Abstract
Swine influenza virus and Streptococcus suis are two important contributors to the porcine respiratory disease complex, and both have significant economic impacts. Clinically, influenza virus and Streptococcus suis co-infections in pigs are very common, which often contribute to severe pneumonia and can increase the mortality. However, the co-infection pathogenesis in pigs is unclear. In the present study, co-infection experiments were performed using swine H1N1 influenza virus and Streptococcus suis serotype 2 (SS2). The H1N1-SS2 co-infected pigs exhibited more severe clinical symptoms, serious pathological changes, and robust apoptosis of lungs at 6 days post-infection compared with separate H1N1 and SS2 infections. A comprehensive gene expression profiling using a microarray approach was performed to investigate the global host responses of swine lungs against the swine H1N1 infection, SS2 infection, co-infection, and phosphate-buffered saline control. Results showed 457, 411, and 844 differentially expressed genes in the H1N1, SS2, and H1N1-SS2 groups, respectively, compared with the control. Noticeably, genes associated with the immune, inflammatory, and apoptosis responses were highly overexpressed in the co-infected group. Pathway analysis indicated that the cytokine–cytokine receptor interactions, MAPK, toll-like receptor, complement and coagulation cascades, antigen processing and presentation, and apoptosis pathway were significantly regulated in the co-infected group. However, the genes related to these were less regulated in the separate H1N1 and SS2 infection groups. This observation suggested that a certain level of synergy was induced by H1N1 and SS2 co-infection with significantly stronger inflammatory and apoptosis responses, which may lead to more serious respiratory disease syndrome and pulmonary pathological lesion.
Introduction
Swine influenza is a highly infectious acute respiratory viral disease of pigs that affects the respiratory tract and has considerable economic impacts [1]. Three main subtypes of swine influenza virus (H1N1, H3N2, and H1N2), with H1N1 as the predominant subtype, have circulated in pigs worldwide [2, 3]. In March 2009, a new swine-origin H1N1 influenza virus became a pandemic [4]. Pig infections with the new H1N1 virus have then been observed in multiple countries, showing that the pandemic H1N1 viruses have become established in swine populations [5–7]. Previous study has showed the new H1N1 viruses have spread from humans to pigs in China [8]. Swine influenza virus replication is mainly restricted to the epithelial cells in the respiratory tract, with the lung being the major target organ. Although it is a highly contagious virus for pigs and has high-morbidity but low-mortality rates, secondary complications would substantially worsen the illness and increase death rate [9]. In fact, swine influenza is one of the several significant contributors to the porcine respiratory disease complex (PRDC), which is caused by infection with more than one pathogen, such as the swine influenza virus and Streptococcus suis (S. suis) co-infection.
S. suis infections have been considered as a major problem worldwide in the swine industry and as a secondary agent of pneumonia, particularly in the past 20 years [10]. Among the 35 serotypes, serotype 2 (SS2) is generally considered as the most prevalent and virulent type [10]. S. suis infections in pigs often cause arthritis, meningitis, pneumonia, endocarditis, and septicemia with or without sudden death. Although S. suis is a major swine pathogen, it has been increasingly detected in wide range of mammalian species. Infections have been observed in humans in 2005 in China, which affected more than 200 people and had approximately 20% mortality [11]. In clinical cases, co-infections of swine influenza virus and S. suis in pigs often contribute to severe pneumonia and can increase the mortality. Co-infection outbreaks have been recently reported in England [12].
Recently, several studies on the pathogenesis of the co-infection of influenza virus and Streptococcus pneumonia have been performed using mouse models [13–15]. Pro- and anti-inflammatory (IL-6, IL-1β, TNF-α, and IL-10) molecules were remarkably elevated in the blood in influenza virus and Streptococcus pneumonia co-infected mouse [16]. However, fewer studies have examined swine influenza and S. suis co-infection in pigs, and its pathogenesis is not yet fully elucidated. In the present study, microarray assay was utilized to explore the global host responses of porcine lungs that suffered from H1N1 influenza virus, SS2, H1N1-SS2 co-infection, and phosphate-buffered saline (PBS) to enhance the understanding of the H1N1 and S. suis co-infection pathogenesis through a pig model. Stronger inflammatory and apoptosis responses were determined to be important contributors to the increased pathogenicity caused by swine H1N1 and SS2 co-infection. Our study would improve the understanding of the pathogenesis of H1N1 and SS2 co-infection in pigs.
Materials and Methods
Ethics statement
The research protocol of animal experiment in this study was approved by the Biological Studies Animal Care and Use Committee in Hubei Province, China.
Viruses, bacteria, and cells
Viruses used in this study were A/swine/Hubei/101/2009 (H1N1) as previously described [17], and quantified as egg infectious dose 50 (EID50) in special pathogen-free chicken eggs. Madin-Darby canine kidney cells (MDCK) and Porcine kidney cells (PK-15) were obtained from the China Center for Type Culture Collection (CTCC, Wuhan, China) and cultured in Dulbecco's modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactived fetal bovine serum (FBS) (HyClone, Logan, UT), 100 units/mL penicillin and 100 μg/mL streptomycin (Invitrogen) at 37°C with 5% CO2. An SS2 strain, 05ZY, was isolated from the brain of a diseased piglet in Sichuan Province, China in 2005. Colony forming units (CFU) were counted through serial dilutions of bacteria on tryptic soy agar (TSA) plates supplemented with 5% (v/v) FBS.
Infection experiment
The pigs used in the present study were raised in isolated facilities and determined to be negative for influenza virus antibody and SS2 antibody by analyzing the pig sera with hemagglutination inhibition test and enzyme-linked immunosorbent assay, respectively. The pigs were randomly divided into four groups with three pigs per group. The assigned groups were as follows: H1N1 infection alone (H1N1), SS2 infection alone (SS2), H1N1 and SS2 co-infection (H1N1-SS2) and PBS control. Considering the animal welfare and avoiding the waste, H1N1 and control groups were the same as those used in our previous study [17], because the two experiments were performed at the same time and under same conditions. Each group was housed in an isolated room. At 35 day of age, the H1N1 and H1N1-SS2 groups were inoculated intranasally with 2 mL of 107.0 EID50/mL of the H1N1 swine influenza virus, and the other pigs were inoculated with PBS. On day 3 of the experiment (3 d after H1N1 challenge), the SS2 and H1N1-SS2 groups were inoculated intranasally with 2 mL of PBS containing 106.0 CFU/mL SS2 strain ZY05. The H1N1 and control groups were treated similarly with PBS. Considering the biological features of swine H1N1 virus and to be much closer to the natural infections, we performed the H1N1 infection first and then SS2 infection. All pigs were monitored daily for rectal temperature and clinical signs. The nasal swabs were obtained from the right nostrils every other day using sterile cotton swabs. On day 6 of the experiment, all pigs were humanely euthanized by intravenous injection of 60 mg/kg body weight sodium pentobarbital. Lungs from all groups were immediately obtained. The macroscopic lesions of lung tissues were estimated visually, and scored blindly by a pathologist. Lesion areas in left caudal lobes of the lungs were immediately frozen and stored in liquid nitrogen after collection until RNA extraction, and another area in the same lobes was used for isolating the virus and bacteria. Lesion areas in right caudal lobes were fixed in formalin and embedded in paraffin, sectioned at 5μm, and stained with hematoxylin and eosin for further histopathologic evaluation. Slides were randomized, and read blindly by an experienced pathologist. At least 6 sections per lung from each group were examined microscopically for tissue damage, necrosis, apoptosis, and inflammatory cellular infiltration.
Virus and bacteria isolation
At specified time points, the collected nasal swabs were inserted into vials containing 1.5 mL of sterile PBS supplemented with 100 U/mL penicillin and streptomycin. After thorough vortex, the vials were centrifuged at 12,000 rpm at 4°C for 10 min. The supernatants were collected, and the virus titer was quantified as EID50. To prevent cross-contamination, separate sterile instruments were used to collect the lungs of pigs from each group, which were homogenized in sterile PBS and then centrifuged to collect the supernatant. Virus titer was quantified as EID50, and SS2 content was determined by incubation on TSA plates supplemented with 5% (v/v) FBS for CFU counting.
Agilent microarray experiment
The same areas in the lungs from each group were sampled for RNA extraction using TRIzol (Invitrogen, Carlsbad, CA) and RNaeasy MiNi Kit (QIAGEN) according to the manufacturer’s instruction. The microarray experiments were performed as previously described [17]. RNA labeling and hybridization were performed using a commercial Agilent array (Shanghaibio, China) that followed the standard one-cycle protocol according to the manufacturer’s instructions. Transcriptional profiles were assessed using Agilent 4x44K Porcine Oligo Microarray. The hybridization and scanning of the arrays were conducted using a G2565BA Scanner (Agilent Technologies, Palo Alto, CA) according to the standard protocol. The hybridization data were normalized using Agilent Feature Extraction Software. ANOVA was used to analyze the data. The raw and processed microarray data of H1N1 and control groups used here were described as our previous study [17], because these two experiments were performed under same conditions. All differentially expressed (DE) genes were then examined using hierarchical cluster (Ver. 3.0) and TreeView (Ver. 1.1.1) analyses [18]. Genes with significant similarities (greater than 70%) to transcripts in the nr database which were functionally annotated based on BLASTX searches were selected for GO and pathway analyses using MAS 3.0 software, which is based on DAVID and KEGG databases (CapitalBio, Beijing, China) [19]. All DE genes were identified by setting a fold change (FC) > 1.5 and p < 0.05 as the criteria.
