Abstract
The occurrence of genes encoding biotechnologically relevant α/β-hydrolases in mangrove soil microbial communities was assessed using data obtained by whole-metagenome sequencing of four mangroves areas, denoted BrMgv01 to BrMgv04, in São Paulo, Brazil. The sequences (215 Mb in total) were filtered based on local amino acid alignments against the Lipase Engineering Database. In total, 5923 unassembled sequences were affiliated with 30 different α/β-hydrolase fold superfamilies. The most abundant predicted proteins encompassed cytosolic hydrolases (abH08; ∼ 23%), microsomal hydrolases (abH09; ∼ 12%) and Moraxella lipase-like proteins (abH04 and abH01; < 5%). Detailed analysis of the genes predicted to encode proteins of the abH08 superfamily revealed a high proportion related to epoxide hydrolases and haloalkane dehalogenases in polluted mangroves BrMgv01-02-03. This suggested selection and putative involvement in local degradation/detoxification of the pollutants. Seven sequences that were annotated as genes for putative epoxide hydrolases and five for putative haloalkane dehalogenases were found in a fosmid library generated from BrMgv02 DNA. The latter enzymes were predicted to belong to Actinobacteria, Deinococcus-Thermus, Planctomycetes and Proteobacteria. Our integrated approach thus identified 12 genes (complete and/or partial) that may encode hitherto undescribed enzymes. The low amino acid identity (< 60%) with already-described genes opens perspectives for both production in an expression host and genetic screening of metagenomes.
Introduction
Mangroves harbour diverse microbial communities which play critical roles in the functioning and maintenance of these sensitive and complex systems (Kathiresan and Bingham, 2001; Sahoo and Dhal, 2006). Given the peculiar factors that drive these systems (salt, anaerobic/aerobic shifts), mangroves offer rich sources of genes for new biotechnological products/enzymes, such as lipases (Couto et al., 2010), cellulases (Thompson et al., 2013) and laccases (Ye et al., 2010). Mangrove soils have already been explored for microbial diversity using a wide range of culture-dependent and culture-independent methods (Dias et al., 2009; dos Santos et al., 2011). In particular, the modern metagenomics-based tools, i.e. high-throughput sequencing of environmental DNA followed by a directed search for target genes, allow a ready access to the metabolic potential of mangrove microbial communities (Andreote et al., 2012). Moreover, genes for important enzymes can be further custom-synthesized and codon-optimized, after which heterologous expression may be achievable in a suitable host, an approach that has been coined ‘synthetic metagenomics’ (Chistoserdova, 2010). To access whole operons, metagenomic libraries can be constructed in large-insert vectors and screened either by functional or genetic approaches. Importantly, functional screening analysis does not depend on prior sequence information to detect the target proteins, provided these become available and active in the novel host. Recently, it was shown that particular sequence/activity incoherencies in databases can be solved using expression detection (Fernández-Arrojo et al., 2010; Jiménez et al., 2012a). However, proper testing requires that the expression conditions in the heterologous host are adequate (Ekkers et al., 2012).
Current classification of metagenomic sequencing data relies strongly on local alignments (e.g. using blast) against public databases (e.g. NCBI, SEED and KEGG) (Montaña et al., 2012). However, completely novel biotechnologically relevant proteins cannot be easily discovered using such approach. Metagenome datasets can be assessed by the use of more specific databases, such as for example the CAZy (Carbohydrate-Active Enzyme) (Cantarel et al., 2008), PeroxiBase (Fawal et al., 2013), Lipase/Laccase Engineering (Fischer and Pleiss, 2003; Sirim et al., 2011), ESTHER (Lenfant et al., 2013), 3DM (Kourist et al., 2010), Epoxide Hydrolases and/or Haloalkane Dehalogenases (Barth et al., 2004). Alternatively, catalytic or structurally conserved domains can be detected using hidden Markov models (HMM). However, most HMM are designed based on protein sequences retrieved from databases and so the true novelty is still questionable.
The α/β-hydrolase fold enzymes, present in the Lipase Engineering Database (LED), constitute a protein family with diverse catalytic and non-catalytic functions. The α/β-hydrolase fold proteins consist of eight β-strands connected by α-helices. These enzymes are characterized by a common catalytic triad formed by a catalytic nucleophile (serine, aspartate or cysteine), a histidine and an acidic residue (aspartate or glutamate). These residues occur on conserved locations in loops and the α/β-hydrolase fold brings them together to form the active site (Lenfant et al., 2013). These proteins encompass several key enzymes for biocatalytic applications, e.g. lipases, esterases, epoxide hydrolases (EHs), C–C breaking enzymes, dehalogenases and hydroxynitrile lyases (Holmquist, 2000).
Interestingly, EHs (Enzyme Commission number-EC 3.3.2.9) from microbial sources have been recently recognized as a versatile group of enzymes that are important for the synthesis of enantiopure oxides and vicinal diols (intermediates in the organic synthesis of chiral pharmaceutical compounds, drugs and agrochemicals) (Lee and Shuler, 2007; Choi, 2009; Sareen and Kumar, 2011). Such EHs are involved in the degradation of several hydrocarbons including 1,3-dihalo-2-propanol, epichlorohydrin, 9,10-epoxy fatty acids, trans-2,3-epoxysuccinate and 2,3- chlorostyrene oxides (van der Werf et al., 1998; Fretland and Omiecinski, 2000). The presence of EHs has been reported in bacteria recovered from gasoline and oil-contaminated marine sediments (Kwon et al., 2007; 2010; Woo et al., 2007; 2013). Moreover, haloalkane dehalogenases (HDs) (EC 3.8.1.5) have attracted considerable attention due to their unique catalytic mechanism, broad substrate specificity, stability, enantioselectivity and catalytic efficiency (Koudelakova et al., 2013). The HDs catalyse the cleavage of carbon–halogen bonds, which is a key step in the aerobic mineralization of many halogenated pollutants, such as oil compounds (Janssen et al., 2005). Previous studies indicated HDs to be important for the preparation of optically pure building blocks for organic synthesis, recycling of by-products from chemical processes, decontamination of chemical warfare agents and for bio-sensing of environmental pollutants and protein tagging for cell imaging and protein analysis (Koudelakova et al., 2013).
