Abstract
Background
BAP1 is a nuclear deubiquitinase that regulates gene expression, transcription, DNA repair, and more. Several findings underscore the apparent “driver” role of BAP1 in malignant mesothelioma (MM). However the reported frequency of somatic BAP1 mutations in MM varies considerably, a discrepancy that appeared related to either methodological or ethnical differences across various studies.
Methods
To address this discrepancy, we carried out comprehensive genomic and immunohistochemical (IHC) analyses to detect somatic BAP1 gene alterations in 22 frozen MM biopsies from US MM patients.
Results
By combining Sanger sequencing, Multiplex Ligation-Dependent Probe Amplification, copy number analysis and cDNA sequencing, we found alteration of BAP1 in 14/22 biopsies (63.6%). No changes in methylation were observed. IHC revealed normal nuclear BAP1 staining in the 8 MM containing wild-type BAP1, while no nuclear staining was detected in the 14 MM biopsies containing tumor cells with mutated BAP1. Thus, IHC results were in agreement with those obtained by genomic analyses. We then extended IHC analysis to an independent cohort of 70 MM biopsies, of which there was insufficient material to perform molecular studies. IHC revealed loss of BAP1 nuclear staining in 47 out of these 70 MM biopsies (67.1%).
Conclusions
Our findings conclusively establish BAP1 as the most commonly mutated gene in MM, regardless of ethnic background or other clinical characteristics. Our data point to IHC as the most accessible and reliable technique to detect BAP1 status in MM biopsies.
Keywords: Sporadic Malignant Mesothelioma, Somatic BAP1 mutation, MLPA
INTRODUCTION
Malignant mesothelioma (MM) is an aggressive tumor that arises from the mesothelial cells that form the lining of the pleural, pericardial, and peritoneal cavities 1. In the US, an estimated 3,200 people are diagnosed with MM each year, with nearly 100,000 new cases expected to occur over the next 40 years 2. The association between exposure to asbestos and to other mineral fibers and MM is well established 3. Only a fraction of asbestos-exposed individuals develop MM, indicating that other factors may contribute to the development of the disease 1. We have shown that in some families an extremely high susceptibility to develop MM is transmitted in an autosomal dominant fashion 4. Recently, we identified germline mutations in the BRCA1-associated protein 1 (BAP1) gene as the cause of a novel cancer syndrome characterized by a very high incidence of MM and uveal melanoma 5, 6, and benign atypical melanocytic lesions7, known as MBAITs (Melanocytic BAP1-mutated Atypical Intradermal Tumors) 8, and also by an elevated risk of other malignancies, such as cutaneous melanoma 7, renal cell carcinoma 9, cholangiocarcinoma 10, basal cell carcinoma 11, and possibly more 5, 6.
BAP1 is a nuclear deubiquitinase, which belongs to the ubiquitin C-terminal hydrolase (UCH) family 12. Among UCH family members, BAP1 is unique because of its long C-terminal tail, which contains two nuclear localization signals (NLS1 at 656–661 and NLS2 at 717–722) 13, 14. It has been postulated that BAP1 functions as a tumor suppressor 6, possibly through three or more mechanisms, all requiring the nuclear localization of the BAP1 protein: 1) as component of the Polycomb repressive deubiquitinase (PR-DUB) complex, BAP1 deubiquitinates histone H2A, leading to transcriptional activation of genes that regulates cell growth 15; 2) BAP1 acts as a transcription coregulator, associating with Host Cell Factor-1 (HCF-1), YY1, and E2F1 to induce transcription of genes involved in cell cycle regulation 12, 16–18; and 3) BAP1 contributes to the process of DNA repair 6, 19, 20. Whether BAP1 also has some presently unknown cytoplasmic activity remains unknown.
Somatic BAP1 mutations are found in sporadic MM (i.e., MMs that occur in individuals that do not carry germline BAP1 mutations), as well as in other malignancies, although the frequency of BAP1 mutations varies widely across different tumor types. Pena-Llopis et al. detected somatic BAP1 mutations in 14% (24/176 tumors) of renal cell carcinoma specimens using Sanger and whole genome sequencing 21, while Harbour et al. detected mutations in 84% (26/31) of metastasizing uveal melanoma biopsies using next generation sequencing 22. In sporadic MM, by using Sanger sequencing, we found somatic BAP1 mutations in 22% (4/18) of US Caucasian MM biopsies 5. These results are in accordance with the results of Bott et al. 23 and Zauderer et al., 24, who found 23% (12/53) and 20% (24/121) BAP1 mutations in sporadic US MM, also by Sanger sequencing. We described loss of BAP1 nuclear staining in MM tumor biopsies containing mutated BAP1 5. We and Bott et al. 23 also noted loss of BAP1 nuclear staining in some MM biopsies that apparently had wild-type BAP1 according to Sanger sequencing. Arzt el al.25 revealed absence of BAP1 nuclear staining in 60% of 123 MM biopsies in a study that was based exclusively on IHC. Yoshikawa et al. found that BAP1 was mutated in 61% (14/23) of cell cultures derived from pleural fluids of Japanese MM patients 26. In this study, Yoshikawa et al. performed Array CGH, Sanger sequencing and real-time PCR on DNA extracted from tumor cells established in tissue culture, after several subcultures. Instead, our study 5, and the studies of Bott et al., 23, and Zauderer et al., 24 were based on Sanger sequencing of tumor DNA from primary frozen tissue biopsies. The significant difference in frequency of BAP1 mutations reported may due to differences in ethnicities of the patients studied, lack of sensitivity and/or reproducibility of Sanger sequencing or IHC in detecting BAP1 mutations, procedure of tumor cell isolation and accumulation of novel BAP1 mutations by MM cells in tissue culture 26, lack of specificity of IHC, or some other differences in methodology. To investigate the basis of the discrepancies between these studies and to try to conclusively address this issue, we performed multiplex molecular analyses to comprehensively identify all possible genetic alterations of the BAP1 gene in sporadic MM biopsies. We found that BAP1 is mutated in over 60% of MM specimens.
MATERIALS AND METHODS
Specimen Collection
Twenty-two MM frozen biopsies and matching normal leukocytes were obtained from the New York University (NYU) Langone Medical Center (HIP). MM samples were harvested during surgery and immediately frozen in liquid nitrogen.
An independent cohort of 70 formalin-fixed paraffin-embedded tissue slides was obtained from the National Mesothelioma Virtual Bank (NMVB) at the University of Pittsburgh). These latter specimens were used exclusively to extend BAP1 IHC analyses, as the amount of tissue available was insufficient to perform molecular studies. Specimens were de-identified prior to analysis. Gender, ethnicity, asbestos exposure, age at diagnosis, histology and stage data were collected and are reported in the Supplemental Table 1A, Supplemental Digital Content 1A (SDC1A).
Germline BAP1 wild type (WT) DNAs were obtained from healthy volunteers. Written and informed consent was obtained from all patients included in these studies according to the guidelines set forth by the Institutional IRBs.
