Abstract
Transposon-based genetic transformation has facilitated insect functional genomics and new strategies of pest management. However, there is a need for alternative, site-specific approaches to overcome limitations of random integration (and associated position-effects) and potential instability of inserted transgenes. Here we describe a transposon-free, site-specific genetic transformation system mediated by transcription activator-like effector nucleases (TALENs) in the silkworm, Bombyx mori, a lepidopteran model insect. We successfully established a site-specific transgenic system with comparable transformation efficiency to transposon-based genetic transformation through microinjection of TALENs mRNA targeting the BmBLOS2 locus and a linearizable donor plasmid encoding an expression cassette of the DsRed2 red fluorescent protein. This system provides a valuable approach for insect transgenesis and will enable future functional gene analysis and generate novel applications in agricultural and medical insect pest-management technologies.
Keywords: Bombyx mori, TALENs, homologous recombination
1. Introduction
Conventional insect transgenesis (IT) technologies for functional genomics and the development of novel applications in agricultural and medical insect pest management have depended until recently on non-autonomous transposable elements such as piggyBac, Hermes and Minos (Robinson et al., 2004). These elements have a number of disadvantages resulting from random integration and insertion-site effects, including mutagenesis and local enhancer influences (Wilson et al., 1990). In addition, some species also have pre-existing autonomously active transposons, and these may contribute to instability of inserted transgenes (Labrador and Corces, 1997). Hence, deletion of transposon terminal sequence is needed to achieve a stable germline transformation (Handler et al., 2004; O'Brochta and Handler, 2008). Site-specific bacteriophage recombinases or integrases for insect genomic manipulation mitigate some of these problems, however, they require prior introduction into chromosomes of ‘docking’ sites using transposon-based genetic transformation (Schetelig et al., 2011; Schetelig et al., 2009). Thus, there is a need in insects for alternative, site-specific approaches to create generalizable IT strategies to advance functional genetics and practical applications.
Recently-established, custom-engineered nuclease technologies such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered regularly-interspaced short-palindromic repeats and crispr-associated protein (CRISPR/Cas) systems provide promising alternatives. These engineered nucleases usually induce double-strand DNA breaks (DSBs) at target genomic sites that are repaired by non-homologous end joining (NHEJ) or homologous recombination (HR) in the presence of homologous donor template DNA(Gaj et al., 2013). If the two broken DNA ends have tandem-repeated nucleotide sequences, DSBs also could be repaired by single-strand annealing (SSA) recombination (Heyer et al., 2010). ZFNs, TALENs or CRISPR/Cas systems have been used to induce targeted integration in several species but not in Bombyx mori yet (Baena-Lopez et al., 2013; Cui et al., 2011; Dickinson et al., 2013; Katsuyama et al., 2013; Yang et al., 2013; Zu et al., 2013). More research is necessary to develop general strategies that can be applied to a variety of agriculturally- and medically-important insect species.
The domesticated silkworm, Bombyx mori, is an economically valuable lepidopteran model insect. ZFNs-, TALENs- and CRISPR/Cas9-mediated targeted gene mutagenesis were achieved recently in this species (Ma et al., 2012; Takasu et al., 2010; Yueqiang Wang, 2013). The BmBLOS2 gene, a B. mori homolog of the human biogenesis of lysosome-related organelles complex 1, subunit 2, is located on the Z chromosome and is responsible for urate granule synthesis in the larval epidermis (Fujii et al., 2010). Mutagenesis of BmBLOS2 results in a larval epidermal color change from opaque to translucent, the so-called ‘oily skin phenotype’ (OSP). OSP is detected easily throughout larval stages, especially in the final larval instar, making BmBLOS2 an excellent marker gene for designing advanced IT strategies. We began addressing this by developing TALENs-mediated, targeted integration at the BmBLOS2 locus in B. mori.
