Abstract
Biphenyl (BP)-degrading bacteria were identified to degrade various polychlorinated BP (PCB) congers in long-term PCB-contaminated sites. Exploring BP-degrading capability of potentially useful bacteria was performed for enhancing PCB bioremediation. In the present study, the bacterial composition of the PCB-contaminated sediment sample was first investigated. Then extracellular organic matter (EOM) from Micrococcus luteus was used to enhance BP biodegradation. The effect of the EOM on the composition of bacterial community was investigated by combining with culture-dependent and culture-independent methods. The obtained results indicate that Proteobacteria and Actinobacteria were predominant community in the PCB-contaminated sediment. EOM from M. luteus could stimulate the activity of some potentially difficult-to-culture BP degraders, which contribute to significant enhancement of BP biodegradation. The potentially difficult-to-culture bacteria in response to EOM addition were mainly Rhodococcus and Pseudomonas belonging to Gammaproteobacteria and Actinobacteria respectively. This study provides new insights into exploration of functional difficult-to-culture bacteria with EOM addition and points out broader BP/PCB degrading, which could be employed for enhancing PCB-bioremediation processes.
Introduction
Polychlorinated biphenyls (PCBs) are toxic persistent pollutants that threaten both natural ecosystem and human health (Zanaroli et al., 2010). In situ PCB bioremediation have aroused increasing concern because they are less expensive and more environmentally sound than conventional methods (Leigh et al., 2006). A large number of bacteria have been identified and shown to cometabolize PCBs through the biphenyl (BP) catabolic pathway (Petrić et al., 2007). Indeed, research on aerobic PCB biodegradation bacteria isolated so far has mainly focused on BP-utilizing bacteria. Numerous phylogenetically diverse BP-utilizing bacteria that have the capability to transform several PCB congeners have been isolated (Abraham et al., 2002; Pieper, 2005). However, until recently, the full-scale bacterial remediation of PCB-contaminated environment performed not very well, because the activity and capability of BP/PCB-degrading strains soon decreased when exposed to natural environment. Although bioaugmentation of sites with degradative bacteria have been unsuccessful application in the field, efforts to stimulate indigenous BP/PCB-degrading bacteria have been promising (Leigh et al., 2006).
In the natural environment, it is very common for bacteria to survive under a wide variety of stress conditions by entering a ‘viable but non-culturable’ (VBNC) state, in which cells are intact and alive but fail to normally grow on the routine bacteriological media (Oliver, 2010). The high toxicity of PCBs and their low bioavailability exerted significant stress on indigenous microorganisms. It should be interesting to find out the survival and activity of bacteria exposed to such adverse conditions (Chávez et al., 2006). Although numerous BP/PCB-degrading bacteria have been isolated and studied, there are more phylogenetically diverse bacteria detected in the environment using molecular tools (Macedo et al., 2007). Undoubtedly, using conventional plate separation methods, only a small fraction of BP/PCB-degrading bacteria existed in nature can be obtained. Furthermore, it is common knowledge that artificial mixed cultures consisting of purified cultivable isolates from enrichment cultures are less efficient in BP/PCB degradation than mixed cultures (Mikesková et al., 2012; Uhlik et al., 2012). One reason for this discrepancy is that there are abundant VBNC or uncultured bacteria possessing BP/PCB-degrading abilities in mixed cultures (Tang et al., 2013). Hence, the resuscitation and stimulation of potential BP/PCB-degrading indigenous bacteria is crucial for in situ bioremediation of PCBs.
The most exciting development in reaction of VBNC bacteria is the role of extracellular protein secreted by Micrococcus luteus, known as a resuscitation-promoting factor (Rpf), which has been shown to promote the resuscitation and growth of high G + C Gram-positive organisms (Mukamolova et al., 1998; Su et al., 2013b). It is worth noting that these organisms contain well-known BP/PCB degraders, including Rhodococcus, Arthrobacter, Bacillus and Microbacterium (Pieper, 2005; Petrić et al., 2007). In addition, Ding and Yokota (2010) indicated that Rpf could also enhance the culturability of several other Gram negative. Hence, Rpf is capable of culturing difficult-to-culture bacteria involved in the BP/PCB degradation process and stimulating the activity of indigenous bacterial communities in PCB-contaminated environment. Mukamolova and colleagues (2006) indicated that Rpf control the culturability of several bacteria because of its muralytic activity. And at least two additional extracellular proteins in the M. luteus culture supernatant were also found to possess the same muralytic activity as Rpf protein. On the other hand, the recombinant Rpf protein presented lower activity than the native Rpf protein (purified from M. luteus culture supernatant), and both of them were prone to lose its activity after storage at 4°C for 1 week (Mukamolova et al., 2006). By analysing on the disadvantage of Rpf protein, extracellular organic matter (EOM) from M. luteus offers an attractive and cost-effective alternative additive for resuscitating and stimulating VBNC or difficult-to-culture bacteria, as well as enhancing the activity and degrading capability of indigenous bacteria in PCB-contaminated area.
The bacterial composition of the PCB-contaminated sediment sample was first investigated in the present study. Above all, to test the hypothesis that EOM from M. luteus was a feasible and effective additive for enhancing bacterial BP-degradation capability, the effect of EOM on BP biodegradation and bacterial community was assessed. Specifically, the degradation abilities, bacterial composition and abundance of enrichment cultures with different experimental treatment were investigated. In this work, we aimed to identify whether EOM could resuscitate the BP-degrading potential of VBNC or difficult-to-culture bacteria by isolating pure cultures unique to the enrichment culture with EOM addition. To our best knowledge, this is the first attempt to reveal the effect of EOM from M. luteus on BP-degrading bacterial communities using the combination of Illumina high-throughput sequencing and culture-dependent methodology.