Real-time quantitative PCR (RT-qPCR)
RT-qPCR was used to validate the expression data. RNA was extracted using TRIzol (Invitrogen). RNA (0.5 μg) was reversely transcribed in a 20μl reaction mixture containing 2 μl avian myeloblastosis virus (AMV) buffer, 50 pM Oligo18T, 0.5 mM dNTPs, 10 U RNase inhibitor and 10 U AMV reverse transcriptase (TAKARA, Japan), and the gene expression was monitored through a Power SYBR Green PCR master mix kit (Applied Biosystems) with corresponding primers. The fluorescence signals were measured using the ABI ViiA7 PCR system (Applied Biosystems). GAPDH was used as the internal control, and all gene expressions were normalized to GAPDH. The primers used are listed in S1 Data.
Microarray Data Accession Number
The raw and processed data in this study have been deposited in NCBI’s Gene expression omnibus and are accessible through GEO Series accession number GSE60172.
Statistical analysis
Standard two-sample t-tests were used to compare virus titers, gene expressions, lung pathological damage score, and the apoptosis rate among infection groups. All data are expressed as the mean ± SEM. A value of P < 0.05 was considered statistically significant.
Results
Clinical signs and pathological evaluation
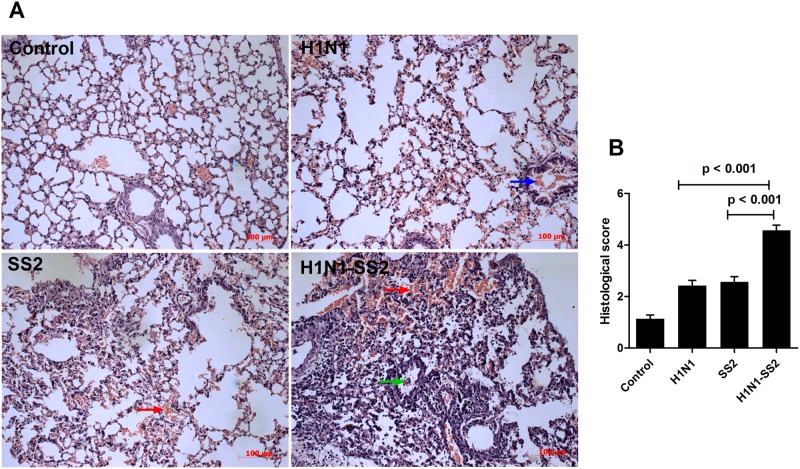
To assess the infection process, clinical signs were observed daily starting from 1 d before challenge (day 0) (Table 1). The H1N1 and H1N1-SS2 groups experienced fever, which started on day 1, reaching to 40.8°C and 40.9°C, respectively, on day 3. In the H1N1 group, coughing, nasal discharge, and anorexia were observed. Clinical signs recovered to normal on day 6. However, in the H1N1-SS2 co-infection group, in addition to the signs observed in H1N1 group, pigs also exhibited high fever (41.5°C, 41.8°C, and 41.5°C) on day 6. Moreover, limp and arthrocele, which are the typical symptoms of SS2 infection, were observed. In the SS2 infection group, all pigs did not show any abnormal sign before inoculating with SS2. However, 3 d after SS2 infection (day 6), the SS2 group exhibited typical SS2 infection symptoms, such as lameness, joint swelling, lethargy to different extents, and one pig exhibited fever over 40.5°C. Notably, the H1N1-SS2 co-infection group exhibited more severe symptoms compared with the SS2 infection alone group. To investigate pathological damage to lungs of infected pigs, pigs were euthanized on day 6, and the macroscopic lesions of lungs tissues were estimated visually (S2 Data). Much more serious lung pathological damage was caused by co-infection, including hyperemia, necrosis, bleeding, and consolidation, compared with separate infections. Microscopic pathological changes in the lungs were also evaluated (Fig 1A, S3 Data (Control), S4 Data (H1N1), S5 Data (SS2), and S6 Data (H1N1-SS2)). All groups except the control exhibited different degrees of pathological damage. Extensive lung damages (that were, extensive cellular infiltrates, bleeding, and cellular debris) were observed in H1N1-SS2 group. However, H1N1 and SS2 groups showed lesser lung damages. These observations were supported by blinded histological scoring (Fig 1B). These results demonstrated that H1N1 and SS2 co-infection increased the pathogenicity.
Table 1. Clinical signs after infection in each group.
| Group | Clinical signs | |
|---|---|---|
| Days 1–3 | Days 4–6 | |
| Control | N. | N. |
| H1N1 | Fever (3/3), cough (1/3), sneeze (1/3), snotty (2/3). | Fever (0/3), cough (1/3), sneeze (0/3), snotty (0/3). |
| SS2 | N. | Fever (2/3), depressed (2/3), limp (2/3), arthrocele (1/3). |
| H1N1-SS2 | Fever (3/3), cough (1/3), sneeze (1/3), snotty (1/3). | High fever (2/3), hypothermia (1/3), limp (2/3), recumbent (1/3), arthrocele (2/3), cough (1/3), sneeze (2/3), snotty (0/3). |
Clinical signs in each infection group were monitored daily. N: No obvious clinical sign was observed. X/X refers to morbidity/total.
Fig 1. Histopathologic changes in lungs of infected pigs.
Pigs were PBS-control infected or infected with H1N1, SS2 and H1N1-SS2 as described in methods. On day 6 of the experiment, lungs were removed and underwent Hematoxylin and eosin stain. (A) The microscopic lesions of lung tissues from each group showed different extent of acute pneumonia with pathological changes: alveolar wall thickening, bleeding (red arrow), debris in the lumen (green arrow), erythrocyte effusion (blue arrow), and the accumulation of inflammatory cells. (B) Histological score of sections of lungs in pigs from each group were showed as mean ± SEM by two-tailed Student’s t-test. P-value less than 0.05 was considered to represent a statistically significant difference.
Virus and bacteria isolation
The swabs, which were used for virus isolation, were collected from all pigs every 2 d after inoculation. As expected, virus were isolated from the pigs in the H1N1 and H1N1-SS2 groups on days 2, 4, and 6 (Fig 2). The virus titer in the H1N1-SS2 groups was slightly but significantly higher than that in the H1N1 group on day 6. On day 6 after inoculation, all pigs were humanely euthanized, and viruses in lungs were also detected in both the H1N1 and H1N1-SS2 groups. The virus titer in the H1N1-SS2 group was also significantly higher than that in H1N1 group (Fig 2). No virus was isolated from the SS2 and control groups. However, no significant difference was observed on the bacterial loads in the lungs between the H1N1-SS2 and SS2 groups (data are not shown). No bacteria were detected in the H1N1 and control groups.
Fig 2. Virus isolation of nasal swabs and lungs in each group.

Swabs were collected from the right nostrils of pigs from each group every other day. The obtained nasal swabs were inserted into vials containing 1.5 mL of sterile PBS. Supernatants were collected and viruses were qualified as LgEID50. At the day 6 of the experiment, all pigs were humanely euthanized and lungs were collected and homogenized in sterile PBS and then centrifuged to collect the supernatant. Virus titer was quantified as LgEID50. Data were showed as mean ± SEM by Student’s t-test. P-value less than 0.05 was noted with a single-asterisk and p-value less than 0.01 was noted with a double-asterik.
