Summary
G protein-coupled receptors (GPCRs) mediate the majority of cellular responses to hormones and neurotransmitters. The majority of GPCR crystal structures have been obtained using a fusion protein strategy, where the flexible third intracellular loop is replaced by T4 lysozyme (T4L). However, wild-type T4L may not be ideally suited for all GPCRs due to its size and the inherent flexibility between N- and C-terminal subdomains. Here we report two modified T4L variants, designed to address flexibility and size, that can be used to optimize crystal quality or promote alternative packing interactions. These variants were tested on the M3 muscarinic receptor (M3). The original M3-T4L fusion protein produced twinned crystals that yielded 3.4 Å structure from a 70-crystal data set. We replaced T4L with the modified T4L variants. Both T4L variants yielded new M3 muscarinic receptor crystals, with alternate lattices that were not twinned, including a 2.8 Å structure.
Introduction
During the past seven years there has been remarkable progress in the structural biology of G protein-coupled receptors (GPCRs). GPCRs are inherently difficult to crystallize because of their low natural abundance, structural instability in detergents and minimal polar surface area. Several strategies have been used to facilitate crystallogenesis including fusion proteins, antibody complexes (Rasmussen et al., 2007), and thermostabilization (Warne et al., 2008). To date, the largest number of published crystal structures has been obtained using the fusion protein strategy. This approach was first applied to the β2–adrenergic receptor (β2AR) where the flexible third intracellular loop was replaced by T4 lysozyme (T4L), a well-folded soluble protein that crystallizes under many different conditions (Baase et al., 2010; Rosenbaum et al., 2007). More recently it was shown that fusing T4L to the amino terminus could also facilitate crystallogenesis (Zou et al., 2012). Subsequently at least 14 other GPCRs have been crystallized using the T4L fusion protein strategy (N terminus or third intracellular loop), and an additional 3 new structures have been obtained using this strategy with another soluble fusion partner, thermostabilized apocytochrome b562 RIL (Chun et al., 2012).
In crystals generated from GPCR-T4L fusion proteins, crystal packing is primarily driven by T4L-T4L interactions or T4L-GPCR interactions. In an effort to increase the number of potential packing interactions for a given GPCR, we explored the use of modified versions of T4L that could be exchanged for the original T4L after the links between T4L and the GPCR have been optimized.
T4L is composed of three structural components: a small N-terminal helix and larger N-terminal and C-terminal lobes which are coupled via a small hinge region; together, the intersection of these lobes defines the substrate binding groove (Matthews et al., 1981) (Figure 1A). The flexibility between the lobes, which is required for enzymatic activity of T4L (Matsumura and Matthews, 1989), can be observed by comparing the structures of the T4L in several previously published GPCR structures (Figure 1B) where the position of the N-terminal lobe differs by as much as 11.8 Å. In some fusion protein structures this flexibility may be important in forming crystal contacts; however for others it may contribute to poor crystal quality. In this study, we investigate the effect of modifying T4L size and flexibility on crystallogenesis of the M3 muscarinic receptor. The M3 receptor was chosen as a test case because it was one of the most challenging to solve due to twinning and relatively low-resolution diffraction (Kruse et al., 2012). Here we show that modifications of T4L used in M3-T4L fusion proteins could produce non-twinned crystals that diffract to higher resolution.
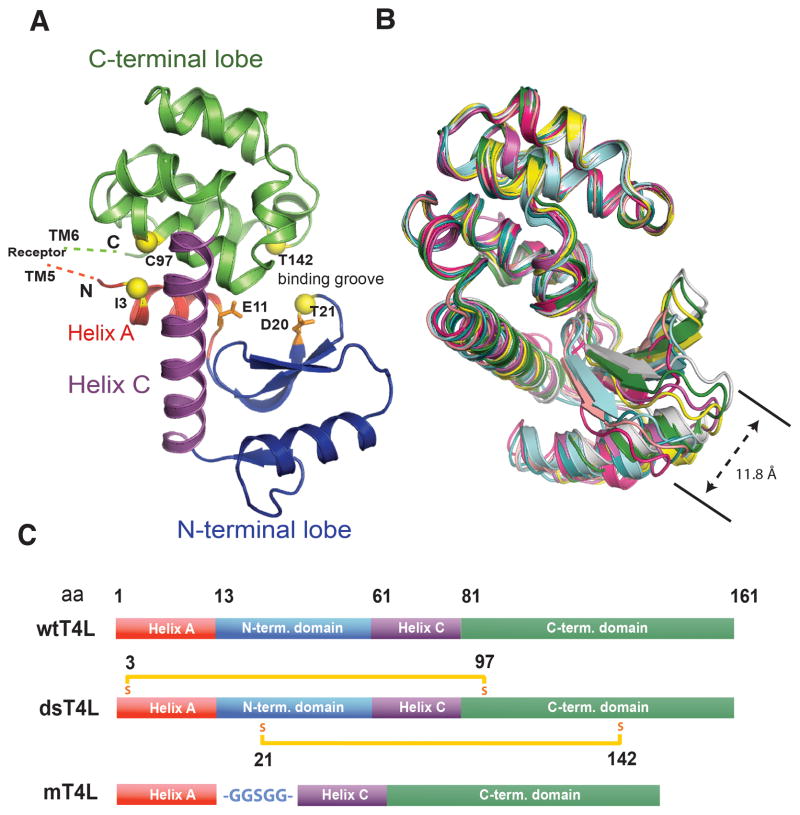
Figure 1. T4 lysozyme is a two-domain protein with a flexible hinge region.
