Summary
Prions are self‐perpetuating amyloid protein aggregates which underlie various neurodegenerative diseases in mammals and heritable traits in yeast. The molecular basis of how yeast and mammalian prions form spontaneously into infectious amyloid‐like structures is poorly understood. We have explored the hypothesis that oxidative stress is a general trigger for prion formation using the yeast [PSI +] prion, which is the altered conformation of the Sup35 translation termination factor. We show that the frequency of [PSI +] prion formation is elevated under conditions of oxidative stress and in mutants lacking key antioxidants. We detect increased oxidation of Sup35 methionine residues in antioxidant mutants and show that overexpression of methionine sulphoxide reductase abrogates both the oxidation of Sup35 and its conversion to the [PSI +] prion. [PSI +] prion formation is particularly elevated in a mutant lacking the Sod1 Cu,Zn‐superoxide dismutase. We have used fluorescence microscopy to show that the de novo appearance of [PSI +] is both rapid and increased in frequency in this mutant. Finally, electron microscopy analysis of native Sup35 reveals that similar fibrillar structures are formed in both the wild‐type and antioxidant mutants. Together, our data indicate that oxidative stress is a general trigger of [PSI +] formation, which can be alleviated by antioxidant defenses.
Introduction
Prions are novel, protein‐only, infectious agents associated with a group of transmissible neurodegenerative diseases typified by human Creutzfeldt Jakob disease. In mammalian cells, conversion of the normal prion protein (PrPC) into its infectious PrPSc conformation underlies the pathogenesis of these transmissible spongiform encephalopathies (Collinge and Clarke, 2007). This ‘protein‐only’ mechanism of infectivity also underpins the unusual inheritance of several novel epigenetic determinants found in the yeast Saccharomyces cerevisiae (Wickner, 1994). Around 10 different proteins are now known to form prions in yeast with many other proteins classified as potential prion candidates (Alberti et al., 2009). The [Het‐s] prion that controls vegetative incompatibility has also been described in Podospora anserina, an unrelated fungal species (Saupe, 2011) illustrating the wider occurrence of prions in fungi. Amongst the most extensively studied of the fungal prions are [PSI +], formed from Sup35, a translation termination factor (Wickner, 1994; Tuite and Cox, 2007) and [URE3], the prion form of the transcriptional regulator, Ure2 (Wickner, 1994). In several cases, the infectious behaviour of the fungal prion associated with a particular phenotype has been directly demonstrated (Maddelein et al., 2002; King and Diaz‐Avalos, 2004; Tanaka et al., 2004), including [RNQ +], the prion form of Rnq1 (Patel and Liebman, 2007), a protein of undetermined function. The [RNQ +] prion is also one of a number of prions generically designated as [PIN +] that are required for the de novo formation of [PSI +] (Derkatch et al., 1996; 2001; Osherovich and Weissman, 2001). Studies on fungal prions have not only provided direct evidence for the ‘protein‐only’ nature of prion‐associated traits but also revealed how prions can contribute to the regulation of a range of cellular processes, some of which may be beneficial to the host cell (Tuite and Serio, 2010).
As with the formation of the mammalian PrPSc prion, fungal prions can also arise de novo; however, the underlying molecular mechanism is poorly understood in both cases. PrPc is thought to adopt an alternative conformational state by spontaneous misfolding event(s) triggered possibly by mutation, mistranslation, environmental stresses and/or by disruption of the chaperone network (DeMarco and Daggett, 2005). The frequency of de novo formation of the [PSI +] prion in yeast is regulated by environmental, cellular and epigenetic factors (Tuite et al., 2007; Tyedmers et al., 2008). Studies of [PSI +] suggest that several misfolded molecules of Sup35 may spontaneously form a catalytically active oligomer that initiates prion formation (Serio et al., 2000). The [PSI +] prion arises de novo at a frequency of ∼ 10−5–10−7 (Lund and Cox, 1981; Lancaster et al., 2010), but this frequency is elevated approximately 1000‐fold by overexpression of either the full length Sup35 or the prion NM domain of Sup35 (Chernoff et al., 1993). Several different environmental stress conditions, including oxidative stress, are capable of increasing the frequency of [PSI +] prion formation (Tyedmers et al., 2008). Previously, we have demonstrated that the de novo formation of the [PSI+] and [PIN +] prions is increased in yeast mutants lacking the peroxiredoxin proteins Tsa1 and Tsa2 (Sideri et al., 2010; 2011). Peroxiredoxins play multiple roles in protecting cells against stress; in particular, they suppress potentially harmful oxidative damage to proteins following oxidative stress. These findings implicated oxidative damage, of Sup35p, as an important trigger for the formation of the heritable [PSI+] prion in yeast.
Oxidative damage following exposure to reactive oxygen species (ROS) is a well‐established trigger of protein misfolding (Dalle‐Donne et al., 2003). Oxidative stress has been implicated in the de novo formation of mammalian prions, where elevated levels of oxidized methionine (Met) residues are detected in misfolded PrPSc relative to the native PrPC form of the protein (Wolschner et al., 2009; Canello et al., 2010; Elmallah et al., 2013). Oxidative damage to Met residues in purified PrPC has been proposed to facilitate the structural conversion underlying the sporadic formation of PrPSc (Younan et al., 2012). Similarly, we have shown that Met oxidation of Sup35 is elevated in tsa1 tsa2 mutants (Sideri et al., 2011). Abrogating Sup35 methionine oxidation in a tsa1 tsa2 mutant by overexpressing methionine sulphoxide reductase (MSRA) prevents [PSI +] formation, indicating that Sup35 oxidation may underlie the switch from a soluble to an aggregated prion form of Sup35 (Sideri et al., 2011). Protein oxidation may therefore be a common mechanism underlying the aggregation of both mammalian and yeast amyloidogenic proteins.
Formation of Sup35 prion aggregates sequesters Sup35 away from its normal function of translation termination resulting in elevated levels of stop‐codon read‐through and the generation of C‐terminally extended polypeptides. It has been suggested that the shift to the [PSI +] prion state, and the associated increase in stop‐codon read‐through, provides a mechanism for generating heritable phenotypic diversity by allowing cells to ‘reprogram’ gene expression, such that new genetic traits become uncovered that potentially aid survival through adverse conditions (Tyedmers et al., 2008). For example, resistance to several environmental stress conditions correlates with the [PSI +] versus [psi −] status of cells (Eaglestone et al., 1999; True and Lindquist, 2000; True et al., 2004). Thus, most phenotypic alterations are thought to arise due to changes in translation termination efficiency (True et al., 2004). In agreement with this idea, the [PSI +] prion has now been found in a number of wild yeast strains and shown to confer diverse phenotypes that are frequently beneficial under selective conditions (Halfmann et al., 2012). In addition, we have previously shown that prion formation provides yeast cells with an adaptive advantage under oxidative stress conditions, since elimination of prions from tsa1 tsa2 mutants renders the cells hypersensitive to hydrogen peroxide (Sideri et al., 2010).