Global gene expression modified by each infection
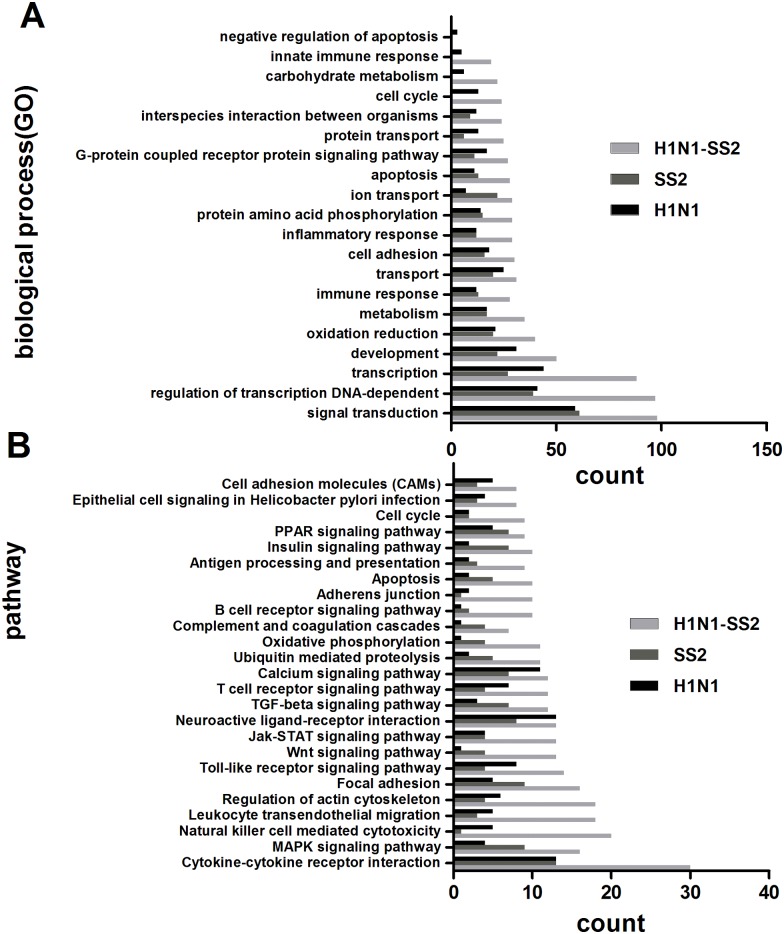
To further understand the H1N1 influenza virus and SS2 co-infection pathogenesis in pigs, Agilent microarray assay was performed to analyze the gene expression in the lungs from each group. The expression profiles of porcine lungs of the infected groups were compared with that in the control group. After quantile normalization and statistical analyses, 1314, 1353, and 2652 transcripts were identified to be different expression in H1N1, SS2, and H1N1-SS2 groups, respectively. These transcripts represented the corresponding 457 genes (230 upregulated and 227 downregulated), 411 genes (199 upregulated and 212 downregulated), and 844 genes (597 upregulated and 247 downregulated), respectively. MAS3.0 was used to analyze the functions of the DE genes. Majority of the DE genes were associated with signal transduction, transcription, development, immune response, cell adhesion, inflammatory response, apoptosis, innate immune response, and oxidation reduction (Fig 3A). Interestingly, inflammatory response and apoptosis were more intensely activated in the H1N1-SS2 group compared with the H1N1 and SS2 groups, indicating that they may have important roles in the infection process. To gain insight into the different biological processes of each infection, pathway analysis was also performed on the DE genes. Those containing more genes were selected for the analysis (Fig 3B), which showed that more DE genes were clustered in the immune and inflammation response (cytokine–cytokine receptor interaction, MAPK signaling, Toll-like receptor (TLR) signaling, JAK-STAT signaling pathway, antigen processing and presentation, and Complement and coagulation cascades) and apoptosis in the H1N1-SS2 group than the two other infection groups.
Fig 3. Characterization of the differential expression of genes.
(A) Categories of genes based on biological process GO term in each group. Only the top20 terms based on the gene numbers are showed. (B) Clustered pathways of the DE genes by KEGG analysis. Only the pathways containing more genes are presented. Many categories shared the same genes.
Genes related to inflammatory response
The DE genes associated with inflammatory responses were examined to further investigate the different gene expression. H1N1-SS2 co-infection increased the expression of more cytokine–cytokine receptor interaction genes compared with the other two infection groups (Fig 3B and Tables 2, 3 and 4). These genes included interferon receptors IFNAR1 (1.74-fold), IFNAR2 (2.27-fold), and IFNGR1 (1.73-fold). Several interleukin receptors were also significantly upregulated, including IL13RA1 (1.65-fold), IL1R2 (4.56-fold), and IL4R (2.15-fold). In addition, co-infection significantly regulated the mRNA expression of several chemokine and chemokine receptor genes, including CCL3 (1.62-fold), CX3CL1 (1.7-fold), CXCL16 (0.56-fold), CCR1 (2.23-fold), and CXCR4 (1.55-fold). Although the pro-inflammatory cytokine TNF-α was not significantly regulated, five tumor necrosis factor receptor superfamily genes were significantly regulated in the co-infection group (Table 4). To describe the inflammatory responses, several genes related to inflammation were tested by RT-PCR (Fig 4A). It indicated that TLR4 and MYD88 were significantly increased in H1N1-SS2 infection group, which play important roles in TLR signaling. IL-8, CCL2, and IL-6 were also remarkably increased in the co-infection group.
Table 2. The DE genes associated with immune and inflammatory responses in H1N1 group.
| Functional classification | Description | Gene symbol | Gene ID | Fold change |
|---|---|---|---|---|
| Immune response | complement component 6 | C6 | 100037952 | 2.66 |
| toll-like receptor 3 | TLR3 | 100037937 | -2.54 | |
| ring finger protein 125, E3 ubiquitin protein ligase | RNF125 | 100517591 | 2.19 | |
| CD59 molecule, complement regulatory protein | CD59 | 397347 | 2.00 | |
| CD1d molecule | CD1D | 100124526 | -1.93 | |
| chemokine (C-C motif) receptor-like 1 | CCRL1 | 100037292 | 1.86 | |
| peroxidasin homolog (Drosophila) | PXDN | 100516076 | 1.72 | |
| Rep:CD274 molecule CD274, mRNA- Homo sapiens | CD247 | 397302 | 1.71 | |
| lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) | LCP2 | 100511843 | 1.70 | |
| toll-like receptor 1 | TLR1 | 396607 | 1.65 | |
| GTP binding protein overexpressed in skeletal muscle | GEM | 404772 | -1.63 | |
| CD4 molecule | CD4 | 404704 | 1.58 | |
| toll-like receptor 6 | TLR6 | 396621 | 1.58 | |
| Inflammatory response | sphingosine-1-phosphate receptor 3 | S1PR3 | 100154607 | 2.29 |
| Rep: transforming growth factor, beta 2- Macaca mulatta | TGFB2 | 397084 | -2.18 | |
| interleukin 17D | IL17D | 100738902 | 2.16 | |
| chemokine (C-C motif) ligand 2 | CCL2 | 397422 | -1.99 | |
| interferon (alpha, beta and omega) receptor 1 | IFNAR1 | 396658 | 1.89 | |
| vascular endothelial growth factor C | VEGFC | 100127470 | -1.87 | |
| CD163 molecule | CD163 | 397031 | -1.84 | |
| interleukin 2 | IL2 | 396868 | 1.71 | |
| toll-like receptor adaptor molecule 2 | TICAM2 | 100520421 | 1.66 | |
| mitogen-activated protein kinase 12 | MAPK12 | 574062 | -1.65 | |
| chemokine (C-C motif) ligand 20 | CCL20 | 553951 | 1.61 | |
| chemokine (C-X-C motif) ligand 2 | CXCL2 | 414904 | 1.59 | |
| tumor necrosis factor receptor superfamily, member 1B | TNFRSF1B | 100037306 | 1.58 | |
| inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | IKBKG | 100127355 | -1.52 | |
| toll-interleukin 1 receptor (TIR) domain containing adaptor protein | TIRAP | 100514174 | -1.71 |
The DE genes associated with immune and inflammatory responses were assigned based on GO term and manual annotation. Manual annotations were listed in italics. Many genes with multiple functions were only listed in one category. Gene ID refers to NCBI gene ID in the study.
Table 3. The DE genes associated with immune and inflammatory responses in SS2 group.
| Functional classification | Description | Gene symbol | Gene ID | Fold change |
|---|---|---|---|---|
| Immune response | protein C receptor, endothelial | PROCR | 654289 | 2.86 |
| chitinase 1 (chitotriosidase) | CHIT1 | 100512552 | 2.50 | |
| C-type lectin domain family 5, member A | CLEC5A | 397050 | -2.21 | |
| chitinase, acidic | CHIA | 100156433 | 1.99 | |
| Fc fragment of IgA | FCAR | 100144539 | 1.99 | |
| cytotoxic T-lymphocyte-associated protein 4 | CTLA4 | 397286 | 1.80 | |
| dipeptzzidyl-peptidase 8 | DPP8 | 100155833 | -1.53 | |
| interleukin 1 receptor, type II | IL1R2 | 100628112 | 14.28 | |
| CD14 molecule | CD14 | 100037938 | 5.23 | |
| CD163 molecule | CD163 | 397031 | 3.60 | |
| A1 adenosine receptor | ADORA1 | 606743 | 3.08 | |
| histone deacetylase 9-like | HDAC9 | 100523163 | -3.07 | |
| interleukin 2 | IL2 | 396868 | -2.50 | |
| sialoadhesin | SIGLEC1 | 397623 | -2.22 | |
| kinase insert domain receptor (a type III receptor tyrosine kinase) | KDR | 397311 | -2.22 | |
| insulin-like growth factor binding protein 4 | IGFBP4 | 100144490 | 2.14 | |
| Inflammatory response | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | KIT | 396810 | 2.10 |
| interleukin 17D | IL17D | 100738902 | 2.07 | |
| erythropoietin receptor | EPOR | 397554 | -2.07 | |
| vascular endothelial growth factor A | VEGFA | 397157 | -1.91 | |
| erythropoietin | EPO | 397249 | 1.85 | |
| chemokine (C-X-C motif) ligand 12 | CXCL12 | 494460 | -1.77 | |
| VCP-interacting membrane protein | SELS | 100151836 | 1.67 | |
| neutrophil cytosolic factor 4, 40kDa | NCF4 | 100152055 | 1.64 | |
| KIT ligand | KITLG | 397509 | -1.60 | |
| Rep: transforming growth factor, beta 2- Macaca mulatta | TGFB2 | 397084 | -1.60 | |
| toll-like receptor 9 | TLR9 | 397007 | 1.58 | |
| vacuolar protein sorting 45 homolog (S. cerevisiae) | VPS45 | 100514766 | 1.56 |
The DE genes associated with immune and inflammatory responses were assigned based on GO term and manual annotation. Manual annotations were listed in italics. Many genes with multiple functions were only listed in one category.