Results and discussion
Bacterial composition in the PCB-contaminated sediment
Illumina high-throughput sequencing was performed to determine the diversity and composition of the bacterial communities in the sediment sample. The top 10 dominant phyla/genera are shown in Fig. 1. As shown in Fig. 1A, of the 10 major phyla, the most predominant phylum was Proteobacteria, which represented 31.3% of the total bacteria. Actinobacteria, Chloroflexi and Firmicutes were the subdominant groups, constituting around 50% of the total population. At the genus level (Fig. 1B), the four most abundant genera, which belonged to Rhodococcus, Achromobacter, Mycobacterium and Dechloromonas, accounted for 18.8% of all sequences. The proportion of sequences assigned as unclassified constituted highly 31.3% of the total population. The composition of the bacterial community of the sediment sample identified in this study is in accordance with previous studies (Fukuda et al., 1998; Macedo et al., 2007; Uhlik et al., 2009), which are rather common abundant bacteria in PCB-contaminated environments adapted for biodegradation of the pollutants. To improve bioremediation practices, it is important to understand the composition of bacterial community in long-term PCB-contaminated sites (Petrić et al., 2011). These results indicate that genera Rhodococcus and Achromobacter with outstanding degradation abilities were widespread in long-term PCB-contaminated sediment, in which the total of 20 PCB congeners (from one to six chlorine atoms) concentration averaged 29.16, and the standard deviation was 0.72 μg g−1. In addition, it would be interesting to find that majority indigenous bacteria cannot be cultivated. Indeed, yet-to-be cultured bacteria probably account for the majority of the degradation activity among indigenous bacteria (Uhlik et al., 2012; Tang et al., 2013).
Fig 1.
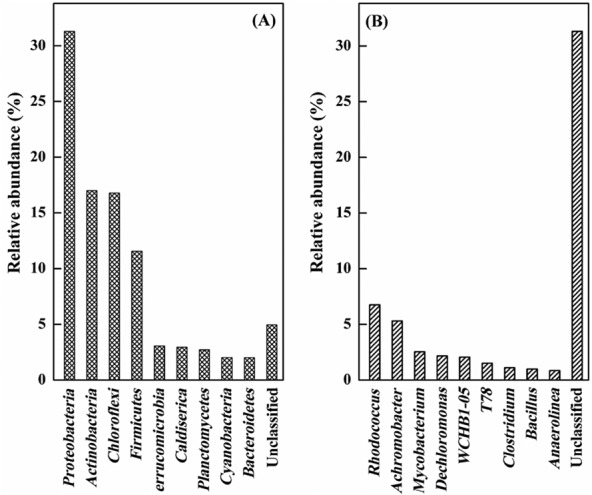
Taxonomic composition of bacterial community in the PCB-contaminated sediment sample. (A) Taxonomic affiliations at the phylum level. (B) Taxonomic affiliations at the genus level.
Effect of EOM on BP degradation
Based on the BP-degradation efficiency and cell growth (OD600), the effect of EOM on the BP tolerance concentration, BP-degrading curve and cell growth was investigated. The degradation of BP by enrichment samples under various BP concentrations (from 500 to 4500 mg l−1) is detailed in Fig. 2. As shown in Fig. 2, the BP-degradation efficiency of all the groups decreased with elevated BP concentrations. When BP concentration was increased from 500 to 3500 mg l−1, the BP-degradation efficiency decreased from 99.9% to 68.3% in treatment group (TG), whereas it decreased from 65.5% to 12.4% in blank group (BG). Under a BP concentration of 4000 mg l−1, BP-degradation efficiency reached 59.5% in TG, whereas the efficiency was 29.6% and 7.6% in CG and BG respectively. Similar to the trend of variations in BP-degradation efficiency, cell growth decreased with elevated BP concentrations in the CG and BG. However, the values of OD600 in TG underwent an increasing trend, reaching its maximum value of 2.63 at 2000 mg l−1, and then decreased to 0.58 at BP concentration of 4500 mg l−1. Meanwhile, under a BP concentration of 4000 mg l−1, the value of OD600 in TG was observed to be around fivefold and ninefold greater than that observed in control group (CG) and BG. Meanwhile, Fig. 3 depicts the changes in the BP-degradation curves and cell growth among the three groups. As shown in Fig. 3, the BP-degradation efficiency and cell growth were significantly enhanced in TG. After 60 h, the BP-degradation efficiency in TG reached up to 97.4%, whereas the efficiency was less than 60.3% in CG and 47.2% in BG. The overall trend in cell growth coincided with the degradation rate, and significant increases in the value of OD600 were recorded. These results indicated that TG with EOM addition maintained higher degradation efficiency and faster cell growth and was more tolerant to increasing BP concentration. Specifically, the BP-degradation efficiencies of TG were higher than those of CG, suggesting that EOM performed better than autoclaved EOM in enhancing BP degradation. The better performance of TG could be attributed to the positive effects of some proteins in EOM on cell growth and BP-degradation efficiency, because autoclaved EOM had the same constituents as EOM except proteins. In addition, the BP-degradation efficiencies of CG were higher than BG, suggesting that some polysaccharides of EOM may also play a role in enhancing BP degradation.
Fig 2.
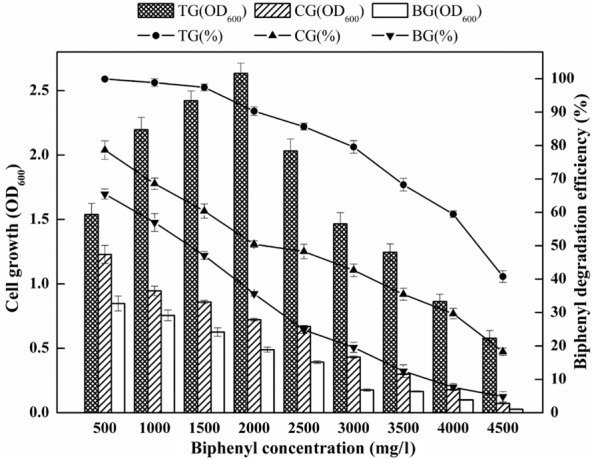
Comparison of tolerance concentration of biphenyl among different groups of enrichment cultures. TG: addition of EOM; CG: addition of autoclaved EOM; BG: without EOM. Error bars indicate the standard deviations of triplicate samples.