2. Materials and methods
2.1. TALENs vector construction and luciferase SSA assay
DNA fragments complementary to selected targeting sites were cloned and sequenced to confirm the absence of single-nucleotide polymorphisms. TALENs targeting the BmBLOS2 exon2 were constructed by Golden-Gate assembly and ligated into the pTD1 vector (Shimazu) containing a T7 promoter sequence, polyA signal and T7 transcription terminator. A human cytomegalovirus (CMV) promoter was sub-cloned into pTD1-TALEN vectors to facilitate the luciferase SSA assay in 293T cells. A firefly luciferase single-strand annealing (SSA) assay reporter vector was constructed following reported methods (Huang et al., 2011). To determine activity in vitro, TALENs vectors (200ng for each TALEN vector), luciferase SSA reporter vector (100ng) and an internal control vector (100ng) expressing the Renilla luciferase were co-transfected with Lipofectamine 2000 (Life Technologies) in triplicate into 293T cells in 24-wells plates. Cells were harvested after 24 h and luciferase activity was determined using a dual-luciferase assay (Promega). For transcription in vitro of TALENs mRNA, TALENs vectors were linearized by restriction endonulease NotI (Fermentas) digestion followed by phenol/chloroform/isoamylalcohol (25:24:1, Sangon) treatment, and used as templates in subsequent transcription experiments in vitro. TALENs mRNA was synthesized with mMESSAGE mMACHINE T7 kit (Ambion).
2.2. Donor construction
Donor A (pMD18T_L-Homo_HR5-IE1-DsRed2-SV40_R-Homo) was constructed as follows: a HR5-IE1-DsRed2-SV40 cassette (with Sa1I and BamHI sites at 5’-end, AscI and ApaI sites at 3’-end), including a HR5-IE1 promoter, DsRed2 ORF and SV40 polyA signal, was sub-cloned into the pMD18T-simple vector (Takara) to generate pMD18T_HR5-IE1-DsRed2-SV40. An 820bp 5’-homologous arm (L-Homo), digested by Sa1I and BamHI, and a 791bp 3’-homologous arm (R-Homo), double-digested by AscI and ApaI, were sub-cloned separately into pMD18T_HR5-IE1-DsRed2-SV40. Donor B has the same structure, except that it has two 49bp TALENs sites flanked with the left and right homologous arms.
2.3. Silkworm rearing and microinjection
A multivoltine, nondiapausing silkworm strain, Nistari, was used for all experiments. TALENs mRNA (300 ng/μl for each) and donor DNA (Donor A or Donor B, 250 ng/μl) were co-injected into preblastoderm embryos that then were incubated at 25 °C in a humidified chamber for 10–12 days until larval hatching. Larvae were reared on fresh mulberry leaves under standard conditions. Putative transgenic adult G0 were sib-mated or crossed with wild-type moths, and G1 progeny were scored for the presence of the marker gene under fluorescence microscopy (Nikon AZ100).
2.4. Transgene integration analysis
Genomic DNA was extracted from DsRed2-positive G1 mutant animals by standard SDS lysis–phenol treatment after incubation with proteinase K, followed by RNase A treatment and purification. 5’- and 3’-end junction fragments at the integration event were cloned separately and sequenced. Amplification conditions were 94°C for 2 min, 30 cycles of 94°C for 10s, 55°C for 10s, and 68°C for 6min, followed by a final extension period 68°C for 10min. Amplified products were cloned and sequenced. Primers used in here were listed in the supplementary information (Supplementary Table 1).
2.5. Off-target sites detection
Off-target integration was investigated by inverse PCR. Genomic DNA was extracted from G1 transgenic moths by standard SDS lysis-phenol treatment after incubation with proteinase K, followed by RNase A treatment and purification. DNA was digested with NlaIII and circularized by ligation for 6 hours at 16 °C. The 3′-end transgene junction:genome sequences were amplified using the primers (Supplementary Table 1) designed to anneal to the 3′ homology arm of the donor vector. The PCR conditions were as follows: 94 °C for 2 min, 30 cycles of 94 °C for 10 s, 50 °C for 10 s, and 65 °C for 2 min, followed by a final extension period of 65 °C for 10 min. Amplified fragments were cloned and sequenced.
3. Results
3.1. TALENs target site and related donor
TALENs were designed to target exon 2 of the BmBLOS2 locus (Fig. 1A). The TALENs activity in vitro was determined in mammalian 293T cells by using the firefly luciferase single strand annealing (SSA) recombination assay (Huang et al., 2011). The luciferase activity of the experimental samples was 4.4-fold higher compared with control, indicating that the TALENs worked efficiently (Supplementary Fig.1).