Table 4. The DE genes associated with immune and inflammatory responses in H1N1-SS2 group.
| Functional classification | Description | Gene symbol | Gene ID | Fold change |
|---|---|---|---|---|
| Immune response | CD1d molecule | CD1D | 100124526 | 3.99 |
| A1 adenosine receptor | ADORA1 | 606743 | 3.37 | |
| chitinase 1 (chitotriosidase) | CHIT1 | 100512552 | 3.02 | |
| PREDICTED: Sus scrofa guanylate binding protein 4 (GBP4) | GBP4 | 100155195 | 2.91 | |
| nuclear factor, interleukin 3 regulated | NFIL3 | 100153822 | 2.53 | |
| SAM domain and HD domain 1 | SAMHD1 | 100625064 | 2.41 | |
| CD72 molecule | CD72 | 100038011 | 2.39 | |
| vav 1 guanine nucleotide exchange factor | VAV1 | 100519821 | 2.41 | |
| bone morphogenetic protein 2 | BMP2 | 100157103 | -1.88 | |
| mitochondrial antiviral signaling protein | MAVS | 100037290 | 1.74 | |
| bone morphogenetic protein receptor, type IB | BMPR1B | 396691 | -1.72 | |
| Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | FCGR3B | 397684 | 1.71 | |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 | DDX46 | 100626833 | -1.69 | |
| hepcidin antimicrobial peptide | HAMP | 397207 | 1.58 | |
| granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) | GZMA | 100526762 | -1.57 | |
| B-cell linker | BLNK | 100152350 | 1.56 | |
| ferritin, heavy polypeptide 1 | FTH1 | 397030 | 1.50 | |
| Inflammatory response | CD163 molecule | CD163 | 397031 | 4.98 |
| CD14 molecule | CD14 | 100037938 | 4.96 | |
| Rep: IL-1 receptor 2-Bos taurus Bovine, partial 98% | IL1R2 | 100628112 | 4.56 | |
| neutrophil cytosolic factor 4, 40kDa | NCF4 | 100152055 | 3.34 | |
| myeloid differentiation primary response gene (88) | MYD88 | 396646 | 2.32 | |
| oncostatin M receptor | OSMR | 100519398 | 2.31 | |
| toll-like receptor 9 | TLR9 | 397007 | 2.27 | |
| chemokine (C-C motif) receptor 1 | CCR1 | 414374 | 2.24 | |
| interferon (alpha, beta and omega) receptor 2 | IFNAR2 | 100533555 | 2.23 | |
| interleukin 4 receptor | IL4R | 397614 | 2.15 | |
| neutrophil cytosolic factor 1 | NCF1 | 100134857 | 2.10 | |
| toll-like receptor adaptor molecule 2 | TICAM2 | 100520421 | 2.00 | |
| kininogen 1 | KNG1 | 396568 | -2.08 | |
| erythropoietin | EPO | 397249 | 1.94 | |
| bone morphogenetic protein 2 | BMP2 | 100157103 | -1.88 | |
| fms-related tyrosine kinase 3 ligand | FLT3LG | 100322867 | -1.85 | |
| lipopolysaccharide binding protein | LBP | 397303 | 1.81 | |
| vascular endothelial growth factor A | VEGFA | 397157 | -1.81 | |
| allograft inflammatory factor 1 | AIF1 | 397271 | 1.80 | |
| toll-like receptor 4 | TLR4 | 399541 | 1.79 | |
| scavenger receptor for phosphatidylserine and oxidized low density lipoprotein | CXCL16 | 396735 | -1.78 | |
| SMAD family member 1 | SMAD1 | 397016 | 1.75 | |
| interferon (alpha, beta and omega) receptor 1 | IFNAR1 | 396658 | 1.74 | |
| nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | NFKBIA | 406188 | 1.74 | |
| interferon gamma receptor 1 | IFNGR1 | 100152238 | 1.73 | |
| bone morphogenetic protein receptor, type IB | BMPR1B | 396691 | -1.72 | |
| chemokine (C-X3-C motif) ligand 1 | CX3CL1 | 100621027 | 1.70 | |
| activin A receptor, type IIA | ACVR2A | 397282 | 1.68 | |
| neutrophil cytosolic factor 2 | NCF2 | 100142665 | 1.68 | |
| toll-interleukin 1 receptor (TIR) domain containing adaptor protein | TIRAP | 100514174 | -1.66 | |
| interleukin 13 receptor, alpha 1 | IL13RA1 | 397615 | 1.65 | |
| chemokine (C-C motif) ligand 3-like 1 | CCL3 | 494459 | 1.62 | |
| anti-Mullerian hormone receptor, type II | AMHR2 | 100154297 | -1.58 | |
| B-cell linker | BLNK | 100152350 | 1.56 | |
| bone morphogenetic protein receptor, type IB | BMPR1B | 396691 | 1.56 | |
| chemokine (C-X-C motif) receptor 4 | CXCR4 | 396659 | 1.55 | |
| interleukin 17D | IL17D | 100738902 | 1.51 | |
| interleukin 6 signal transducer (gp130, oncostatin M receptor) | IL6ST | 100037294 | 1.50 | |
| NADPH oxidase heavy chain subunit | CYBB | 397108 | 1.50 | |
| signal transducer and activator of transcription 5B | STAT5B | 397340 | 1.50 |
The DE genes associated with immune and inflammatory responses were assigned based on GO term and manual annotation. Manual annotations were listed in italics. Many genes with multiple functions were only listed in one category.
Fig 4. Expression of genes related to inflammation and apoptosis by qRT-PCR.
RNA was extracted from lungs of each infection group, and was reversely transcribed. mRNA levels were measured by real-time PCR. (A) Changes of genes related to proinflammatory response. (B) Changes of genes related to apoptosis. Data were showed as mean ± SEM and were representatives of two independent experiments. Significant levels were analyzed by T-test.
Robust apoptosis response to H1N1-SS2 co-infection
The microarray analysis results showed that a number of genes involved in apoptosis were markedly upregulated to various extents in the H1N1-SS2 group (Table 5). These genes included the tumor necrosis factor receptor (TNFR) superfamily members (TNFRSF10A, TNFRSF1B, and TNFRSF8), genes involved in the activation cascade of caspase that is responsible for apoptosis (CASP2, CASP3, and CASP4) and BCL2-like genes (BCL2L11 and BCL2L14). qRT-PCR also demonstrated co-infection significantly increased expressions of CASP2, CASP3, BC2L11, FASLG, and TNFRSF8 (Fig 4B). The markedly expressed apoptosis-associated genes in H1N1 and SS2 groups were relatively lesser in the H1N1-SS2 group. Thus, apoptosis in each group was further investigated. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) was used to detect cell apoptosis in the lungs from each group. Fig 5A showed that the H1N1 and SS2 infections caused significant apoptosis, compared with the control. However, H1N1-SS2 co-infection notably triggered the significantly more cell apoptosis rate compared with the two other infections (Fig 5B). This result was consistent with the modification of the significantly regulated genes involved in apoptosis by each infection (Table 5). The result suggested that abundant cell apoptosis induced by H1N1 and SS2 co-infection would contribute to the enhanced virulence.