Fig 3.
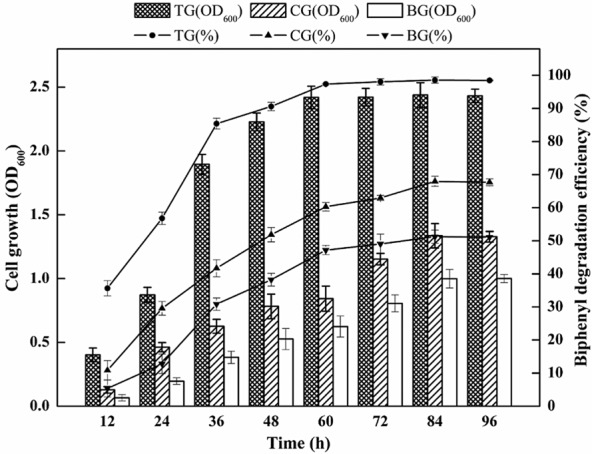
Comparison of biphenyl-degrading curve among different enrichment cultures at a concentration of 1500 mg l−1. TG: addition of EOM; CG: addition of autoclaved EOM; BG: without EOM. Error bars indicate the standard deviations of triplicate samples.
In previous studies, the resuscitation and stimulation function of proteins in the culture supernatants of M. luteus has been tentatively verified (Ding, 2004; Su et al., 2013b). This study further indicated that EOM from M. luteus as an efficient additive could significantly enhance the performance of BP biodegradation. Recent advances in PCB bioremediation processes have focused on selecting BP-degrading enrichment cultures to obtain BP/PCB-degrading community and identifying the key members involved in this process using culture-independent approaches (Cámara et al., 2004; Uhlik et al., 2012). Nevertheless, most of bacteria and enrichment cultures, which were obtained with high removal rates in the laboratory, showed exceptionally slow removal rates in situ (Abraham et al., 2002). Therefore, the challenge for PCB-bioremediation is to enhance the activities of bacterial communities involved in biodegradative processes and develop approaches to realize the full potential of BP/PCB degraders (Gomes et al., 2013). Clearly, culture-dependent studies can focus only on a limited number of bacteria that are not likely to represent the bacterial diversity in the environment, as there are abundant of VBNC or difficult-to-culture bacteria that are important players in bioremediation existed ubiquitously in PCB-contaminated (de Cárcer et al., 2007; Zanaroli et al., 2010). Although culture-independent molecular techniques can investigate the diversity of bacteria potentially responsible for ecologically relevant processes, to elucidate bacteria related function and genotype, it will be necessary to recover them and study their microbiology in pure cultures (Uhlik et al., 2012; Su et al., 2013b). Thus, EOM from M. luteus can be employed for culturing difficult-to-culture bacteria and for exploiting the potential environmental functions of VBNC or difficult-to-culture bacteria, which will demonstrate highly valuable in future bioremediation strategies.
Effect of EOM on bacterial diversity and composition
Illumina high-throughput sequencing was adopted to investigate bacterial community diversity and richness among sediment sample and enrichment cultures (TG, CG and BG) (Table 1). In order to compare the bacterial species richness, operational taxonomic units (OTUs) were estimated by Chao1 estimator at a distance level of 3%. The total number of OTUs in TG (2272) with EOM addition was the largest among the enrichment cultures. And BG (1981) contained the least OTUs amount, indicating that TG with EOM addition exhibited the greatest bacterial richness. Furthermore, the bacterial phylotype richness can also be reflected using Shannon diversity index, which is generally used to demonstrate species diversity in a bacterial community, and it accounts for both evenness and abundance of the species present (Ma et al., 2013). Considering the fact that Shannon index of TG (5.60) was much higher than that of CG (5.02) and BG (4.87), it could be inferred that the enriched OTUs in TG community distributed more evenly than those in CG and BG. These results suggested that bacterial community diversity and richness increased with EOM addition.
Table 1.
Alpha diversity of samples
| ` | OTUs | Chao 1 richness estimation | Shannon diversity |
|---|---|---|---|
| Sediment sample | 5623 | 23 187 | 9.86 |
| Enrichment sample TG | 2272 | 10 561 | 5.60 |
| Enrichment sample CG | 2175 | 9803 | 5.02 |
| Enrichment sample BG | 1981 | 9259 | 4.87 |
The effective bacterial sequences in the TG, CG and BG were all assigned to corresponding taxonomies. Figure 4 shows the taxonomic compositions at the class level for the three enrichment cultures, demonstrating a relative abundance greater than 0.1% in at least one enrichment culture. In total, 10 identified class were observed; Bataproteobacteria, Actinobacteria and Gammaproteobacteria were the dominant class, which were consistent with previous studies on bacterial community composition in polycyclic aromatic hydrocarbon and PCB exposed environments (Stach and Burns, 2002; Petrić et al., 2011). However, the relative abundance of each dominant class among the three enrichment cultures was distinct. Especially, the relative abundance of Bataproteobacteria, Actinobacteria and Gammaproteobacteria were 27.6%, 39.3% and 24.3% in TG, and 58.0%, 15.6% and 18.1% in BG. Actinobacteria were the most dominant bacterial community in TG instead of Bataproteobacteria in CG and BG. Thus, it could be inferred that after EOM addition, Actinobacteria and Gammaproteobacteria were greatly enriched. Moreover, a small fraction of class Bacteroidia accounting for less than 0.1% of total community showed a similar trend, which the order of abundance was TG > CG > BG. The Bacteroidia class is believed to be involved in the degradation of aromatic compounds (Xu et al., 2012). Petrić and colleagues (2011) had previously reported that Actinobacteria and Bacteroides were the predominant phyla in bioremediation of PCB-contaminated soil. In view of these results, it could be concluded that EOM from M. luteus greatly affects the composition and abundance of bacterial communities closely related to BP/PCB degradation.
