Google Scholar]
- 135.Preuss I, Kurig B, Nurnberg B, Orth JH, Aktories K. Pasteurella multocida toxin activates Gβγ dimers of heterotrimeric G proteins. Cell Signal. 2009;21(4):551–558. doi: 10.1016/j.cellsig.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 136.Katada T, Oinuma M, Ui M. Two guanine nucleotide-binding proteins in rat brain serving as the specific substrate of islet-activating protein, pertussis toxin. Interactior of the α-subunits with β γ-subunits in development of their biological activities. J. Biol. Chem. 1986;261(18):8182–8191. [PubMed] [Google Scholar]
- 137.Orth JH, Lang S, Taniguchi M, Aktories K. Pasteurella multocida toxin-induced activatior of RhoA is mediated via two families of G{α} proteins, G{α}q and G{α}12/13. J. Biol. Chem. 2005;280(44):36701–36707. doi: 10.1074/jbc.M507203200. [DOI] [PubMed] [Google Scholar]
- 138.Buhl AM, Johnson NL, Dhanasekaran N, Johnson GL. Gα 12 and G α 13 stimulate Rho-dependent stress fiber formation and focal adhesion assembly. J. Biol. Chem. 1995;270(42):24631–24634. doi: 10.1074/jbc.270.42.24631. [DOI] [PubMed] [Google Scholar]
- 139.Goulimari P, Knieling H, Engel U, Grosse R. Larg and Mdia1 link Gct12/13 to cell polarity and microtubule dynamics. Mol Biol. Cell. 2008;19(1):30–40. doi: 10.1091/mbc.E06-11-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jin S, Exton JH. Activation of RhoA by association of Gα(13) with DBL. Biochem. Biophys. Res. Commun. 2000;277(3):718–721. doi: 10.1006/bbrc.2000.3744. [DOI] [PubMed] [Google Scholar]
- 141.Tanabe S, Kreutz B, Suzuki N, Kozasa T. Regulation of RGS-RhoGEFs by Gα12 and Gα13 proteins. Methods Enzymol. 2004;390:285–294. doi: 10.1016/S0076-6879(04)90018-3. [DOI] [PubMed] [Google Scholar]
- 142.Yamaguchi Y, Katoh H, Negishi M. N-terminal short sequences of α subunks of the G12 family determine selective coupling to receptors. J. Biol. Chem. 2003;278(17):14936–14939. doi: 10.1074/jbc.M301409200. [DOI] [PubMed] [Google Scholar]
- 143.Orth JH, Lang S, Aktories K. Action of Pasteurella multocida toxin depends on the helical domain of Gαq. J. Biol. Chem. 2004;279(33):34150–34155. doi: 10.1074/jbc.M405353200. [DOI] [PubMed] [Google Scholar]
- 144.Kashimoto T, Katahira J, Cornejo WR, et al. Identification of functional domains of Bordetella dermonecrotizing toxin. Infect. Immun. 1999;67(8):3727–3732. doi: 10.1128/iai.67.8.3727-3732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lemichez E, Flatau G, Bruzzone M, Boquet P, Gauthier M. Molecular localization of the Escherichia coli cytotoxic necrotizing factor CNF1 cell-binding and catalytic domains. Mol. Microbiol. 1997;24(5):1061–1070. doi: 10.1046/j.1365-2958.1997.4151781.x. [DOI] [PubMed] [Google Scholar]
- 146.Pullinger GD, Sowdhamini R, Lax AJ. Localization of functional domains of the mitogenic toxin of Pasteurella multocida . Infect. Immun. 2001;69(12):7839–7850. doi: 10.1128/IAI.69.12.7839-7850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Aminova LR, Luo S, Bannai Y, Ho M, Wilson BA. The C3 domain of Pasteurella multocida toxin is the minimal domain responsible for activation of Gq-dependent calcium and mitogenic signaling. Protein Sci. 2008;17(5):945–949. doi: 10.1110/ps.083445408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kitadokoro K, Kamitani S, Miyazawa M, et al. Crystal structures reveal a thiol protease-like catalytic triad in the C-terminal region of Pasteurella multocida toxin. Proc. Natl Acad. Sci. USA. 2007;104(12):5139–5144. doi: 10.1073/pnas.0608197104. ▪▪ Crystal structure of Pasteurella multocida toxin-C3 active fragment and its structural comparison to other members of the Cys-His-Asp triad-containing cysteine protease family.
