Abstract
Killer cell immunoglobulin-like receptors (KIRs) are a diverse family of activating and inhibitory receptors expressed on natural killer (NK) cells and T cells, the genes of which show extreme polymorphism. Some KIRs bind to human leucocyte antigen (HLA) class I subgroups, and genetic interactions between KIR genes and their ligand HLA have been shown to be associated with several autoimmune diseases. The present study aimed to investigate whether the combinations of KIR genes and HLA-Cw ligands associate with the susceptibility of systemic lupus erythematosus (SLE). Polymerase chain reaction using sequence-specific primers was used to determine the genotypes of KIR genes and HLA-Cw alleles. We found that the frequencies of HLA-Cw07 were statistically significantly higher in the patient group than those in the control group (P = 0·009). KIR2DS1+HLA−CwLys was more common in subjects with SLE compared to control subjects (P = 0·015). In addition, the frequency of KIR2DS1 was increased in SLE when KIR2DL1/HLA-Cw are absent, and the difference was significant (P = 0·001). KIR genotype and HLA ligand interaction may potentially influence the threshold for NK (and/or T) cell activation mediated through activating receptors, thereby contributing to the pathogenesis of SLE.
Keywords: genes, killer cell immunoglobulin-like receptors, systemic lupus erythematosus
Introduction
Systemic lupus erythematosus (SLE) is a systemic autoimmune disorder characterized by the production of a broad variety of autoantibodies and subsequent immune complex deposition, which results in chronic inflammation in multiple organ systems. Although the pathogenesis of SLE has still not been clarified, the involvement of genetic factors is supported by twin concordance studies as well as by numerous reports on individual associations with candidate genes 1–5. The immunological abnormalities of SLE are thought to be complex, and involve multiple genes encoding different molecules with significant functions in the regulation of the immune system.
Immunological disturbance is of importance, as SLE exhibits myriad aberrations in the immune system that involve both natural killer (NK) and T cells, resulting in the activation of polyclonal B cells and the production of autoantibodies. As a result, the chronic inflammation of multiple organs occurs. A number of studies 6,7 suggested recently that impaired NK cell differentiation in SLE could contribute to immune system dysregulation. As our understanding of NK cell biology has improved, it has become clear that NK cell responses are dictated by the balance between inhibitory and activating signals originating from cell surface receptors 8. Of these receptors, killer cell immunoglobulin-like receptors (KIRs), which are expressed by NK cells and subsets of T lymphocytes, have generated considerable interest in recent years. In fact, the functions of NK and T cells are modulated, in part, by KIRs that recognize HLA class I molecules on target cells.
KIRs are encoded by a family of tightly clustered 14 KIR genes and two pseudogenes on the leucocyte receptor complex at chromosome 19q13·4 9,10. These multi-gene KIRs interact with their polymorphic HLA-A, HLA-B and HLA-C ligands to diversify and individualize the human immune system 11. The binding of inhibitory KIR (designated 2DL and 3DL) to specific HLA molecules has been demonstrated clearly and correlates well with their ability to inhibit NK cytolysis of target cells bearing those HLA allotypes. The major ligands for inhibitory KIRs are HLA-C molecules 12.
There are many allelic variants of HLA-C, but in terms of KIR recognition these can be reduced to two groups: HLA-C1 molecules (characterized by asparagines at position 80) are recognized by inhibitory KIRs, KIR2DL2 and KIR2DL3, whereas HLA-C2 molecules (characterized by lysine at position 80) are recognized by KIR2DL1 10,13. It has been suggested that the ligands for activating KIRs recognize the same HLA-B or HLA-Cw molecules as are recognized by their related inhibitory KIRs, but with poorly binding affinities 14. Overall, upon interaction with HLA class I ligands, KIR genes provide inhibitory or activating signals to regulate the activation of NK cells and T cells, which contributes to the pathogenesis of diverse types of disease. Because both HLA-C and KIR genes are polymorphic and are encoded on different chromosomes, an individual may have both KIRs and the corresponding HLA-C ligands, may have only KIRs and no corresponding HLA-C ligands or may have only the HLA-C ligand but no KIR for a certain HLA–KIR interaction. Such diversity of KIR and HLA-C genes among individuals might be related to the heterogeneity of the immune response to infectious agents or susceptibility to autoimmune or inflammatory diseases 10. In recent years, many studies in different populations have shown associations of specific KIR alleles with HLA-C alleles and autoimmune diseases 15–18.