Laser Capture Micro-dissection
Laser capture micro-dissection was performed to enrich tumor cells from frozen tissue specimens for DNA and RNA isolation. The detailed procedure is described in the Supplemental Material SDC2.
DNA and RNA Extraction
DNA was extracted using DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany), or QiAamp DNA Micro Kit (Qiagen 56304) as per manufacturer’s instructions. Total RNA was extracted using mirVana miRNA isolation kit (Ambion, Austin, TX) as per manufacturer’s instructions. Sample quality was evaluated by nanodrop. The 260/280 ratio was between 1.66 and 2.07 for DNA samples, and between 1.94 and 2.15 for RNA samples.
Real Time-Polymerase Chain Reaction (RT-PCR)
Total RNA was converted to cDNA using ImProm-II™ Reverse Transcription System (Promega, Madison, WI), following the manufacturer’s instructions. A double-strand cDNA library was synthesized using the LD PCR kit (SMART cDNA Library Construction Kit, Clontech Laboratory, Mountain View, CA) according to the manufacturer’s instructions.
DNA and RNA Sequencing Analysis
Conventional Sanger sequencing was performed as described 5. Genomic BAP1 PCR product sizes ranged from 560–670 bp with 100–150 bp overlap between primer sets. A similar primer design was employed for cDNA sequencing. Sequencing was conducted using the ABI 3730XL DNA Sequencer, at the Advanced Studies in Genomics, Proteomics and Bioinformatics facility at the University of Hawai’i at Manoa.
Multiplex Ligation-Dependent Probe Amplification Assay
MLPA is a reliable technique extensively validated for detection of medium and large, gene alterations, which cannot be detected by conventional Sanger sequencing 27, 28. MLPA assay was performed according to the manufacturer’s protocol (SALSA MLPA probemix P417-B1 BAP1, MRC-Holland, Amsterdam, The Netherlands). The detailed procedure is described in the Supplemental Material SDC2 and Supplemental Figure SDC3.
Immunohistochemistry (IHC)
IHC analysis of BAP1 protein expression was performed as described using a mouse monoclonal anti-BAP1 antibody (C-4: Santa Cruz Biotechnology, Dallas, TX) 5. This antibody recognizes the epitope between aa 430 and 739, therefore it is predicted to detect BAP1 wild-type and mutant forms that retain the nuclear localization signal. We had previously reported that BAP1 is expressed in normal pleural cells 8. We extensively validated this antibody for the detection of nuclear BAP1 on a number of normal human pleural samples –all showing nuclear staining in 100% of pleural cells, as well as on MM-derived cell lines in which BAP1 gene status has been comprehensively characterized in our laboratory. Histologically normal pleura and liver biopsies from autopsies were used as positive and negative controls, respectively, for BAP1 nuclear staining. Liver biopsies were chosen because they were shown to contain negligible amounts of BAP1 (http://www.proteinatlas.org/ENSG00000163930-BAP1/tissue/liver). We confirmed that BAP1 does not stain the nuclei of liver cells.
Two US Board certified pathologists with extensive experience in MM diagnosis, M.C and A.P., independently reviewed all IHC results and concurred with their interpretation. The patients characteristics of the control materials are not known and this information cannot be obtained due to HIPAA (Health Insurance Portability and Accountability Act) constraints.
Promoter Methylation Analysis
Tumor DNA was modified with sodium bisulfide by using EZ DNA methylation kit (ZYMO research, Orange, CA) according to the manufacturer’s protocol. The detailed procedure is described in the Supplemental Material SDC2.
Whole gene methylation Analysis
BAP1 gene body methylation analysis was performed Using the Methyl Primer Express® Software v1.0 (Applied Biosystems, Grand Island, NY), as described in the Supplemental Material SDC2.
TaqMan Copy Number Assay
TaqMan copy number assay was performed at the Genomics Shared Resource, University of Hawai’i Cancer Center, according to the manufacturer’s instructions (see Supplemental Material SDC2).
Statistical analysis
We studied the relationship between BAP1 mutation status and age, gender, ethnicity, asbestos exposure and histology using Fisher’s exact test. Epithelioid MMs were compared to all other histological types; MM from patients older than 65 years were compared to all the others; patients exposed to asbestos were compared to all the others. Overall survival was estimated using the Kaplan–Meier method. Patients alive at the end of the study were censored at the time of the last available follow-up. Using the log-rank test we compared survival between MM patients with altered BAP1 versus patients with wild-type BAP1.
RESULTS
Sanger Sequencing and IHC for BAP1
Sanger sequencing was performed on DNA extracted from laser-micro-dissected tumor cells and from matching germline DNA isolated from blood leukocytes of each of 22 MM patients. Matching germline DNA was found to contain the wild-type BAP1 gene in 22/22 samples. Sequencing analysis detected BAP1 alterations in a total of 6 of the 22 MM biopsies tested (27.3%, Table 1), which is in accordance with our previous findings 5. The 22 MM biopsies included 15 samples that had been tested previously by Sanger sequencing, whose DNA was re-extracted from a separate vial containing frozen tumor tissue, and 7 samples not previously analyzed yet. All 22 samples were analyzed blindly to test for reproducibility. The results obtained using Sanger sequencing were very reproducible as the same 4 samples that had tested positive for BAP1 mutations, NYU647, NYU658, NYU866, and NYU937 previously 5, retested positive, and the 11 samples that had tested negative 5, re-tested negative (Table 1). These findings indicated that Sanger sequencing of DNA from tumor cells laser microdissected from frozen MM biopsies is very sensitive and specific in detecting at least certain types of BAP1 DNA alterations.
Table 1.