- 149.Sugai M, Hatazaki K, Mogami A, et al. Cytotoxic necrotizing factor type 2 produced by pathogenic Escherichia coli deamidates a Gln residue in the conserved G-3 domain of the Rho family and preferentially inhibits the GTPase activity of RhoA and Rac1. Infect. Immun. 1999;67(12):6550–6557. doi: 10.1128/iai.67.12.6550-6557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kamtiani S, Kitadokoro K, Miyazawa M, et al. Characterization of the membrane-targeting C1 domain in Pasteurella multocida toxin. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.102285. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Busch C, Orth J, Djouder N, Aktories K. Biological activity of a C-terminal fragment of Pasteurella multocida toxin. Infect. Immun. 2001;69(6):3628–3634. doi: 10.1128/IAI.69.6.3628-3634.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Orth JH, Blocker D, Aktories K. His1205 and His1223 are essential for the activity of the mitogenic Pasteurella multocida toxin. Biochemistry. 2003;42(17):4971–4977. doi: 10.1021/bi0272959. [DOI] [PubMed] [Google Scholar]
- 153. Ginalski K, Kinch L, Rychlewski L, Grishin NV. BTLCP proteins: a novel family of bacterial transglutaminase-like cysteine proteinases. Trends Biochem. Sci. 2004;29(8):392–395. doi: 10.1016/j.tibs.2004.06.001. ▪▪ Reviews the family of transglutaminase-like cysteine proteases.
- 154.Yee VC, Pedersen LC, Bishop PD, Stenkamp RE, Teller DC. Structural evidence that the activation peptide is not released upon thrombin cleavage of factor XIII. Thromb. Res. 1995;78(5):389–397. doi: 10.1016/0049-3848(95)00072-y. [DOI] [PubMed] [Google Scholar]
- 155.Noguchi K, Ishikawa K, Yokoyama K, Ohtsuka T, Nio N, Suzuki E. Crystal structure of red sea bream transglutaminase. J. Biol. Chem. 2001;276(15):12055–12059. doi: 10.1074/jbc.M009862200. [DOI] [PubMed] [Google Scholar]
- 156.Yamaguchi S, Jeenes DJ, Archer DB. Protein-glutaminase from Chryseobacterium proteolyticum, an enzyme that deamidates glutaminyl residues in proteins. Purification characterization and gene cloning. Eur. J. Biochem. 2001;268(5):1410–1421. doi: 10.1046/j.1432-1327.2001.02019.x. [DOI] [PubMed] [Google Scholar]
- 157.Sinclair JC, Sandy J, Delgoda R, Sim E, Noble ME. Structure of arylamine N-acetyltransferase reveals a catalytic triad. Nat. Struct. Biol. 2000;7(7):560–564. doi: 10.1038/76783. [DOI] [PubMed] [Google Scholar]
- 158.Kashiwagi T, Yokoyama K, Ishikawa K, et al. Crystal structure of microbial transglutaminase from Streptoverticillium mobaraense . J. Biol. Chem. 2002;277(46):44252–44260. doi: 10.1074/jbc.M203933200. [DOI] [PubMed] [Google Scholar]
- 159.Yokoyama K, Nio N, Kikuchi Y. Properties and applications of microbial transglutaminase. Appl. Microbiol. Biotechnol. 2004;64(4):447–454. doi: 10.1007/s00253-003-1539-5. [DOI] [PubMed] [Google Scholar]
- 160.Chao X, Muff TJ, Park SY, et al. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell. 2006;124(3):561–571. doi: 10.1016/j.cell.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 161.Kim Y, Maltseva N, Dementieva I, Collart F, Holzle D, Joachimiak A. Crystal structure of hypothetical protein YfiH from Shigella flexneri at 2 Å resolution. Proteins. 2006;63(4):1097–1101. doi: 10.1002/prot.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Lax AJ. Opinion: bacterial toxins and cancer – a case to answer? Nat. Rev. Microbiol. 2005;3(4):343–349. doi: 10.1038/nrmicro1130. ▪ Proposal for the connection between toxin exposure and cancer development.