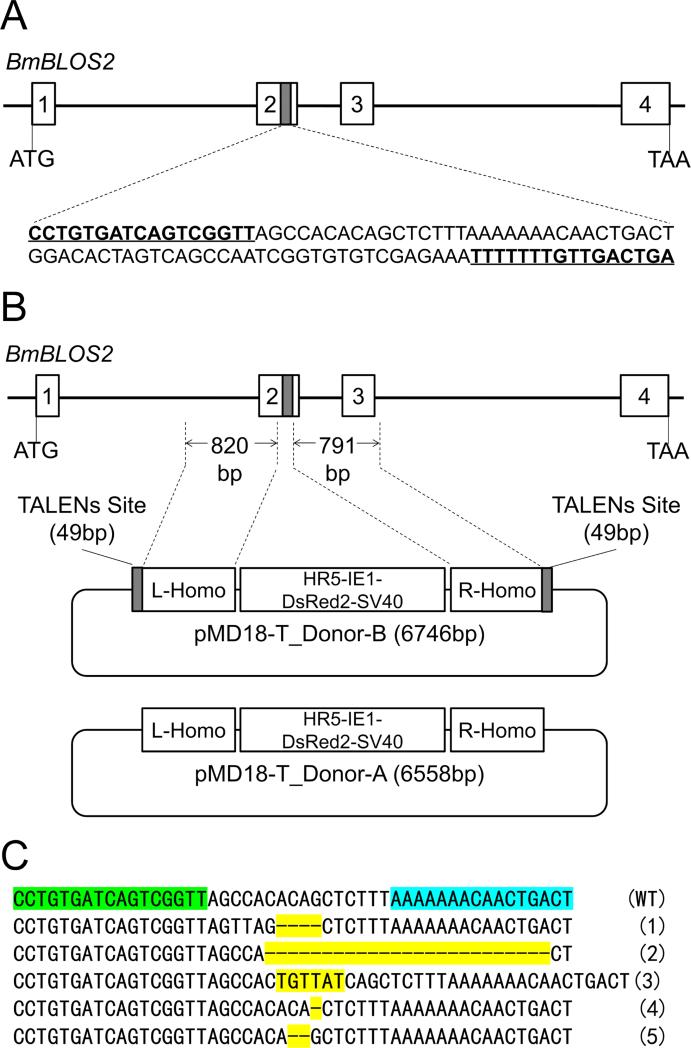
Fig.1.
Schematic representation of TALENs targeting site and donor B. (A) Schematic representation of the TALENs targeting site. The thin black line represents the BmBLOS2 genome locus with the open boxes signifying the four exons. A 49bp fragment (gray box) located in exon-2 is the targeting site. The sequences in bold and underlined are the TALENs targeting sequence. A 16bp sequence serves as a spacer between the two sites. (B) Schematic representation of the donor B. A cassette expressing a DsRed2 marker (with a HR5 enhancer sequence, driven by a baculovirus IE1 promoter) was cloned into the pMD18T- simple vector. Two homologous DNA fragments (L-homo and R-homo), 820bp and 791bp at the 5’- and 3’-ends, respectively, of the TALEN site were cloned separately into the left and right sides of the DsRed2 cassette. The DsRed2 marker with two homologous fragments was flanked by two of the 49bp TALENs sites (donor B). (C) Mutations detected in G0 mutants with oily skin phenotype (OSP). Genomic DNA was extracted from G0 mutants with severe OSP. Fragments covering the target site were amplified and sequenced. Small deletions or insertions resulting from non-homologous end joining repair were detected. Sequences highlight in green and blue are TALENs target sequence. Mutations are highlight in yellow.
The design of an efficient homologous donor DNA is crucial for site-specific transgene integration in custom-engineered nuclease technologies. Here we prepared two kinds of donor DNAs, the first of which, designated ‘donor A’, is a circular construct that uses the baculovirus immediate early one (IE1) gene promoter to express the red fluorescent protein,DsRed2, ubiquitously throughout the insect. Two genomic fragments flanking the TALENs targeting site in BmBLOS2 locus (820bp and 791bp at the 5’- and 3’-ends, respectively) were amplified and served as homologous left and right arms. The second donor construct, designated ‘donor B’, has the same structure as donor A except that it includes two 49bp TALENs site sequences flanking the left and right homologous arms that can be digested in vivo by the corresponding endonuclease, releasing a linear donor homologous template (Fig. 1B).