In this study, we show that oxidation of Sup35 is a common response to oxidative stress and results in Sup35 conversion to the [PSI +] state, presumably due to structural transitions favouring conversion to the propagatable [PSI+] form. We found the frequency of [PSI+] prion formation is elevated both in mutants lacking key antioxidants, and in response to the addition of exogenous oxidants. The increased frequency of [PSI+] formation in multiple antioxidant mutants correlates with elevated levels of methionine oxidation in Sup35, and the switch from a soluble to an aggregated form of Sup35 is suppressed by overexpressing methionine sulphoxide reductase. We have used fluorescence microscopy in conjunction with Sup35NM‐GFP and electron microscopy (EM) analysis of native Sup35 to characterize Sup35 aggregate formation and show that similar fibrillar structures are formed in wild‐type and antioxidant mutants. To our knowledge, this is the first systematic investigation of the role of oxidative stress in yeast prion formation.
Results
[PSI +] prion formation is increased in response to oxidative stress
To assay for [PSI +] formation, we used a ura3–14 allele containing the ade1–14 nonsense mutation engineered into the wild‐type URA3 gene (Manogaran et al., 2006). The ura3–14 allele allows [PSI +] prion formation to be scored by growth on medium lacking uracil, indicative of decreased translational termination efficiency in [PSI +] cells. [PSI +] formation was differentiated from nuclear gene mutations which give rise to uracil prototrophy by their irreversible elimination in 3 mM guanidine hydrochloride (GdnHCl) (Tuite et al., 1981). This simple diagnostic assay has been widely used to identify prion‐associated traits including [PSI +] and [PIN +] (e.g. Derkatch et al., 2001; Ganusova et al., 2006; Tyedmers et al., 2008; Halfmann et al., 2012). Using this assay, the frequency of de novo [PSI +] prion formation in a [PIN +][psi −] strain was estimated to be approximately 5 × 10−5 (Fig. 1A) comparable with previously reported frequencies ranging between ∼ 10−5 and 10−7 (Lund and Cox, 1981; Lancaster et al., 2010).
Figure 1.
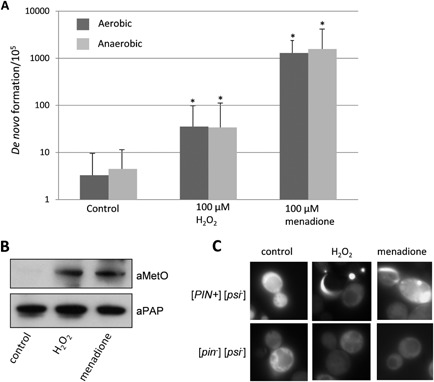
Oxidative stress conditions increase the frequency of [PSI +] prion formation.
A. [PSI +] prion formation was quantified using an engineered ura3–14 allele, which contains the ade1–14 nonsense mutation inserted into the wild‐type URA 3 gene (Manogaran et al., 2006). [PSI +] prion formation was scored by growth on media lacking uracil, indicative of decreased translational termination efficiency. [PSI +] formation was also scored by growth on media lacking uracil under anaerobic conditions. [PSI +] formation was differentiated from nuclear gene mutations which give rise to uracil prototrophy by their irreversible elimination in GdnHCl. The control [PIN +] [psi −] strain was grown in the presence of 100 μM hydrogen peroxide or 100 μM menadione for 16 h prior to scoring [PSI +] prion formation (in the absence of exogenous oxidants). Data shown are the means of at least three independent experiments expressed as the frequency of [PSI +] prion colonies per 105 viable cells. Error bars denote the standard deviation. *P < 0.05.
B. Western blot analysis of Sup35 oxidation. Sup35 was affinity purified using TAP chromatography from a control [PIN +] [psi −] strain following growth in the presence of 100 μM hydrogen peroxide or 100 μM menadione for 16 h. Western blots were probed with α‐PAP to confirm that similar amounts of Sup35 were purified from each strain. Sup35 oxidation was detected using antibodies that recognize methionine sulphoxide (αMetO).
C. Representative fluorescence micrographs are shown for the [PIN +] [psi +] control strain containing the Sup35NM‐GFP plasmid following growth in the presence of 100 μM hydrogen peroxide or 100 μM menadione for 16 h. The Sup35NM‐GFP plasmid was induced for 2 h using copper prior to visualizing aggregate formation.
We tested whether oxidative stress caused by the addition of exogenous oxidants increases the frequency of [PSI +] prion formation. The control [PIN +][psi −] strain was grown in the presence of 100 μM hydrogen peroxide or the superoxide anion generator menadione for 16 h prior to scoring [PSI +] prion formation. These concentrations of oxidants are quite mild and only moderately slow the growth of a wild‐type strain. Growth in the presence of 100 μM hydrogen peroxide increased the frequency of [PSI +] prion formation by approximately 10‐fold (Fig. 1A). Stronger induction of [PSI +] prion formation was observed in response to the superoxide anion, generated by exposure to menadione, which increased the frequency of formation by approximately 300‐fold in response to 100 μM menadione exposure. To confirm that de novo [PSI +] prion formation occurs during the 16 h exposure to the oxidant rather than during the subsequent growth on selective plates used to score for [PSI +] prion formation, plates were also incubated under anaerobic conditions. No differences in the frequency of [PSI +] prion formation following exposure to hydrogen peroxide or the superoxide anion were observed when cells were grown on plates under aerobic compared with anaerobic conditions (Fig. 1A). These data indicate that the oxidant‐induced de novo [PSI +] prion formation scored in these experiments occurs during exposure to the oxidants in the initial 24 h growth period.
Increased frequency of de novo [PSI +] prion formation is a common feature of antioxidant mutants
The ura3–14 reporter plasmid was introduced into yeast mutants lacking major antioxidants, including superoxide dismutases (sod1, sod2), catalases (ctt1, cta1) and peroxiredoxins (tsa1, tsa2). The frequency of de novo [PSI +] prion formation was significantly increased in tsa1 tsa2 mutants (Fig. 2A), as previously described (Sideri et al., 2010; 2011). Similarly, the frequency of [PSI +] prion formation was elevated by approximately 10‐fold in mutants lacking CTT1, encoding cytosolic catalase. In contrast, loss of peroxisomal CTA1 did not significantly affect [PSI +] prion formation and no further elevation was observed in a double ctt1 cta1 mutant. Loss of SOD1, encoding cytosolic copper–zinc superoxide dismutase, caused the most dramatic effect, increasing the frequency of de novo [PSI +] prion formation almost 200‐fold. Mutants lacking the Sod2 mitochondrial manganese superoxide dismutase showed an increased frequency of [PSI +] prion formation of approximately fourfold, but only a modest further increase was observed in a double sod1 sod2 mutant.
Figure 2.
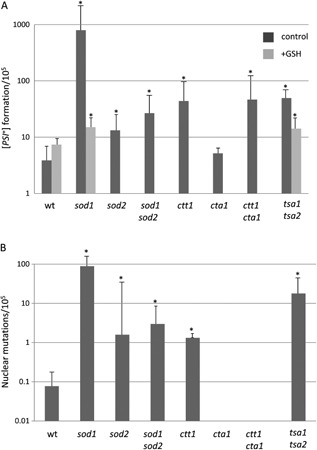
The frequency of [PSI +] formation is increased in antioxidant mutants.
A. [PSI +] prion formation was quantified in [PIN +] [psi −] versions of the indicated antioxidant mutant strains as described for Fig. 1. The addition of 1 mM GSH during the initial 16 h growth period reduced the high frequency of [PSI +] prion formation observed in the sod1 and tsa1 tsa2 mutants to a level approaching that of the wild‐type strain.