(A) T4L is composed of a small N-terminal helix (helix A) (red) which extends into a larger N-terminal lobe including helix B (blue). The N-terminal lobe couples via its larger helix C (purple) to the C-terminal domain (green), which forms the core of the protein. Together the N- and C-terminal domains form a binding groove in which hydrolysis is catalyzed by E11 and D20 (orange). The positions of the cysteines introduced to make dsT4L are shown as yellow spheres. (B) T4Ls of GPCR fusion proteins previously crystalized in our laboratory (PDB IDs; 3UON, 4DKL, 4EJ4, 3VW7, 4DAJ, 3SN6 and 2RH1) were superimposed by their C-terminal lobes illustrating the flexibility of the N-terminal lobes relative to the C-terminal lobes. (C) Schematic representation of the primary structure of wtT4L compared with the structure of the two recombinant versions of T4L, dsT4L and mT4L used in the current study. The two disulfide bridges introduced in dsT4L between position 3 and 97 and 21 and 142 are represented by orange bars. The glycine/serine linker (-GGSGG-) replacing the N-terminal domain in mT4L is shown in blue. See also figure S3.
Results
The M3 receptor was crystallized using modified T4 lysozymes
In our first approach to address the issue of flexibility in T4L, we introduced two disulfide bridges to stabilize the interactions between the N- and C-terminal domains. Based on the work of Matsumura et al (Matsumura et al., 1989b), cysteines were introduced by the following mutations: I3C, T21C, A97C and T142C to create “disulfide stabilized T4L” (dsT4L) (Figure 1C). Note that natural T4L has cysteines at positions 54 and 97, which have been mutated to threonine and alanine respectively in the protein used for all published GPCR-T4L fusion proteins. We will refer to this C54T/C97A variant as “wild type T4L” (wtT4L). In dsT4L, Cys3 forms a disulfide bond with Cys97, and Cys21 forms a disulfide bond with Cys142. Introduction of disulfide bridges at these and other positions has previously been shown to significantly increase the thermal stability of T4L, and stabilizes a more closed conformation (Jacobson et al., 1992; Matsumura et al., 1989a; Matsumura and Matthews, 1989; Matsumura et al., 1989b). The insertion site for dsT4L in the M3 receptor third intracellular loop was identical to that used for the original M3-wtT4L structure. Tiotropium bound M3-dsT4L crystallized in lipid cubic phase in 100 mM Tris pH 8.1, 113.5 mM lithium citrate, 110 mM ammonium sulfate and 45% PEG 300 (Figure S1A–B). The M3-dsT4L crystallized in a higher symmetry space group (P41212) compared to (P1) for the original M3-wtT4L crystals (PDB 4DAJ), and the crystals were not twinned. However, the resolution of 3.6 Å (Table 1) was slightly lower than the original P1 structure (3.4 Å).
Table 1.
M3-dsT4L collection statistics. Crystals collected can be viewed in figure S1.
| Data collectiona | |
| Number of crystals | 15 |
| Space group | P 41 21 2 |
| Cell dimensions | |
| a, b, c (Å) | 54.95, 54.95, 348.0 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 31.6 - 3.57 (3.70 - 3.57) |
| Rmerge (%) | 20.4 (82.7) |
| <I>/<σI> | 4 (1.1) |
| Completeness | 89.4 (89.3) |
| Redundancy | 3.1 (3.0) |
| CC1/2 | 0.978 (0.634) |
| Refinement | |
| Resolution (Å) | 31.6 - 3.57 |
| No of unique reflections | 9628 (489) |
| RWork/Rree (%) | 0.274 / 0.325 |
| Average B-factor (Å2) | |
| M3 muscarinic receptor | 88.8 |
| Tiotropium | 78.5 |
| T4 lysozyme | 85.8 |
| R.m.s. deviation from ideality | |
| Bond length (Å) | 0.007 |
| Bond angles (°) | 1.1 |
| Ramachandran statistics | |
| Favored regions (%) | 92.4 |
| Allowed regions (%) | 7.1 |
| Outliers (%) | 0.5 |
Highest shell statistics are in parentheses.
In a second approach, we deleted the smaller flexible N-terminal lobe of wtT4L and inserted a short -GGSGG-linker to connect wtT4L helices A and C to create a “minimal T4L” (mT4L). Tiotropium bound M3-mT4L crystallized in 100 mM Tris pH 7.5, 44% PEG 300 and 400 mM ammonium tartrate. The crystals were in space group C2, with a block-like morphology, and were strongly birefringent (Figure S1C–E). Data were collected giving a final resolution of 2.8 Å (Table 2).
Table 2.
M3-mT4L Tiotropium collection statistics. Crystals collected can be viewed in figure S1.
| Data collectiona | |
| Number of crystals | 37 |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 152.3, 184.6 52.6 |
| α, β, γ (°) | 90 98.5 90 |
| Resolution (Å) | 29.6-2.8 (2.90-2.80) |
| Rmerge (%) | 0.131 (0.972) |
| <I>/<σI> | 7.0 (1.8) |
| Completeness | 96.8 (93.9) |
| Redundancy | 5.2 (4.6) |
| CC1/2 | 0.996 (0.649) |
| Refinement | |
| Resolution (Å) | 28.5-2.8 |
| No of unique reflections | 60975 (3022 in test set) |
| RWork/Rree (%) | 23.0/26.0 |
| Average B-factor (Å2) | |
| M3 muscarinic receptor (chain A/B) | 100/92 |
| Tiotropium (chain A/B) | 84/80 |
| T4 lysozyme (chain A/B) | 75/72 |
| monoolein | 112 |
| PEG300 | 100 |
| Tartrate | 104 |
| Water | 57 |
| R.m.s. deviation from ideality | |
| Bond length (Å) | 0.003 |
| Bond angles (°) | 0.65 |
| Ramachandran statistics | |
| Favored regions (%) | 96.8 |
| Allowed regions (%) | 3.2 |
| Outliers (%) | 0 |
Highest shell statistics are in parentheses.