Previously, we have demonstrated that the total carriage frequency of KIR2DL2 and KIR2DS1 and the number of activating KIRs increased in SLE patients compared with that in healthy subjects 19. The aim of this study was to investigate further the role of the HLA-Cw/KIR pairs in SLE in our population.
Patients and methods
Patients and control
We recruited 236 patients (210 females, 26 males) with SLE from Shandong Qianfoshan Hospital. All patients fulfilled the American College of Rheumatology revised criteria for the disease 20, and all patients gave informed consent. Their mean age was 28·78 ± 11·31 years (range = 13–55 years). The control samples were obtained from 230 random healthy Chinese individuals. The control population consisted of unrelated ethnicity-, age- and sex-matched individuals. Subjects with other autoimmune disease were excluded from our study.
Genome DNA extraction
Genomic DNA was extracted from 5 ml ethylenediamine tetraacetic acid (EDTA)-treated peripheral blood using the standard salting-out procedure.
KIR genotyping
Similar to MHC loci, KIR sequences are highly polymorphic. A typical feature of KIR haplotypes is the high variability in the number and type of genes they contain. The most extreme aspect of KIR polymorphism is the varying composition of the genes, which is based on the presence or absence of KIR genes in the genomes of different individuals. We present here a refined polymerase chain reaction (PCR) sequence-specific primer (PCR–SSP) method for KIR genotyping in all the recruited subjects. KIR genotyping was performed by KIR locus typing to detect the presence or absence of a total of 14 KIR loci and one pseudogene, KIRZ. Among them, eight KIR genes (2DLI, 2DL2, 2DL3, 2DL4, 2DL5, 3DL1, 3DL2 and 3DL3) were responsible for inhibitory functions and six KIR genes (2DS1, 2DS2, 2DS3, 2DS4, 2DS5 and 3DS1) for conveying activating functions. The PCR–SSP primers used for the detection of KIR loci were based on primer sites that have been described previously 21. Among the 29 pairs of primers (synthesized by Shanghai BoYa Biotechnology Co. Ltd, Shanghai, China), one pair of primers was used to amplify the 2DS5 gene and two pairs of primers were used to amplify each of the other 14 genes, in order to ensure a high detectable rate of positive genes. The framework genes (2DL4, 3DL2 and 3DL3) served as positive markers of PCR. PCR reaction was carried out on a 9700 thermal cycler (PE Applied Biosystems, Foster City, CA, USA). Specifically, 20–50 ng DNA was amplified in a 20 ml reaction containing 0·2 mM deoxyribonucleotide triphosphate (dNTP), 2 mM MgCl2, 214·3 nM primer and 0·5 U Taq polymerase. After the initial denaturation for 1 min at 96°C, the samples were amplified under the following conditions: five cycles of 25 s at 96°C, 45 s at 65°C and 30 s at 72°C; 21 cycles of 25 s at 96°C, 45 s at 60°C and 30 s at 72°C; five cycles of 25 s at 96°C, 1 min at 55°C, 2 min at 72°C; and a prolongation of 10 min at 72°C. PCR products were separated on 1·5% agarose gels containing ethidium bromide. After electrophoresis, the agarose gel was scanned and imaged by Alphaimager TM 2200 instrument (Alpha Innotech Corporation, San Leandro, CA, USA). All typing was repeated at least once.