Fig 4.
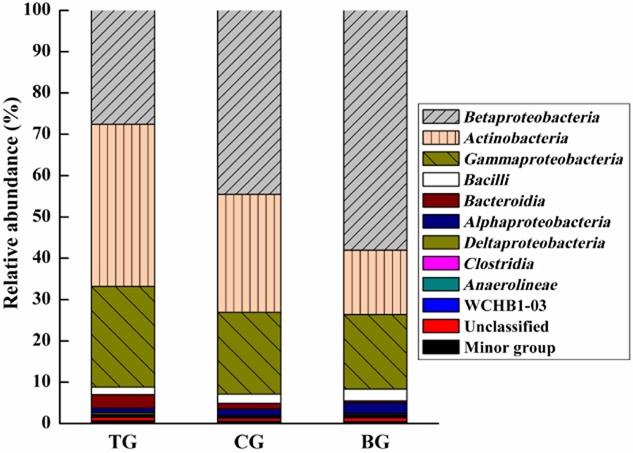
Taxonomic composition of bacterial communities at the class level in enrichment cultures. TG: addition of EOM; CG: addition of autoclaved EOM; BG: without EOM.
To further explore the variation in bacterial community with EOM addition, bacterial abundance was also analysed more specifically at the genus level. As Fig. 5 illustrates, a relative abundance greater than 0.1% in at least one enrichment culture is summarized. Other genera were grouped into minor groups. At the genus level, the majority of sequences were affiliated to five types, such as Achromobacter, Rhodococcus, Pseudomonas, Stenotrophomonas and unclassified bacteria. And the composition of TG with EOM addition was also reflected to be dramatically different. The genus Rhodococcus dominated TG (26.4% of total reads) community, followed by Achromobacter (18.2%), Pseudomonas (6.1%) and Stenotrophomonas (3.1%). However, the genus Achromobacter was the most dominant bacterial community, accounting for 30.8% and 37.69% of the total reads in CG and BG community respectively. Notably, the order of abundance for the genera Rhodococcus, Pseudomonas and Stenotrophomonas was TG > CG > BG. The discrepancy between TG and CG could be attributed to the function of some proteins in EOM. Compared with BG, CG with autoclaved EOM had higher relative abundance of the three genera which may be attributable to the stimulation function of polysaccharides in EOM. Overall, well-known PCB degraders (Rhodococcus, Pseudomonas and Stenotrophomonas) were greatly abundant after EOM addition (Macedo et al., 2007; Correa et al., 2010). The results in this study is consistent with earlier reports that Rpf could promote the resuscitation and growth of not only high G + C Gram-positive organisms, but also several other Gram-negative organisms (Mukamolova et al., 2006; Oliver, 2010; Su et al., 2013a).
Fig 5.
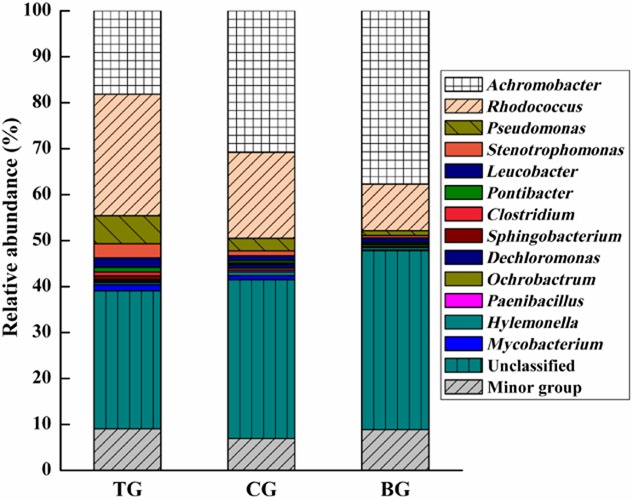
Taxonomic composition of bacterial communities at the genus level in enrichment cultures. TG: addition of EOM; CG: addition of autoclaved EOM; BG: without EOM.
It was interesting to observe that a large number of unclassified sequences (30.1–38.9% of total reads) were found in the enrichment cultures, suggesting that a wide variety of novel species or yet-to-be cultured bacteria may inhabit complex BP enrichment cultures communities. The results indicated that some uncultured or VBNC bacteria frequently detected in PCB-contaminated sites and BP enrichment cultures may be highly correlated to the functions and performances of BP/PCB biodegradation process. This is in agreement with previous investigation on degradative bacterial communities in BP/PCB-contaminated environments (Abraham et al., 2002; Uhlik et al., 2012). Indeed, the traditionally isolated genera of bacteria possessing the capability for pollutants degradation represent only a small fraction of the total diversity in nature. In this study, bacteria in the enrichment culture were exposed to high concentrations of BP and progressive nutrient depletion, both of which may initiate the VBNC state. Tang and colleagues (2013) found that many clones related to BP/PCB degradation were uncultured bacteria. Furthermore, Actinobacteria as the predominance of the phylum in the BP/PCB-degrading community process was prone to enter VBNC state in which they have significantly reduced metabolic activity and lose culturability (Keep et al., 2006; Mukamolova et al., 2006). In addition, Mukamolova and colleagues (2006) had previously shown that the culturability of several Actinobacteria is controlled by Rpf. And Rpf homologues are widespread throughout the Actinobacteria. Meanwhile, Schroeckh and Martin (2006) found that Rpf could resuscitate Actinobacteria and was a useful tool for isolating new actinobacterial species. Typically, Rhodococcus is known to enter into a VBNC state quite rapidly under adverse conditions, such as R. rhodochrous and R. fascians (Shleeva et al., 2002). The results supported the hypothesis that the positive effects of EOM on BP-degrading capability enhancement was mainly caused by its ability to resuscitate and stimulate VBNC bacteria, especially for BP/PCB-degrader Rhodococcus-like populations. Therefore, it may be inferred that EOM from M. luteus containing the Rpf and Rpf-like proteins provides a useful approach for isolating novel species or yet-to-be cultured bacteria and realizing the full potential of BP/PCB degraders.