The effect of modified T4Ls on M3 receptor and T4L packing interactions
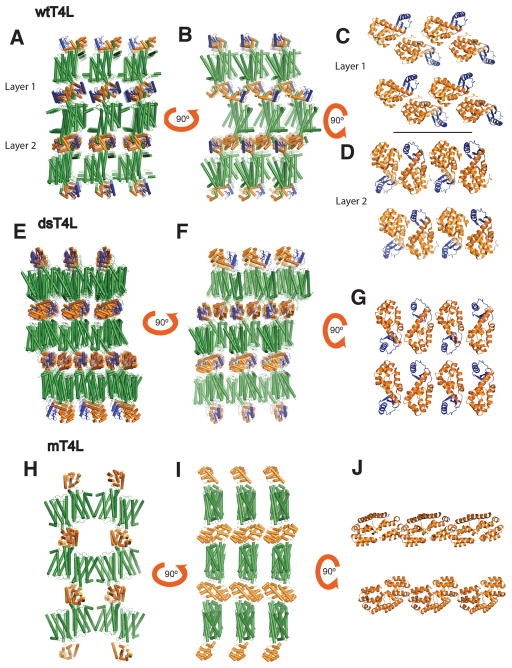
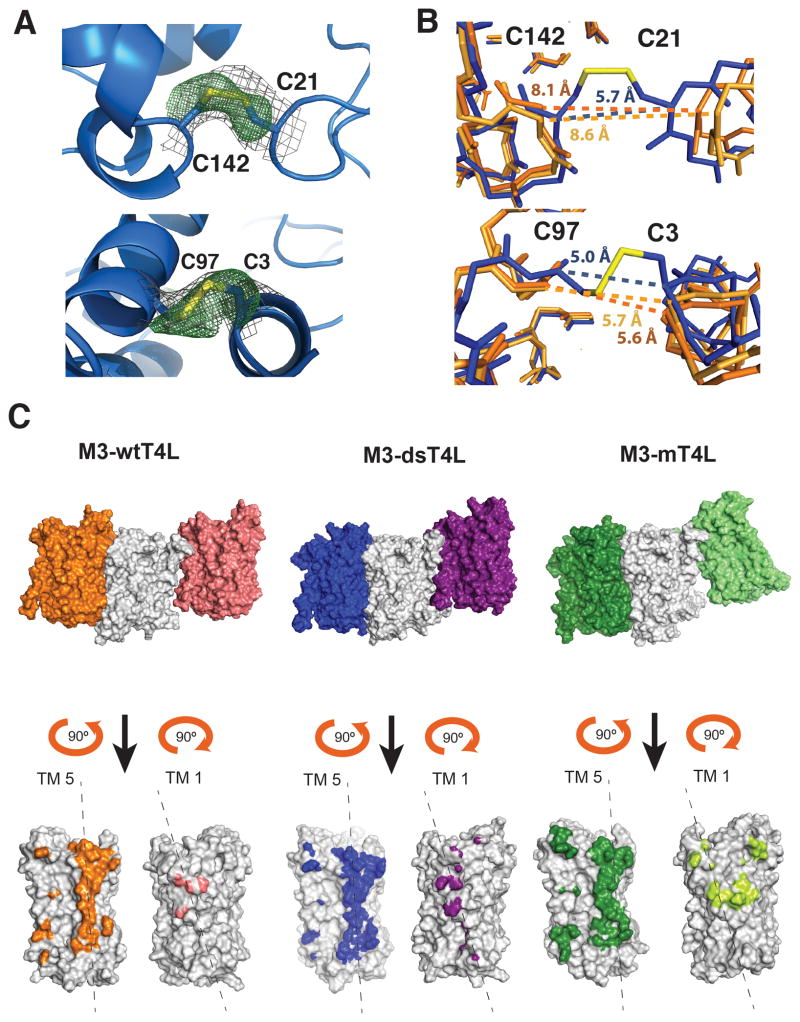
Figure 2 shows the packing arrangement of M3-wtT4L, M3-dsT4L and M3-mT4L crystals. All three are type 1 crystals with alternating layers of T4L and M3. The M3-wtT4L crystal has four molecules in the asymmetric unit, the M3-mT4L has two and the M3-dsT4L has one. In M3-wtT4L, there are two different arrangements of T4L packing interactions, which gives rise to the observed epitaxial twinning (Kruse et al., 2012) (Figure 2A–D). In contrast, the M3-dsT4L and M3-mT4L crystals have only one packing arrangement (Figure 2E–J). In layer 1 of M3-wtT4L crystals the amino terminal lobe of T4L (colored blue) is not involved in packing interactions, whereas in layer 2 the amino terminal lobe packs against the C-terminal lobe of the adjacent T4L (Figure 2C–D). The packing in layer 2 of M3-wtT4L is similar to that observed in M3-dsT4L. Both T4L molecules in M3-wtT4L have a larger distance between their N- and C-terminal lobes than is observed in M3-dsT4L, where we observe clear electron density for the two disulfides introduced between residues 21 and 142 and residues 3 and 97 (Figure 3A). As a result, interactions between the N- and C-terminal lobes were stabilized, with a distance between the alpha carbons of the disulfide bonded cysteines of 5.7 Å between C21 and C142 (Figure 3B top panel). In contrast, the crystal structure of M3-wtT4L (4DAJ) showed larger distances of 8.1 and 8.6 Å between the alpha carbons at homologous positions (Figure 3B top panel).
Figure 2. Crystal packing interactions for M3-wtT4L, M3-dsT4L and M3-mT4L.
(A and B) M3-wtT4L was previously crystallized in a P1 crystal in which the receptor (green) forms arrays of anti parallel dimers stacked in between layers of T4L (orange with the N-terminal domain shown in blue). The packing of T4L alternates in every second layer (panels C and D). (E and F) Packing interactions in M3-dsT4L are similar to M3 wtT4L with the receptor forming arrays of anti parallel dimers. But unlike M3 wtT4L, dsT4L has only one T4L packing arrangement (panel G). (H and I) In M3-mT4L receptor formed two oligomerization interfaces such that the receptor positions in a linear arrangement in the crystal. The first weaker interaction is mediated by helix 1 and 2. A second interface is formed by an antiparallel interaction between helix 4 and 5 on each receptor. The dimeric packing of mT4L is mediated by the C-terminal surface of T4L that is exposed upon removal of the N-terminal domain. (Figure J).
Figure 3. Features of the M3 mT4L and dsT4L structures.