B. The nuclear mutation rate was quantified by the formation of Ura + colonies which are not curable with GdnHC. Data shown are the means of at least three independent experiments expressed as the number of colonies per 105 viable cells. Error bars denote the standard deviation. *P < 0.05.
Given the increased frequency of [PSI +] prion formation observed in antioxidant mutants, we tested whether the addition of exogenous glutathione (GSH) could prevent prion formation in antioxidant mutants. GSH is an essential metabolite that protects cells against oxidative stress (Schafer and Buettner, 2001). Exogenous GSH can enter yeast cells and provide protection against oxidative stress (Grant et al., 1996). GSH addition reduced the high frequency of [PSI +] prion formation observed in the sod1 and tsa1 tsa2 mutants to a level approaching that of the wild type, indicating that the increased frequency of prion formation caused by oxidative stress could be abrogated by addition of an exogenous antioxidant.
The nuclear mutation rate was scored in these antioxidant mutants by the formation of Ura+ colonies that are not curable with GdnHCl (Fig. 2B). This is to control for any mutations that stabilize nonsense mRNAs, or any mutations in termination factors (Sup35, Sup45) or the translation apparatus itself that might also cause read‐through at the ura3–14 stop codon. The mutants that showed the highest frequencies of [PSI +] prion formation also tended to have higher frequencies of nuclear mutations. This ranged from approximately 1 × 10−3 in the sod1 mutant to 1 × 10−5 in the ctt1 mutant. However, nuclear mutations were formed at significantly lower frequencies compared with the frequency of de novo [PSI +] prion formation. Taken together, these data indicate that increased [PSI +] prion formation is a common feature of antioxidant mutants.
Sup35 methionine oxidation is increased in antioxidant mutants
We have previously shown that loss of TSA1 and TSA2 results in elevated levels of Sup35 methionine oxidation (Sideri et al., 2011). Abrogating Sup35 methionine oxidation by overexpressing MSRA prevented [PSI +] formation, indicating that Sup35 oxidation may underlie the switch from a soluble to an aggregated prion form of Sup35 in tsa1 tsa2 mutants. We therefore examined whether Sup35 is similarly oxidized in other antioxidant mutants by immunoblot analysis using an antibody that recognizes methionine sulfoxide (MetO). The basal levels of MetO were significantly elevated in antioxidant mutants, which showed an increased rate of [PSI +] prion formation, including sod1, sod2, sod1 sod2, ctt1, ctt1 cta1 and tsa1 tsa2 mutants (Fig. 3A). MetO formation was not detected in Sup35 in the wild‐type strain, nor in a cta1 mutant strain which did not significantly increase the frequency of [PSI +] prion formation.
Figure 3.
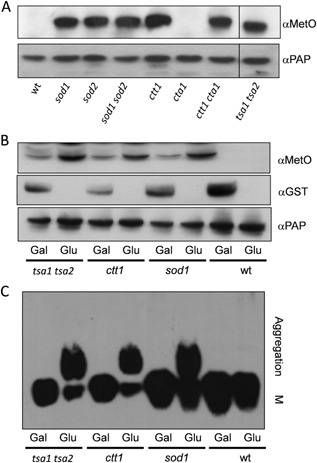
Overexpression of MSRA protects Sup35 against methionine oxidation and prion formation.
A. Sup35 was affinity purified using TAP chromatography from a wild‐type strain and the indicated antioxidant mutants. Western blots were probed with α‐PAP to confirm that similar amounts of Sup35 were purified from each strain. Sup35 oxidation was detected using antibodies that recognize methionine sulphoxide (αMetO).
B. Methionine sulphoxide reductase (MSRA) was overexpressed using plasmid GAL1 ‐ MSRA ‐ GST in wild‐type and antioxidant mutant strains. Overexpression was confirmed under inducing (Gal) versus repressing (Glu) conditions using an anti‐GST antibody (αGST). MSRA expression prevented methionine oxidation of Sup35 detected using the α‐MetO antibody.
C. SDS‐resistant Sup35 aggregates were detected in the same strains using SDD‐AGE. Aggregate and monomer (M) forms are indicated.
If methionine oxidation underlies the switch to the [PSI +] prion, we reasoned that MetO should also be detected in Sup35 from cells exposed to oxidative stress condition that promote prion formation. We therefore examined Sup35 from a control [PIN +][psi −] strain grown in the presence of 100 μM hydrogen peroxide or the superoxide anion generator menadione for 16 h as described in Fig. 1A. These oxidative stress conditions increased the frequency of [PSI +] prion formation (Fig. 1A) and also significantly increased MetO levels.
To determine whether methionine oxidation correlates with Sup35 aggregation, we examined whether overexpression of MSRA could protect against methionine oxidation and prevent [PSI +] prion formation. Wild‐type, ctt1, sod1 and tsa1 tsa2 mutant strains were first transformed with a GAL1‐MSRA‐GST plasmid and then [pin −][psi −] derivatives generated by growth with 3 mM GdnHCl. The resulting [pin −][psi −] transformed colonies were grown in SD or SGal media for approximately 40 generations in liquid culture to allow for the formation of new prions. As expected, no MSRA expression was observed in glucose grown cells and elevated MetO formation was detected in the antioxidant mutants (Fig. 3B). Growth on galactose was confirmed to induce the expression of MSRA. Elevated MSRA correlated with decreased levels of methionine oxidation in the ctt1, sod1 and tsa1 tsa2 mutants (Fig. 4B). Semi‐denaturing detergent–agarose gel electrophoresis (SDD–AGE) was used to examine whether methionine oxidation influences the formation of high molecular weight sodium dodecyl sulfate (SDS)‐resistant Sup35 aggregates, diagnostic of [PSI +] prion formation. High‐molecular weight SDS‐resistant aggregates of Sup35 were observed in ctt1, sod1 and tsa1 tsa2 mutants where MSRA expression was repressed by growth on glucose (Fig. 3C). In contrast, these aggregates did not form when MSRA was overexpressed by growth on galactose (Fig. 3C). These data indicate that methionine oxidation in Sup35 plays a critical role in the de novo formation of the [PSI +] prion in antioxidant mutants.
Figure 4.
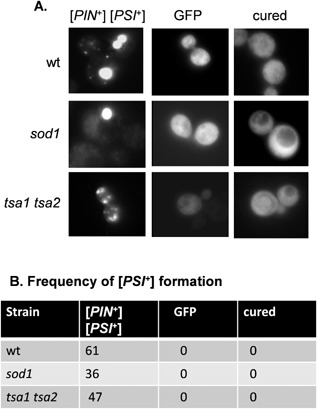
Visualization of [PSI +] prion formation in antioxidant mutants.
A. Representative fluorescence micrographs are shown for [PIN +] [PSI +] versions of the wild‐type, sod1 and tsa1 tsa2 mutant strains containing the Sup35NM‐GFP plasmid. The Sup35NM‐GFP plasmid was induced for 2 h using copper prior to visualizing aggregate formation. Cured strains were analyzed following growth with 3 mM GndHCl. A control GFP plasmid (GFP) resulted in diffuse cytoplasmic fluorescence in all strains examined ruling out any non‐specific effects on protein aggregation in these mutants.