The current study aimed at bioprospection of shotgun sequence datasets generated from four mangrove soils for α/β-hydrolase fold proteins by using specific LED. In addition, a metagenomic fosmid library constructed from one oil-impacted mangrove site (BrMgv02) was used for sequence-based screening. The prevalence [relative abundance (RA)] of EHs and HDs was addressed, with special reference to the oil contamination, biodegradation and future potential industrial application.
Results and discussion
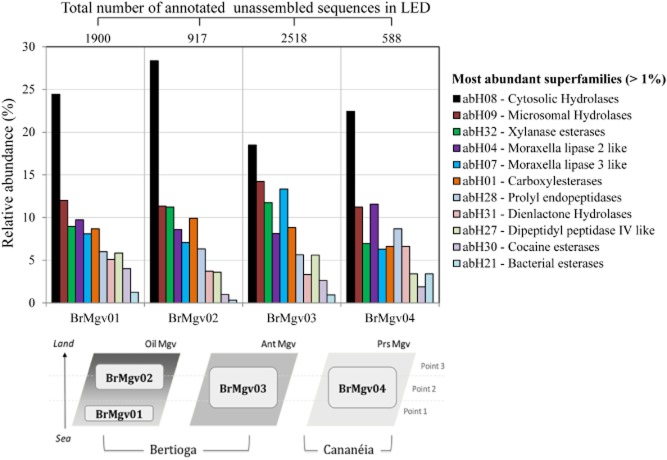
In this study, an analysis of the composition and diversity of metagenomic sequences encoding α/β-hydrolase fold proteins in four distinct mangrove soils was performed. Briefly, mangrove soil samples were collected in July 2008 in three distinct mangroves in the state of São Paulo (Brazil). Samples were divided in four groups: BrMgv01 and BrMgv02 (23°53′49″S; 46°12′28″W – Bertioga city) are two sites in the same mangrove separated by a small stream. This area has been affected by oil contamination (petroleum). Sample BrMgv03 (23°54′06″S; 45°15′03″W – Bertioga city) was taken from a site adjacent to BrMgv01-02 that had not been affected by oil, but by household waste. Finally, BrMgv04 (25°05′02″S; 47°57′42″W – Cananéia city) represents a sample from a pristine mangrove (Fig. 1) as detailed in Andreote and colleagues (2012). In total, 1.8 g (six samples per site) of soil from each area was subjected to total genomic DNA extraction, after which the DNAs were subjected to shotgun sequencing using the 454 GS-FLX titanium technology (Indianapolis, IN, USA). The sequences obtained (905 521 unassembled sequences with an average length of 236 bp) were sorted and trimmed based on length and quality, using an in-house python script (Jiménez et al., 2012b). The total numbers of trimmed sequences obtained for each mangrove area were 249 993 for BrMgv01 (average read length ∼ 235 bp), 231 233 for BrMgv02 (∼ 238 bp), 214 921 for BrMgv03 (∼ 248 bp) and 217 605 for BrMgv04 (∼ 223 bp). These sequences were uploaded to the metagenomic RAST (MG-RAST) server and made publically accessible under the project codes 4451033.3, 4451034.3, 4451035.3 and 4451036.3 for mangroves BrMgv01, BrMgv02, BrMgv03, and BrMgv04 respectively. As a complement to a previous study (Andreote et al., 2012), we performed BLASTX against LED (Fischer and Pleiss, 2003) using a cut-off e-value of 1e-5, as in other studies in which moderately rigid criteria were used to find genetic novelty and to evaluate functional and taxonomic profiles (Jung et al., 2011; Jiménez et al., 2012b; Mendes et al., 2014). It is important to note that BLASTX has been successfully used against LED in other studies (Kim et al., 2009; Damon et al., 2012). However, we are aware of the fact that these parameter settings may result in spurious hits and thus data need to be carefully re-examined. To address this critical issue, we performed manual annotation in the best hits. With this strategy, the information retrieved was compared across the four datasets. Thus, totals of 1900 (0.8% RA), 917 (0.4% RA), 2518 (1.1% RA) and 588 (0.2% RA) unassembled sequences were found to match 30 different α/β-hydrolase superfamilies, for mangrove samples BrMgv01, BrMgv02, BrMgv03 and BrMgv04 respectively (Fig. 1). Collectively, these sequences matched 22 superfamilies in the class GGGX. In the LED, proteins were assigned to the classes GX, GGGX and Y, in accordance with their amino acid sequences and the structures of the oxy anion holes (Pleiss et al., 2000). The oxy anion hole helps to stabilize the negatively charged transition state that occurs during enzymatic hydrolysis (Nardini and Dijkstra, 1999).
Fig 1.
Relative abundance (%) of the most abundant α/β-hydrolase fold protein superfamilies in the four mangrove metagenomes (BrMgv01 to BrMgv04). Oil Mgv: oil-polluted mangrove site; Ant Mgv: anthropogenic polluted site; Prs: pristine mangrove site. The database was downloaded from the LED ftp website – http://www.led.uni-stuttgart.de/ – release 3.0, last update on 10 December 2009. Normalization was performed using the total number of annotated sequences (in LED) in each dataset.
The most abundant superfamilies across all metagenomes were annotated as cytosolic hydrolases (abH08) (∼ 23% RA) and microsomal hydrolases (abH09) (∼ 12% RA) (except in BrMgv04), followed by Moraxella lipase 2-like sequences (abH04) (9.7% RA in BrMgv01 and 11.5% RA in BrMgv04), xylanase esterases (abH32) (11.7% RA in BrMgv02) and Moraxella lipase 3-like (abH07) (13.3% RA in BrMgv03) (Fig. 1). Superfamily abH08 consists of 15 homologous families that include 3188 protein entries in LED. Into the abH08 superfamily, we can found a large group of bacterial EHs, non-heme peroxidases and HDs. In addition, superfamily abH09 contains three families, namely microsomal EHs, BioH protein like (biotin biosynthesis) and proline iminopeptidases (Fischer and Pleiss, 2003). Furthermore, proline iminopeptidases, propyl endopeptidases and dipeptidyl peptidases make part of the bacterial proteolytic system, which has been reported as being key for bacterial nitrogen utilization (nitrogen from amino acids), especially under nitrogen-limiting conditions (Kunji et al., 1996; Li et al., 2010). Nitrogen limitation may reign in anoxic soil mangroves, in which denitrification leading to gaseous nitrogen effluxes is rampant (Fernandes et al., 2012).