Table 5. The DE genes associated with apoptosis in each infection group.
| Group | Description | Gene symbol | Gene ID | Fold change |
|---|---|---|---|---|
| H1N1 | C1D nuclear receptor corepressor | C1D | 100515656 | 4.23 |
| cytoplasmic FMR1 interacting protein 2 | CYFIP2 | 100523290 | 1.93 | |
| Rep: baculoviral IAP repeat-containing protein 2 | BIRC2 | 100622859 | -1.85 | |
| RING1 and YY1 binding protein | RYBP | 100526249 | -1.71 | |
| lymphotoxin beta (TNF superfamily, member 3) | LTB | 100155581 | 1.63 | |
| lipopolysaccharide-induced TNF factor | LITAF | 100518302 | -1.62 | |
| tumor necrosis factor receptor superfamily, member 1B | TNFRSF1B | 100037306 | 1.58 | |
| sulfatase 1 | SULF1 | 100152427 | -1.53 | |
| SS2 | cellular FLICE-like inhibitory protein | C-FLIP | 414381 | 2.24 |
| tumor necrosis factor receptor superfamily, member 8 | TNFRSF8 | 574055 | 2.08 | |
| death-associated protein kinase 2 | DAPK2 | 100155578 | -1.94 | |
| Rep: Mdm4, transformed 3T3 cell double minute 4, p53 binding protein isoform 1 | MDM4 | 100512731 | -1.94 | |
| programmed cell death 4 (neoplastic transformation inhibitor) | PDCD4 | 100157112 | -1.76 | |
| tumor necrosis factor receptor superfamily, member 1B | TNFRSF1B | 100037306 | 1.64 | |
| H1N1-SS2 | caspase 10, apoptosis-related cysteine peptidase | CASP10 | 100154896 | 1.55 |
| granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | GZMB | 100233184 | 4.29 | |
| Fas ligand (TNF superfamily, member 6) | FASLG | 396726 | 3.33 | |
| BCL2-like 11 (apoptosis facilitator) | BCL2L11 | 396632 | 2.40 | |
| caspase 4, apoptosis-related cysteine peptidase | CASP4 | 100522887 | 2.36 | |
| tumor necrosis factor receptor superfamily, member 10a | TNFRSF10A | 100134832 | 2.11 | |
| cell death-inducing DFFA-like effector c | CIDEC | 100127161 | 1.96 | |
| BCL2-like 14 (apoptosis facilitator) | BCL2L14 | 100514901 | 1.81 | |
| caspase 2, apoptosis-related cysteine peptidase | CASP2 | 100521118 | 1.81 | |
| adenomatous polyposis coli | APC | 100517932 | -1.81 | |
| tumor necrosis factor receptor superfamily, member 8 | TNFRSF8 | 574055 | 1.75 | |
| Rep: baculoviral IAP repeat-containing protein 2 | BIRC2 | 100622859 | 1.75 | |
| Rep: HECT, UBA and WWE domain containing 1 | HUWE1 | 100517442 | 1.65 | |
| PREDICTED: Sus scrofa tumor necrosis factor alpha-induced protein 8-like | TNFAIP8 | 100524014 | 1.64 | |
| tumor necrosis factor receptor superfamily, member 11b | TNFRSF11B | 100049688 | -1.63 | |
| DNA-damage-inducible transcript 3 | DDIT3 | 100240743 | 1.62 | |
| tumor necrosis factor receptor superfamily, member 1B | TNFRSF1B | 100037306 | 1.54 | |
| caspase 3, apoptosis-related cysteine peptidase | CASP3 | 397244 | 1.51 |
The DE genes associated with apoptosis were based on GO term and manual annotation. Manual annotations were listed in italics. Many genes with multiple functions were only listed in one category.
Fig 5. Apoptosis induced by H1N1 virus, SS2, H1N1-SS2 and PBS control.
Histological sections of lungs from each group were evaluated for apoptosis by TUNEL test. (A) The infected groups showed varying degrees of apoptosis. Brown ovals (red arrows) indicate apoptotic nucleus. Blue ovals (blue arrows) indicate normal nucleus. (B) Statistical analysis of apoptosis rate in each infection group. Data was showed as mean ± SEM. T-test was used to analyze the significance.
Validation of microarray data by qRT-PCR
qPCR was performed to validate the expression patterns during infections for specific genes on the same samples used in the microarray analysis. Eight genes (NCF4, CD14, IL17D, CD1D, CD163, IL1R2, TLR9, and TLR2) with different expression in each infection group were selected for qPCR analysis (Table 6). All selected genes were regulated with the same pattern as in the microarray assay through qPCR analysis, although a variation in the fold changes was observed between qPCR and microarray. This phenomenon may arise from the different sensitivities and accuracies between the two techniques. The coincident expression pattern indicated the reliability of the microarray analysis.
Table 6. Verification of differentially expressed genes via qRT-PCR.
| Genes | H1N1 group | SS2 group | H1N1-SS2 group | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Microarray fold change | qRT-PCR fold change | qRT-PCR p-value | Microarray fold change | qRT-PCR fold change | qRT-PCR p-value | Microarray fold change | qRT-PCR fold change | qRT-PCR p-value | |
| NCF4 | NS | -0.21 | < 0.0010 | 1.64 | 6.73 | < 0.0010 | 3.34 | 17.56 | < 0.0010 |
| CD14 | NS | 2.70 | < 0.0010 | 5.23 | 13.65 | < 0.0010 | 4.96 | 6.37 | < 0.0010 |
| IL17D | NS | 2.21 | < 0.0010 | 2.07 | 4.54 | 0.019 | 1.50 | 15.25 | 0.0050 |
| CD1D | -1.93 | -4.29 | < 0.0010 | -1.84 | -2.16 | < 0.0010 | 3.99 | 38.90 | < 0.0010 |
| CD163 | -1.93 | -3.62 | 0.777 | 3.60 | 6.01 | 0.0060 | 4.98 | 14.53 | 0.060 |
| IL1R2 | NS | 1.49 | 0.221 | 14.28 | 24.73 | < 0.0010 | 4.56 | 4.12 | 0.018 |
| TLR9 | NS | 1.44 | 0.127 | 1.58 | 4.l9 | 0.0050 | 2.27 | 2.16 | 0.022 |
| TLR2 | NS | -3.95 | 0.042 | NS | 53.34 | 0.0070 | NS | -1.66 | 0.83 |
Fold changes of gene expression in infected group compared with the PBS control. mRNA expressions were measured by real-time RT-PCR. The data represent means ± SEM of triplicate reactions for each gene transcript. The expression level of GAPDH was assayed for normalization. Statistical analysis was performed by T-test. NS: No significant change.
Discussion
Clinically, influenza virus and S. suis co-infections are common. We also investigated the co-infections in pig farms in the Hubei Province of China using serological test, which exhibited a high co-infection rate (S7 Data). The altered host response after swine influenza virus infection predisposes to secondary bacterial infection because of the complicated mechanisms that would significantly increase the mortality rate. Secondary infection by S. suis is an example of such case. However, the molecular mechanisms underlying this high mortality rate have not yet been established. The augmented host response is considered as a critical factor for the severe pathogenicity of influenza virus infection in humans [20]. Therefore, a genome expression analysis was performed to obtain comprehensive information on the host responses in the lungs of infected pigs through H1N1, SS2, and H1N1-SS2 infections. The clinical and pathological findings demonstrated that H1N1 and SS2 co-infection enhanced the virulence in pigs. In addition, the transcriptional profiles indicated that co-infection resulted in augmented inflammatory and apoptosis responses, which could account for the increased pathogenecity. The increased virulence of co-infection might also be attributed to the enhanced viral replication or SS2 growth. Increased viral load was observed in lungs and nasal swabs in co-infection group, which might play a role in the virulence. However, the SS2 content in lungs was not significantly different between SS2 and H1N1-SS2 groups in the present study. It suggested that the H1N1 infection might not contribute to the enhanced SS2 growth in pig lungs. Yet, in spite of this, H1N1 infection could damage the epithelia lining of the bronchi and lungs, allowing the SS2 a foothold, and therefore strengthened and disordered the host responses, especially the immune responses in a synergetic manner.
Inflammatory responses were enhanced in response to H1N1-SS2 co-infection
TLRs are families of pattern-recognition receptors that sense the invasion of pathogenic microorganisms and trigger innate immune responses. Our data showed that several genes in the TLR signaling were significantly but differently regulated in each infection group. For instance, CD14 was markedly upregulated in SS2 and H1N1-SS2 infection groups. CD14 has been identified as an LPS receptor [21], which can bind to the LPS-binding protein (LBP) [22]. LBP catalyzes the transfer of LPS to CD14, which then mediates the transfer of LPS to the membrane, where LPS binds with the TLR4-MD-2 complex, leading to the nuclear factor-kappa B (NF-κB) activation [23, 24]. TLR4 is necessary in this LPS signal transduction process. In fact, CD14 is an essential co-receptor of influenza virus recognition, which is necessary for inflammatory cytokine production induced by influenza virus [25]. TLR4 signaling has also been reported as a key pathway of the acute lung injury induced by H5N1 infection and can be blocked by the TLR4 antagonist, thus protecting the mouse from influenza virus [26, 27]. Based on the microarray data, TLR4 and LBP were slightly but markedly upregulated (1.79- and 1.8-fold, respectively) in the H1N1-SS2 co-infection group (Table 4). However, these genes were not significantly changed in the H1N1 or SS2 infection alone group. MYD88 is an important signaling adapter that contains the toll/IL-1R domain, which can mediate the TLR4 signaling response [28], to elicit a pro-inflammatory response [29]. The results showed that MYD88 was significantly upregulated in the H1N1-SS2 group at 2.32-fold compared with that in the other two infection groups. These results demonstrated that TLR4 signaling have some important roles in the co-infection pathogenesis. Several studies have previously determined that S. suis was mainly recognized by TLR2, which is associated with CD14 and activates the proinflammatory cytokines. However, the present data showed that TLR2 was not significantly changed in each infection group compared with the control, which suggested that TLR2 may be not a key contributor to the increased virulence by co-infection and this result is consistent with that of a previous study [30]. Thus, the role of TLR4 and its downstream signaling in co-infection are worthy of further investigations.