Summary of Somatic BAP1 Alterations in Sporadic Malignant Mesotheliomas
| Sample ID | 2011 Nat Gen IDa | Genomic Sequencing | MLPA Analysis | DNA Copy Number Assay |
cDNA Sequencing | BAP1 Nuclear localization |
Prediction of Amino Acid Change |
Comprehensive Interpretation |
Promoter Methylation |
|---|---|---|---|---|---|---|---|---|---|
| NYU-047 | SP-024 | WT | WNL | - | WT | (+) | Normal | 1.7% | |
| NYU-207 | WT | Ex 7 to 13 deletion | Ex7 to 13 deletion | Ex 7 to 13 deletion | (−) | p.Pro147fsX63c | Mutant | 1.3% | |
| NYU-217 | SP-019 | WT | WNL | - | WT | (+) | Normal | 1.6% | |
| NYU-269 | SP-017 | WT | WNL | - | WT | (+) | Normal | 1.3% | |
| NYU-517 | SP-016 | WT | WNL | - | WT | (+) | Normal | 1.7% | |
| NYU-524 | Int 14 splicing site (52,436,887 G>T) | WNL | - | 70nt insertion at Ex 14 and 15 | (−) | p.Glu630fsX23 | Mutant | 1.5% | |
| NYU-540 | SP-025 | WT | Entire gene deletion | Entire BAP1 deletion | - | (−) | Mutant | 1.4% | |
| NYU-559 | SP-022 | WT | Ex4-17 deletion | Ex4-17 deletion | - | (−) | Mutant | 3.5% | |
| NYU-647 | SP-001 | Ex 11 (52,439,219 C deletion) | WNL | - | Ex 11 C deletion | (−) | p.Ser341fsX21 c | Mutant | 1.8% |
| NYU-658 | SP-018 | Ex 17 (52,436,398- 399 CG deletion) | WNL | - | Ex 17 CG deletion | (−) | p.Arg699fsX17 | Mutant | 1.4% |
| NYU-809 | SP-011 | WT | WNL | - | WT | (+) | Normal | 1.5% | |
| NYU-851 | SP-012 | WT | Ex 1,4,15 deletion | WT | Ex11 to 17 deletion | (−) | p.Val335fsx27 d | Mutant | 1.3% |
| NYU-866 | SP-013 | Ex16 (52,436,599–627 29bpdeletion) | Entire deletion of other allele | - | insertion of Int 16 | (−) | p.Ala683fsX3 | Mutant | 1.2% |
| NYU-929 | SP-023 | WT | WNL | - | WT | (+) | Normal | 1.1% | |
| NYU-937 | SP-015 | Ex 9 (52,440,352 G deletion) b | Ex9 deletion | - | Ex 9 G deletion | (−) | p.Val234fsX15 c | Mutant | 1.7% |
| Ex 13 (52,437,664 C deletion) b | WNL | - | Ex13 C deletion | p.Ile499fsX72 | |||||
| NYU-966 | WT | Ex 1&4 deletion | WNL | Ex 12–15 deletion | (−) | Deletion between 375–633 | Mutant | 1.8% | |
| NYU-1017 | SP-009 | WT | WNL | - | WT | (+) | Normal | 1.5% | |
| NYU-1024 | SP-026 | EX14 (52,437,281-52,437,282 G insertion) | Ex 10, 12, 14, 16, and 17 deletion | - | EX14 G insertion | (−) | p.Pro588fsx55 | Mutant | 2.1% |
| NYU-1250 | WT | WNL | - | Deletion in Ex 13 | (−) | p.Tyr418fsx113 | Mutant | 1.5% | |
| NYU-1306 | WT | Triplication | Increase of Ex2,4,7,12,15 and 17 | Ex 12,13,14 deletion | (−) | p.Lys368fsx33 d | Mutant | 1.4% | |
| NYU-1359 | WT | WNL | - | WT | (+) | Normal | 1.6% | ||
| NYU-1419 | WT | Random exon deletion | Decrease of Ex 2,4,7,15 and 17 | WT | (−) | Mutant | 1.4% |
None of these point mutations and alterations were detected in germline DNA samples from these patients.
Average promoter methylation % is shown.
Sample ID from Testa et al., Nature Genetics 43:10, 1022–26 (2011), Supplementary Data Table 1
Point mutations were detected in each allele.
WT: wild type sequence
WNL: within normal limit (duploid, 2 copies)
- : not tested
BAP1 protein nuclear expression: (+) positive, (−) negative
Triplication: gain of gene copy number (3 copies)
Putative BAP1 mutant form may not be detected by BAP1 antibody.
Putative BAP1 mutant form may be detected by BAP1 antibody.
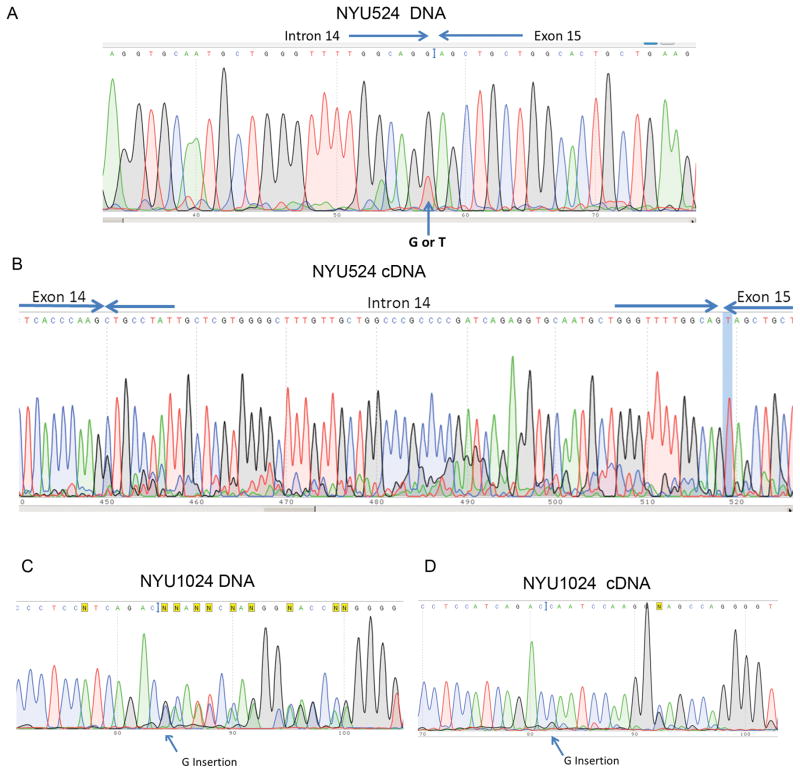
In addition, we identified 2 new BAP1 mutations among the 7 new MM biopsies, while 5 tested negative (BAP1 wild type). A point mutation (G → T) was found at the end of intron 14 on the genomic DNA of NYU524 (Figure 1A). cDNA sequencing of this region revealed a 70 nucleotide insertion of an extra sequence from intron 14 (Figure 1B). A single G insertion in exon 14 was detected in MM NYU1024 by sequencing both genomic DNA and cDNA (Figure 1C). This variant encodes a presumed truncated BAP1 protein (642aa) missing the nuclear localization signal (NLS) as a result of the frameshift mutation (Figure 1D). Overall, Sanger sequencing detected BAP1 mutations in 6 out of 22 samples (27.3%) all predicted to result in truncated proteins lacking the NLS.
Figure 1. Point mutations of BAP1 genes detected by Sanger sequencing.
(A) Tumor NYU524. Genomic DNA sequencing shows point mutation (G → T) at the splicing site at the end of intron 14. (B) cDNA sequencing revealed a 70 nt insertion from part of intron 14 (c.1891_1892Ins 70, p.Glu630fsX23). (C) Tumor NYU1024. Genomic sequencing displayed G insertion in exon 14. (c.1762_1763 Ins G) (D) cDNA sequence of the corresponding region, (p.Pro588fsx55).