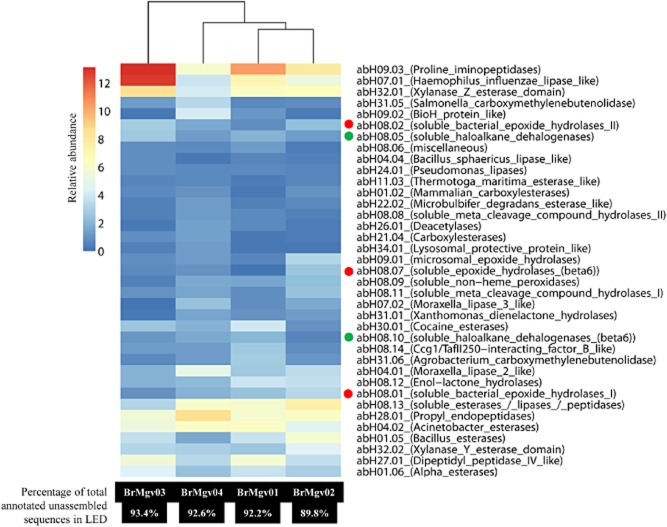
We then assessed the distribution of the unassembled sequences over the predicted protein families per mangrove sample. The prevalences of proline iminopeptidases (abH09.03), Haemophilus influenzae lipase-like (abH07.01), xylanase Z esterase domain (abH32.01) and dipeptidyl peptidases (abH27.01) were highest in BrMgv01 (10.3%, 7.2%, 6.4% and 5.8% RA respectively) and BrMgv03 (13.1%, 12.7%, 8.5% and 5.5% RA respectively). The families encompassing Acinetobacter esterases (abH04.02) and propyl endopeptidases (abH28.01) showed highest prevalences (6.3 and 8.6% RA respectively) in the unpolluted mangrove site (BrMgv04). Conversely, EHs, belonging to families abH08.01, abH09.01, abH08.07 and abH08.02, were abundant in BrMgv02 (3.3%, 3.1%, 2.6% and 2.3% RA respectively) compared with the unpolluted mangrove BrMgv04 (1.8%, 1.1%, 1.0% and 1.7% RA respectively) (Fig. 2). On another matter, genes encoding xylanases/esterases were very prevalent in all mangrove samples (between 4% and 8% RA), irrespective of pollution. This suggests that the conversion of hemicellulose is likely to occur in these environments, which are anaerobic most of the time (Benner et al., 1984; Ye et al., 2010).
Fig 2.
Relative abundance (%) of the most abundant α/β-hydrolase fold protein families in four mangrove metagenomes (BrMgv01-BrMgv04). Hierarchical dendrogram and heat-mapping were performed using cluster V3.0 (Eisen et al., 1998) software. Red and green dots represent the epoxide hydrolase (EHs) and haloalkane dehalogenase (HDs) families respectively.
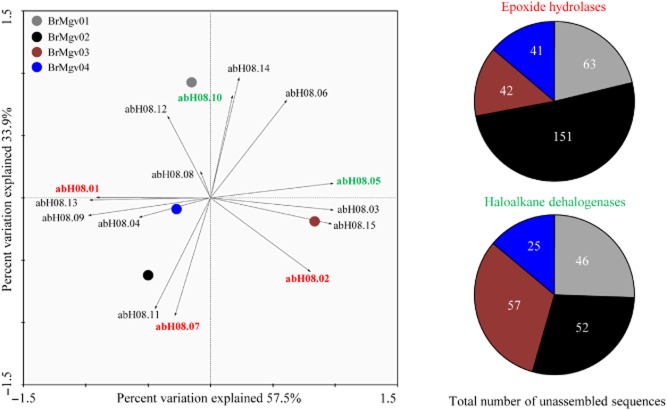
To shed light on the α/β-hydrolase fold protein superfamilies detected in our dataset, we focused on the protein superfamily that was most abundant across the board, i.e. abH08. Principal component analyses showed that EHs, families abH08.07 and abH08.02, were preferentially present in BrMgv02 and BrMgv03 respectively (Fig. 3). In terms of numbers of annotated sequences, the prevalences of these predicted enzymes (abH08.01, abH08.07 and abH08.02) were highest in BrMgv02 site (151 unassembled sequences – 30% RA based on the number of annotated sequences within abH08 family) comparatively to BrMgv04 (41 unassembled sequences – 17% RA). Regarding HDs (abH08.10 and abH08.05), highest numbers of annotated genes were observed in the polluted mangroves BrMgv02 (52 unassembled sequences – 11% RA) and BrMgv03 (57 unassembled sequences – 22% RA) (Fig. 3). Interestingly, van Loo and colleagues (2006) reported the screening of various genomic databases for the presence of EHs to find ways to express these proteins in different bacterial hosts. In addition, putative open reading frames for EHs and subsequent expression have been investigated in Cupriavidus metallidurans CH34 (Kumar et al., 2011). Clearly, novel EHs, provided they offer features such as enhanced activity, substrate specificity and/or stability in the face of chemicals, are useful in the production of a range of compounds (e.g. β3-adrenergic receptor agonists, anti-obesity and anti-inflammatory drugs, nematicides, anticancer agents, anti-fungal chemicals). In addition, they may also serve the detoxification of xenobiotics such as polyaromatic hydrocarbons and the production of enantiopure epoxides and vicinal diols from cheap racemic epoxides (Lee and Shuler, 2007).
Fig 3.
Left: principal component analysis (PCA) within the family abH08 using Canoco software v4.52 (Wageningen, the Netherlands). Right: Number of unassembled sequences annotated in the abH08.02 [epoxide hydrolases (EHs)] and abH08.05 [haloalkane dehalogenases (HDs)] families in BrMgv samples.