B. The percentage of cells containing visible puncta is shown for each strain from an average of 200 cells counted.
Visualization of Sup35 aggregation in antioxidant mutants
We used a SUP35NM‐GFP fusion protein to visualize Sup35 aggregate formation as previously described (Patino et al., 1995). This construct contains the amino‐terminal prion forming domain of Sup35 under the control of the copper regulatable CUP1 promoter (SUP35NM‐GFP). Our studies focused on the sod1 mutant since it has the highest rate of prion formation detected in any of the antioxidant mutants examined. Following induction with copper, diffuse cytoplasmic fluorescence is observed in [psi −] cells, whereas coalescence of newly made Sup35‐GFP with pre‐existing Sup35 aggregates in [PSI +] allows the detection of [PSI +] foci. The Sup35‐GFP reporter construct was introduced into [PIN +][PSI +] versions of the sod1 and tsa1 tsa2 mutant strains, which were grown for 16 h prior to inducing the expression of SUP35‐GFP for 2 h (Fig. 4A). Many large aggregates of Sup35 were detected in the majority of [PIN +][PSI +] control cells following the 2 h induction period (Fig. 4). Similarly, Sup35‐GFP was detected predominantly as large bright fluorescent foci in approximately 36% of sod1 mutant cells, comparable with the [PIN +][PSI +] control strain. In contrast, the foci were of varying sizes in tsa1 tsa2 mutant cells. We ruled out any non‐specific effects on protein aggregation in these mutants since expression of a control GFP protein resulted in diffuse cytoplasmic fluorescence in all strains examined (Fig. 4). One genetic criterion for a yeast prion is reversible curability (Wickner, 1994). GdnHCl blocks the propagation of yeast prions by inhibiting Hsp104, a molecular chaperone that is absolutely required for yeast prion propagation (Ferreira et al., 2001; Jung and Masison, 2001). Treatment of all the [PIN +][PSI +] strains with GdnHCl resulted in diffuse cytoplasmic Sup35‐GFP fluorescence, with no detectable foci, confirming the requirement for Hsp104 to propagate [PSI+] prion formation in these strains (Fig. 4).
Overexpression of Sup35 in [PIN +][psi −] cells leads to the de novo appearance of the [PSI+] prion since the excess Sup35 increases the possibility for prion seed formation (Wickner, 1994). We found that overexpression of Sup35‐GFP in a [PIN +][psi −] control strain resulted in detectable protein aggregates following 28 h of expression induced by copper addition (Fig. 5), similar to previous reports (Bradley and Liebman, 2004; Mathur et al., 2010). The detectable aggregates included large fluorescent aggregates that arise due to the ‘decoration’ of existing aggregates, as well as rod‐ and ribbon‐like aggregate characteristics of the de novo formation of [PSI+]. We examined the formation of aggregates in antioxidant mutants to determine whether fluorescent foci are detectable at earlier time points. Multiple large foci were detected in approximately 1% of tsa1 tsa2 mutant cells examined within 4 h, but again rod‐ and ribbon‐like aggregates could only be detected within 28 h (Fig. 5). In comparison, small foci were detected in the sod1 mutant following as little as 30 min expression of the Sup35‐GFP construct (Fig. 5). By 4 h, more than 3% of sod1 mutant cells examined contained large fluorescent foci. Rod‐ and ribbon‐like aggregate characteristics of the de novo formation of [PSI+] could also be detected within as little as 4 h in the sod1 mutant. These aggregates were confirmed to be [PSI+] prions since they were curable with GdnHCl. Loss of antioxidants, therefore, both increases the frequency of [PSI+] appearance as well as shortens the time in which detectable aggregates are formed.
Figure 5.
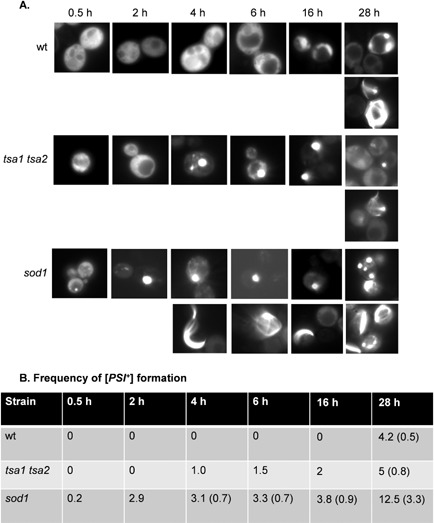
A [PIN +] [psi −] sod1 mutant shows increased frequency and rate of [PSI +] prion formation.
A. Fluorescence micrographs are shown for [PIN +] [psi −] versions of the wild‐type, sod1 and tsa1 tsa2 mutant strains containing the Sup35NM‐GFP plasmid induced with copper for the indicated times. Representative images are shown where puncta formation was first detected in the wild‐type (28 h), sod1 (0.5 h) and tsa1 tsa2 (4 h) strains (upper rows of images). Representative images are shown where rod‐ and ribbon‐like aggregates, indicative of de novo prion formation, were detected in the wild‐type (28 h), sod1 (4 h) and tsa1 tsa2 (28 h) strains (lower rows of images).
B. The percentage of cells containing visible puncta and rod‐ and ribbon‐like aggregates (in parentheses) is shown for each strain from an average of 500 cells counted. The numbers indicate the percentage of cells containing puncta at each time point. Numbers in parentheses are the percentage of cells containing rod‐ or ribbon‐like aggregates.
To further confirm oxidative stress conditions promote [PSI+] formation, we used the Sup35‐GFP reporter to visualize Sup35 aggregate formation following exposure to oxidants. The control [PIN +][psi −] strain was grown in the presence of 100 μM hydrogen peroxide or the superoxide anion generator menadione for 16 h as described in Fig. 1A. Following induction of SUP35‐GFP for 2 h, both puncta and rod‐ and ribbon‐like aggregates could be detected in the cells exposed to hydrogen peroxide or menadione, indicative of [PSI+] formation (Fig. 1C).
Formation of Sup35 fibrils in antioxidant mutants
Prion formation in sod1 mutants has some unusual features relative to wild type including a high frequency of de novo prion formation and a fast appearance of rod‐ and ring‐like aggregate structures. We therefore used EM to further visualize the aggregate structures formed by Sup35 in antioxidant mutants to determine whether they show any differences to the structures formed in control [PSI +] strains. Overexpression of NM‐GFP has previously been used to facilitate visualization of the fibrillar structures of [PSI +] formed inside live yeast cells (Kawai‐Noma et al., 2010; Tyedmers et al., 2010; Saibil et al., 2012). We were able to detect similar structures formed in the [PSI +] control strain and [PSI +] versions of tsa1 tsa2 and sod1 mutants (Fig. 6). General protein aggregation would be expected to be detected as amorphous, irregular structures using EM. Instead, we observed the formation of spherical structures with ordered parallel fibrils in the control and antioxidant mutant strains, suggesting the presence of a similar amyloid fiber organization in these mutants. In all cases, curing with GdnHCl generated [pin −][psi −] cells which contained no visible fibrillar structures (data not shown).
Figure 6.