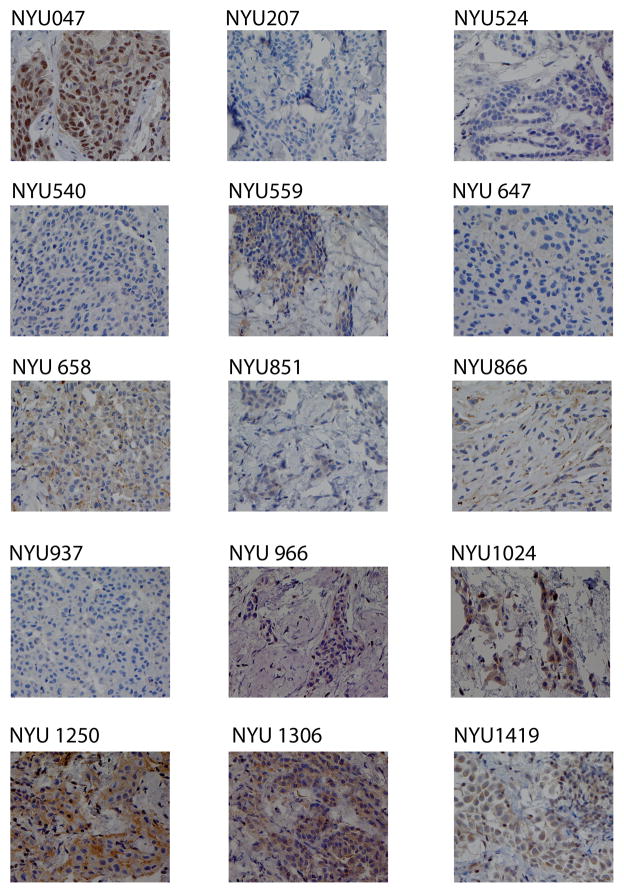
IHC showed complete lack of BAP1 nuclear staining in 100% of tumor cells of these 6 mutated samples. Unexpectedly, in 8 additional biopsies that appeared to contain wild-type BAP1 by Sanger sequencing, no nuclear staining was detected. Overall, 14/22 MM biopsies (63.6%) contained tumor cells lacking BAP1 nuclear staining. Instead, the nuclei of the tumor cells of the additional 8 MM biopsies carrying wild-type BAP1, as detected by Sanger sequencing, uniformly (all tumor cells) stained for BAP1 (Figure 2).
Figure 2. Detection of BAP1 protein expression in sporadic MM tumor samples by immunohistochemistry.
Wild-type BAP1 tumor sample NYU047 shows strong nuclear staining in all tumor cells. All samples carrying alteration of BAP1 gene are shown (Photomicrographs magnification: 400x).
MLPA, TaqMan Copy Number Analysis & cDNA sequencing detected a number of previously unidentified gross alterations of BAP1 in somatic MM
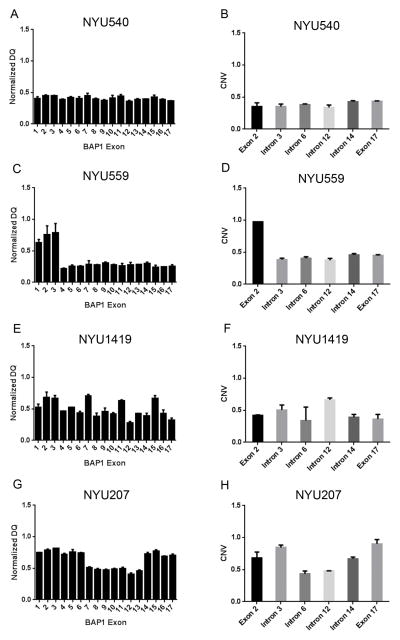
Tumor DNA from NYU540, NYU559, NYU1419, NYU207, NYU851, NYU966, NYU1306 and NYU1250, appeared to contain wild-type BAP1 according to Sanger sequencing, while none of the tumor cells displayed nuclear BAP1 staining (Figure 2). NYU540 tumor DNA displayed homozygous deletion of the entire BAP1 gene by MLPA analysis (average NDQ of all exon = 0.387; Figure 3A; NDQ, Normalized Dosage Quotient). This deletion was confirmed by TaqMan copy number analysis (Figure 3B). NYU559 tumor DNA showed a homozygous deletion from exon 4 to exon 17 by MLPA analysis (exon 4–17 average NDQ = 0.267; Figure 3C and D). MLPA also revealed a homozygous deletion of the BAP1 gene in NYU1419, with the lowest NDQ value of 0.27 for exon 12 (Figure 3E). These results were confirmed by TaqMan copy number assay (Figure 3F). A 3919bp deletion spanning exon 7 to 13 was detected in NYU207 tumor DNA by MLPA analysis and TaqMan copy number assay (NDQ value of 0.407 at exon 12; Figure 3G and H), which was further confirmed by genomic DNA sequencing using primers specifically designed to detect this deletion (Supplemental Figure SDC4). cDNA sequencing (c.437_1729del 1293) revealed that this deletion would result in a putative truncated protein (210 aa) lacking the NLS.
Figure 3. Gross BAP1 gene alterations detected by MLPA and TaqMan copy number assays.
MLPA assay is measured at 17 points, one each on 17 exons of BAP1 gene. TaqMan copy number assay is measured at six points on BAP1 gene; one each on exon 2, 3, 6, 12, 14 & 17. (A, B) NYU540 MLPA and TaqMan data (C, D) NYU559 MLPA and TaqMan data (E, F) NYU1419 MLPA and TaqMan data (G, H) NYU207 MLPA and TaqMan data.