Cytokine is an important part of the host defensive system that helps eliminate pathogens by recruiting inflammatory cells. However, overabundant cytokine production could cause pathology. In the present study, several cytokines were significantly regulated. For instance, CXCL2, CCL20, and IL2 were significantly upregulated in the H1N1 group (Table 3). In the SS2 infection, IL17D was elevated by 2.07-fold (Table 3), which can stimulate production of IL6, IL8, and GM-CSF production in endothelial cells [31]. In the H1N1-SS2 group, CCL3, CCR1, CXCR4, CX3CL1, IL6ST, and IL17D were differently upregulated (Table 4). Besides, MYD88, NFKB1, TIRAP, TLR4, and TLR9 were upregulated from 1.63- to 2.32-fold in this group. These genes are all involved in the TLR signaling pathway, which can induce and then release pro-inflammatory factors. MAPK, a key pathway that regulates the synthesis of numerous cytokines, chemokines, and other inflammatory mediators [32], was also significantly regulated in the three infection groups. Expectedly, co-infection modified the most number of regulated genes related to MAPK signalling (Fig 3B). The MAPK signaling pathway has been found to be greatly induced and may be one of the key pathways that led to the immune response against S. suis [33]. Thus, it might suggest that the significant activation of the MAPK pathway plays some important roles in the pro-inflammatory responses caused by co-infection. Interestingly, CD163 was significantly upregulated in the SS2 (3.6-fold) and H1N1-SS2 (4.98-fold) infection groups, however, it was downregulated in H1N1 infection group. CD163 is a macrophage receptor for the bacterial binding [34, 35] and can induce the pro-inflammatory cytokines during infection [35, 36]. In addition, CD163 was associated with SS2 infection [37]. The CD163 upregulation demonstrated that CD163 may have some important roles in inflammatory responses, especially in H1N1-SS2 co-infection. The CD163 function in the co-infection pathogenesis needs further studies. Furthermore, genes related to ROS production were also identified to be upregulated. ROS is involved in many physiological processes, including cell proliferation, apoptosis, and immune and pro-inflammatory responses [38, 39]. Excessive ROS production could cause acute lung injury induced by H5N1 [26, 40]. NADPH oxidase mainly causes the ROS production in inflammatory cells [41, 42], and CYBB is the primary component. CYBB is related to lung inflammation caused by influenza virus [43]. NCF1, NCF2, and NCF4 are all NADPH oxidase components. In the present study, CYBB, NCF1, NCF2, and NCF4 were significantly up-regulated in the H1N1-SS2 group. The increased NADPH oxidase expression implied more ROS production and would lead to oxidative stress, which caused more severe inflammatory damage in H1N1-SS2 group.
To maintain the development and metabolism homeostasis, the host must express anti-inflammatory factors to prevent infection-induced excessive inflammation. The PPAR pathway has an important role in anti-inflammatory response because it can reduce several pro-inflammatory cytokines and chemokines [44]. In the current study, several genes involved in this pathway were upregulated in all three infection groups. In addition, other regulated negative regulators, such as IL13RA1, IL1R2, and IL4R, were detected. This result indicated that the host tried to terminate the over-induced inflammatory responses. TGF-β is another interesting gene, which can act as a global regulator of immunity by controlling the initiation and resolution of inflammatory responses [45, 46]. Previous studies have proven that human H1N1 could induce TGF-β elevation, however, H5N1 downregulated the TGF-β secretion in a mouse model [47, 48]. TGF-β can also protect the host from the lethal influenza virus in a mouse model [49]. Therefore, TGF-β can function as a protective agent against virus invasion. Surprisingly, in our data, TGF-β was downregulated in H1N1 and SS2 group, and no significant change was observed in H1N1-SS2 groups. This result may be attributed to the different animal models used here. The role of TGF-β in immunopathology of infection in pigs remains to be further studied. Based from these results and with the lung pathological lesions, the H1N1-SS2 co-infection is proposed to induce stronger inflammatory responses, which caused more severe inflammatory damage in the lungs.
Stronger immune responses were induced by H1N1-SS2 co-infection
Cellular immune responses are critical for clearing the influenza virus [50]. NK cells, CD4+ and CD8+ T cells, and neutrophils increased upon influenza virus infection [51]. The increase of the immune cells is not only beneficial for clearing the virus, but may also contribute to pulmonary inflammation, which could cause immune damage and enhance the virulence. In the present study, several genes, as immune cell markers, were moderately upregulated, which contained CD14, CD1D, and CD163, in the SS2 and H1N1-SS2 groups. However, CD163 and CD1D were both downregulated, and CD14 was not significantly changed in H1N1 group. It indicated that the following SS2 infection might enhance or maintain the activation of immune response. However, the composition of the infiltrating immune cells was unknown, and investigating the frequency of immune cells in the lungs and BAL fluid from the H1N1 and SS2 co-infected pigs is highly attractive, because of the important contributions of their inflammatory and immune effect. Antigen processing and presentation is critical for the T cell activation and the adaptive immune response. However, T cells can also contribute to pathology when presented and activated in excess. In the present study, H1N1-SS2 co-infection enhanced several genes expression responsible for the antigen processing and presentation (S8 Data). For instance, SLA-DQA1, which belongs to the MHC class II histocompatibility antigen, was upregulated in H1N1-SS2 group. MHC class II molecules have significant functions in many virus infections in pigs. LGMN was upregulated by 3.47-fold. LGMN, a cysteine protease, was reported to be implicated in human regulatory T cell function [52], and may be involved in the processing of proteins for MHC class II antigen presentation. Specially, KLRC1, an inhibitory receptor of Natural killer (NK) cell, was also upregulated in H1N1-SS2 group. NK cells play an important role in the immune response to viral infections, which can activate dendritic cells and also secret T helper type 1 (Th1) cytokines to augment cytotoxic T cell responses [53]. Previous studies showed that KLRC1 was upregulated on NK cells in HIV and HCV infection [54, 55], and KLRC1 expressing NK cells correlated inversely with HCV RNA [56]. It indicated that KLRC1 might play roles in the H1N1-SS2 co-infection.
Complement is a major effector system of innate immunity, and it also influences the host adaptive immune response to micro-organisms or potential autoantigens [57, 58]. However, accumulating data have suggested that excessive activation of complement is involved in pathogenesis of inflammatory disease, inflammation responses, and acute respiratory distress syndrome [59, 60]. Our data indicated that more genes related to complement system were upregulated, which included C1R, C1S, C5AR1, C6, and KNG1 (S9 Data). It was possible that H1N1-SS2 co-infection triggered much stronger complement response to induce more intense immune responses.
Robust apoptosis induced by H1N1-SS2 co-infection
Influenza virus has been demonstrated to induce apoptosis, and swine streptococcus has also been reported to induce apoptosis in cultured porcine choroid plexus epithelia cells [61]. The modulation of host cell death pathways may eliminate key immune cells or evade the host defence through pathogen infection. Moreover, the activation or prevention of cell death could be a critical factor for an infection outcome [62]. In the present study, apoptosis was observed in the lungs from all infection groups, and increased robust apoptosis was induced by the H1N1-SS2 co-infection. The microarray analysis results also indicated that more genes involved in apoptosis pathway were regulated. In the H1N1-SS2 group, members of the TNFR superfamily (TNFRSF10A, TNFRSF1B, and TNFRSF8), caspase family (CASP2, CASP3, and CASP4), BCL2-like genes (BCL2L11 and BCL2L14), FASLG, and GZME were upregulated (Table 5). TUNEL test also demonstrated that co-infection led to much higher apoptosis rate compared with separate infections (Fig 5). The impacts of apoptosis on the viral replication vary depending on different virus. Infected cells possibly release the infectious viral particles into micro-environment, which then infect the nearby cells and trigger apoptosis. In this case, the infected nearby cells can host additional virus replication. In the present study, abundant cell apoptosis did not only cause serious lung damage, but also contribute to the dissemination of viruses, which in turn induced stronger pro-inflammatory responses with increased viral load. Therefore, co-infection by H1N1 and SS2 could potentiate virulence by causing severe apoptosis.
In this study, we co-infected SS2 on 3 days after H1N1 inoculation, and killed the pigs on day 6 of the experiment for microarray analysis. The choice of the time was depend on the biological characteristic of H1N1 and SS2. H1N1 infection could cause high-morbidity, but low mortality. The H1N1 infection with 107.0 EID50/mL could cause obvious flu symptoms within 3 days. However, SS2 was highly virulent to pigs. SS2 challenge with 106.0 CFU/mL could lead to severe typical symptoms within 24 hours. In the microarray analysis, many genes reported to play important roles in inflammatory response were not fund to be significantly changed, such as TNF-α, IFN-β, and IL-10. It might be due to the time point selected in our study was relatively later, or the sensitivity of microarray hybridization. The clinical co-infections of influenza virus and SS2 are highly complex, which makes the mechanism of the increased virulence much complex. The factors contributing to the complex may include infection doses of pathogens, the secondary infection time, different pathogens with various virulence, and even the environment factors. Based on this, more studies (studies with more infection times, more infection models, and more pathogens models) are needed to be performed to make the pathogenicity of co-infection much clearer.