Isolation and phylogenetic analysis of bacteria
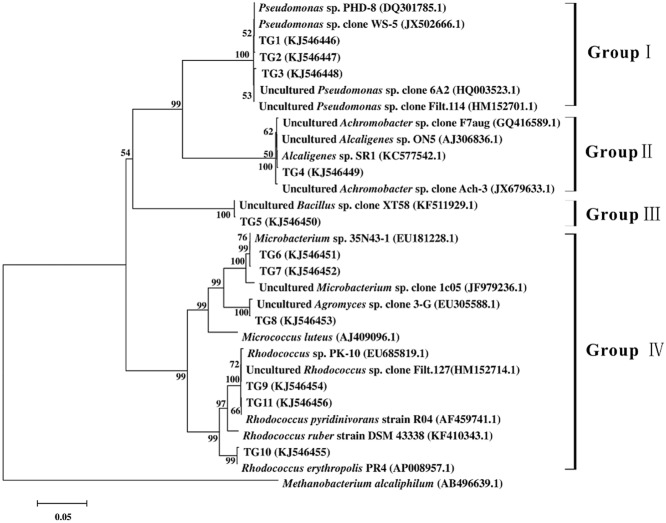
In order to further verify the resuscitating function of EOM, the three enrichment cultures (TG, CG and BG) were also investigated using a culture-dependent method. Colonies that were unique in TG, with no counterpart in CG and BG, were isolated using a modified most probable number method. Eleven unique strains in TG (TG1–TG11) were isolated on mineral salts agar plates. Phylogenetic analysis was based on 16S rDNA sequences. Closely matching representatives were determined by a Blast search at GenBank. Figure 6 presents an overview of the 11 unique strains based phylogenetic tree, generated by including representative members. Overall, at the class level, strains belonging exclusively to TG can be broadly divided into four subgroups. The most dominant classified subgroup was Actinobacteria (subgroups IV), accounting for 54.5% (6/11) of the total strains. Gammaproteobacteria (subgroups I) was the subdominant group, which constituted 27.2% of the total strains. Bataproteobacteria (subgroups II) and Bacilli (subgroups III) only contributed to about 18% of the total population. The results are consistent with the investigations on variation of community composition in TG analysed by Illumina high-throughput sequencing. And the analyses based on the culture-dependent method further confirmed that EOM could resuscitate functional bacteria for BP degradation, especially the Actinobacteria.
Fig 6.
Phylogenetic tree of bacterial 16S rRNA gene sequences, including eleven isolates unique to enrichment culture with EOM addition and 18 of their most similar GenBank sequences. Bootstrap values (> 50%) are showed at branch points. The bar represents a sequence distance of 0.1.
Moreover, it was worth noting that strains TG6–TG11 belonging to genera Microbacterium, Agromyces and Rhodococcus are highly homologous with M. luteus. Furthermore, it is evident that strain TG9 represents a novel species of the genus Rhodococcus, for which the name Rhodococcus biphenylivorans sp. nov. is proposed (Su et al., 2015). In addition, VBNC state of the strain TG9T in response to low temperature and oligotrophic nutrients was verified (data not shown). This is agreement with previous reports that Rpf could promote the resuscitation and growth of Gram-positive organisms (Keep et al., 2006; Su et al., 2013a). By contrast, strains TG1–TG4 belonging to genera Pseudnomonas and Achromobacter are Gram-negative bacteria, which were well supported by the observation that Rpf also resuscitated the growth of several other Gram-negative bacteria (Ding and Yokota, 2010; Su et al., 2013b). Additionally, these bacteria unique to TG were closely related to uncultured bacteria, which are known for their catabolic diversity in degradation of aromatic compounds (Zanaroli et al., 2010; Xu et al., 2012). In view of these results, it could be inferred that after EOM addition, functional difficult-to-culture bacteria belonging to genera Microbacterium, Agromyces, Rhodococcus, Pseudnomonas and Achromobacter recovered their culturability.
Application prospect of EOM from M. luteus
In the present study, the influence of EOM from M. luteus on the BP-degradation capability and composition of bacterial community in PCB-contaminated sediment was first evaluated by combining with culture-dependent and culture-independent methods. The obtained results provided evidence that EOM from M. luteus could significantly enhance the BP-degrading capability, which could be attributed to enrichment of some potentially VBNC or difficult-to-culture BP/PCB degraders. In addition, it was evident that the mainly functional bacteria in response to EOM addition were Actinobacteria, which were prone to enter into VBNC state and showed a low degrading activity in long-term PCB-contaminated sediment.