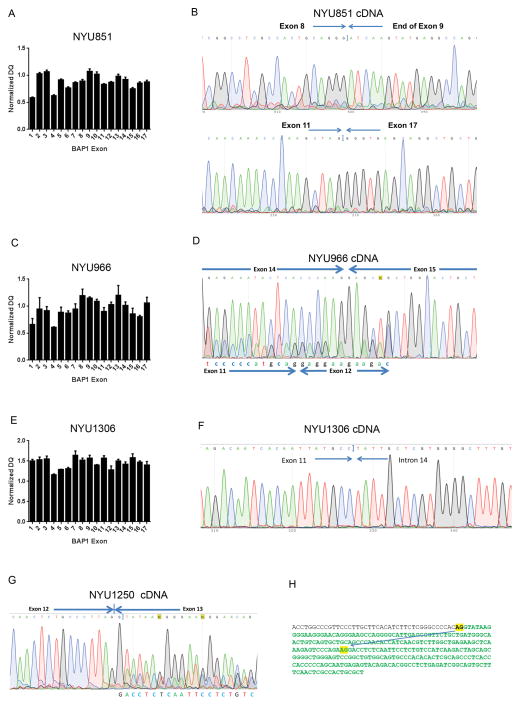
Three additional MM biopsies (NYU851, NYU966, and NYU1306) appeared to contain wild-type BAP1 by Sanger sequencing, yet they lacked nuclear BAP1 staining (Figure 2). These 3 tumors displayed various types of copy number alterations in BAP1 exons by MLPA. As shown in Figure 4, NYU851 tumor DNA showed low NDQ values in exon 1, 4, and 15 (Figure 4A). Aberrant splicing was found between exon 8 and exon 9 and cDNA sequencing identified the deletion of 1820 nucleotides from 1003 to 2822 (c.1003_2822 del 1820, Figure 4B). NYU966 showed decreased copy number of exon 1 and exon 4 (Figure 4C) while cDNA sequencing identified a transcript with deletion of 769 nucleotides from 1128 to 1896 (c.1128_1896del 769, Figure 4D). An increase in exon copy number was detected in multiple BAP1 exons using MLPA in tumor NYU1306, particularly exons 1–3, 7–9, and 10–17 showing a NDQ value above 1.4. (Figure 4E). This copy number increase was confirmed by TaqMan Assay (Supplemental Figure SDC5). Moreover, NYU1306 cDNA sequencing identified a deletion of 791 nucleotides from 1101 to 1891 leading to a putative truncated BAP1 (c.1101_1891 del 791, 400aa, Figure 4F). NYU1250 also lacked nuclear BAP1 staining (Figure 2). However, in this sample, neither Sanger sequencing nor MLPA could identify BAP1 alterations. cDNA sequencing detected a splicing mutant (c.1250_1371 del 122) in which the end of exon 12 was juxtaposed to the middle of exon 13 (Figure 4G). A cryptic splicing site AG was found at the junction, which may generate a putative BAP1 protein of only 531 amino acids lacking the NLS (p.Tyr418fsx113, Figure 4H). This splicing variant has not been reported previously (National Center for Biotechnology Information, and GeneCards from the Weizmann Institute of Science).
Figure 4. Detection of aberrant forms of BAP1 by MLPA and cDNA sequencing.
(A) Tumor NYU851. MLPA assay detected deletions of exons 1, 4, and 15. (B) NYU851 cDNA sequencing revealed two aberrant splicing at Exon 8–9 and Exon 11–17 in one allele. (C) MLPA on NYU966 showed deletion of exon 1 and 4. (D) Tumor NYU966 cDNA sequencing revealed a cDNA with skipping of most of exon 12, 13, 14, and 6 bp of exon 15 as well as a wild type cDNA (exons 14 and 15 shown). (E). MLPA on NYU1306 detected triplication, copy number increase to 3 copies, in most exons. (F) NYU1306 cDNA sequencing revealed a deletion of exon 12 and 13. (G). NYU1250 cDNA sequencing revealed a new splicing isoform of BAP1 due to a 121bp deletion in exon 13. (H) Aberrant usage of criptic splicing site AG, found in NYU1250, is shown by arrow on reference DNA sequence at intron 12 and exon 13. Nucleotides in green indicate the exon sequence.
Three samples showed BAP1 alterations using both, Sanger sequencing and MLPA. NYU937 showed a reduced NDQ of exon 9. Sanger sequencing revealed a point mutation in the region of the MLPA probe for exon 9. (Supplemental Figure, SDC3). MLPA also detected allele deletions in NYU tumor biopsies 866 and 1024 (Supplemental Figure SDC6), which were shown by Sanger sequencing to contain DNA mutations in the other allele and lacked BAP1 nuclear staining (Figure 1C and 2). Together these analyses revealed loss of heterozygosity (LOH) for these two samples.
No deletions were detected by MLPA, or by other techniques, in MM samples NYU047, NYU0217, NYU269, NYU517, NYU809, NYU929, NYU1017 and NYU1359. IHC revealed positive BAP1 nuclear staining in 100% of the tumor cells in these 8 biopsies (Figure 2 and Table 1).
BAP1 promoter and body Methylation are not altered in Malignant Mesothelioma
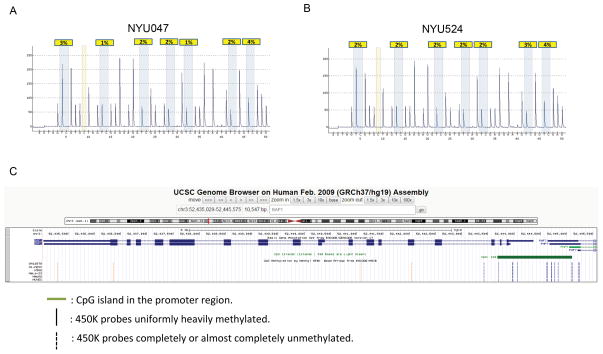
We sought to determine whether DNA methylation could be an additional mechanism leading to BAP1 inactivation in sporadic MM. Transcriptional silencing may occur through methylation around the transcriptional start site, therefore, seven CpG loci, located about 450 bp upstream of ATG start codon, were used to check the methylation status of BAP1 (Supplemental Figure SDC7A). Bisulfite-converted pyrosequencing revealed 86–98% methylation in a human high methylation genomic DNA control (Supplemental Figure SDC7B), while 2–4% methylation was detected in the low methylation control (Supplemental Figure SDC7C). We analyzed all 22 sporadic MM biopsies and found a consistently low level (1–4%) of methylation in all the samples. Methylation analysis of patient samples NYU047 and N524 are shown as an example (Figure 5 and Table 1). These results indicate that hypermethylation is not a significant mechanism for BAP1 inactivation in MM. Recently, gene body methylation was found positively correlated with gene expression 29. Therefore, we investigated whether BAP1 body methylation may influence BAP1 gene expression. Using The Cancer Genome Atlas (TCGA) database we analyzed BAP1 methylation in several tumor types, at nineteen sites spanning the entire BAP1 promoter, gene and 3′ untranslated region (Figure 5C). Four probes located in the body region and 3′ untranslated region were uniformly heavily methylated in all non-malignant and malignant tissues examined. The remaining 15 probes including all the ones located in the promoter/exon 1 region were completely or nearly completely unmethylated in all tissue samples examined (Figure 5C and Supplemental Tables SDC8 and 9), support the interpretation that methylation plays no role in the inactivation of the BAP1 gene.
Figure 5. BAP1 promoter and body methylation analysis.
(A) Sample NYU047 (B) Sample NYU524. Low BAP1 promoter methylation was detected by pyrosequencing in all sporadic MM samples tested. Pyrosequencing values detected in MM tumor samples are as low as the normal control DNA. Two representative samples are shown, where yellow highlights indicate the percentage of methylation at each site. (C) The Cancer Genome Atlas portal database (https://tcga-data.nci.nih.gov/tcga/) was examined for methylation at BAP1 gene, which was covered by nineteen probes (data generated by use of the Infinium HumanMethylation450 BeadChip).