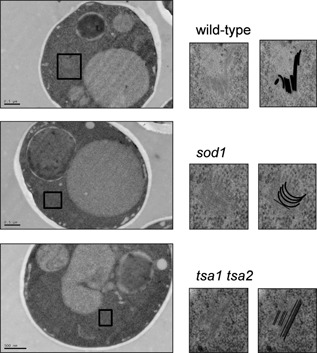
EM images of [PSI +] aggregates in antioxidant mutants. Representative EM micrographs are shown for [PIN +] [PSI +] versions of the wild‐type, sod1 and tsa1 tsa2 mutant strains. The boxed areas in the main images are magnified on the right along with images containing line drawings of each aggregate.
Discussion
Peroxiredoxins (Prx) have multiple roles in stress protection, acting as antioxidants, molecular chaperones and in the regulation of signal transduction (Wood et al., 2003). Tsa1 is the major yeast 2‐Cys Prx and acts as an antioxidant in the detoxification of hydroperoxides (Garrido and Grant, 2002; Wong et al., 2004), particularly as a result of endogenous ROS generated during normal aerobic metabolism (Iraqui et al., 2009). The Tsa2 peroxiredoxin is highly homologous to Tsa1 (86% identity) and possesses similar peroxidase activity, although it is normally expressed at significantly lower levels compared with Tsa1 (Jang et al., 2004; Wong et al., 2004). We previously showed that a tsa1 tsa2 mutant of 74D‐694 gives rise to [PSI+] prion strains at a high frequency (Sideri et al., 2010). In this current study, we have quantified prion formation using a ura3–14 allele‐based assay and show that [PSI+] prion formation is elevated by approximately 11‐fold in a tsa1 tsa2 mutant compared with a wild‐type strain. Given the multiple roles that Prx's play in stress defense, it was unclear if elevated prion formation is a common response to oxidative stress conditions. Our current data indicate that prion formation is similarly increased in a range of antioxidant mutants, as well as in response to the addition of exogenous oxidants suggesting that protein oxidative damage may be a common mechanism underlying the switch from a normal soluble protein to the amyloid prion form of Sup35.
We found that oxidative stress induced by exposure to hydrogen peroxide or menadione increased the frequency of [PSI+] prion formation. Exposure to 100 μM hydrogen peroxide increased the frequency of [PSI+] prion formation by approximately 10‐fold compared with a 300‐fold increase in response to menadione. Menadione is a redox‐cycling agent which transfers electrons to molecular oxygen to generate the superoxide anion, and hence it is difficult to directly compare its dose–response effects on prion formation with that of hydrogen peroxide. H2O2 is a ubiquitous molecule formed as a byproduct of aerobic respiration and following exposure to diverse biological and environmental factors. It must be removed from cells to avoid Fenton and Haber–Weiss reactions, leading to the formation of highly reactive hydroxyl radicals (OH·). The superoxide anion (O2 −) is generated by one electron reduction of oxygen and can be produced from the mitochondrial electron transport chain. The superoxide anion can be dismutated to hydrogen peroxide by superoxide dismutases and can generate hydroxyl radicals via metal‐catalyzed reactions. Hence, hydrogen peroxide and the superoxide anion would be expected to oxidize amino acid residues through the generation of the hydroxyl radical (Berlett and Stadtman, 1997).
Superoxide dismutases (SODs) are ubiquitous antioxidants which convert the superoxide anion to hydrogen peroxide. Yeast contains a cytoplasmic Cu,Zn‐SOD (Sod1) and a mitochondrial matrix Mn‐SOD (Sod2) which play distinct roles during oxidative stress conditions (Culotta et al., 2006). Cells deleted for SOD1 are hypersensitive to the superoxide anion and display a number of oxidative stress‐related phenotypes including vacuole damage and increased free iron concentrations (Culotta, 2000). Mutants deleted for SOD2 are less affected in growth and stress sensitivity compared with sod1 mutants but do show a reduced ability to grow under respiratory conditions (van Loon et al., 1986). Interestingly, loss of SOD1 caused a dramatic increase in [PSI+] prion formation, which was elevated by approximately 200‐fold. In contrast, a modest fourfold increase in [PSI+] prion formation was detected in a sod2 mutant. This difference in prion formation may reflect the difference in superoxide sensitivity between the sod1 and sod2 mutants, with the Sod1 playing the predominant role in protection against endogenous superoxide. It is unclear why sod1 sod2 mutants display reduced [PSI+] prion formation rates compared with a sod1 single mutant, but this may indicate that compensatory changes occur in the double mutant which reduce prion formation.
Catalases are heme‐containing enzymes that catalyze the dismutation of H2O2 into H2O and O2. Yeast expresses a peroxisomal catalase A encoded by CTA1 and a cytosolic catalase T encoded by CTT1. Ctt1 is thought to play a more general role as an antioxidant during exposure to oxidative stress, whereas Cta1 functions in the detoxification of H2O2 generated from fatty acid β‐oxidation (Martinez‐Pastor et al., 1996). [PSI+] prion formation was elevated approximately 10‐fold in ctt1 and ctt1 cta1 mutants, but loss of CTA1 did not appear to affect prion formation. This is in agreement with Ctt1 protecting against endogenous hydrogen peroxide and therefore required to suppress [PSI +] during normal growth conditions, compared with Cta1 which plays a more specialized role in peroxisomes. Thus loss of both superoxide dismutases and catalases, which are major antioxidants in eukaryotic cells, both increased the frequency of de novo formation of the [PSI+] prion, suggesting that prion formation is a common response in cells subjected to oxidative stress.
Reactive oxygen species are toxic molecules that can potentially oxidize all amino acids in proteins, including Met residues, which are particularly susceptible to oxidation by ROS (Dean et al., 1997). We previously detected elevated levels of methionine oxidation in tsa1 tsa2 mutants (Sideri et al., 2011), and similarly, MetO levels were found to be increased in sod1, sod2, sod1 sod2, ctt1 and ctt1 cta1 mutants, which all display increased [PSI+] prion formation. A similar increase in the levels of Mets–SO formation was observed in all of these mutants, despite the finding that they showed significant differences in the frequency of de novo [PSI+] prion formation. This may indicate that methionine oxidation is not the most important form of protein oxidative damage for prion formation. Alternatively, the Western blots used to detect MetO levels may not provide a quantitative measure of methionine oxidation. Nevertheless, no increase in MetO levels was detected in cta1 mutants, which was unaffected in prion formation. We previously showed that overexpression of MSRA prevents MetO formation in a tsa1 tsa2 mutant and largely prevented the conformational shift from soluble Sup35 to the insoluble aggregated form (Sideri et al., 2011). Similarly, overexpression of MSRA in antioxidant mutants protected against methionine oxidation in these mutants and abrogated Sup35 aggregation. These data confirm that methionine oxidation of Sup35 plays a critical role in the de novo formation of [PSI+] during oxidative stress conditions.