Recent advances in the bioremediation of PCB-contaminated sites have focused on the development of ways to stimulate the activities of indigenous BP/PCB-degrading community (Leigh et al., 2006; Gomes et al., 2013). However, the application of PCB bioremediation is still inefficient and not well established up to now. It has been known that diverse BP-utilizing bacteria have the capability to transform several PCB congeners by bph encoded BP pathway (Cámara et al., 2004). And numerous studies have focused on the BP-degrading bacterial community and BP degradation pathway in order to explore a competitive advantage of PCB degraders and establish optimized PCB-bioremediation processes (Pieper, 2005; Uhlik et al., 2009). However, highly efficient BP/PCB-degrading community in the laboratory experiments showed lower efficiency and survived poorly when these cultures were inoculated in PCB-contaminated sites. Thus, it will be of critical importance to stimulate functional bacteria to enhance their BP/PCB degrading capabilities for in situ bioremediation. The present study demonstrated that EOM from M. luteus is an efficient additive which can significantly enhance BP biodegradation by recovering and stimulating the potentially functional BP-degrading community. It provided new insight into the exploration of potentially functional bacteria for enhancing in situ bioremediation, which could be inferred that the addition of EOM to PCB-contaminated areas holds great potential for the efficient and cost-effective bioremediation of PCB-contaminated environments.
Conclusions
The obtained results suggest that Proteobacteria and Actinobacteria were two predominant classes in long-term PCB-contaminated sediment. EOM from M. luteus enhanced the performance of BP biodegradation, which could be attributed to stimulation of the growth and activity of some potentially BP/PCB-degraders. Illumina high-throughput sequencing and culture-dependent methods indicated that the genera of Rhodococcus and Pseudomonas which were related to BP/PCB-degradation were greatly abundant after EOM addition, and potentially difficult-to-culture bacteria in response to EOM addition were mainly Actinobacteria. This study provides new insights into the identity of as-yet uncultured and unclassified bacteria actively metabolizing BP/PCB with EOM addition, and points out broader BP/PCB-degrading community which could be employed in bioremediation of sites.
Experimental procedures
Preparation of EOM from M. luteus
Micrococcus luteus IAM 14879 (= NCIMB 13267) used in this study had previously been described (Mukamolova et al., 1998; Ding, 2004). The pure culture was inoculated at 30°C on a rotary shaker at 160 r.p.m. for 36 h in modified lactate minimal medium (LMM) of Kaprelyants and Kell (1992). Then, the pre-culture was grown under the same culture conditions until the cells reached the stationary phase. The obtained fermentation broth was centrifuged (8000 r.p.m., 15 min) to separate the cells and then filtered through a 0.22 μm filter to remove floating cells. Finally, EOM was obtained and stored at −20°C for further experiments. The pH and redox potential (Eh) of EOM were 8.8 and −106 mV respectively. The concentration of total carbon, nitrogen, phosphorus and sulphur of EOM were 867.8, 279.3, 99.8 and 42.1 mg l−1 respectively. The main components of EOM were polysaccharides and proteins, and their concentrations were 405.7 and 25.1 mg l−1 respectively. Especially, Rpf protein was dominated in the proteins of EOM (Su et al., 2015).
Enrichment and cultivation
A PCB-contaminated sediment sample was obtained from a river very close to the e-waste recycling site in Luqiao Town of Taizhou City, which has been involved in e-waste disassembly for nearly 30 years (Shen et al., 2008; Chen et al., 2010; 2012). The sediment (4%, w/v) was incubated in a mineral salts medium (Su et al., 2013b), which BP (reagent grade, Sigma-Aldrich) was added as the sole carbon and energy source at an initial concentration of 500 mg l−1. Cultures were cultivated in conical flasks at 30°C on a rotary shaker at 180 r.p.m. Initial enrichments were transferred into fresh medium with 5% (v/v) inoculum after 5 days of cultivation. The BP concentration was increased in steps of 500 mg l−1 until a final concentration of 2000 mg l−1 was reached. Throughout the entire enrichment period, an equal amount EOM (10%, v/v), autoclaved EOM (sterilized at 121°C for 15 min) and LMM were added to TG, CG and BG respectively. Finally, the obtained enrichment samples were subjected to three different treatments, including TG, CG and BG.
BP degradation of enrichment cultures
The BP degradation capability of the enrichment samples (TG, CG and BG) were assessed by investigating the BP-tolerance concentrations and BP-degradation curve of the samples described elsewhere (Su et al., 2013b). Briefly, after cultivating for 64 h at 30°C on a rotary shaker, BP-tolerance concentrations were investigated with the concentration of BP varying from 500 to 4500 mg l−1 in steps of 500 mg l−1. And the BP-degradation and cell growth curve was depicted at 12 h intervals within 96 h under a BP concentration of 1500 mg l−1. Lastly, the cell growth (OD600) and BP-degradation efficiency of each experimental culture were measured as previously described (Su et al., 2013b). All of the experiments were performed in triplicate, and standard deviation (SD) was calculated by SPSS software (version 18.0, Chicago, IL, USA) for analysis of statistical significance of the triplicate samples.
DNA extraction
After lyophilization, DNA extraction of sediment sample was carried out using a beating method (FastDNATM SPIN Kit for Soil, Bio101, USA) following the manufacturer's protocol. For the enrichment samples and pure bacterial cultures, DNA extraction was performed using the EZ-10 spin column genomic DNA miniprep kit (Bio Basic, Canada) according to the manufacturer's instructions. The DNA extracts were stored at −20°C for further analysis.
Illumina high-throughput sequencing
Illumina high-throughput sequencing was performed to determine the diversity and composition of the bacterial communities in the sediment and enrichment samples (Capodicasa et al., 2009). PCR amplifications were conducted with the 515F/806R primer set that amplifies the V4 region of the 16S rRNA gene. The reverse primer contained a 6 bp error-correcting barcode unique to each sample. DNA was amplified following the protocol described by (Magoč and Salzberg, 2011). Sequencing was conducted on an Illumina MiSeq platform. Sequences were analysed with the Quantitative Insights Into Microbial Ecology software package and UPARSE pipeline (Wang et al., 2007), and were assigned to OTUs at 97% similarity. All sequences have been deposited in GenBank short-read archive (SRA: SRS632100).