Extended IHC analyses on a separate MM cohort
Our genetic results were very consistent with BAP1 IHC results, as all BAP1 gene alterations detected resulted in loss of BAP1 nuclear staining by IHC. Thus, IHC appeared a reliable method to identify MM biopsies containing mutated BAP1. To extend these results, we performed BAP1 IHC on unstained paraffin embedded glass slides from an independent, annotated cohort from the NMVB (Supplemental Table SDC1B) consisting of a total of 70 cases of US MM biopsies, of which we did not have additional material to conduct genetic analyses. This cohort included 63 Caucasian and 7 non-Caucasian cases. We found loss of BAP1 nuclear staining in 47 out of all 70 MM (67.1%), and in 44 out of the 63 Caucasian MM biopsies (69.8%) (Supplemental Table SDC1B).
Patients’ demographics and possible clinical correlations
Overall, the MM patients studied appeared to have received similar treatments as 45% of BAP1-mutated and 44% of patients with normal BAP1 had surgery, while 35% of BAP1-altered and 31% of patients with normal BAP1 received chemotherapy (Supplemental Table SDC1); thus it is unlikely that treatment influenced some of the results. We did not find significant relationships between frequency of BAP1 alterations and gender, age of diagnosis, ethnicity or history of asbestos exposure (Supplemental Table SDC1). Patients with altered BAP1 had similar survivals than those with normal BAP1 (Supplemental Figure SDC10). This was expected as each cancer contains many genetic alterations and it would be highly unlikely that mutations of a single gene influence prognosis. There was an apparent increased frequency of BAP1 alterations in patients with epithelial type MM, versus other histological subtypes. However, the possible significance of all of these clinical correlates is tempered by the small sample size.
DISCUSSION
Using an integrated molecular approach that included Sanger sequencing, MLPA, TaqMan copy number analysis, mRNA sequencing as well as IHC (Supplemental Figure SDC11), we conclusively demonstrate that the majority (14/22, 63.6%) of sporadic MMs contain somatic BAP1 mutations. Our study underscores the need of using distinct technical approaches to identify the nature of the different types of BAP1 mutations possibly causing lack of BAP1 nuclear staining.
Our results indicate that the use of a single molecular technique is insufficient to detect all types of BAP1 alterations. While Sanger sequencing is reliable to detect single point mutations, which are various and frequent in MM and other cancer types 5, 8, 21, 26, 30, 31, the MLPA assay is much more reliable to detect large deletions 32. We found that Sanger sequencing was very reliable and reproducible in detecting BAP1 point mutations and small deletions in preparations of DNA from laser dissected MM biopsies containing over 80% purity of tumor cells. However, this technique could not detect large DNA deletions. Conversely, MLPA was reliable to detect large exon gains and losses, even under conditions of relatively low sample purity (~ 50% of tumor cells). In this setting, MLPA is cost-efficient compared to array CGH, fluorescence in situ hybridization, and gene copy number assay 33. However, MLPA could not detect single nucleotide changes unless they fall within the MLPA probe target regions. In our study, 6/14 mutations were detected using conventional Sanger sequencing. Of these 6 mutations, 5 were not detected by MLPA because the mutated regions were small and not covered by the MLPA probe. On the other hand, 7 additional aberrant forms of the BAP1 gene were identified by the MLPA assay. Since the regions of exon copy number changes in these seven biopsies are large, Sanger sequencing could not detect these mutations. For accurate detection of deletions of large exons, such as exon 13 of BAP1, several MLPA probes had to be used. Abnormal splicing forms were detected in one MM biopsy –in which both Sanger sequencing and MLPA had not found any alteration- by RNA sequencing. Introns contain splicing enhancers and silencers 34–36, which may partially explain the presence of some aberrant splicing variants in these MM samples.
Six of 14 MM biopsies had homozygous BAP1 deletions as determined by both genomic analyses and IHC. For the remaining 8 additional mutated samples, lack of nuclear staining supports LOH. However, molecular studies could not definitely establish LOH, possibly because traces of contaminant normal cell DNA were present in the samples.
There was 100% concurrence between detection of some molecular alterations in 14/22 biopsies and absence of BAP1 nuclear staining (Table 1 and Figure 2). Strong nuclear and weaker cytoplasmic BAP1 IHC reliably identified the 8 MM biopsies containing wild-type BAP1 from 14 biopsies containing mutated BAP1, whose nuclei did not stain for BAP1. Among the 14 mutated biopsies, 7 contained tumor cells lacking BAP1 staining entirely (Figure 2). Cytoplasmic BAP1 tumor cell staining was detected in 7 biopsies with genetically mutated BAP1 (Figure 2). Cytoplasmic staining can be expected when mutations result in a truncated BAP1 protein lacking the nuclear localization signal or bearing mutations in the catalytic subunit that prevent BAP1 auto-deubiquitination which is required for nuclear localization 37. All mutated forms of BAP1 detected here and in previous publications fall in this category, and therefore are expected to lead to similar biological effects, as they all cause complete BAP1 protein loss or BAP1 isoforms sequestered in the cytoplasm and thus loss of BAP1 nuclear activity.
These results suggest that IHC may be the most reliable and accessible method to identify MM biopsies that harbor BAP1 genetic alterations, while much more expensive and time consuming genetic approaches can be used to: 1) confirm the IHC findings, 2) detect heterozygosity, and 3) understand the mechanisms leading to loss of BAP1 expression.
To this date all BAP1 activities have been related to nuclear localization 10, 13, 14. Analysis of a separate cohort of MM biopsies from the NMVB revealed lack of BAP1 nuclear staining in 47/70 biopsies (67.1%), confirming the high frequency of BAP1 mutations in US MMs. Of note, in this cohort, six specimens contained a mixed tumor cell population, some with and some without BAP1 nuclear staining, possibly a result related to intra-tumoral heterogeneicity and/or polyclonality 38, 39.
Promoter methylation is a common mechanism of tumor suppressor gene inactivation in several MM and other tumor types 40, 41. For example, RASSF1 (Ras association -RalGDS/AF-6- domain family member 1) and E-cadherin promoter hypermethylation was detected in MM patients 42, 43. High expression of DNA methyltransferases, DNMT1, was detected in pleural MM 44. However, we observed no differences in methylation of the BAP1 promoter or gene body. Similarly, no BAP1 methylation alterations were found in clear cell renal carcinoma 45. Whole body methylation was also examined, considering the increasing volume of publications documenting a role of body methylation in gene expression 29. While methylation data on MM are not available yet within the TCGA database, the data collected from multiple other tumor types indicate that methylation plays no role in the inactivation of the BAP1 gene. In summary, DNA methylation does not appear to be a mechanism that contributes to BAP1 inactivation in sporadic MM.
Although the information of demographics, histology and survival is presented, we are not elaborating about possible clinical correlation with BAP1 status as the relatively small size of this cohort may bias interpretation.