(A) Close up showing the formation of a disulfide bridge between dsT4L position C21 and C142 (top panel) and position C3 and C97 (bottom panel). The 2Fo-Fc electron density map around the disulfide bridges contoured at 1.0σ is shown in grey. Shown in green is a Fo-Fc omit map, refined with the disulfide bond omitted, contoured at 3.0σ. Maps are carved at 2.0 A around the disulfide atoms. (B, top panel) Side view of the dsT4L and wtT4L showing that the disulfide bridge between dsT4L position C21 and C142 stabilizes a more closed conformation of dsT4L (blue) compared with wtT4L (orange and light orange). (B, bottom panel) In contrast, the distance between positions C3 and C97 in dsT4L is nearly the same as in wtT4L. (C) Surfaces of M3 involved in packing interactions for M3- wtT4L, M3-dsT4L and M3-mT4L. Note that the right light green monomer in M3- mT4L is parallel to the center grey monomer. In M3-wtT4L and M3-dsT4L this interface is antiparallel.
We observe more extensive packing interactions between adjacent M3 molecules in M3-mT4L compared to M3-wtT4L and M3-dsT4L. In M3-mT4L there is a 2504 Å2 antiparallel dimer interface involving TM 4 and TM 5 and a 914 Å2 parallel dimer interface involving TM 1 and 2 (Figure 2H and Figure 3C). Packing interactions between adjacent M3 molecules in M3-wtT4L primarily involve a single antiparallel interface of 1967 Å2 involving TM4 and 5, although in every second receptor layer there is a second weak 278 Å2 antiparallel interface involving TM1. For M3-dsT4L there is an antiparallel interface of 2228 Å2 involving TM4 and TM5 and a second antiparallel interface of 738 Å involving TM1. The more extensive interactions between M3 protomers in M3-mT4L are likely due to the smaller size of mT4L. The packing arrangement observed in M3-mT4L would not be possible with wtT4L (Figure 2H–J).
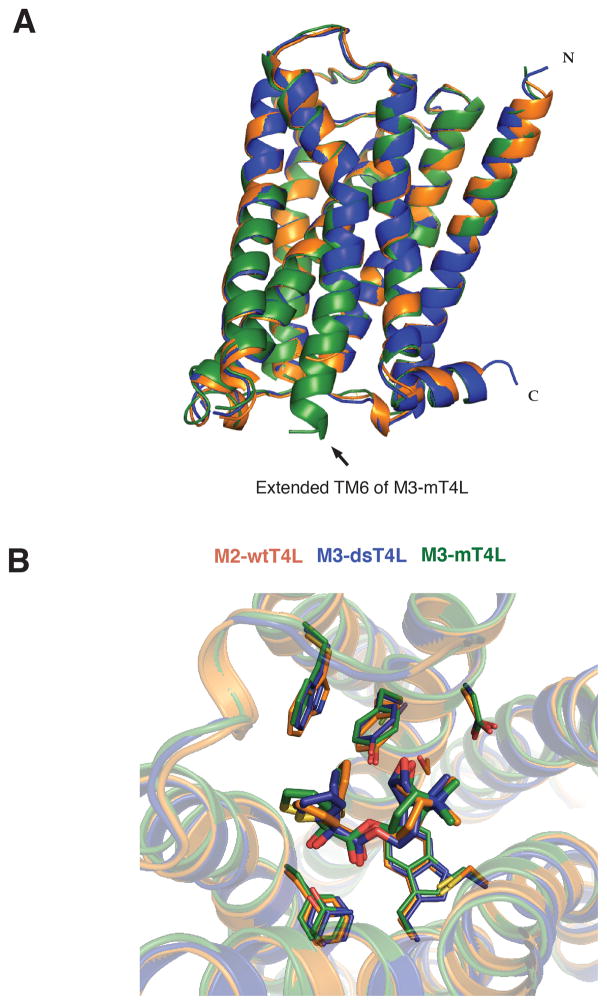
The structure of M3 in the three crystals is nearly identical (RMSD = 0.44 comparing M3-T4L and M3-dsT4L, RMSD = 0.63 comparing M3-T4L and M3-mT4L) (Figure 4A). The most notable difference among the three structures is at the cytoplasmic end of TM6, where an extended alpha helix is observed in the M3-mT4L structure (Figure 4A), providing insight into the structure of the AAxxLS motif that plays a role in G-protein selectivity (Blin et al., 1995; Liu et al., 1995). An overlay of the three structures confirms the previously reported binding mode of the tiotropium ligand, which has a nearly identical pose in the three receptors (Figure 4B).
Figure 4. Differences in T4L have only small effects on M3 structure.
(A) Alignment of the M3-mT4L (green) (PDB 4U15), M3-dsT4L (blue) (PDB 4U15) and M3-wtT4L (orange) (PDB 4DAJ) structures showing that the M3 structures are highly similar. (B) Alignment of the binding pockets of M3-mT4L, M3-dsT4L and M3-wtT4L reveals nearly identical interactions with the antagonist tiotropium. The binding mode of methylscopolamine and a PEG molecule occupying the alloteric binding pocket are shown in figure S2.
The M3 receptor was crystallized with N-methylscopolamine and PEG300
The M3-mT4L was additionally prepared and crystallized in the presence of the antagonist N-methyl scopolamine (NMS) under the same conditions as with tiotropium (Figure S1F). The poorer diffraction of 3.7 Å (Table 3) is likely due to the lower affinity and more rapid dissociation rate of NMS (t1/2 of approx. 40 min compared with tiotropium t1/2 of 462–1620 min) (Casarosa et al., 2009; Dowling and Charlton, 2006; Sykes et al., 2012; Tautermann et al., 2013; Trankle et al., 2001). We observed NMS binds in a similar manner to tiotropium (Figure S2A). For both tiotropium and NMS bound M3 receptor, a clear density was found in the extracellular vestibule above the orthosteric binding pocket, which has been shown to constitute the allosteric binding site of this class of receptors (Dror et al., 2013; Lazareno and Birdsall, 1995; Prilla et al., 2006). The electron density in the M3-mT4L was consistent with a single PEG 300 molecule in chain B (Figure S2B and Supplementary Methods). This was supported by ligand binding studies in which PEG 300 slowed the dissociation of radioactive NMS from the receptor (Figure S2C).