Isolation and phylogenetic analysis of bacteria
The enrichment cultures TG, CG and BG were diluted respectively in 10-fold steps. Then serial dilutions (from 102-fold to 108-fold) was plated on mineral-salts agar plates with BP as the carbon source. Colonies that were unique in the TG, with no counterpart in the CG and BG, were selected. All isolates were purified and stored at 4°C for further identification. Genomic DNA was extracted from the pure bacterial cultures and then was amplified by PCR using primers 8F/1541R as described previously (Su et al., 2013b). Phylograms were constructed by the neighbour-joining method with 1000 replicate trees in the MEGA6 computer software program (Tamura et al., 2013). All sequences reported in this study had been submitted to NCBI GenBank under accession numbers KJ546446-KJ546456.
Conflict and interest
None declared.
References
- Abraham WR, Nogales B, Golyshin PN, Pieper DH. Timmis KN. Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr Opin Microbiol. 2002;5:246–253. doi: 10.1016/s1369-5274(02)00323-5. [DOI] [PubMed] [Google Scholar]
- Cámara B, Herrera C, González M, Couve E, Hofer B. Seeger M. From PCBs to highly toxic metabolites by the biphenyl pathway. Environ Microbiol. 2004;6:842–850. doi: 10.1111/j.1462-2920.2004.00630.x. [DOI] [PubMed] [Google Scholar]
- de Cárcer DA, Martín M, Karlson U. Rivilla R. Changes in bacterial populations and in biphenyl dioxygenase gene diversity in a polychlorinated biphenyl-polluted soil after introduction of willow trees for rhizoremediation. Appl Environ Microbiol. 2007;73:6224–6232. doi: 10.1128/AEM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capodicasa S, Fedi S, Carnevali M, Caporali L, Viti C, Fava F. Zannoni D. Terminal-restriction fragment length polymorphism analysis of biphenyl dioxygenase genes from a polychlorinated biphenyl-polluted soil. Res Microbiol. 2009;160:742–750. doi: 10.1016/j.resmic.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Chávez FP, Gordillo F. Jerez CA. Adaptive responses and cellular behaviour of biphenyl-degrading bacteria toward polychlorinated biphenyls. Biotechnol Adv. 2006;24:309–320. doi: 10.1016/j.biotechadv.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Chen L, Yu C, Shen C, Zhang C, Liu L, Shen K, et al. Study on adverse impact of e-waste disassembly on surface sediment in East China by chemical analysis and bioassays. J Soil Sediment. 2010;10:359–367. [Google Scholar]
- Chen L, Yu C, Shen C, Cui J, Chen C. Chen Y. Occurrence of (anti) estrogenic effects in surface sediment from an E-waste disassembly region in East China. Bull Environ Contam Toxicol. 2012;89:161–165. doi: 10.1007/s00128-012-0632-9. [DOI] [PubMed] [Google Scholar]
- Correa PA, Lin L, Just CL, Hu D, Hornbuckle KC, Schnoor JL. Van Aken B. The effects of individual PCB congeners on the soil bacterial community structure and the abundance of biphenyl dioxygenase genes. Environ Int. 2010;36:901–906. doi: 10.1016/j.envint.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding LX. 2004. Tokyo: The University of Tokyo Studies on the isolation of viable but non-culturable bacteria and the phylogenetic analysis of the genus Aquaspirillum.
- Ding LX. Yokota A. Curvibacter fontana sp. nov., a microaerobic bacteria isolated from well water. J Gen Appl Microbiol. 2010;56:267–271. doi: 10.2323/jgam.56.267. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Shimizu S, Okita N, Seto M. Masai E. Structural alteration of linear plasmids encoding the genes for polychlorinated biphenyl degradation in Rhodococcus strain RHA1. Antonie Van Leeuwenhoek. 1998;74:169–173. doi: 10.1023/a:1001732718159. [DOI] [PubMed] [Google Scholar]
- Gomes HI, Dias-Ferreira C. Ribeiro AB. Overview of in situ and ex situ remediation technologies for PCB-contaminated soils and sediments and obstacles for full-scale application. Sci Total Environ. 2013;445–446:237–260. doi: 10.1016/j.scitotenv.2012.11.098. [DOI] [PubMed] [Google Scholar]
- Kaprelyants A. Kell D. Rapid assessment of bacterial viability and vitality by rhodamine 123 and flow cytometry. J Appl Microbiol. 1992;72:410–422. [Google Scholar]
- Keep NH, Ward JM, Robertson G, Cohen-Gonsaud M. Henderson B. Bacterial resuscitation factors: revival of viable but non-culturable bacteria. Cell Mol Life Sci. 2006;63:2555–2559. doi: 10.1007/s00018-006-6188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh MB, Prouzová P, Macková M, Macek T, Nagle DP. Fletcher JS. Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl Environ Microbiol. 2006;72:2331–2342. doi: 10.1128/AEM.72.4.2331-2342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Wang Z, Yang Y, Mei X. Wu Z. Correlating microbial community structure and composition with aeration intensity in submerged membrane bioreactors by 454 high-throughput pyrosequencing. Water Res. 2013;47:859–869. doi: 10.1016/j.watres.2012.11.013. [DOI] [PubMed] [Google Scholar]
- Macedo AJ, Timmis KN. Abraham WR. Widespread capacity to metabolize polychlorinated biphenyls by diverse microbial communities in soils with no significant exposure to PCB contamination. Environ Microbiol. 2007;9:1890–1897. doi: 10.1111/j.1462-2920.2007.01305.x. [DOI] [PubMed] [Google Scholar]