HLA-C genotyping
We performed HLA-C genotyping on the DNA of all samples. Genomic DNA was amplified using sequence-specific primers, as described previously 22,23. The final volume PCR mixture was 20 μl, including 20–50 ng genomic DNA, 10 μl of 2 × GC PCR Buffer (TaKaRa Bio Inc., Shiga, Japan), 0·2 mM dNTP, 1 μM allele-specific primer, 0·1 μM of DRB1 control primers and Taq DNA polymerase 1 U. Cycling was carried out in a 9700 thermal cycler (PE Applied Biosystems) under the following conditions: 1 min at 96°C, five cycles of 96°C for 30 s, 68°C for 45 s, 72°C for 60 s; 21 cycles of 96°C for 30 s, 63°C for 50 s, 72°C for 60 s; four cycles of 96°C for 30 s, 55°C for 1 min, 72°C for 2 min, followed by a final extension step for 10 min at 72°C. PCR products were electrophoresed in 1·5% agarose gels containing ethidium bromide, and predicted size products were visualized under ultraviolet light. All typing was repeated at least once.
Statistical analysis
Frequency differences of HLA-Cw allelic gene and KIR/HLA-Cw combinations were tested for significance by the two-tailed Fisher's test or χ2 test. To correct for incidental significance, the P-value (if <0·05) was multiplied by the number of comparisons, and a corrected P-value (Pc) ≤ 0·05 was considered statistically significant. Analyses were performed using the sas version 9·0 statistical package.
Results
Distribution of HLA-Cw*01–08 alleles in patients with SLE
We studied HLA-Cw01-08 alleles which are specific for the KIR locus. The frequency of HLA-Cw alleles in 236 individuals with SLE compared with 230 control subjects is shown in Table 1. HLA-Cw07 was identified in 41·10% of patients, but in only 29·13% of the control subjects (P = 0·009; Table 1).
Table 1.
The frequencies of human leucocyte antigen (HLA)-Cw*01–08 genes in patients and control subjects
| HLA-Cw | Control (n = 230) | SLE (n = 236) | P |
|---|---|---|---|
| Cw01 | }43 (18·70%) | 35 (14·81%) | 0·286 |
| Cw 02 | 4 (1·74%) | 3 (1·27%) | 0·486 |
| Cw 03 | 86 (37·39%) | 74 (31·35%) | 0·174 |
| Cw 04 | 30 (13·04%) | 22 (9·32%) | 0·239 |
| Cw 05 | 5 (2·17%) | 7 (2·96%) | 0·772 |
| Cw 06 | 37 (16·08%) | 49 (20·76%) | 0·232 |
| Cw 07 | 67 (29·13%) | 97 (41·10%) | 0·009 |
| Cw 08 | 102 (44·35%) | 96 (40·68%) | 0·454 |
Significant differences between patients and controls in genotype frequency are shown in bold type. P-values < 0·05 were considered statistically significant.
KIR/HLA-Cw combinations
We next analysed combinations of activating/inhibitory KIR-2D and their HLA-Cw ligands for a possible association with SLE. Four combinations, KIR2DL1/HLA-CwLys, 2DS1/HLA-CwLys, KIR2DL2/2DL3/HLA-CwAsn and 2DS2/HLA-CwAsn, were analysed. The genotype KIR2DS1+HLA-CwLys was more common in subjects with SLE compared to control subjects (P = 0·015; Table 2).
Table 2.
The frequency of killer cell immunoglobulin-like receptor–human leucocyte antigen (KIR-HLA)-Cw in systemic lupus erythematosus (SLE) and control subjects
| KIR-HLA-Cw | Control (n = 230) | SLE (n = 236) | P |
|---|---|---|---|
| KIR2DL1+HLA-CwLys | 65 (28·26%) | 78 (33·91%) | 0·271 |
| KIR2DL2/3+HLA-CwAsn | 198 (86·08%) | 207 (87·71%) | 0·681 |
| KIR2DS1+HLA-CwLys | 36 (15·65%) | 59 (25·00%) | 0·015 |
| KIR2DS2+HLA-CwAsn | 40 (17·39%) | 49 (20·76%) | 0·410 |
P-values < 0·05 were considered statistically significant.