Table 3.
M3-mT4L NMS collection statistics. Crystals collected can be viewed in figure S1.
| Data collectiona | |
| Number of crystals | 12 |
| Space group | C2 |
| Cell dimensions | |
| A, b, c (Å) | 153.4 187.2 53.5 |
| α, β, γ (°) | 90 99.8 90 |
| Resolution (Å) | 33.7-3.7 (3.83-3.70) |
| Rmerge (%) | 0.219 (0.884) |
| <I>/<σI> | 8.9 (2.2) |
| Completeness | 93.7 (89.1) |
| Redundancy | 2.6 (2.5) |
| CC1/2 | 0.509 (0.964) |
| Refinement | |
| Resolution (Å) | 34-3.7 |
| No of unique reflections | 14787 (749 in test set) |
| RWork/Rree (%) | 23.9/28.5 |
| Average B-factor (Å2) | |
| M3 muscarinic receptor (chain A/B) | 155/149 |
| NMS (chain A/B) | 138/130 |
| T4 lysozyme (chain A/B) | 145/131 |
| Tartrate | 177 |
| R.m.s. deviation from ideality | |
| Bond length (Å) | 0.003 |
| Bond angles (°) | 0.60 |
| Ramachandran statistics | |
| Favored regions (%) | 95.1 |
| Allowed regions (%) | 4.9 |
| Outliers (%) | 0 |
Highest shell statistics are in parentheses.
Discussion
The fusion protein strategy to crystallize GPCRs was initially demonstrated using T4L inserted between TM5 and TM6 (Rosenbaum et al., 2007), and subsequently it was shown that crystals of the β2AR could be grown by inserting T4L immediately before TM1 (Zou et al., 2012). More recently it was shown that a thermostabilized apocytochrome b562 RIL could be used as the fusion partner (Chun et al., 2012). The investigators also tried removing the amino terminal domain of T4L by generating a circular-permutated T4L in which the N-terminal helix A was moved to the C-terminal end of the protein and the amino terminal domain was deleted. In contrast to the approach described here, the use of a circular-permutated T4L required changing the positions where T4L is fused to the GPCR (Figure S3). Fusion of the circular permutated T4L to the third intracellular loop of A2a and β2AR receptors resulted in dramatically reduced expression and no crystals were obtained (Chun et al., 2012).
In conclusion, the GPCR fusion protein strategy, initially employing T4L as the fusion partner, has been used to obtain the majority of GPCR structures. However, the size and flexibility of T4L may not be ideal for all GPCRs. We describe alternate forms to T4L that can be used to facilitate GPCR crystallogenesis. dsT4L reduces the intrinsic flexibility of T4L, while mT4L reduces the size. The modified T4L proteins do not require further optimization of the insertion site. Recently mT4L was used to obtain crystals of the M4 muscarinic receptor that diffract to 2.1Å (data to be published elsewhere), showing that the approach can be applied to other receptors.
Experimental Procedures
The T4L mutant dsT4L was generated by a short overlap extension (SOE) PCR from wtT4L in a previous M3 construct and one piece of synthetic DNA (gBlocks, Integrated DNA technologies).
Synthetic gBlock DNA;
CGCATCGACGAAGGCCTGCGTCTCAAGATTTACAAGGACTGCGAAGGTTATTACACGATTGGCATCGGCCACCTCCTGACAAAGAGCCCATCACTCAACGCTGCCAAGTCTGAACTGGACAAAGCCATTGGTCGCAACACCAACGGTGTCATTACAAAGGACGAGGCGGAGAAACTCTTCAACCAAGATGTAGATGCGGCTGTCCGTGGCATCCTGCGTAATGCCAAGTTGAAGCCCGTGTATGACTCCCTTGATGCTGTTCGCCGTTGCGCCTTGATCAACATGGTTTTCCAAATGGGTGAGACCGGAGTGGCTGGTTTTACGAACTCCCTGCGCATGCTCCAGCAGAAGCGCTGGGACGAGGCCGCAGTGAATTTGGCTAAATCTCGCTGGTACAATCAGTGCCCTAACCGTGCCAAGCGTGTCATCACTACCTTCCGTACTGGAACTTGGGACGCTTACCTCATCAAGGAGAAGAAGGCCGCCCAGACGCTCAGT
SOE was performed using Phusion Polymerase (Finnzymes cat. nr. F-530S) with 20 sec. elongation in the first round and 40 sec. in the second. The following primers were used:
Primers;
RecepF; CTTGTCATTGTGGCCTTCAAGG, RecepR; GCGCAGCATCTCGAAGCAGTTC ATCTTCTCAGTTTCCTTATAGATCC, GenebF; GAGAAGATGAACTGCTTCGAGA TGCTGCGCATCGACGAAGGCCTG and GenebR; GATGAAGGCTAGCAAGATG GCACTGAGCGTCTGGGCGGC
The obtained PCR fragment was subcloned into an existing pVL 1393 M3 construct using AflII and NheI enzymes. This M3 construct is essentially the same as the one used for the previously published M3 structure (Kruse et al., 2012). Briefly, Rat M3 receptor had a cleavable signal sequence followed by a Flag epitope fused to its N-terminus. Four potential N-glycosylation site asparagine residues in the N-terminus were mutated to glutamine. Residues 50–56 were replaced with a TEV protease site. The third intracellular loop was replaced by the inserted T4L between residues 259 and 482. The only difference between the M3 used here and that used to obtain the previous M3 structure (Kruse et al., 2012) is that an additional 20 amino acids were truncated from the C-terminal tail as these were not observed in the original M3 structure. Therefore the receptor ends at residue 569. Both versions of the M3 receptor had a His tag in the C-terminus (6 histidines in the previous version and 8 in the current truncated version). Four cysteines were introduced into T4L at positions 3, 21, 97 and 142 giving rise to two disulfide bridges as described by Matsumura et al (Matsumura et al., 1989b). The third bridge described in the paper by Matsumura et al was omitted since it links position 9 with 164, of which the latter residue is truncated for compactness in our constructs. mT4L was generated by a single quick change of the same M3 construct in a PrsetA vector deleting amino acids 13 to 60 of T4L and inserting the linker sequence – GGSGG-instead. This was done using the primers: T4Ldel_F; GACGAAGGCGGCGGCAGTGGCGGCGACGAGGCGGAGAAACTCTTC and T4Ldel_R; CCTCGTCGCCGCCACTGCCGCCGCCTTCGTCGATGCGCAG. The polymerase used was Pfu Turbo Hotstart (Aglient Technologies cat. nr. 600322-51). The QC program used employed 24 cycles with 30 sec. annealing at 49°C and 6 min extension at 72C.