Since the spontaneous frequency of [PSI+] prion formation is so low, de novo formation is often visualized by overexpression of Sup35, as overexpression of Sup35 causes very high rates of [PSI+] formation (Derkatch et al., 1997). Overexpression of SUP35NM‐GFP in [PIN +][psi −] cells facilitates the detection of ring‐, rod‐ and ribbon‐like aggregates. These ring‐like structures are found in the cell periphery or surrounding the vacuole and mature into an infectious prion state detected as large dot‐like aggregates (Zhou et al., 2001; Ganusova et al., 2006). Overexpression of SUP35NM‐GFP in [PIN +][PSI +] cells can give rise to cells with multiple large fluorescent puncta, where Sup35‐GFP decorates pre‐existing aggregates. We found that overexpression of SUP35NM‐GFP for 2 h in [PIN +][PSI +] versions of sod1 and tsa1 tsa2 mutant cells gave rise to multiple large fluorescent foci. The foci detected in the tsa1 tsa2 mutant were more variable in size relative to the control [PIN +][PSI+] strain and sod1 mutant. Overexpression of SUP35NM‐GFP in [PIN +][psi −] versions of sod1 and tsa1 tsa2 mutant cells gave rise to detectable fluorescent foci at much earlier time points than in the control [PIN +][psi −] strain. This was most apparent in the sod1 mutant, where small foci were detected within 30 min and larger foci were detected in more than 3% of cells examined following 4 h of overexpression of SUP35NM‐GFP. Large foci were also detected in approximately 1% of tsa1 tsa2 mutant cells following overexpression of SUP35NM‐GFP for 4 h. In contrast, large fluorescent puncta were only detected in the control [PIN +][psi −] strain within 28 h of overexpression of SUP35NM‐GFP. Rare, ring‐ and rod‐like structures, which are thought to represent de novo [PSI +] formation, were also detected at this time point. In contrast, ring‐ and rod‐like structures could be detected within 4 h in the sod1 mutant. Thus, the high frequency of [PSI +] prion formation in sod1 mutants correlates with the earlier de novo appearance of [PSI +] prion aggregates.
Overexpression of SUP35NM‐GFP in [PIN +][PSI +] cells gives rise to cells with multiple large aggregates that have been characterized using EM (Kawai‐Noma et al., 2010; Tyedmers et al., 2010; Saibil et al., 2012). Analysis of native Sup35 in sod1 and tsa1 tsa2 mutants revealed the presence of similar spherical structures with ordered parallel fibrils, which are comparable with a control strain. Additionally, similar SDS‐resistant high molecular weight aggregates were detected using SDD–AGE analysis in wild‐type and antioxidant mutants. Thus, although the frequency of [PSI +] formation is elevated during oxidative stress conditions, there does not appear to be any obvious structural differences in the protein aggregates formed under these conditions. Oxidative stress is a major biological problem that has proposed causal relationships with many disease processes including cancer, neurodegenerative disease and cardiovascular disease (Gutteridge, 1993). All organisms are exposed to ROS during the course of normal aerobic metabolism or following exposure to radical‐generating compounds. In addition, there are many established links between oxidative stress and aging, and, for example, ROS may play a causal role in cellular decline during aging (Harman, 1972). The molecular basis by which mammalian and fungal prions arise spontaneously is poorly understood at present. Our data are therefore important since they indicate a potential causal link for oxidative protein damage, in the switch from a normal soluble protein to the amyloid form.
Loss of yeast SOD1 had the most dramatic effect on [PSI +] formation, increasing the frequency of prion formation by approximately 200‐fold when compared with the wild‐type strain. Additionally, overexpression of SUP35NM‐GFP in a [PIN +][psi −] strain resulted in the detection of prion‐like structures immediately after expression. The observed puncta were most likely aggregates that had already formed in cells, and were decorated with newly expressed Sup35 following induction of SUP35NM‐GFP. Moreover, rod‐ and ring‐like structures, which are thought to represent the initial stages in de novo prion formation (Mathur et al., 2010), were detected in sod1 mutants after 4 h of SUP35NM‐GFP expression, compared with 28 h in other strains. In other work, we have shown that the frequency of de novo formation of the [PIN +] prion is similarly increased in a sod1 mutant strain (data not shown). Thus, two sequence‐unrelated yeast prions, [PSI +]/Sup35 and [PIN +]/Rnq1, are similarly formed in a sod1 mutant, suggesting that prion formation may be a common phenomenon in this mutant. Mutations in human SOD1 have been associated with amyotrophic lateral sclerosis (ALS), which is a fatal neurodegenerative disease (Sreedharan and Brown, 2013). Hence, our data may also indicate an unexpected link between ALS and prion diseases, which both involve protein aggregation. The pathology associated with ALS is not believed to arise as a consequence of the inactivation of Sod1, but misfolding and aggregation of the Sod1 protein itself are thought to cause downstream neurotoxic events. Loss of Sod1 activity may not directly contribute to the pathology of ALS, but it may have additional pathological effects via increasing the frequency of the sporadic de novo formation of spongiform neuropathies.
Experimental procedures
Yeast strains and plasmids
The wild‐type strain 74D‐694 (MATa ade1–14 ura3–52 leu2–3, 112 trp1–289 his3–200) and its isogenic derivatives deleted for TSA1 (tsa1::LEU2) and TSA2 (tsa2::kanMX) have been described previously (Sideri et al., 2010). Strains deleted for SOD1 (sod1::TRP1), SOD2 (sod2::HIS), CTT1 (ctt1::TRP1) and CTA1 (cta1::HIS3) were constructed in 74D‐694 using standard yeast methodology. Sup35 was tagged at its C‐terminus in with a tandem affinity purification (TAP) tag and has been described previously (Sideri et al., 2011). Overexpression of methionine sulphoxide reductase (MSRA) was achieved using a GAL1‐MSRA‐GST plasmid (pEGH) supplied by Open Biosystems. This plasmid expresses the MXR1 gene encoding methionine S‐sulphoxide reductase.
Growth and stress condition
Strains were grown at 30°C with shaking at 180 rpm in rich yeasts extract peptone dextrose (YEPD) medium (2% w/v glucose, 2% w/v bactopeptone, 1% w/v yeast extract) or minimal SD (0.67% w/v yeast nitrogen base without amino acids, 2% w/v glucose) supplemented with appropriate amino acids and bases. SGal media contained 2% w/v galactose and SRaf media contained 2% w/v raffinose in place of glucose. Media were solidified by the addition of 2% (w/v) agar. Strains were cured by three rounds of growth on YEPD agar plates containing 3 mM guanidine hydrochloride (GdnHCl).
Determination of spontaneous [PSI +] prion formation
A plasmid containing an engineered ura3–14 allele, which contains the ade1–14 nonsense mutation in the wild‐type URA3 gene, was used to score the frequency of [PSI +] prion formation (Manogaran et al., 2006). Briefly, freshly grown cells were inoculated into 25 ml of media selective for the plasmid (SD minus Leu) in 250 ml flasks and incubated with vigorous aeration for 16 h. Cells were washed and appropriate dilutions were plated on selective plates. A number of viable colonies were scored on plates selective for the plasmid (SD minus Leu) after 3 day incubation at 30°C. [PSI +] prion formation was scored by growth on SD minus uracil following incubation at room temperature for 3 weeks (Manogaran et al., 2006). For anaerobic conditions, plates were incubated in an anaerobic jar (Oxoid) containing a gas generating kit (anaerobic system BR38; Oxoid). Each colony was spotted and scored as [PSI+] if it was able to grow on plates selective for [PSI+] but not in the presence of GdnHCl. The number of non‐prion derivatives was estimated by growing 200 of the resulting colonies on selective plates containing GdnHCl. The absence of growth indicates that it was caused by prion formation. Each experiment was performed at least in triplicate.