Activating KIR2D while absence of ligands for corresponding inhibitory KIRs
Martin et al. 21 believed that individuals expressing only activating KIR, while HLA ligands for the corresponding inhibitory KIRs were missing, would allow a greater impact of stimulatory KIRs on NK cell function, enhancing activation. We determined the frequency of activating KIR2D with the absence of the KIR2DL/HLA-Cw combination; the frequency of KIR2DS1 was increased when KIR2DL1/HLA-Cw are absent in SLE, and the difference was significant (P = 0·001; Table 3, Fig. 1).
Table 3.
Frequency of activating killer cell immunoglobulin-like receptor (KIR)2D in the absence of ligands for corresponding inhibitory KIRs
| KIR-HLA-Cw | Control | SLE | P |
|---|---|---|---|
| KIR2DS1(+)2DL1-HLA-CwLys(–) | 72 (31·30%) | 110 (46·61%) | 0·001 |
| KIR2DS2(+)2DL2-HLA-CwAsn(–) | 7 (3·04%) | 4 (1·69%) | 0·376 |
P-values < 0·05 were considered statistically significant.
Fig 1.
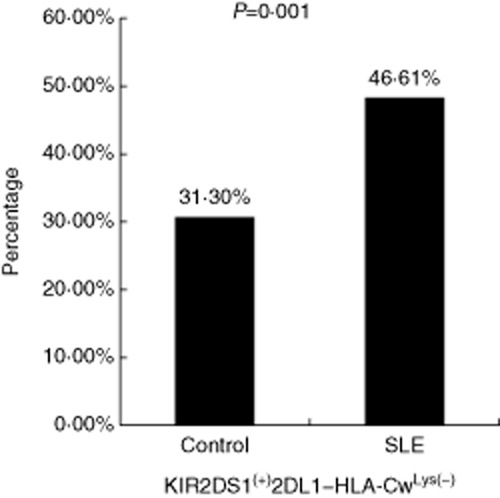
The frequency of killer cell immunoglobulin-like receptor (KIR)2DS1(+)2DL1–human leucocyte antigen (HLA)-CwLys(–) in systemic lupus erythematosus (SLE) and control group. The frequency of KIR2DS1 was increased when KIR2DL1/HLA-Cw were absent in SLE, and the difference was significant (P = 0·001).
Discussion
The extent to which KIRs act as inhibitory self-receptors depends upon an individual's HLA type. KIR recognition of specific HLA class I allotypes contributes to the array of receptor–ligand interactions that determine the response of an NK cell to its target. If the polymorphism of both HLA and the KIR–gene complex is considered together, then dissimilar numbers and qualities of KIR/HLA pairs appear to function in different individuals. In addition, the independent segregation of HLA and KIR genes raises the possibility that any given individual can express the receptor or the ligand only, or both receptor and ligand. Several studies on autoimmune or inflammatory diseases, including psoriatic arthritis 24, type 1 diabetes mellitus 25, rheumatoid arthritis 26 and scleroderma 27, unequivocally demonstrated an association of ‘less inhibitory’ KIR2DS and 2DL profiles, either alone or in combination, with HLA-Cw groups.
We have shown previously that the frequency of KIR2DL2 and KIR2DS1 increased in SLE patients compared with that in healthy subjects. It is well known that the function of KIR is highly dependent upon the HLA molecules expressed on target cells. HLA-C alleles, the ligands of KIR2D genes, are highly polymorphic; these loci segregate to KIR genes, and NK cells can express KIRs for which there is no known HLA ligand present. In this study, we showed that HLA-Cw*07 was present more frequently in SLE patients than in healthy subjects, which is consistent with the findings reported by Kong et al. 28. HLA-C is the classic HLA-I gene, which was on the short arm of human chromosome 6, between HLA-A and B. This was found in 1970, and the C seat was identified in 1975. According to loci recognized by KIR2D, we divided HLA-C into group 1 (Cw*01/3/7/8, Asn80) and group 2 (Cw*02/4/5/6/, Lys80). We found that the HLA-Cw group 1 was more common in both SLE and control, and HLA-Cw07 was increased in patients with SLE. The HLA-C imbalance between groups 1 and 2 will impact more easily upon the interplay between HLA-C/KIR genes, leading to a genetic susceptibility to inappropriate or dysregulated NK cell surveillance.