- Magoč T. Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikesková H, Novotný Č. Svobodová K. Interspecific interactions in mixed microbial cultures in a biodegradation perspective. Appl Microbiol Biotechnol. 2012;95:861–870. doi: 10.1007/s00253-012-4234-6. [DOI] [PubMed] [Google Scholar]
- Mukamolova GV, Kaprelyants AS, Young DI, Young M. Kell DB. A bacterial cytokine. Proc Natl Acad Sci USA. 1998;95:8916–8921. doi: 10.1073/pnas.95.15.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamolova GV, Murzin AG, Salina EG, Demina GR, Kell DB, Kaprelyants AS. Young M. Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation. Mol Microbiol. 2006;59:84–98. doi: 10.1111/j.1365-2958.2005.04930.x. [DOI] [PubMed] [Google Scholar]
- Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev. 2010;34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- Petrić I, Hršak D, Fingler S, Vončina E, Ćetković H, Begonja Kolar A. Udiković Kolić N. Enrichment and characterization of PCB-degrading bacteria as potential seed cultures for bioremediation of contaminated soil. Food Technol Biotech. 2007;45:11–20. [Google Scholar]
- Petrić I, Bru D, Udiković-Kolić N, Hršak D, Philippot L. Martin-Laurent F. Evidence for shifts in the structure and abundance of the microbial community in a long-term PCB-contaminated soil under bioremediation. J Hazard Mater. 2011;195:254–260. doi: 10.1016/j.jhazmat.2011.08.036. [DOI] [PubMed] [Google Scholar]
- Pieper DH. Aerobic degradation of polychlorinated biphenyls. Appl Microbiol Biotechnol. 2005;67:170–191. doi: 10.1007/s00253-004-1810-4. [DOI] [PubMed] [Google Scholar]
- Schroeckh V. Martin K. Resuscitation-promoting factors: distribution among actinobacteria, synthesis during life-cycle and biological activity. Antonie Van Leeuwenhoek. 2006;89:359–365. doi: 10.1007/s10482-005-9039-5. [DOI] [PubMed] [Google Scholar]
- Shen CF, Huang SB, Wang ZJ, Qiao M, Tang XJ, Yu CN, et al. Identification of Ah receptor agonists in soil of e-waste recycling sites from Taizhou area in China. Environ Sci Technol. 2008;42:49–55. doi: 10.1021/es071162z. [DOI] [PubMed] [Google Scholar]
- Shleeva M, Bagramyan K, Telkov M, Mukamolova GV, Young M, Kell DB. Kaprelyants AS. Formation and resuscitation of ‘non-culturable’ cells of Rhodococcus rhodochrous and Mycobacterium tuberculosis in prolonged stationary phase. Microbiology. 2002;148:1581–1591. doi: 10.1099/00221287-148-5-1581. [DOI] [PubMed] [Google Scholar]
- Stach JE. Burns RG. Enrichment versus biofilm culture: a functional and phylogenetic comparison of polycyclic aromatic hydrocarbon-degrading microbial communities. Environ Microbiol. 2002;4:169–182. doi: 10.1046/j.1462-2920.2002.00283.x. [DOI] [PubMed] [Google Scholar]
- Su XM, Chen X, Hu JX, Shen CF. Ding LX. Exploring the potential environmental functions of viable but non-culturable bacteria. World J Microbiol Biotechnol. 2013a;29:2213–2218. doi: 10.1007/s11274-013-1390-5. [DOI] [PubMed] [Google Scholar]
- Su XM, Ding LX, Hu JX, Shen CF. Chen YX. A novel approach to stimulate the biphenyl-degrading potential of bacterial community from PCBs-contaminated soil of e-waste recycling sites. Bioresour Technol. 2013b;146:27–34. doi: 10.1016/j.biortech.2013.07.028. [DOI] [PubMed] [Google Scholar]
- Su XM, Liu YD, Hashmi MZ, Hu JX, Ding LX, Wu M. Shen CF. Rhodococcus biphenylivorans sp. nov., a polychlorinated biphenyl-degrading bacterium. Antonie Van Leeuwenhoek. 2015;107:55–63. doi: 10.1007/s10482-014-0303-4. [DOI] [PubMed] [Google Scholar]
- Su XM, Liu YD, Hu JX, Ding LX. Shen CF. Optimization of protein production by Micrococcus luteus for exploring pollutant-degrading uncultured bacteria. Springerplus. 2014;3:117. doi: 10.1186/2193-1801-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A. Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XJ, Chen C, Chen LT, Cui JL, Yu CN, Chen L, et al. Bacterial communities of polychlorinated biphenyls polluted soil around an e-waste recycling workshop. Soil Sediment Contam. 2013;22:562–573. [Google Scholar]
- Uhlik O, Jecna K, Mackova M, Vlcek C, Hroudova M, Demnerova K, et al. Biphenyl-metabolizing bacteria in the rhizosphere of horseradish and bulk soil contaminated by polychlorinated biphenyls as revealed by stable isotope probing. Appl Environ Microbiol. 2009;75:6471–6477. doi: 10.1128/AEM.00466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlik O, Wald J, Strejcek M, Musilova L, Ridl J, Hroudova M, et al. Identification of bacteria utilizing biphenyl, benzoate, and naphthalene in long-term contaminated soil. PLoS ONE. 2012;7:e40653. doi: 10.1371/journal.pone.0040653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM. Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Chen X, Qiu M, Zeng X, Xu J, Deng D, et al. Bar-coded pyrosequencing reveals the responses of PBDE-degrading microbial communities to electron donor amendments. PLoS ONE. 2012;7:e30439. doi: 10.1371/journal.pone.0030439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanaroli G, Balloi A, Negroni A, Daffonchio D, Young LY. Fava F. Characterization of the microbial community from the marine sediment of the Venice lagoon capable of reductive dechlorination of coplanar polychlorinated biphenyls (PCBs) J Hazard Mater. 2010;178:417–426. doi: 10.1016/j.jhazmat.2010.01.097. [DOI] [PubMed] [Google Scholar]