In the light of the high prevalence of sequences related to EHs (family abH08.02) and HDs (family abH08.05) in highly oil-impacted mangrove BrMgv02 (73 and 36 unassembled sequences respectively), this specific dataset and the best BLAST hits (based on e-values) were further analysed. For the former unassembled sequences, we found some putative novel enzymes (based on relatively low amino acid identity: < 60%) that were mostly affiliated with similar proteins (amino acid identity between 50% and 73%) from Streptomyces, Hyphomonas, Bradyrhizobium and Phenylobacterium. For the abH08.05 family, HDs were affiliated to proteins from Salinispora, Alcanivorax, Photobacterium, Moritella and Chloroflexus (Table 1 – approach 1). It is important to stress that the taxonomic affiliation is only an indication of identity as it is based on function of the most similar or homologous proteins, and thus may not directly reflect the true microbial source. Thus a whole new suite of partial genes encoding novel enzymes within these two classes was unlocked. These are hypothesized to function best under the environmental conditions of the habitat, i.e. the local salt, oxic/anoxic conditions as well as the presence of hydrocarbons (Arulazhagan and Vasudevan, 2011; Arfi et al., 2013). Such enzymes might well suit industrial and environmental (pollutant removal) needs, as will be explored in future work. According to Andreote and colleagues (2012), the bacterial community at the BrMgv02 mangrove site were dominated by Proteobacteria belonging to the classes Delta < Gamma < Alpha. Moreover, Alphaproteobacteria, next to Actinobacteria were significantly raised compared with BrMgv04. Hence, we surmised that such bacterial groups were carriers of the biodegradative functions in the oil-polluted BrMgv02, which is consistent with findings in other estuarine ecosystems (Greer, 2010).
Table 1.
Putative epoxide hydrolases (EHs) retrieved from the BrMgv02 dataset
| Approach (Mangrove) | Best-hit protein (NCBI Accession No.) | Putative microbial source based on homology (Best hit protein) | Hit-length (aa) | Query-length (bp) | e-value | Amino acid identity (%) | ESTHERc |
|---|---|---|---|---|---|---|---|
| 1 (BrMgv02) | Putative epoxide hydrolase (YP_761108) | Hyphomonas neptunium | 320 | 345 | 7e-48 | 73.04 | XE |
| Epoxide hydrolase (NP_823281) | Streptomyces avermitilis | 328 | 394 | 2e-28 | 48.46a | XE | |
| Epoxide hydrolase (YP_002203910) | Streptomyces sviceus | 330 | 393 | 8e-28 | 47.69a | XE | |
| Epoxide hydrolase (NP_767754) | Bradyrhizobium diazoefficiens | 330 | 345 | 7e-24 | 50a | XE | |
| Epoxide hydrolase (YP_002129963.1) | Phenylobacterium zucineum | 321 | 347 | 4e-23 | 51a | XE | |
| 2 (BrMgv02) | Alpha/beta hydrolase fold protein (WP_008687080.1) | Rhodopirellula sallentina | 319 | 850 | 1e-65 | 40ab | XE |
| Epoxide hydrolase – like protein (YP_003705817.1) | Frankia sp. | 398 | 351 | 4e-32 | 51ab | XE | |
| Alpha/beta hydrolase fold protein (YP_002544714.1) | Truepera radiovictrix | 292 | 504 | 3e-38 | 69b | XE | |
| Alpha/beta hydrolase fold protein (YP_003336243.1) | Streptosporangium roseum | 307 | 1459 | 3e-75 | 55ab | XE | |
| Epoxide hydrolase domain protein (WP_007534516.1) | Rhizobium mesoamericanum | 199 | 345 | 3e-21 | 49ab | XE | |
| Epoxide hydrolase (WP_009025691) | Bradyrhizobium sp. | 334 | 802 | 8e-74 | 46ab | XE | |
| Alpha/beta hydrolase (YP_727726.1) | Rhodospirillum rubrum | 305 | 1039 | 4e-116 | 64b | XE |
Putative novel protein based on low amino acid identity (< 60%).
Complete genes.
Classification based on BLASTX against ESTHER database (Lenfant et al., 2013).
The table was constructed using two different approaches: (1) Whole metagenome sequencing – and BLASTX against the LED; (2) Fosmid library sequencing – and BLASTX against the NCBI using the MG-RAST pipeline.
XE, Block_X, Family Epoxide_hydrolase.