As any effect of KIR on disease susceptibility might depend upon the presence of putative HLA ligands within an individual, we analysed combinations of activating/inhibitory KIR2D and their HLA-Cw ligands for a possible association with SLE. In the current study, we found that individuals positive for a combination of KIR2DS1+HLA-CwLys were more frequently patients than control subjects. This may explain that, in SLE patients, more activating KIRs sent more activating signals to NK cells or to T cells, resulting in the recruitment of other cells of the immune system, and this suggested that there were excess immune states in SLE. This suggested that KIR2DS1+HLA-CwLys might contribute to the pathogenesis of SLE by influencing NK or T cell activity.
In addition, a significant difference (P = 0·001) was observed between the SLE and control groups that the frequency of KIR2DS1 was increased when KIR2DL1/HLA-Cw are absent in the SLE group. Martin et al. 21 reported that subjects with activating KIR2DS1 and/or KIR2DS2 genes were susceptible to developing psoriatic arthritis, but only when HLA ligands for their homologous inhibitory receptors, KIR2DL1 and KIR2DL2/3, are missing. The absence of ligands for inhibitory KIRs could potentially lower the threshold for NK (and/or T) cell activation mediated through activating receptors, thereby contributing to the pathogenesis of psoriatic arthritis. They suggested that, in the presence of their HLA ligands, corresponding inhibitory KIR may neutralize the effect of the activating KIR 21. Activating KIR molecules are known to bind poorly to HLA molecules compared with that observed for inhibitory KIR, perhaps explaining the observed dominance of inhibition over activation of NK or T cells. KIR tetramer-binding studies suggest that activating and inhibitory receptors recognize the same set of HLA class I molecules, differing in their binding affinities, such that the stimulatory KIR is not always sufficient to trigger an NK cell response to ligands 29. This allows fine control during cellular activation. There is support for the notion that non-HLA molecules (such as foreign or microbial antigens, aberrantly expressed normal cell surface proteins or complexes of pathogen-derived peptides bound to MHC class I molecules) may behave as ligands for activating KIRs 21.
Some studies have been conducted on the KIR association with SLE, and these studies have investigated the frequencies of KIR genes in SLE patients, reporting discrepant results. Pellett et al. showed that the frequency of KIR2DS1 in the absence of KIR2DS2 was increased significantly in SLE patients compared with controls 30. Kimoto et al. reported that the KIR2DL5 gene was associated significantly with a reduced risk of SLE, as well as with an increased risk of infectious events in general in patients with SLE 15. In 2011, a study suggested that the predisposition to SLE is associated with the presence of the KIR2DS2+/KIR2DS5+/KIR3DS1+ profile and with the GTGT deletion at the SLC11A1 3′-untranslated region (3′-UTR) 16. Taken together, those results could mean that individuals with more stimulatory receptors or fewer inhibitory receptors, under hypomethylation, could be more susceptible to develop lupus. This conclusion is also in agreement with a general tendency for the association of stimulatory KIR genes with autoimmune disorders 31.
In conclusion, our data indicate that the imbalance of HLA-C groups 1 and 2 may impact upon the interplay between HLA-C/KIR genes, leading to genetic susceptibility to dysregulated NK cell surveillance. In addition, the study found that not only were the frequencies of KIR2DS1 in combination with its ligand HLA-C increased statistically significantly in the patient group, but also when their homologous inhibitory receptor-ligands were absent. Our results suggest that KIR genotype and HLA ligand interaction may contribute to the genetic pathogenesis of SLE.