Expression and purification of M3-T4L fusion proteins
The expression and purification of M3-T4L fusion proteins was essentially the same as previously described (Kruse et al., 2012). A notable exception was that M3-dsT4L was not treated with iodoacetamide until after Ni NTA purification to allow time for disulfide bond formation. The fusion proteins were expressed for 60 hours in Sf9 cells using the baculovirus expression system. 8 L of cells were harvested by centrifugation and frozen. Cells were thawed in 800 ml lysis buffer containing 10 mM Tris pH 7.5, 1 mM EDTA, 2.5 mg/L leupeptin, 1 mM benzamidine, and 10 uM antagonist, either tiotropium (pharmaChem) or N-methylscopolamine (Sigma cat. no. 2250). For the mT4L fusion, 1 mg/ml iodoacetamide (sigma cat. nr. I1149) was added at this stage to alkylate free cysteines. Cells were pelleted by centrifugation at 18,000 RPM for 15 min. The pellet was then solubilized in 800 ml buffer containing 20 mM HEPES pH 7.5, 1% DDM, 10 mM Tris pH 7.5, 0.2% sodium cholate, 0.03% cholesteryl hemisuccinate, 750 mM NaCl, 30% glycerol, 2.5 mg/L leupeptin, 1 mM benzamidine, 1 ul/800 ml benzonase (Sigma cat. nr. B6506) and 10 uM either tiotropium or N-methylscopolamine. Again for mT4L fusion only, 1 mg/ml iodoacetamide was added to the solubilization buffer. Cells were homogenized 20 times with a dounce tissue grinder and stirred for 15 min at 4 °C. Next, 24 ml nickel chelating sepharose resin beads (GE Healthcare) were added to the solution, which was then gently shaken for 2 h at 4 °C. The beads were recovered by centrifugation and washed in batch mode with three 100 ml volumes of nickel wash buffer containing 20 mM HEPES pH 7.5, 0.1% DDM, 10 mM Tris pH 7.5, 0.02% sodium cholate, 0.03% cholesteryl hemisuccinate, 750 mM NaCl, 30% glycerol, 5 mM imidazole, and 10 uM tiotropium or N-methylscopolamine. Beads were then loaded into a column and washed with an additional 100 ml of nickel was buffer. The protein eluted under gravity flow in the same buffer supplemented with 300 mM imidazole. At this stage 1 mg/ml iodoacetamide was added to the eluted dsT4L fusion protein. The nickel-pure protein was stored at −20 °C overnight before further purification. Next the purified protein was supplemented with 2 mM CaCl2 and loaded onto 5 ml M1 antibody resin by gravity flow and washed with 30 ml nickel wash buffer supplemented with 2 mM CaCl2. While on the M1 antibody resin the protein was exchanged into lauryl maltose neopentyl glycol (MNG) detergent-containing buffer composed of 20 mM HEPES pH 7.5, 0.1% MNG-14, 0.01% cholesteryl hemisuccinate, 100 mM NaCl, 10 uM tiotropium or N-methylscopolamine and 2 mM CaCl2. The detergent exchange was performed by washing the column over 60 min with a series of 6 buffers (10 ml each) made up of the following ratios (v:v) of MNG exchange buffer and nickel wash buffer: 1:1, 4:1, 9:1, 20:1, 99:1 and MNG exchange buffer alone. The column was then washed with 10 ml of 10x CMC MNG buffer containing 20 mM HEPES pH 7.5, 0.01% MNG-14, 0.001% cholesteryl hemisuccinate, 100 mM NaCl, 10 uM tiotropium or N-methylscopolamine and 2 mM CaCl2. Elution was performed with the 10X CMC buffer supplemented with 5 mM EDTA and 0.2 mg/ml FLAG peptide. This procedure typically yielded 3.5–4 mg protein in a volume of 10–15 ml. 0.3 mg of TEV protease was added for each mg of purified receptor and incubated for 4 hr on ice to remove the flexible amino terminal tail. The digested protein was concentrated to 500 ul using 50 kDa spin filters and purified further on a Sephadex-200 size exclusion column (GE Healthcare) in 10x CMC MNG buffer. The pooled fractions were concentrated in 50 kDa spin columns to Abs280 = 40–80 and flash frozen in 8 ul aliquots.
Crystallization
The protein was reconstituted in a 2:3 ratio by weight into a 10:1 mixture of monoolein (Sigma cat. Nr. M7765) and cholesterol (Sigma cat. Nr. C3045) and spotted on glass plates in 15–30 nl drops using a Gryphon LCP robot (Art Robbins Instruments). Crystals were harvested after 4–5 days growth in a 20 °C incubator. Crystal diffraction was collected at APS beamline 23D, at the Argonne National Laboratory, IL, USA.
Four rounds of optimization were performed on the M3-dsT4L construct before bright rhombic and flat rhombic crystals approximately 15–20 μm were obtained. The optimal crystallization condition comprised 100 mM Tris pH 8.1, 113.5 mM lithium citrate, 110 mM ammonium sulfate and 45% PEG 300. The lipid was in most cases sponge phase, with cubic phase obtained by varying the PEG or ammonium sulfate concentration in a gradient. The crystals diffracted to 2.9 Å. The space group is P41212, with unit cell dimensions a= 54.95 Å, c= 348.0 Å. Data were collected from 15 crystals.