In summary, our results demonstrate that: 1) BAP1 inactivation in MM is achieved primarily by DNA mutation and changes in exon copy number; 2) different types of mutations lead to BAP1 lacking nuclear localization as shown by absence of BAP1 nuclear staining in all of the mutated samples and; 3) ethnicity does not appear to influence the frequency of BAP1 alterations. Previously CDKN2A/2B (cyclin-dependent kinase inhibitor 2A/2B) and NF2 (Neuriofibromin 2) have been considered the most commonly mutated tumor suppressor genes in MM as they are found mutated in 30–50%, and 35–40% of human MM biopsies, respectively 46, 47. However, the high percentage of BAP1 mutations that we found in sporadic MM (>60%) now identifies BAP1 as the most commonly mutated gene in this malignancy.
Our findings are supported by two recent next generation sequencing studies (NGS) of the MM genome by Guo et al. 48 and lo Iacono et al. 49. These two NGS studies revealed that various inactivating mutations occur randomly and are rarely shared among MM biopsies, with the exception of BAP1 that was found mutated in 41% 48 and 58% 49 of MMs respectively, and to a lesser extent NF2, CDKN2A (Refs48,49) and possibly CUL1 (Cullin 1)48. Lo Iacono et al. also performed IHC studies and found that 52% of 116 MM biopsies from 116 patients stained for nuclear BAP1 – an indication of normal BAP1 activity – while 48% did not, an indication of mutated BAP1 49. BAP1 staining correlated with presence/absence of DNA mutations (p=0.001) - of note in the same study NF2 staining did not -, independently supporting our findings that IHC is a reliable technology to identify BAP1 mutations 49.
Guo et al. performed whole exome sequencing of 22 MM biopsies from 22 MM patients and found 490 mutated genes of which 447 were mutated only in one biopsy. BAP1 alterations were the most frequent mutations as they were detected in 41% of the 22 biopsies they studied, followed by CDKN2A and NF2. Six of these 22 MM biopsies were also included in our study (NYU 269, 517, 647, 658, 929, and 937), allowing a direct comparison of the sensitivity of the technology used to detect BAP1 mutations. In accordance with our results, Guo et al. found that samples NYU269, 517, and 929 contained wild-type BAP148. They also detected the identical BAP1 mutation in sample NYU937 (p.Ile499fsX15). However, while we detected homozygous mutations in samples NYU647 and 658 by Sanger sequencing–further confirmed by lack of nuclear staining by IHC-, Guo et al. did not (Table 1). This discrepancy may be due to the limitations of the NGS approach, which can produce base substitution errors, and to the fact that, differently from Guo et al. we laser microdissected tumor cells from the biopsy, thus decreasing/eliminating the background of contaminating normal cells that contain wild-type BAP1. Alternatively, this discrepancy may be due to intra-tumor heterogeneity 38 and/or polyclonality 39.
Our results and those from these two recent NGS MM studies, underscore the apparent “driver” role of BAP1 in MMs and point at BAP1 as a potentially useful therapeutic target. In addition, somatic mutations in BAP1 are present in other malignancies 6, thus therapies to restore BAP1 activity are of potential relevance to many cancer patients.
Supplementary Material
Acknowledgments
Funding:
This work was supported by National Institute of Health [grant numbers R01CA106567, P01CA114047, P30CA071789 to MC and R01CA160715-0A to HY]; the DoD CDMRP PRCRP Career Development Award to HY, and the V Foundation to MC and HY, the P30CA071789 (UHCC Pathology Shared Resource and UHCC GSR); the Mesothelioma Applied Research Foundation to HY, the United-4 A Cure, the Hawai’i Community Foundation to HY, the National Mesothelioma Virtual Bank, CDC NIOSH 2U24-OH009077-08 to MB and HP and the University of Hawai’i Foundation, which received donations to support mesothelioma research from Honeywell International Inc., to MC.
We thank Mr. Hiroyasu Nishioka for helping our sequencing work during his internship. We also wish to thank Ms. Min-Ae Song at the University of Hawaii Cancer Center Genomics Shared Resource (supported by NCI P30CA071789 to MC), Dr. Shaobin Hou and Dr. Xuehua Wan at the Advanced Studies in Genomics, Proteomics and Bioinformatics (ASGPB) of UH Manoa. We thank Dr. Waqas Amin, at the University of Pittsburg, for his precious help in the process of collecting the NMVB cohort specimens and all related clinical information.
Footnotes
Conflict of Interest Statement:
M. Carbone has pending patent applications on BAP1, and both Dr. Carbone and Dr. Gazdar provide consultation for mesothelioma expertise and diagnosis. The remaining authors declare no competing financial interests.
References
- 1.Carbone M, Ly BH, Dodson RF, et al. Malignant mesothelioma: facts, myths, and hypotheses. J Cell Physiol. 2012;227:44–58. doi: 10.1002/jcp.22724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ismail-Khan R, Robinson LA, Williams CC, Jr, et al. Malignant pleural mesothelioma: a comprehensive review. Cancer Control. 2006;13:255–263. doi: 10.1177/107327480601300402. [DOI] [PubMed] [Google Scholar]
- 3.Baumann F, Ambrosi JP, Carbone M. Asbestos is not just asbestos: an unrecognised health hazard. Lancet Oncology. 2013;14:576–578. doi: 10.1016/S1470-2045(13)70257-2. [DOI] [PubMed] [Google Scholar]
- 4.Roushdy-Hammady I, Siegel J, Emri S, et al. Genetic-susceptibility factor and malignant mesothelioma in the Cappadocian region of Turkey. Lancet. 2001;357:444–445. doi: 10.1016/S0140-6736(00)04013-7. [DOI] [PubMed] [Google Scholar]
- 5.Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carbone M, Yang H, Pass HI, et al. BAP1 and cancer. Nature reviews Cancer. 2013;13:153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiesner T, Obenauf AC, Murali R, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carbone M, Korb Ferris L, Baumann F, et al. BAP1 cancer syndrome: malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J Transl Med. 2012;10:179. doi: 10.1186/1479-5876-10-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popova T, Hebert L, Jacquemin V, et al. Germline BAP1 mutations predispose to renal cell carcinomas. American journal of human genetics. 2013;92:974–980. doi: 10.1016/j.ajhg.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wadt KA, Aoude LG, Johansson P, et al. A recurrent germline BAP1 mutation and extension of the BAP1 tumor predisposition spectrum to include basal cell carcinoma. Clinical genetics. 2014 doi: 10.1111/cge.12501. [DOI] [PubMed] [Google Scholar]