Supporting Information
(DOCX)
On day 6, all pigs were humanely euthanized, and lungs were immediately removed. (A) lungs from each group exhibited different extent lesions. Black arrow indicated bleeding and hyperaemia, Blue arrow indicated necrosis, and Red arrow indicated hyperaemia and pulmonary consolidation. (B) Lung pathology scores were evaluated blindly.
(TIF)
Lungs were removed on day 6, and were fixed in formalin and embedded in paraffin, sectioned at 5μm, and stained with hematoxylin and eosin for further histopathologic evaluation.
(TIF)
Lungs were removed on day 6, and were fixed in formalin and embedded in paraffin, sectioned at 5μm, and stained with hematoxylin and eosin for further histopathologic evaluation.
(TIF)
Lungs were removed on day 6, and were fixed in formalin and embedded in paraffin, sectioned at 5μm, and stained with hematoxylin and eosin for further histopathologic evaluation.
(TIF)
Lungs were removed on day 6, and were fixed in formalin and embedded in paraffin, sectioned at 5μm, and stained with hematoxylin and eosin for further histopathologic evaluation.
(TIF)
A total of 376 serum samples from 4 different pig farms were tested for the H1N1 and SS2 antibody by HI and ELISA test respectively.
(DOCX)
The DE genes associated with antigen processing and presentation were assigned based on GO term and manual annotation. Manual annotations were listed in italics. Many genes with multiple functions were only listed in one category.
(DOCX)
The DE genes associated with Complement and coagulation cascades were assigned based on GO term and manual annotation. Manual annotations were listed in italics. Many genes with multiple functions were only listed in one category.
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files. The raw and processed data of microarray files are available from the NCBI’s Gene expression omnibus database (accession number GSE60172).
Funding Statement
The study was supported by China National Basic Research Program (China "973" Program 2012CB518805). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jung K, Ha Y, Chae C. Pathogenesis of swine influenza virus subtype H1N2 infection in pigs. J Comp Pathol. 2005; 132: 179–184. [DOI] [PubMed] [Google Scholar]
- 2. Gatherer D. The 2009 H1N1 influenza outbreak in its historical context. J Clin Virol. 2009; 45: 174–178. 10.1016/j.jcv.2009.06.004 [DOI] [PubMed] [Google Scholar]
- 3. Brown IH. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol. 2000; 74: 29–46. [DOI] [PubMed] [Google Scholar]
- 4. AVIT, Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009; 360: 2605–2615. 10.1056/NEJMoa0903810 [DOI] [PubMed] [Google Scholar]
- 5. Weingartl HM, Berhane Y, Hisanaga T, Neufeld J, Kehler H, Emburry-Hyatt C, et al. Genetic and pathobiologic characterization of pandemic H1N1 2009 influenza viruses from a naturally infected swine herd. J Virol. 2010; 84: 2245–2256. 10.1128/JVI.02118-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hofshagen M, Gjerset B, Er C, Tarpai A, Brun E, Dannevig B, et al. Pandemic influenza A(H1N1)v: human to pig transmission in Norway? Euro Surveill. 2009; 14. [DOI] [PubMed] [Google Scholar]
- 7. Pasma T, Joseph T. Pandemic (H1N1) 2009 infection in swine herds, Manitoba, Canada. Emerg Infect Dis. 2010; 16: 706–708. 10.3201/eid1604.091636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou H, Wang C, Yang Y, Guo X, Kang C, Chen H, et al. Pandemic (H1N1) 2009 virus in swine herds, People's Republic of China. Emerg Infect Dis. 2011; 17: 1757–1759. 10.3201/eid1709.101916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loving CL, Brockmeier SL, Vincent AL, Palmer MV, Sacco RE, Nicholson TL. Influenza virus coinfection with Bordetella bronchiseptica enhances bacterial colonization and host responses exacerbating pulmonary lesions. Microb Pathog. 2010; 49: 237–245. 10.1016/j.micpath.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 10. Gottschalk M, Xu J, Calzas C, Segura M. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 2010; 5: 371–391. 10.2217/fmb.10.2 [DOI] [PubMed] [Google Scholar]
- 11. Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang H, et al. Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis. 2006; 12: 914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Williamson SM, Tucker AW, McCrone IS, Bidewell CA, Brons N, Habernoll H, et al. Descriptive clinical and epidemiological characteristics of influenza A H1N1 2009 virus infections in pigs in England. Vet Rec. 2012; 171: 271 10.1136/vr.100673 [DOI] [PubMed] [Google Scholar]
- 13. Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, Liu SY, et al. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest. 2009; 119: 1910–1920. 10.1172/JCI35412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seki M, Yanagihara K, Higashiyama Y, Fukuda Y, Kaneko Y, Ohno H, et al. Immunokinetics in severe pneumonia due to influenza virus and bacteria coinfection in mice. Eur Respir J. 2004; 24: 143–149. [DOI] [PubMed] [Google Scholar]
- 15. Kudva A, Scheller EV, Robinson KM, Crowe CR, Choi SM, Slight SR, et al. Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. J Immunol. 2011; 186: 1666–1674. 10.4049/jimmunol.1002194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith MW, Schmidt JE, Rehg JE, Orihuela CJ, McCullers JA. Induction of pro- and anti-inflammatory molecules in a mouse model of pneumococcal pneumonia after influenza. Comp Med. 2007; 57: 82–89. [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y, Zhou H, Wen Z, Wu S, Huang C, Jia G, et al. Transcription analysis on response of swine lung to H1N1 swine influenza virus. BMC genomics. 2011; 12: 398 10.1186/1471-2164-12-398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008; 24: 719–720. [DOI] [PubMed] [Google Scholar]
- 19. Li R, Zhang A, Chen B, Teng L, Wang Y, Chen H, et al. Response of swine spleen to Streptococcus suis infection revealed by transcription analysis. BMC genomics. 2010; 11: 556 10.1186/1471-2164-11-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peiris JS, Cheung CY, Leung CY, Nicholls JM. Innate immune responses to influenza A H5N1: friend or foe? Trends Immunol. 2009; 30: 574–584. 10.1016/j.it.2009.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990; 249: 1431–1433. [DOI] [PubMed] [Google Scholar]
- 22. Schumann RR, Leong SR, Flaggs GW, Gray PW, Wright SD, Mathison JC, et al. Structure and function of lipopolysaccharide binding protein. Science. 1990; 249: 1429–1431. [DOI] [PubMed] [Google Scholar]
- 23. Triantafilou K, Triantafilou M, Ladha S, Mackie A, Dedrick RL, Fernandez N, et al. Fluorescence recovery after photobleaching reveals that LPS rapidly transfers from CD14 to hsp70 and hsp90 on the cell membrane. J Cell Sci. 2001. 114: 2535–2545. [DOI] [PubMed] [Google Scholar]
- 24. da Silva Correia J, Soldau K, Christen U, Tobias PS, Ulevitch RJ. Lipopolysaccharide is in close proximity to each of the proteins in its membrane receptor complex transfer from CD14 to TLR4 and MD-2. J Bio Chem. 2001; 276: 21129–21135. [DOI] [PubMed] [Google Scholar]
- 25. Pauligk C, Nain M, Reiling N, Gemsa D, Kaufmann A. CD14 is required for influenza A virus-induced cytokine and chemokine production. Immunobiology. 2004; 209: 3–10. [DOI] [PubMed] [Google Scholar]
- 26. Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, Nicholls J, et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008; 133: 235–249. 10.1016/j.cell.2008.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shirey KA, Lai W, Scott AJ, Lipsky M, Mistry P, Karp CL, et al. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature. 2013; 497: 498–502. 10.1038/nature12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fitzgerald KA, Palsson-McDermott EM, Bowie AG, Jefferies CA, Mansell AS, Brady G, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001; 413: 78–83. [DOI] [PubMed] [Google Scholar]
- 29. Fan J, Frey RS, Malik AB. TLR4 signaling induces TLR2 expression in endothelial cells via neutrophil NADPH oxidase. J Clin Invest. 2003; 112: 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dessing MC, van der Sluijs KF, Florquin S, Akira S, van der Poll T. Toll-like receptor 2 does not contribute to host response during postinfluenza pneumococcal pneumonia. Am J Respir Cell Mol Biol. 2007; 36: 609–614. [DOI] [PubMed] [Google Scholar]
- 31. Starnes T, Broxmeyer HE, Robertson MJ, Hromas R. Cutting edge: IL-17D, a novel member of the IL-17 family, stimulates cytokine production and inhibits hemopoiesis. J Immunol. 2002; 169: 642–646. [DOI] [PubMed] [Google Scholar]
- 32. Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002; 20: 55–72. [DOI] [PubMed] [Google Scholar]