In a second stage, we studied selected sequences retrieved from a fosmid library constructed using the CopyControl Fosmid Library Production Kit – Epicentre which contains the pCC2FOS as a vector and Escherichia coli EPI 300 T1R Phage T1 resistant [F– mcrA Δ(mrr-hsdRMS-mcrBC) (StrR) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139Δ(ara, leu)7697 galU galK λ– rpsL nupG trfA tonA dhfr] as host cells. Environmental DNA extracted from the oil-impacted mangrove soil BrMgv02 was used to construct the metagenomic library, with insert sizes ranging from 25 to 40 kb. A total of 12 900 clones was obtained and further subjected to 454 pyrosequencing (GS FLX Titanium technology), yielding a total of 1 380 509 unassembled sequences with average length of 484 bp (approximately 624 Mb). The sequences were assembled into contigs using CLC Genomics Workbench (version 6.5.1; CLC Bio, Cambridge, MA, USA) (under default parameters) resulting in 118 882 sequences (average length 913 bp), totalling 108 Mb. The data were annotated using the MG-RAST pipeline (cut-off e-value of 1e-5) (Meyer et al., 2014). This allowed the detection of seven putative EHs in fosmids (fosmid library metagenomic data were deposited at MG-RAST under the ID No. 4555913.3). These were further compared and were found to be affiliated with EHs in genomes of Rhodopirellula, Bradyrhizobium (family abH08.02), Truepera, Streptosporangium (family abH08.01), Rhizobium, Frankia (family abH09.01) and Rhodospirillum (family abH08.07) (Table 1 – approach 2). Moreover, five HDs sequences were detected, and these matched those present in the genomes of Anaeromyxobacter, Oceanicaulis (family abH08.05), Mycobacterium (families abH08.05 and abH08.10) and Aeromicrobium (abH08.10). In some cases (five EHs and two HDs), the sequence identity with already described genes (database) was low (< 60%), suggesting that the sequences could represent putative new proteins. The hierarchical classification of these 22 (complete and/or partial) genes, based on the ESTHER database, was done (Tables 1 and 2). Importantly, the activity of such enzymes may not be detected in E. coli host cells, specially due to the different expression systems present in this species when compared with those of Actinobacteria, Alphaproteobacteria, Deinococcus-Thermus and Planctomycetes members (Gabor et al., 2004). On the other hand, the use of degenerate primers has been applied as a sequence-driven approach to identify enzymes directly from metagenomes. For example, Kotik and colleagues (2009) amplified fragments of EH genes using degenerate primers targeted to conserved motifs, followed by assembly by genome walking. These results highlight the importance of using both sequence and function-based approaches in metagenomic library screenings, thus circumventing problems inherent to either lack of heterologous expression efficiency or limited sequence information based on known protein sequences available at public databases. Our fosmid-based approach opens up the possibility of finding whole operons, start/stop codons and expression signals on the basis of the genetic information gathered in this study. The EHs and HDs encoding genes from the oil-impacted mangrove soils can thus be further expressed in appropriate expression vectors, for future practical use in industrial or biotechnological processes. Comparison of the EHs genes between the BrMgv02 metagenome and the fosmid library datasets revealed protein families (abH08.01, abH09.01, abH08.07 and abH08.02) to be coincident. Similar results were found with HDs, as the main protein families found within the BrMgv02 metagenome were also observed in the fosmid library dataset (abH08.05 and abH08.10). Thus, the metagenomic library represented to a considerable extent the diversity of EHs and HDs present in the analysed mangrove sample.
Table 2.
Putative haloalkane dehalogenases (HDs) retrieved from the BrMgv02 dataset
| Approach (Mangrove) | Best-hit protein (NCBI Accession No.) | Putative microbial source based on homology (Best hit protein) | Hit-length (aa) | Query-length (bp) | e-value | Amino acid identity (%) | ESTHERc |
|---|---|---|---|---|---|---|---|
| 1 (BrMgv02) | Haloalkane dehalogenase (YP_001537995) | Salinispora arenicola | 310 | 412 | 1e-29 | 65.8 | X1 |
| Haloalkane dehalogenase (YP_694135) | Alcanivorax borkumensis | 296 | 410 | 5e-29 | 63.5 | X1 | |
| Putative haloalkane dehalogenase (ZP_01221858) | Photobacterium profundum | 303 | 242 | 1e-27 | 62.5 | X1 | |
| Putative haloalkane dehalogenase (ZP_01897865) | Moritella sp. | 292 | 190 | 2e-25 | 75.8 | X1 | |
| Haloalkane dehalogenase (ZP_02986139) | Chloroflexus sp. | 293 | 306 | 5e-19 | 45a | X1 | |
| 2 (BrMgv02) | Haloalkane dehalogenase (YP_001377212.1) | Anaeromyxobacter sp. | 304 | 840 | 8e-37 | 49ab | X1 |
| Haloalkane dehalogenase (WP_020727623.1) | Mycobacterium marinum | 325 | 1752 | 1e-146 | 80ab | X2 | |
| Haloalkane dehalogenase (WP_022699531.1) | Oceanicaulis alexandrii | 301 | 249 | 1e-46 | 64ab | X1 | |
| Haloalkane dehalogenase (YP_003336243.1) | Mycobacterium vanbaalenii | 298 | 607 | 5e-81 | 61ab | X1 | |
| Haloalkane dehalogenase (WP_007078410.1) | Aeromicrobium marinum | 290 | 1241 | 3e-65 | 52ab | X1 |
Putative novel protein based on low amino acid identity (< 60%).
Complete genes.
Classification based on BLASTX against ESTHER database (Lenfant et al., 2013).
The table was constructed using two different approaches: (1) Whole metagenome sequencing – and BLASTX against the LED; (2) Fosmid library sequencing – and BLASTX against the NCBI using the MG-RAST pipeline.
X1, Block_X, Family Haloalkane_dehalogenase-HLD1; X2, Block_X, Family Haloalkane_dehalogenase-HLD2.
Clearly, Actinobacteria and Alphaproteobacteria might serve as genetic sources for EHs bio-exploration. For instance, analysis of the genome of the actinobacterium Mycobacterium tuberculosis revealed an unusually large number of potential EHs, i.e. nine EHs genes occurred scattered on the genome (Johansson et al., 2005). Also, the presence of EHs in Agrobacterium radiobacter AD1 has been reported (Rink et al., 1997). These were further engineered towards an increasing activity for industrial purposes (Rui et al., 2005). Protein engineering proved to be an efficient method to tailor α/β-hydrolase fold enzymes towards a desired property. Moreover, enzymes with completely new catalytic activities have been generated, for instance the conversion of an esterase from Pseudomonas fluorescens into an EH (Jochens et al., 2009). The EHs, HDs and haloperoxidases have a typical lipase catalytic triad (G-X-S-X-G) and share approximately 25% of amino acid identity with lipases belonging to the family V (Arpigny and Jaeger, 1999; Tirawongsaroj et al., 2008). In the catalytic triad, the nucleophilic aspartate carries out an attack on the carbon atom of the epoxide ring, thus displacing the oxygen and producing a covalent intermediate compound (de Vries and Janssen, 2003). Soluble EHs have recently been found in an Andean forest soil metagenome, in this case affiliated to the bacterium Streptomyces scabies (Montaña et al., 2012). In addition, Procópio and colleagues (2013) reported the presence of five putative genes encoding EHs in the genome of Dietzia cinnamea (a common soil Actinobacterium). These studies are consistent with the notion that members of the Actinobacteria can produce EHs in the environment, which supports their potential use as bioremediation agents, for instance in oil-contaminated systems. Moreover, we also found HDs belonging to Alcanivorax and Phenylobacterium. These microorganisms are known as ‘hydrocarbonoclastic’ based on their capacity to degrade an exceptionally broad range of haloalkane hydrocarbons (Sabirova et al., 2006; dos Santos et al., 2011). The genus Phenylobacterium (a facultatively anaerobic bacterium) has a unique preference for phenyl moieties from heterocyclic compounds such as chloridazon, antipyrine and pyramidon (Oh and Roh, 2012). Conversely, Alcanivorax species has also been reported as a key bacterial group present in crude oil enrichments based on mangrove soils as the microbial source (Brito et al., 2006).