- 12.Misaghi S, Ottosen S, Izrael-Tomasevic A, et al. Association of C-Terminal Ubiquitin Hydrolase BRCA1-Associated Protein 1 with Cell Cycle Regulator Host Cell Factor 1. Mol Cell Biol. 2009;29:2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen DE, Proctor M, Marquis ST, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 14.Ventii KH, Devi NS, Friedrich KL, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Research. 2008;68:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheuermann JC, Alonso AGD, Oktaba K, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–U138. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machida YJ, Machida Y, Vashisht AA, et al. The Deubiquitinating Enzyme BAP1 Regulates Cell Growth via Interaction with HCF-1. J Biol Chem. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eletr ZM, Wilkinson KD. An emerging model for BAP1’s role in regulating cell cycle progression. Cell Biochem Biophys. 2011;60:3–11. doi: 10.1007/s12013-011-9184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu H, Mashtalir N, Daou S, et al. The Ubiquitin Carboxyl Hydrolase BAP1 Forms a Ternary Complex with YY1 and HCF-1 and Is a Critical Regulator of Gene Expression. Mol Cell Biol. 2010;30:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ismail IH, Davidson R, Gagne JP, et al. Germ-line Mutations in BAP1 Impair its Function in DNA Double-Strand break Repair. Cancer research. 2014 doi: 10.1158/0008-5472.CAN-13-3109. [DOI] [PubMed] [Google Scholar]
- 20.Yu H, Pak H, Hammond-Martel I, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:285–290. doi: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harbour JW, Onken MD, Roberson EDO, et al. Frequent Mutation of BAP1 in Metastasizing Uveal Melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bott M, Brevet M, Taylor BS, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21. 1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–U681. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zauderer MG, Bott M, McMillan R, et al. Clinical Characteristics of Patients with Malignant Pleural Mesothelioma Harboring Somatic BAP1 Mutations. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2013;8:1430–1433. doi: 10.1097/JTO.0b013e31829e7ef9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arzt L, Quehenberger F, Halbwedl I, et al. BAP1 protein is a progression factor in malignant pleural mesothelioma. Pathol Oncol Res. 2014;20:145–151. doi: 10.1007/s12253-013-9677-2. [DOI] [PubMed] [Google Scholar]
- 26.Yoshikawa Y, Sato A, Tsujimura T, et al. Frequent inactivation of the BAP1 gene in epithelioid-type malignant mesothelioma. Cancer Science. 2012;103:868–874. doi: 10.1111/j.1349-7006.2012.02223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerutti R, Sahnane N, Carnevali I, et al. Identification of the first case of germline duplication of BRCA1 exon 13 in an Italian family. Fam Cancer. 2010;9:275–282. doi: 10.1007/s10689-009-9315-z. [DOI] [PubMed] [Google Scholar]
- 28.Yamada H, Shinmura K, Ito H, et al. Germline alterations in the CDH1 gene in familial gastric cancer in the Japanese population. Cancer Sci. 2011;102:1782–1788. doi: 10.1111/j.1349-7006.2011.02038.x. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Han H, De Carvalho DD, et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48:856–859. doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Njauw CN, Kim I, Piris A, et al. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PloS one. 2012;7:e35295. doi: 10.1371/journal.pone.0035295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herman S, Varga D, Deissler HL, et al. Medium-sized deletion in the BRCA1 gene: Limitations of Sanger sequencing and MLPA analyses. Genetics and molecular biology. 2012;35:53–56. doi: 10.1590/s1415-47572012005000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Homig-Holzel C, Savola S. Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagnostic molecular pathology : the American journal of surgical pathology, part B. 2012;21:189–206. doi: 10.1097/PDM.0b013e3182595516. [DOI] [PubMed] [Google Scholar]
- 34.Gamazon ER, Stranger BE. Genomics of alternative splicing: evolution, development and pathophysiology. Human genetics. 2014;133:679–687. doi: 10.1007/s00439-013-1411-3. [DOI] [PubMed] [Google Scholar]
- 35.Adamia S, Haibe-Kains B, Pilarski PM, et al. A genome-wide aberrant RNA splicing in patients with acute myeloid leukemia identifies novel potential disease markers and therapeutic targets. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:1135–1145. doi: 10.1158/1078-0432.CCR-13-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandao RD, Tserpelis D, Gomez Garcia E, et al. Detection of exon skipping events in BRCA1 RNA using MLPA kit P002. Molecular biology reports. 2012;39:7429–7433. doi: 10.1007/s11033-012-1575-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mashtalir N, Daou S, Barbour H, et al. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Molecular cell. 2014;54:392–406. doi: 10.1016/j.molcel.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Comertpay S, Pastorino S, Tanji M, et al. Evaluation of clonal origin of malignant mesothelioma. J Transl Med. 2014;12:301. doi: 10.1186/s12967-014-0301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandermeers F, Neelature Sriramareddy S, Costa C, et al. The role of epigenetics in malignant pleural mesothelioma. Lung cancer. 2013;81:311–318. doi: 10.1016/j.lungcan.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 41.Christensen BC, Houseman EA, Godleski JJ, et al. Epigenetic Profiles Distinguish Pleural Mesothelioma from Normal Pleura and Predict Lung Asbestos Burden and Clinical Outcome. Cancer Research. 2009;69:227–234. doi: 10.1158/0008-5472.CAN-08-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christensen BC, Godleski JJ, Marsit CJ, et al. Asbestos exposure predicts cell cycle control gene promoter methylation in pleural mesothelioma. Carcinogenesis. 2008;29:1555–1559. doi: 10.1093/carcin/bgn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer JR, Ohnmacht U, Rieger N, et al. Promoter methylation of RASSF1A, RARbeta and DAPK predict poor prognosis of patients with malignant mesothelioma. Lung cancer. 2006;54:109–116. doi: 10.1016/j.lungcan.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 44.Amatori S, Papalini F, Lazzarini R, et al. Decitabine, differently from DNMT1 silencing, exerts its antiproliferative activity through p21 upregulation in malignant pleural mesothelioma (MPM) cells. Lung cancer. 2009;66:184–190. doi: 10.1016/j.lungcan.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Ibragimova I, Maradeo ME, Dulaimi E, et al. Aberrant promoter hypermethylation of PBRM1, BAP1, SETD2, KDM6A and other chromatin-modifying genes is absent or rare in clear cell RCC. Epigenetics : official journal of the DNA Methylation Society. 2013;8:486–493. doi: 10.4161/epi.24552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirao T, Bueno R, Chen CJ, et al. Alterations of the p16(INK4) locus in human malignant mesothelial tumors. Carcinogenesis. 2002;23:1127–1130. doi: 10.1093/carcin/23.7.1127. [DOI] [PubMed] [Google Scholar]
- 47.Ladanyi M, Zauderer MG, Krug LM, et al. New strategies in pleural mesothelioma: BAP1 and NF2 as novel targets for therapeutic development and risk assessment. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:4485–4490. doi: 10.1158/1078-0432.CCR-11-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo G, Chmielecki J, Goparaju C, et al. Whole-Exome Sequencing Reveals Frequent Genetic Alterations in BAP1, NF2, CDKN2A, and CUL1 in Malignant Pleural Mesothelioma. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-14-1008. [DOI] [PubMed] [Google Scholar]
- 49.Lo Iacono M, Monica V, Righi L, et al. Targeted next-generation sequencing of cancer genes in advanced stage malignant pleural mesothelioma: a retrospective study. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2014 doi: 10.1097/JTO.0000000000000436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.