Using the mT4L version of the receptor bound to tiotropium we were able to obtain several initial hits and after a single round of optimization we obtained strongly birefringent, block-like crystals approximately 25 μm on a side. Even larger crystals were obtained by elevating the pH slightly (Figure S1C–E). The best crystallization condition was 100 mM Tris pH 7.5, 44% PEG 300, and 400 mM ammonium tartrate. The lipid was in sponge phase under all conditions. These crystals diffracted to 2.3 Å. The space group is C2 with dimensions a=152.5 Å, b=184.6 Å, c=52.5 Å, = 99°. Data were collected from 37 crystals giving a final resolution of 2.8 Å.
Indexing and scaling of the obtained data was done using HKL2000 (Otwinowski and Minor, 1997) and CCP4 software (Winn et al., 2011). Molecular replacement phasing was done using Phaser MR (Mccoy et al., 2007) in Phenix with the M3 structure from PDB ID 4DAJ and wtT4L (4LZM) or triple cysteine T4L (152L) as ensembles. Refinement was done in Phenix Refine (Adams et al., 2010).
Buried surface calculation
To evaluate the tightness of the M3 receptor packing interfaces within the crystal structures, the command “get_area” was used in PyMOL, as described in pymolwiki.org to calculate the buried area. The total exposed area of the dimer was subtracted from the sum of the surface areas of the two individual monomers.
Calculation of interaction surfaces
To display the surface of a receptor interacting with a neighboring protomer, the receptor chain with hydrogens displayed was selected in PyMOL. Using the “modify around” function, atoms on the receptor in question within 4 A of this were selected and colored for visualization.
Supplementary Material
Figure S1, related to Tables 1–3. M3-dsT4L and M3-mT4L crystalized in lipidic cubic phase.
(A) Image of a 20 nl lipid drop containing plate like M3-dsT4L crystals. (B) Similar crystals as shown in panel a viewed thorough a polarizing filter. (C) 20 nl lipid drop containing block-shaped M3-mT4L crystals. (D) M3-mT4L crystals viewed thorough a polarizing filter. (E) Larger M3-mT4L crystals could be grown at pH 8.0. Though more irregular in shape, they diffracted equally well. (F) The M3 mT4L was also crystalized using NMS as the ligand giving similar appearing crystals as observed with tiotropium.
Figure S2, related to figure 4. Methylscopolamine binding to M3-mT4L
(A) N-methylscopolamine (NMS) (raspberry) was crystallized bound in M3 mT4L (brown) (PDB 4U16). The 2Fo-Fc Map around the NMS contoured at 2.0 σ is shown in grey (left panel). Although the low resolution does not allow for a very exact position of the molecule, it can be seen that NMS binds in the same manner as tiotropium (green) (right panel). The structure of NMS and a lig plot of its interaction with the receptor are also shown.
(B) In the proposed allosteric site of our receptor structures in vicinity of tryptophan 525 a clear density was always present. In the tiotropium bound mT4L M3 structure chain B, the most well resolved chain, a PEG 300 molecule could be fit. The M3 receptor is shown in green and the 2Fo-Fc Map around the PEG 300 (blue) contoured at 1.0 σ is shown in grey. (C) Dissociation experiments show that PEG 300 slows 3H-NMS dissociation from the M3 receptor to a t1/2 of approx. 240 min compared to a t1/2 of approx. 50 min for untreated membranes, supporting a likely binding of PEG in the allosteric site. The data represent the mean +/− s.e.m. of three independent experiments performed in duplicate.
Figure S3, related to figure 1. Comparison of the wt and mT4L with the previously reported circular permutated CP T4L.
A circular permutated version of wtT4L (CPT4L) (PDB: 2O7A) was made by moving the N-teminal helix A of wtT4L to the end of the C-terminal domain, and deleting the N-terminal domain. A short SerGly4Ala linker inserted between the C-terminal domain and N-terminal helix is shown in yellow (Chun et al., 2012). A β2AR fusion protein in which this CPT4L was inserted between TMs 5 and 6 expressed poorly and failed to yield crystals (Chun et al., 2012).
Highlights.
We present new versions of T4 lysozyme to be applied in GPCR crystallography.
We present a new 2.8 Å structure of the M3 muscarinic receptor bound to tiotropium.
We present a structure of the M3 receptor bound with N-methylscopolamine.
We present data showing that PEG300 binds in the M3 allosteric binding site.
Acknowledgments
We acknowledge support from National Institutes of Health grant GM08311806 (B.K.K), National Science Foundation Grant CHE-1223785 (B.K.K.), the Mathers Foundation (B.K.K.), and the Benzon Foundation (T.S.T.). We would also like to thank Andrew Kruse for technical advice.