Acknowledgments
Supported by grants from the Promotive Research Fund for Excellent Young and Middle-Aged Scientists of Shandong Province (BS2010YY054) and Shandong Medical and Health Technology Development (2014WSB04020).
Disclosure
There are no financial or commercial conflicts of interest.
References
- Wong M, Tsao BP. Current topics in human SLE genetics. Springer Semin Immunopathol. 2006;28:97–107. doi: 10.1007/s00281-006-0031-6. [DOI] [PubMed] [Google Scholar]
- Miyagawa H, Yamai M, Sakaguchi D, et al. Association of polymorphisms in complement component C3 gene with susceptibility to systemic lupus erythematosus. Rheumatology (Oxf) 2008;47:158–164. doi: 10.1093/rheumatology/kem321. [DOI] [PubMed] [Google Scholar]
- Koyama T, Tsukamoto H, Masumoto K, et al. A novel polymorphism of the human APRIL gene is associated with systemic lupus erythematosus. Rheumatology (Oxf) 2003;42:980–985. doi: 10.1093/rheumatology/keg270. [DOI] [PubMed] [Google Scholar]
- Tao K, Fujii M, Tsukumo S, et al. Genetic variations of Toll-like receptor 9 predispose to systemic lupus erythematosus in Japanese population. Ann Rheum Dis. 2007;66:905–909. doi: 10.1136/ard.2006.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T, Kiyohara C, Tsukamoto H, et al. A functional M196R polymorphism of tumour necrosis factor receptor type 2 is associated with systemic lupus erythematosus: a case–control study and a meta-analysis. Ann Rheum Dis. 2007;66:320–324. doi: 10.1136/ard.2006.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YW, Kee SJ, Cho YN, et al. Impaired differentiation and cytotoxicity of natural killer cells in systemic lupus erythematosus. Arthritis Rheum. 2009;60:1753–1763. doi: 10.1002/art.24556. [DOI] [PubMed] [Google Scholar]
- Hervier B, Beziat V, Haroche J, et al. Phenotype and function of natural killer cells in systemic lupus erythematosus: excess interferon-γ production in patients with active disease. Arthritis Rheum. 2011;63:1698–1706. doi: 10.1002/art.30313. [DOI] [PubMed] [Google Scholar]
- Leibson PJ. Signal transduction during natural killer cell activation: inside the mind of a killer. Immunity. 1997;6:655–661. doi: 10.1016/s1074-7613(00)80441-0. [DOI] [PubMed] [Google Scholar]
- Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- Williams AP, Bateman AR, Khakoo SI. Hanging in the balance: KIR and their role in disease. Mol Interv. 2005;5:226–240. doi: 10.1124/mi.5.4.6. [DOI] [PubMed] [Google Scholar]
- Moesta AK, Parham P. Diverse functionality among human NK cell receptors for the C1 epitope of HLA-C: KIR2DS2, KIR2DL2, and KIR2DL3. Front Immunol. 2012;3:336. doi: 10.3389/fimmu.2012.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam R. Human diversity of killer cell immunoglobulin-like receptors and disease. Korean J Hematol. 2011;46:216–228. doi: 10.5045/kjh.2011.46.4.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D, Gonzelez F. The extensive polymorphism of KIR genes. Immunology. 2010;129:8–19. doi: 10.1111/j.1365-2567.2009.03208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Phil Trans R Soc Lond B Biol Sci. 2012;367:800–811. doi: 10.1098/rstb.2011.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto Y, Horiuchi T, Tsukamoto H, et al. Association of killer cell immunoglobulin-like receptor 2DL5 with systemic lupus erythematosus and accompanying infections. Rheumatology (Oxf) 2010;49:1346–1353. doi: 10.1093/rheumatology/keq050. [DOI] [PubMed] [Google Scholar]