Visualization of aggregation of prion proteins
De novo [PSI +] prion formation was visualized as described previously using CUP1‐SUP35NM‐GFP (Sideri et al., 2011). Cells were grown for 16 h in SD minimal media, diluted to OD600 0.1–0.2 and 50 μM copper sulphate added for induction of the CUP1 promoter and visualized using an Olympus wide‐field microscope and MetaVue software (Bioimaging Facility, Faculty of Life Science, University of Manchester).
Protein analysis
The analysis of Sup35 amyloid polymers by SDD–AGE was performed as described previously (Alberti et al., 2010). Sup35‐TAP was performed as described by Sideri et al. (2011). Methionine oxidation of Sup35 was detected using αMetO antibodies (Novus Biologicals).
Electron microscopy
Samples for EM were prepared using the super quick freeze substitution method described previously (McDonald and Webb, 2011). High pressure freezing was performed using a Baltec HPM‐010 high‐pressure freezer. Samples were embedded in LR White resin, 70 nm sections cut on a Reichert Ultracut microtome and observed using an FEI Tecnai 12 Biotwin microscope at 100 kV acceleration voltage. Images were acquired using a GATAN Orius SC1000 CCD camera.
Acknowledgements
This work was funded by BBSRC grants BB/J000183/1 (to CG) and BB/J000191/1 (to MFT). The authors wish to thank Dr. Aleksandr Mironov in the Faculty of Life Sciences EM Facility for his assistance and the Wellcome Trust for equipment grant support to the EM Facility.
References
- Alberti, S. , Halfmann, R. , King, O. , Kapila, A. , and Lindquist, S. (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137: 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti, S. , Halfmann, R. , and Lindquist, S. (2010) Biochemical, cell biological, and genetic assays to analyze amyloid and prion aggregation in yeast. Methods Enzymol 470: 709–734. [DOI] [PubMed] [Google Scholar]
- Berlett, B.S. , and Stadtman, E.R. (1997) Protein oxidation in aging, disease, and oxidative stress. J Biol Chem 272: 20313–20316. [DOI] [PubMed] [Google Scholar]
- Bradley, M.E. , and Liebman, S.W. (2004) The Sup35 domains required for maintenance of weak, strong or undifferentiated yeast [PSI+] prions. Mol Microbiol 51: 1649–1659. [DOI] [PubMed] [Google Scholar]
- Canello, T. , Frid, K. , Gabizon, R. , Lisa, S. , Friedler, A. , Moskovitz, J. , and Gasset, M. (2010) Oxidation of Helix‐3 methionines precedes the formation of PK resistant PrP. PLoS Pathog 6: e1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff, Y.O. , Derkach, I.L. , and Inge‐Vechtomov, S.G. (1993) Multicopy SUP35 gene induces de‐novo appearance of psi‐like factors in the yeast Saccharomyces cerevisiae . Curr Genet 24: 268–270. [DOI] [PubMed] [Google Scholar]
- Collinge, J. , and Clarke, A.R. (2007) A general model of prion strains and their pathogenicity. Science 318: 930–936. [DOI] [PubMed] [Google Scholar]
- Culotta, V.C. (2000) Superoxide dismutase, oxidative stress, and cell metabolism. Curr Top Cell Regul 36: 117–132. [DOI] [PubMed] [Google Scholar]
- Culotta, V.C. , Yang, M. , and O'Halloran, T.V. (2006) Activation of superoxide dismutases: putting the metal to the pedal. Biochim Biophys Acta 1763: 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle‐Donne, I. , Giustarini, D. , Colombo, R. , Rossi, R. , and Milzani, A. (2003) Protein carbonylation in human diseases. Trends Mol Med 9: 169–176. [DOI] [PubMed] [Google Scholar]
- Dean, R.T. , Fu, S. , Stocker, R. , and Davies, M.J. (1997) Biochemistry and pathology of radical‐mediated protein oxidation. Biochem J 324: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco, M.L. , and Daggett, V. (2005) Local environmental effects on the structure of the prion protein. C R Biol 328: 847–862. [DOI] [PubMed] [Google Scholar]
- Derkatch, I.L. , Chernoff, Y.O. , Kushnirov, V.V. , Inge‐Vechtomov, S.G. , and Liebman, S.W. (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae . Genetics 144: 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch, I.L. , Bradley, M.E. , Zhou, P. , Chernoff, Y.O. , and Liebman, S.W. (1997) Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147: 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch, I.L. , Bradley, M.E. , Hong, J.Y. , and Liebman, S.W. (2001) Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 106: 171–182. [DOI] [PubMed] [Google Scholar]
- Eaglestone, S.S. , Cox, B.S. , and Tuite, M.F. (1999) Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion‐mediated mechanism. EMBO J 18: 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmallah, M.I. , Borgmeyer, U. , Betzel, C. , and Redecke, L. (2013) Impact of methionine oxidation as an initial event on the pathway of human prion protein conversion. Prion 7: 404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, P.C. , Ness, F. , Edwards, S.R. , Cox, B.S. , and Tuite, M.F. (2001) The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol 40: 1357–1369. [DOI] [PubMed] [Google Scholar]
- Ganusova, E.E. , Ozolins, L.N. , Bhagat, S. , Newnam, G.P. , Wegrzyn, R.D. , Sherman, M.Y. , and Chernoff, Y.O. (2006) Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol 26: 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido, E.O. , and Grant, C.M. (2002) Role of thioredoxins in the response of Saccharomyces cerevisiae to oxidative stress induced by hydroperoxides. Mol Microbiol 43: 993–1003. [DOI] [PubMed] [Google Scholar]
- Grant, C.M. , MacIver, F.H. , and Dawes, I.W. (1996) Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae . Curr Genet 29: 511–515. [DOI] [PubMed] [Google Scholar]
- Gutteridge, J.M.C. (1993) Free radicals in disease processes: a compilation of cause and consequence. Free Radic Res Commun 19: 141–158. [DOI] [PubMed] [Google Scholar]
- Halfmann, R. , Jarosz, D.F. , Jones, S.K. , Chang, A. , Lancaster, A.K. , and Lindquist, S. (2012) Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482: 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman, D. (1972) The biologic clock: the mitochondria? J Am Geriatr Soc 20: 145–147. [DOI] [PubMed] [Google Scholar]
- Iraqui, I. , Kienda, G. , Soeur, J. , Faye, G. , Baldacci, G. , Kolodner, R.D. , and Huang, M.E. (2009) Peroxiredoxin Tsa1 is the key peroxidase suppressing genome instability and protecting against cell death in Saccharomyces cerevisiae. PLoS Genet 5: e1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, H.H. , Lee, K.O. , Chi, Y.H. , Jung, B.G. , Park, S.K. , Park, J.H. , et al (2004) Two enzymes in one; two yeast peroxiredoxins display oxidative stress‐dependent switching from a peroxidase to a molecular chaperone function. Cell 117: 625–635. [DOI] [PubMed] [Google Scholar]
- Jung, G. , and Masison, D.C. (2001) Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol 43: 7–10. [DOI] [PubMed] [Google Scholar]