- 33. de Greeff A, Benga L, Wichgers Schreur PJ, Valentin-Weigand P, Rebel JM, Smith HE. Involvement of NF-kappaB and MAP-kinases in the transcriptional response of alveolar macrophages to Streptococcus suis. Vet Microbiol. 2010; 141: 59–67. 10.1016/j.vetmic.2009.07.031 [DOI] [PubMed] [Google Scholar]
- 34. Van den Heuvel MM, Tensen CP, van As JH, Van den Berg TK, Fluitsma DM, Dijkstra CD, et al. Regulation of CD 163 on human macrophages: cross-linking of CD163 induces signaling and activation. J Leukoc Biol. 1999; 66: 858–866. [DOI] [PubMed] [Google Scholar]
- 35. Fabriek BO, van Bruggen R, Deng DM, Ligtenberg AJ, Nazmi K, Schornagel K, et al. The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood. 2009; 113: 887–892. 10.1182/blood-2008-07-167064 [DOI] [PubMed] [Google Scholar]
- 36. Polfliet MM, Fabriek BO, Daniels WP, Dijkstra CD, van den Berg TK. The rat macrophage scavenger receptor CD163: expression, regulation and role in inflammatory mediator production. Immunobiology. 2006; 211: 419–425. [DOI] [PubMed] [Google Scholar]
- 37. Wilson SM, Norton P, Haverson K, Leigh J, Bailey M. Interactions between Streptococcus suis serotype 2 and cells of the myeloid lineage in the palatine tonsil of the pig. Vet Immunol Immunopathol. 2006; 117: 116–123. [DOI] [PubMed] [Google Scholar]
- 38. Galecka E, Mrowicka M, Malinowska K, Galecki P. Role of free radicals in the physiological processes. Pol Merkur Lekarski. 2008; 24: 446–448. [PubMed] [Google Scholar]
- 39. Nauseef WM. Biological roles for the NOX family NADPH oxidases. J Bio Chem. 2008; 283: 16961–16965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oda T, Akaike T, Hamamoto T, Suzuki F, Hirano T, Maeda H. Oxygen radicals in influenza-induced pathogenesis and treatment with pyran polymer-conjugated SOD. Science. 1989; 244: 974–976. [DOI] [PubMed] [Google Scholar]
- 41. Selemidis S, Sobey CG, Wingler K, Schmidt HH, Drummond GR. NADPH oxidases in the vasculature: molecular features, roles in disease and pharmacological inhibition. Pharmacol Ther. 2008; 120: 254–291. 10.1016/j.pharmthera.2008.08.005 [DOI] [PubMed] [Google Scholar]
- 42. Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007; 87: 245–313. [DOI] [PubMed] [Google Scholar]
- 43. Vlahos R, Stambas J, Bozinovski S, Broughton BR, Drummond GR, Selemidis S. Inhibition of Nox2 oxidase activity ameliorates influenza A virus-induced lung inflammation. PLoS Pathog. 2011; 7: e1001271 10.1371/journal.ppat.1001271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Straus DS, Glass CK. Anti-inflammatory actions of PPAR ligands: new insights on cellular and molecular mechanisms. Trends Immunol. 2007; 28: 551–558. [DOI] [PubMed] [Google Scholar]
- 45. Koli K, Myllarniemi M, Keski-Oja J, Kinnula VL. Transforming growth factor-beta activation in the lung: focus on fibrosis and reactive oxygen species. Antioxid Redox Signal. 2008; 10: 333–342. [DOI] [PubMed] [Google Scholar]
- 46. Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006; 24: 99–146. [DOI] [PubMed] [Google Scholar]
- 47. Lam WY, Yeung AC, Chu IM, Chan PK. Profiles of cytokine and chemokine gene expression in human pulmonary epithelial cells induced by human and avian influenza viruses. Virol J. 2010; 7: 344 10.1186/1743-422X-7-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dybing JK, Schultz-Cherry S, Swayne DE, Suarez DL, Perdue ML. Distinct pathogenesis of hong kong-origin H5N1 viruses in mice compared to that of other highly pathogenic H5 avian influenza viruses. J Virol. 2000; 74: 1443–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carlson CM, Turpin EA, Moser LA, O'Brien KB, Cline TD, Jones JC, et al. Transforming growth factor-beta: activation by neuraminidase and role in highly pathogenic H5N1 influenza pathogenesis. PLoS Pathog. 2010; 6: e1001136 10.1371/journal.ppat.1001136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Graham MB, Braciale TJ. Resistance to and recovery from lethal influenza virus infection in B lymphocyte-deficient mice. J Exp Med. 1997; 186: 2063–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heinen PP, de Boer-Luijtze EA, Bianchi AT. Respiratory and systemic humoral and cellular immune responses of pigs to a heterosubtypic influenza A virus infection. The J Gen Virol. 2001; 82: 2697–2707. [DOI] [PubMed] [Google Scholar]
- 52. Probst-Kepper M, Geffers R, Kroger A, Viegas N, Erck C, Hecht HJ, et al. GARP: a key receptor controlling FOXP3 in human regulatory T cells. J Cell Mol Med. 2009;13: 3343–3357. 10.1111/j.1582-4934.2009.00782.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moretta L, Ferlazzo G, Bottino C, Vitale M, Pende D, Mingari MC, et al. Effector and regulatory events during natural killer-dendritic cell interactions. Immunol Rev. 2006; 214: 219–228. [DOI] [PubMed] [Google Scholar]
- 54. Golden-Mason L, Bambha KM, Cheng L, Howell CD, Taylor MW, Clark PJ, et al. Natural killer inhibitory receptor expression associated with treatment failure and interleukin-28B genotype in patients with chronic hepatitis C. Hepatology. 2011; 54: 1559–1569. 10.1002/hep.24556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang R, Xu J, Hong K, Yuan L, Peng H, Tang H, et al. Increased NKG2A found in cytotoxic natural killer subset in HIV-1 patients with advanced clinical status. Aids. 2007; 21 Suppl 8: S9–17. 10.1097/01.aids.0000304691.32014.19 [DOI] [PubMed] [Google Scholar]
- 56. Bonorino P, Ramzan M, Camous X, Dufeu-Duchesne T, Thelu MA, Sturm N, et al. Fine characterization of intrahepatic NK cells expressing natural killer receptors in chronic hepatitis B and C. J Hepatol. 2009; 51: 458–467. 10.1016/j.jhep.2009.05.030 [DOI] [PubMed] [Google Scholar]
- 57. Fearon DT. Innate immunity and the biological relevance of the acquired immune response. QJM. 1999; 92: 235–237. [DOI] [PubMed] [Google Scholar]
- 58. Garcia CC, Weston-Davies W, Russo RC, Tavares LP, Rachid MA, Alves-Filho JC, et al. Complement C5 activation during influenza A infection in mice contributes to neutrophil recruitment and lung injury. PloS One. 2013; 8: e64443 10.1371/journal.pone.0064443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Martinez-Vila E, Sieira PI. Current status and perspectives of neuroprotection in ischemic stroke treatment. Cerebrovasc Dis. 2001; 11 Suppl 1: 60–70. [DOI] [PubMed] [Google Scholar]
- 60. Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003; 361: 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tenenbaum T, Essmann F, Adam R, Seibt A, Janicke RU, Novotny GE, et al. Cell death, caspase activation, and HMGB1 release of porcine choroid plexus epithelial cells during Streptococcus suis infection in vitro. Brain Res. 2006; 1100: 1–12. [DOI] [PubMed] [Google Scholar]
- 62. Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005; 73: 1907–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
On day 6, all pigs were humanely euthanized, and lungs were immediately removed. (A) lungs from each group exhibited different extent lesions. Black arrow indicated bleeding and hyperaemia, Blue arrow indicated necrosis, and Red arrow indicated hyperaemia and pulmonary consolidation. (B) Lung pathology scores were evaluated blindly.
(TIF)
Lungs were removed on day 6, and were fixed in formalin and embedded in paraffin, sectioned at 5μm, and stained with hematoxylin and eosin for further histopathologic evaluation.
(TIF)
Lungs were removed on day 6, and were fixed in formalin and embedded in paraffin, sectioned at 5μm, and stained with hematoxylin and eosin for further histopathologic evaluation.
(TIF)
Lungs were removed on day 6, and were fixed in formalin and embedded in paraffin, sectioned at 5μm, and stained with hematoxylin and eosin for further histopathologic evaluation.
(TIF)
Lungs were removed on day 6, and were fixed in formalin and embedded in paraffin, sectioned at 5μm, and stained with hematoxylin and eosin for further histopathologic evaluation.
(TIF)
A total of 376 serum samples from 4 different pig farms were tested for the H1N1 and SS2 antibody by HI and ELISA test respectively.
(DOCX)
The DE genes associated with antigen processing and presentation were assigned based on GO term and manual annotation. Manual annotations were listed in italics. Many genes with multiple functions were only listed in one category.
(DOCX)
The DE genes associated with Complement and coagulation cascades were assigned based on GO term and manual annotation. Manual annotations were listed in italics. Many genes with multiple functions were only listed in one category.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. The raw and processed data of microarray files are available from the NCBI’s Gene expression omnibus database (accession number GSE60172).