Footnotes
Author contribution
T.S.T. designed all T4L variants and purified these fused to the M3 receptor. T.S.T. crystallized M3 mT4L bound to tiotropium and NMS and conducted all binding experiments. R.M. purified and crystallized the M3 dsT4L bound to tiotropium. W.W. contributed technical advice. B.K. supervised the project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin N, Yun J, Wess J. Mapping of single amino acid residues required for selective activation of Gq/11 by the m3 muscarinic acetylcholine receptor. J Biol Chem. 1995;270:17741–17748. doi: 10.1074/jbc.270.30.17741. [DOI] [PubMed] [Google Scholar]
- Baase WA, Liu L, Tronrud DE, Matthews BW. Lessons from the lysozyme of phage T4. Protein Sci. 2010;19:631–641. doi: 10.1002/pro.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa P, Bouyssou T, Germeyer S, Schnapp A, Gantner F, Pieper M. Preclinical evaluation of long-acting muscarinic antagonists: comparison of tiotropium and investigational drugs. J Pharmacol Exp Ther. 2009;330:660–668. doi: 10.1124/jpet.109.152470. [DOI] [PubMed] [Google Scholar]
- Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, Katritch V, Kunken J, Xu F, Cherezov V, Hanson MA, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling MR, Charlton SJ. Quantifying the association and dissociation rates of unlabelled antagonists at the muscarinic M3 receptor. Br J Pharmacol. 2006;148:927–937. doi: 10.1038/sj.bjp.0706819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror RO, Green HF, Valant C, Borhani DW, Valcourt JR, Pan AC, Arlow DH, Canals M, Lane JR, Rahmani R, et al. Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature. 2013;503:295–299. doi: 10.1038/nature12595. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Matsumura M, Faber HR, Matthews BW. Structure of a stabilizing disulfide bridge mutant that closes the active-site cleft of T4 lysozyme. Protein Sci. 1992;1:46–57. doi: 10.1002/pro.5560010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazareno S, Birdsall NJ. Detection, quantitation, and verification of allosteric interactions of agents with labeled and unlabeled ligands at G protein-coupled receptors: interactions of strychnine and acetylcholine at muscarinic receptors. Mol Pharmacol. 1995;48:362–378. [PubMed] [Google Scholar]
- Liu J, Conklin BR, Blin N, Yun J, Wess J. Identification of a receptor/G-protein contact site critical for signaling specificity and G-protein activation. Proc Natl Acad Sci U S A. 1995;92:11642–11646. doi: 10.1073/pnas.92.25.11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura M, Becktel WJ, Levitt M, Matthews BW. Stabilization of phage T4 lysozyme by engineered disulfide bonds. Proc Natl Acad Sci U S A. 1989a;86:6562–6566. doi: 10.1073/pnas.86.17.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura M, Matthews BW. Control of enzyme activity by an engineered disulfide bond. Science. 1989;243:792–794. doi: 10.1126/science.2916125. [DOI] [PubMed] [Google Scholar]
- Matsumura M, Signor G, Matthews BW. Substantial increase of protein stability by multiple disulphide bonds. Nature. 1989b;342:291–293. doi: 10.1038/342291a0. [DOI] [PubMed] [Google Scholar]
- Matthews BW, Remington SJ, Grutter MG, Anderson WF. Relation between hen egg white lysozyme and bacteriophage T4 lysozyme: evolutionary implications. J Mol Biol. 1981;147:545–558. doi: 10.1016/0022-2836(81)90399-5. [DOI] [PubMed] [Google Scholar]
- Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Prilla S, Schrobang J, Ellis J, Holtje HD, Mohr K. Allosteric interactions with muscarinic acetylcholine receptors: complex role of the conserved tryptophan M2422Trp in a critical cluster of amino acids for baseline affinity, subtype selectivity, and cooperativity. Mol Pharmacol. 2006;70:181–193. doi: 10.1124/mol.106.023481. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- Sykes DA, Dowling MR, Leighton-Davies J, Kent TC, Fawcett L, Renard E, Trifilieff A, Charlton SJ. The Influence of receptor kinetics on the onset and duration of action and the therapeutic index of NVA237 and tiotropium. J Pharmacol Exp Ther. 2012;343:520–528. doi: 10.1124/jpet.112.194456. [DOI] [PubMed] [Google Scholar]
- Tautermann CS, Kiechle T, Seeliger D, Diehl S, Wex E, Banholzer R, Gantner F, Pieper MP, Casarosa P. Molecular Basis for the Long Duration of Action and Kinetic Selectivity of Tiotropium for the Muscarinic M3 Receptor. J Med Chem. 2013 doi: 10.1021/jm401219y. [DOI] [PubMed] [Google Scholar]
- Trankle C, Kostenis E, Mohr K. Muscarinic allosteric modulation: M2/M3 subtype selectivity of gallamine is independent of G-protein coupling specificity. Naunyn Schmiedebergs Arch Pharmacol. 2001;364:172–178. doi: 10.1007/s002100100441. [DOI] [PubMed] [Google Scholar]
- Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Weis WI, Kobilka BK. N-terminal T4 lysozyme fusion facilitates crystallization of a G protein coupled receptor. PLoS One. 2012;7:e46039. doi: 10.1371/journal.pone.0046039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Tables 1–3. M3-dsT4L and M3-mT4L crystalized in lipidic cubic phase.
(A) Image of a 20 nl lipid drop containing plate like M3-dsT4L crystals. (B) Similar crystals as shown in panel a viewed thorough a polarizing filter. (C) 20 nl lipid drop containing block-shaped M3-mT4L crystals. (D) M3-mT4L crystals viewed thorough a polarizing filter. (E) Larger M3-mT4L crystals could be grown at pH 8.0. Though more irregular in shape, they diffracted equally well. (F) The M3 mT4L was also crystalized using NMS as the ligand giving similar appearing crystals as observed with tiotropium.
Figure S2, related to figure 4. Methylscopolamine binding to M3-mT4L
(A) N-methylscopolamine (NMS) (raspberry) was crystallized bound in M3 mT4L (brown) (PDB 4U16). The 2Fo-Fc Map around the NMS contoured at 2.0 σ is shown in grey (left panel). Although the low resolution does not allow for a very exact position of the molecule, it can be seen that NMS binds in the same manner as tiotropium (green) (right panel). The structure of NMS and a lig plot of its interaction with the receptor are also shown.
(B) In the proposed allosteric site of our receptor structures in vicinity of tryptophan 525 a clear density was always present. In the tiotropium bound mT4L M3 structure chain B, the most well resolved chain, a PEG 300 molecule could be fit. The M3 receptor is shown in green and the 2Fo-Fc Map around the PEG 300 (blue) contoured at 1.0 σ is shown in grey. (C) Dissociation experiments show that PEG 300 slows 3H-NMS dissociation from the M3 receptor to a t1/2 of approx. 240 min compared to a t1/2 of approx. 50 min for untreated membranes, supporting a likely binding of PEG in the allosteric site. The data represent the mean +/− s.e.m. of three independent experiments performed in duplicate.
Figure S3, related to figure 1. Comparison of the wt and mT4L with the previously reported circular permutated CP T4L.
A circular permutated version of wtT4L (CPT4L) (PDB: 2O7A) was made by moving the N-teminal helix A of wtT4L to the end of the C-terminal domain, and deleting the N-terminal domain. A short SerGly4Ala linker inserted between the C-terminal domain and N-terminal helix is shown in yellow (Chun et al., 2012). A β2AR fusion protein in which this CPT4L was inserted between TMs 5 and 6 expressed poorly and failed to yield crystals (Chun et al., 2012).