Conclusions
Current bottlenecks in high-throughput metagenome analysis are mostly due to problems related to sequence annotation. This factor can drastically affect the interpretation of a given dataset, especially in the case of enzyme annotation (Hoff, 2009; Schnoes et al., 2009). In this sense, it becomes important to use specific and curated databases, which – in combination with manual annotation – can improve our capability of data mining. In this study, we make use of metagenomics datasets from mangrove soils to investigate the prevalence and diversity of genes for α/β-hydrolase fold related proteins, using a specific database. Sequences, predicted to belong to Actinobacteria, Chloroflexi, Deinococcus-Thermus, Planctomycetes and Proteobacteria EHs or HDs codifying genes are described and analysed. Moreover, the description of the EHs and HDs will be further explored in the context of the bioconversion of hydrocarbons in oil-contaminated environments. Our results might represent a first step towards the development of a totally synthetic metagenomics approach (synthesis-cloning-expression), to be broadly applied in mangrove tropical ecosystems. Finally, we conclude that the presence of hydrocarbons in mangrove soils has an effect on the abundance and diversity of α/β-hydrolase fold proteins, which were mostly heightened in EHs and HDs.
Acknowledgments
We thank C. Guzman, D. Riaño and S. Restrepo from Andes University (Bogotá D.C, Colombia) for their support through bioinformatics analysis.
Conflict of interest
The authors declare no conflict of interest.
References
- Andreote FD, Jiménez DJ, Chaves D, Dias AC, Luvizotto DM, Dini-Andreote F, et al. The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS ONE. 2012;7:e38600. doi: 10.1371/journal.pone.0038600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfi Y, Chevret D, Henrissat B, Berrin JG, Levasseur A. Record E. Characterization of salt-adapted secreted lignocellulolytic enzymes from the mangrove fungus Pestalotiopsis sp. Nat Commun. 2013;4:1810. doi: 10.1038/ncomms2850. [DOI] [PubMed] [Google Scholar]
- Arpigny JL. Jaeger KE. Bacterial lipolytic enzymes: classification and properties. Biochem J. 1999;343:177–183. [PMC free article] [PubMed] [Google Scholar]
- Arulazhagan P. Vasudevan N. Biodegradation of polycyclic aromatic hydrocarbons by a halotolerant bacterial strain Ochrobactrum sp. VA1. Mar Pollut Bull. 2011;62:388–394. doi: 10.1016/j.marpolbul.2010.09.020. [DOI] [PubMed] [Google Scholar]
- Barth S, Fischer M, Schmid RD. Pleiss J. The database of epoxide hydrolases and haloalkane dehalogenases: one structure, many functions. Bioinformatics. 2004;20:2845–2847. doi: 10.1093/bioinformatics/bth284. [DOI] [PubMed] [Google Scholar]
- Benner R, Maccubbin AE. Hodson RE. Anaerobic biodegradation of the lignin and polysaccharide components of lignocellulose and synthetic lignin by sediment microflora. Appl Environ Microbiol. 1984;47:998–1004. doi: 10.1128/aem.47.5.998-1004.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito EM, Guyoneaud R, Goñi-Urriza M, Ranchou-Peyruse A, Verbaere A, Crapez MA, et al. Characterization of hydrocarbonoclastic bacterial communities from mangrove sediments in Guanabara Bay, Brazil. Res Microbiol. 2006;157:752–762. doi: 10.1016/j.resmic.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V. Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2008;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. Recent progress and new challenges in metagenomics for biotechnology. Biotechnol Lett. 2010;32:1351–1359. doi: 10.1007/s10529-010-0306-9. [DOI] [PubMed] [Google Scholar]
- Choi WJ. Biotechnological production of enantiopure epoxides by enzymatic kinetic resolution. Appl Microbiol Biotechnol. 2009;84:239–247. doi: 10.1007/s00253-009-2110-9. [DOI] [PubMed] [Google Scholar]
- Couto GH, Glogauer A, Faoro A, Chubatsu LS, Souza EM. Pedrosa FO. Isolation of a novel lipase from a metagenomic library derived from mangrove sediment from the south Brazilian coast. Genet Mol Res. 2010;9:514–523. doi: 10.4238/vol9-1gmr738. [DOI] [PubMed] [Google Scholar]
- Damon C, Lehembre F, Oger-Desfeux C, Luis P, Ranger J, Fraissinet-Tachet L. Marmeisse R. Metatranscriptomics reveals the diversity of genes expressed by eukaryotes in forest soils. PLoS ONE. 2012;7:e28967. doi: 10.1371/journal.pone.0028967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias ACF, Andreote FD, Dini-Andreote F, Lacava PT, Sá ALB, Melo IS, et al. Diversity and biotechnological potential of culturable bacteria from Brazilian mangrove sediment. World J Microbiol Biotechnol. 2009;25:1305–1311. [Google Scholar]
- Eisen MB, Spellman PT, Brown PO. Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekkers DM, Cretoiu MS, Kielak AM. van Elsas JD. The great screen anomaly-a new frontier in product discovery through functional metagenomics. Appl Microbiol Biotechnol. 2012;93:1005–1020. doi: 10.1007/s00253-011-3804-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawal N, Li Q, Savelli B, Brette M, Passaia G, Fabre M, et al. PeroxiBase: a database for large-scale evolutionary analysis of peroxidases. Nucleic Acids Res. 2013;41(Database issue):D441–D444. doi: 10.1093/nar/gks1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Arrojo L, Guazzaroni ME, López-Cortés N, Beloqui A. Ferrer M. Metagenomic era for biocatalyst identification. Curr Opin Biotechnol. 2010;21:725–733. doi: 10.1016/j.copbio.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Fernandes SO, Michotey VD, Guasco S, Bonin PC. Bharathi PA. Denitrification prevails over anammox in tropical mangrove sediments (Goa, India) Mar Environ Res. 2012;74:9–19. doi: 10.1016/j.marenvres.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Fischer M. Pleiss J. The Lipase Engineering Database: a navigation and analysis tool for protein families. Nucleic Acids Res. 2003;31:319–321. doi: 10.1093/nar/gkg015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fretland AJ. Omiecinski CJ. Epoxide hydrolases: biochemistry and molecular biology. Chem Biol Interact. 2000;129:41–59. doi: 10.1016/s0009-2797(00)00197-6. [DOI] [PubMed] [Google Scholar]