- Kawai‐Noma, S. , Pack, C.G. , Kojidani, T. , Asakawa, H. , Hiraoka, Y. , Kinjo, M. , et al (2010) In vivo evidence for the fibrillar structures of Sup35 prions in yeast cells. J Cell Biol 190: 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, C.Y. , and Diaz‐Avalos, R. (2004) Protein‐only transmission of three yeast prion strains. Nature 428: 319–323. [DOI] [PubMed] [Google Scholar]
- Lancaster, A.K. , Bardill, J.P. , True, H.L. , and Masel, J. (2010) The spontaneous appearance rate of the yeast prion [PSI +] and its implications for the evolution of the evolvability properties of the [PSI +] system. Genetics 184: 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon, A.P.G.M. , Pesold‐Hurt, B. , and Schatz, G. (1986) A yeast mutant lacking mitochondrial manganese‐superoxide dismutase is hypersensitive to oxygen. Proc Natl Acad Sci USA 83: 3820–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, P.M. , and Cox, B.S. (1981) Reversion analysis of [psi−] mutations in Saccharomyces cerevisiae. Genet Res 37: 173–182. [DOI] [PubMed] [Google Scholar]
- McDonald, K.L. , and Webb, R.I. (2011) Freeze substitution in 3 hours or less. J Microsc 243: 227–233. [DOI] [PubMed] [Google Scholar]
- Maddelein, M.L. , Dos Reis, S. , Duvezin‐Caubet, S. , Coulary‐Salin, B. , and Saupe, S.J. (2002) Amyloid aggregates of the HET‐s prion protein are infectious. Proc Natl Acad Sci USA 99: 7402–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manogaran, A.L. , Kirkland, K.T. , and Liebman, S.W. (2006) An engineered nonsense URA3 allele provides a versatile system to detect the presence, absence and appearance of the [PSI+] prion in Saccharomyces cerevisiae. Yeast 23: 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Pastor, M.T. , Marchler, G. , Schuller, C. , Marchler‐Bauer, A. , Ruis, H. , and Estruch, F. (1996) The Saccharomyces cerevisae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress‐response element. EMBO J 15: 2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Mathur, V. , Taneja, V. , Sun, Y. , and Liebman, S.W. (2010) Analyzing the birth and propagation of two distinct prions, [PSI+] and [Het‐s](y), in yeast. Mol Biol Cell 21: 1449–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich, L.Z. , and Weissman, J.S. (2001) Multiple Gln/Asn‐rich prion domains confer susceptibility to induction of the yeast [PSI +] prion. Cell 106: 183–194. [DOI] [PubMed] [Google Scholar]
- Patel, B.K. , and Liebman, S.W. (2007) Prion‐proof' for [PIN+]: infection with in vitro‐made amyloid aggregates of Rnq1p‐(132–405) induces [PIN+]. J Mol Biol 365: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino, M.M. , Liu, J.J. , Glover, J.R. , and Lindquist, S. (1995) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273: 622–626. [DOI] [PubMed] [Google Scholar]
- Saibil, H.R. , Seybert, A. , Habermann, A. , Winkler, J. , Eltsov, M. , Perkovic, M. , et al (2012) Heritable yeast prions have a highly organized three‐dimensional architecture with interfiber structures. Proc Natl Acad Sci USA 109: 14906–14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe, S.J. (2011) The [Het‐s] prion of Podospora anserina and its role in heterokaryon incompatibility. Semin Cell Dev Biol 22: 460–468. [DOI] [PubMed] [Google Scholar]
- Schafer, F.Q. , and Buettner, G.R. (2001) Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med 30: 1191–1212. [DOI] [PubMed] [Google Scholar]
- Serio, T.R. , Cashikar, A.G. , Kowal, A.S. , Sawicki, G.J. , Moslehi, J.J. , Serpell, L. , et al (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289: 1317–1321. [DOI] [PubMed] [Google Scholar]
- Sideri, T.C. , Stojanovski, K. , Tuite, M.F. , and Grant, C.M. (2010) Ribosome‐associated peroxiredoxins suppress oxidative stress‐induced de novo formation of the [PSI+] prion in yeast. Proc Natl Acad Sci USA 107: 6394–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideri, T.C. , Koloteva‐Levine, N. , Tuite, M.F. , and Grant, C.M. (2011) Methionine oxidation of Sup35 protein induces formation of the [PSI+] prion in a yeast peroxiredoxin mutant. J Biol Chem 286: 38924–38931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan, J. , and Brown, R.H.J. (2013) Amyotrophic lateral sclerosis: problems and prospects. Ann Neurol 74: 309–316. [DOI] [PubMed] [Google Scholar]
- Tanaka, M. , Chien, P. , Naber, N. , Cooke, R. , and Weissman, J.S. (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428: 323–328. [DOI] [PubMed] [Google Scholar]
- True, H.L. , and Lindquist, S.L. (2000) A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483. [DOI] [PubMed] [Google Scholar]
- True, H.L. , Berlin, I. , and Lindquist, S.L. (2004) Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431: 184–187. [DOI] [PubMed] [Google Scholar]
- Tuite, M. , Byrne, L. , Josse, L. , Ness, F. , Koloteva‐Levine, N. , and Cox, B. (2007) Yeast prions and their analysis in vivo . Methods in Microbiology 36: 491–526. [Google Scholar]
- Tuite, M.F. , and Cox, B.S. (2007) The genetic control of the formation and propagation of the [PSI+] prion of yeast. Prion 1: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite, M.F. , and Serio, T.R. (2010) The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Biol 11: 823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite, M.F. , Mundy, C.R. , and Cox, B.S. (1981) Agents that cause a high frequency of genetic change from [psi+] to [psi‐] in Saccharomyces cerevisiae . Genetics 98: 691–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers, J. , Madariaga, M.L. , and Lindquist, S. (2008) Prion switching in response to environmental stress. PLoS Biol 6: e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers, J. , Treusch, S. , Dong, J. , McCaffery, J.M. , Bevis, B. , and Lindquist, S. (2010) Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci USA 107: 8633–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, R.B. (1994) URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264: 566–5699. [DOI] [PubMed] [Google Scholar]
- Wolschner, C. , Giese, A. , Kretzschmar, H.A. , Huber, R. , Moroder, L. , and Budisa, N. (2009) Design of anti‐ and pro‐aggregation variants to assess the effects of methionine oxidation in human prion protein. Proc Natl Acad Sci USA 106: 7756–7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, C.M. , Siu, K.L. , and Jin, D.Y. (2004) Peroxiredoxin‐null yeast cells are hypersensitive to oxidative stress and are genomically unstable. J Biol Chem 22: 23207–23213. [DOI] [PubMed] [Google Scholar]
- Wood, Z.A. , Schroder, E. , Harris, J.R. , and Poole, L.B. (2003) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28: 32–40. [DOI] [PubMed] [Google Scholar]
- Younan, N.D. , Nadal, R.C. , Davies, P. , Brown, D.R. , and Viles, J.H. (2012) Methionine oxidation perturbs the structural core of the prion protein and suggests a generic misfolding pathway. J Biol Chem 287: 28263–28275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Derkatch, I.L. , and Liebman, S.W. (2001) The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion‐like elements [PSI(+)] and [PIN(+)]. Mol Microbiol 39: 37–46. [DOI] [PubMed] [Google Scholar]