- Pedroza LS, Sauma MF, Vasconcelos JM, et al. Systemic lupus erythematosus: association with KIR and SLC11A1 polymorphisms, ethnic predisposition and influence in clinical manifestations at onset revealed by ancestry genetic markers in an urban Brazilian population. Lupus. 2011;20:265–273. doi: 10.1177/0961203310385266. [DOI] [PubMed] [Google Scholar]
- Jiao YL, Zhang BC, You L, et al. Polymorphisms of KIR gene and HLA-C alleles: possible association with susceptibility to HLA-B27-positive patients with ankylosing spondylitis. J Clin Immunol. 2010;30:840–844. doi: 10.1007/s10875-010-9444-z. [DOI] [PubMed] [Google Scholar]
- Ramírez-De los Santos S, Sánchez-Hernández PE, Muñoz-Valle JF, et al. Associations of killer cell immunoglobulin- like receptor genes with rheumatoid arthritis. Dis Markers. 2012;33:201–206. doi: 10.3233/DMA-2012-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YF, Zhang YC, Jiao YL, et al. Disparate distribution of activating and inhibitory killer cell immunoglobulinlike receptor genes in patients with systemic lupus erythematosus. Lupus. 2010;19:20–26. doi: 10.1177/0961203309345779. [DOI] [PubMed] [Google Scholar]
- Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- Martin MP, Nelson G, Lee JH, et al. Cutting edge: susceptibility to psoriatic arthritis: influence of activating killer Ig-like receptor genes in the absence of specific HLA-C alleles. J Immunol. 2002;169:2818–2822. doi: 10.4049/jimmunol.169.6.2818. [DOI] [PubMed] [Google Scholar]
- Bunce M, O'Neill CM, Barnardo MC, et al. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP) Tissue Antigens. 1995;46:355–367. doi: 10.1111/j.1399-0039.1995.tb03127.x. [DOI] [PubMed] [Google Scholar]
- Lu XZ, Hong KX, Qin GM, et al. Genotyping of HLA-Cw locus in Chinese Yi ethnic group by PCR–SSP. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2003;17:62–65. [PubMed] [Google Scholar]
- Chandran V, Bull SB, Pellett FJ, Ayearst R, Pollock RA, Gladman DD. Killer-cell immunoglobulin-like receptor gene polymorphisms and susceptibility to psoriatic arthritis. Rheumatology (Oxf) 2014;53:233–239. doi: 10.1093/rheumatology/ket296. [DOI] [PubMed] [Google Scholar]
- Mehers KL, Long AE, van der Slik AR, et al. An increased frequency of NK cell receptor and HLA-C group 1 combinations in early-onset type 1 diabetes. Diabetologia. 2011;54:3062–3070. doi: 10.1007/s00125-011-2299-x. [DOI] [PubMed] [Google Scholar]
- Yen JH, Moore BE, Nakajima T, et al. Major histocompatibility complex class I-recognizing receptors are disease risk genes in rheumatoid arthritis. J Exp Med. 2001;193:1159–1167. doi: 10.1084/jem.193.10.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim PH, Jobim M, Bredemeier M, et al. Killer cell immunoglobulin-like receptor (KIR) genes in systemic sclerosis. Clin Exp Immunol. 2010;160:325–330. doi: 10.1111/j.1365-2249.2010.04095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong NC, Nasruruddin BA, Murad S, Ong KJ, Sukumaran KD. HLA antigens in Malay patients with systemic lupus erythematosus. Lupus. 1994;3:393–395. doi: 10.1177/096120339400300505. [DOI] [PubMed] [Google Scholar]
- Stewart CA, Laugier-Anfossi F, Vély F, et al. Recognition of peptide–MHC class I complexes by activating killer immunoglobulin like receptors. Proc Natl Acad Sci USA. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett F, Siannis F, Vukin I, Lee P, Urowitz MB, Gladman DD. KIRs and autoimmune disease: studies in systemic lupus erythematosus and scleroderma. Tissue Antigens. 2007;69(Suppl. 1):106–108. doi: 10.1111/j.1399-0039.2006.762_6.x. [DOI] [PubMed] [Google Scholar]
- Kulkarni S, Martin MP, Carrington M. The yin and yang of HLA and KIR in human disease. Semin Immunol. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