- Gabor EM, Alkema WB. Janssen DB. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ Microbiol. 2004;6:879–886. doi: 10.1111/j.1462-2920.2004.00640.x. [DOI] [PubMed] [Google Scholar]
- Greer CW. Bacterial diversity in hydrocarbon-polluted rivers, estuaries and sediments. In: Timmis KN, editor. Handbook of Hydrocarbon and Lipid Microbiology. Berlin Heidelberg, Germany: Springer-Verlag; 2010. pp. 2329–2338. [Google Scholar]
- Hoff KJ. The effect of sequencing errors on metagenomic gene prediction. BMC Genomics. 2009;10:520. doi: 10.1186/1471-2164-10-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist M. Alpha/Beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr Protein Pept Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- Janssen DB, Dinkla IJ, Poelarends GJ. Terpstra P. Bacterial degradation of xenobiotic compounds: evolution and distribution of novel enzyme activities. Environ Microbiol. 2005;7:1868–1882. doi: 10.1111/j.1462-2920.2005.00966.x. [DOI] [PubMed] [Google Scholar]
- Jiménez DJ, Montaña JS, Alvarez D. Baena S. A novel cold active esterase derived from Colombian high Andean forest soil metagenome. World J Microbiol Biotechnol. 2012a;1:361–370. doi: 10.1007/s11274-011-0828-x. [DOI] [PubMed] [Google Scholar]
- Jiménez DJ, Andreote FD, Chaves D, Montaña JS, Osorio-Forero C, Junca H, et al. Structural and functional insights from the metagenome of an acidic hot spring microbial planktonic community in the Colombian Andes. PLoS ONE. 2012b;7:e52069. doi: 10.1371/journal.pone.0052069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochens H, Stiba K, Savile C, Fujii R, Yu JG, Gerassenkov T, et al. Converting an esterase into an epoxide hydrolase. Angew Chem Int Ed Engl. 2009;48:3532–3535. doi: 10.1002/anie.200806276. [DOI] [PubMed] [Google Scholar]
- Johansson P, Unge T, Cronin A, Arand M, Bergfors T, Jones TA. Mowbray SL. Structure of an atypical epoxide hydrolase from Mycobacterium tuberculosis gives insights into its function. J Mol Biol. 2005;351:1048–1056. doi: 10.1016/j.jmb.2005.06.055. [DOI] [PubMed] [Google Scholar]
- Jung JY, Lee SH, Kim JM, Park MS, Bae JW, Hahn Y, et al. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl Environ Microbiol. 2011;77:2264–2274. doi: 10.1128/AEM.02157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan K. Bingham BL. Biology of mangroves and mangrove ecosystems. Adv Mar Biol. 2001;40:81–251. [Google Scholar]
- Kim EY, Oh KH, Lee MH, Kang CH, Oh TK. Yoon JH. Novel cold-adapted alkaline lipase from an intertidal flat metagenome and proposal for a new family of bacterial lipases. Appl Environ Microbiol. 2009;75:257–260. doi: 10.1128/AEM.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotik M, Stepanek V, Maresova H, Kyslik P. Archelas A. Environmental DNA as a source of novel epoxide hydrolase reacting with aliphatic terminal epoxides. J Mol Catal B-Enzym. 2009;56:288–293. [Google Scholar]
- Koudelakova T, Bidmanova S, Dvorak P, Pavelka A, Chaloupkova R, Prokop Z. Damborsky J. Haloalkane dehalogenases: biotechnological applications. Biotechnol J. 2013;8:32–45. doi: 10.1002/biot.201100486. [DOI] [PubMed] [Google Scholar]
- Kourist R, Jochens H, Bartsch S, Kuipers R, Padhi SK, Gall M, et al. The alpha/beta-hydrolase fold 3DM database (ABHDB) as a tool for protein engineering. Chembiochem. 2010;11:1635–1643. doi: 10.1002/cbic.201000213. [DOI] [PubMed] [Google Scholar]
- Kumar R, Wani SI, Chauhan NS, Sharma R. Sareen D. Cloning and characterization of an epoxide hydrolase from Cupriavidus metallidurans-CH34. Protein Expr Purif. 2011;79:49–59. doi: 10.1016/j.pep.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Kunji ERS, Mierau I, Hagfing A, Poolman B. Konings WN. The proteolytic systems of lactic acid bacteria. A van Leeuw J Microb. 1996;70:187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- Kwon KK, Woo JH, Yang SH, Kang JH, Kang SG, Kim SJ, et al. Altererythrobacter epoxidivorans gen. nov., sp. nov., an epoxide hydrolase-active, mesophilic marine bacterium isolated from cold-seep sediment, and reclassification of Erythrobacter luteolus Yoon et al. 2005 as Altererythrobacter luteolus comb. nov. Int J Syst Evol Microbiol. 2007;57(Part 10):2207–2211. doi: 10.1099/ijs.0.64863-0. [DOI] [PubMed] [Google Scholar]
- Kwon TH, Kim JT. Kim JS. Application of a modified sublimation method to screen for PAH-degrading microorganisms. Korean J Microbiol. 2010;46:109–111. [Google Scholar]
- Lee EY. Shuler ML. Molecular engineering of epoxide hydrolase and its application to asymmetric and enantioconvergent hydrolysis. Biotechnol Bioeng. 2007;98:318–327. doi: 10.1002/bit.21444. [DOI] [PubMed] [Google Scholar]
- Lenfant N, Hotelier T, Velluet E, Bourne Y, Marchot P. Chatonnet A. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 2013;41(Database issue):D423–D429. doi: 10.1093/nar/gks1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wu JM, Zhang LF, Zhang YZ. Feng H. Characterization of a unique proline iminopeptidase from white-rot basidiomycetes Phanerochaete chrysosporium. Biochimie. 2010;92:779–788. doi: 10.1016/j.biochi.2010.02.022. [DOI] [PubMed] [Google Scholar]
- van Loo B, Kingma J, Arand M, Wubbolts MG. Janssen DB. Diversity and biocatalytic potential of epoxide hydrolases identified by genome analysis. Appl Environ Microbiol. 2006;72:2905–2917. doi: 10.1128/AEM.72.4.2905-2917.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes LW, Kuramae EE, Navarrete AA, van Veen JA. Tsai SM. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 2014;8:1577–1587. doi: 10.1038/ismej.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, Souza MD, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaña JS, Jiménez DJ, Hernández M, Ángel T. Baena S. Taxonomic and functional assignment of cloned sequences from high Andean forest soil metagenome. A van Leeuw J Microb. 2012;101:205–215. doi: 10.1007/s10482-011-9624-8. [DOI] [PubMed] [Google Scholar]
- Nardini M. Dijkstra BW. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Oh YS. Roh DH. Phenylobacterium muchangponense sp. nov., isolated from beach soil, and emended description of the genus Phenylobacterium. Int J Syst Evol Microbiol. 2012;62(Part 4):977–983. doi: 10.1099/ijs.0.028902-0. [DOI] [PubMed] [Google Scholar]
- Pleiss J, Fischer M, Peiker M, Thiele C. Schmid RD. Lipase Engineering Database—understanding and exploiting sequence-structure-function relationships. J Mol Catal B-Enzym. 2000;10:491–508. [Google Scholar]
- Procópio L, Macrae A, van Elsas JD. Seldin L. The putative α/β-hydrolases of Dietzia cinnamea P4 strain as potential enzymes for biocatalytic applications. A van Leeuw J Microb. 2013;103:635–646. doi: 10.1007/s10482-012-9847-3. [DOI] [PubMed] [Google Scholar]
- Rink R, Fennema M, Smids M, Dehmel U. Janssen DB. Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J Biol Chem. 1997;272:14650–14657. doi: 10.1074/jbc.272.23.14650. [DOI] [PubMed] [Google Scholar]
- Rui L, Cao L, Chen W, Reardon KF. Wood TK. Protein engineering of epoxide hydrolase from Agrobacterium radiobacter AD1 for enhanced activity and enantioselective production of (R)-1-phenylethane-1,2-diol. Appl Environ Microbiol. 2005;71:3995–4003. doi: 10.1128/AEM.71.7.3995-4003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirova JS, Ferrer M, Regenhardt D, Timmis KN. Golyshin PN. Proteomic insights into metabolic adaptations in Alcanivorax borkumensis induced by alkane utilization. J Bacteriol. 2006;188:3763–3773. doi: 10.1128/JB.00072-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo K. Dhal N. Potential microbial diversity in mangrove ecosystems: a review. IJMS. 2009;38:249–256. [Google Scholar]
- dos Santos HF, Cury JC, do Carmo FL, dos Santos AL, Tiedje J, van Elsas JD, et al. Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: bacterial proxies for oil pollution. PLoS ONE. 2011;6:e16943. doi: 10.1371/journal.pone.0016943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D. Kumar R. Prospecting for efficient enantioselective epoxide hydrolases. Indian J Biotech. 2011;10:161–177. [Google Scholar]
- Schnoes AM, Brown SD. Babbitt PC. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirim D, Wagner F, Wang L, Schmid RD. Pleiss J. The Laccase Engineering Database: a classification and analysis system for laccases and related multicopper oxidases. Database (Oxford) 2011;2011:bar006. doi: 10.1093/database/bar006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CE, Beys-da-Silva WO, Santi L, Berger M, Vainstein MH, Guima Rães JA. Vasconcelos AT. A potential source for cellulolytic enzyme discovery and environmental aspects revealed through metagenomics of Brazilian mangroves. AMB Express. 2013;3:65. doi: 10.1186/2191-0855-3-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirawongsaroj P, Sriprang R, Harnpicharnchai P, Thongarama T, Champreda V, Tanapongpipat S, et al. Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J Biotechnol. 2008;133:42–49. doi: 10.1016/j.jbiotec.2007.08.046. [DOI] [PubMed] [Google Scholar]
- de Vries EJ. Janssen DB. Biocatalytic conversion of epoxides. Curr Opin Biotechnol. 2003;14:414–420. doi: 10.1016/s0958-1669(03)00102-2. [DOI] [PubMed] [Google Scholar]
- van der Werf MJ, Overkamp KM. de Bont JA. Limonene-1,2-epoxide hydrolase from Rhodococcus erythropolis DCL14 belongs to a novel class of epoxide hydrolases. J Bacteriol. 1998;180:5052–5057. doi: 10.1128/jb.180.19.5052-5057.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JH, Hwang YO, Kang SG, Lee HS, Cho JC. Kim SJ. Cloning and characterization of three epoxide hydrolases from a marine bacterium, Erythrobacter litoralis HTCC2594. Appl Microbiol Biotechnol. 2007;76:365–375. doi: 10.1007/s00253-007-1011-z. [DOI] [PubMed] [Google Scholar]
- Woo JH, Kwon TH, Kim JT, Kim CG. Lee EY. Identification and characterization of epoxide hydrolase activity of polycyclic aromatic hydrocarbon-degrading bacteria for biocatalytic resolution of racemic styrene oxide and styrene oxide derivatives. Biotechnol Lett. 2013;35:599–606. doi: 10.1007/s10529-012-1114-1. [DOI] [PubMed] [Google Scholar]
- Ye M, Li G, Liang WQ. Liu YH. Molecular cloning and characterization of a novel metagenome-derived multicopper oxidase with alkaline laccase activity and highly soluble expression. Appl Microbiol Biotechnol. 2010;87:1023–1031. doi: 10.1007/s00253-010-2507-5. [DOI] [PubMed] [Google Scholar]