3.2. TALENs induced high efficient germline transformation with donor B
A total of 1040 preblastoderm Nistari eggs were injected with a mixture of TALENs mRNA and donor A. Of these, 601 (57.8%) hatched and most of the larvae exhibited the OSP, demonstrating that the TALENs worked efficiently in vivo. Small deletions or insertions were detected at the target site (Fig. 1C). A total of 173 animals survived to adulthood and these were sib-mated or out-crossed to wild-type moths, creating 73 egg batches. Unfortunately, fluorescent microscopic screening at late embryonic and larval stages did not recover any DsRed2-positive G1 individuals.
Previous reports indicated that a circular homologous donor may not be efficient enough in some systems for enhancing germline integration in vivo (Cui et al., 2011; Zu et al., 2013). Therefore, we repeated the experiments with the linearizable donor B construct. Of the injected 720 preblastoderm eggs, 374 (51.9%) hatched and 206 animals survived to the pupal stage. Chimeric expression of DsRed2 was detected in 72 pupas which indicated high somatic integration efficiency (Supplementary Fig. 2). Surviving moths were sib-mated or out-crossed to wild-type insects and 76 egg batches were examined for fluorescence. Screening at the late embryonic and larval stages detected six DsRed2-positive batches (broods) and one or more DsRed2-positive individuals were recovered in each brood. Red fluorescence was detectable in the whole body throughout all developmental stages (Fig. 2). The transformation efficiency, 7.9%, was defined as the proportion of fertile injection survivors producing one or more transgenic offspring (Tan et al., 2013).
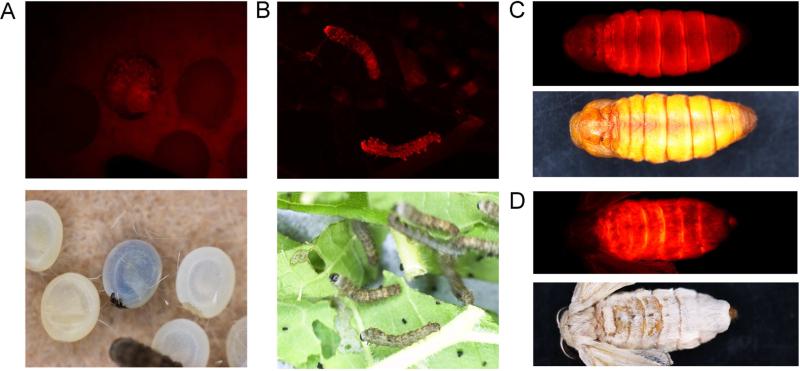
Fig.2.
Red fluorescence could be detected in positive individuals at embryonic, larval, pupal and adult stages. DsRed2 expression was driven by a baculovirus IE1 promoter in a ubiquitous manner. Red fluorescence detected in embryos (A), the first instar larvae (B), pupa (C) and adult (D).
3.3. Site-specific integration was achieved in BmBLOS2 locus
A total of 30 DsRed2-positive G1 individuals were recovered from the six broods. Four of these had the OSP and the remaining 26 were wild-type. Since the BmBLOS2 is located on the Z chromosome, these G1 transgenic animals with wild-type skin color were likely heterozygous males (ZZ type) and polymerase chain reaction (PCR) based analyses verified the presence of diagnostic fragments consistent with the precise integrity at BmBLOS2 locus in three distinct broods 1, 3 and 6 (Fig. 3A and 3B). PCR-based analyses of these broods failed to amplify the complete integrated fragment, most likely due to complex rearrangements arising from the DNA repair mechanism involved in homologous recombination (Dickinson et al., 2013). However, we confirmed the precise integration of the complete HR5-IE1-DsRed2-SV40 expression cassette by gene amplification and sequencing separately the 5’- and 3’-end junction (Supplementary Sequences). Furthermore, identical site-specific transgene integration was observed in all of the three broods. These data support the conclusion that we recovered three independent mutations in the BmBLOS2 locus.
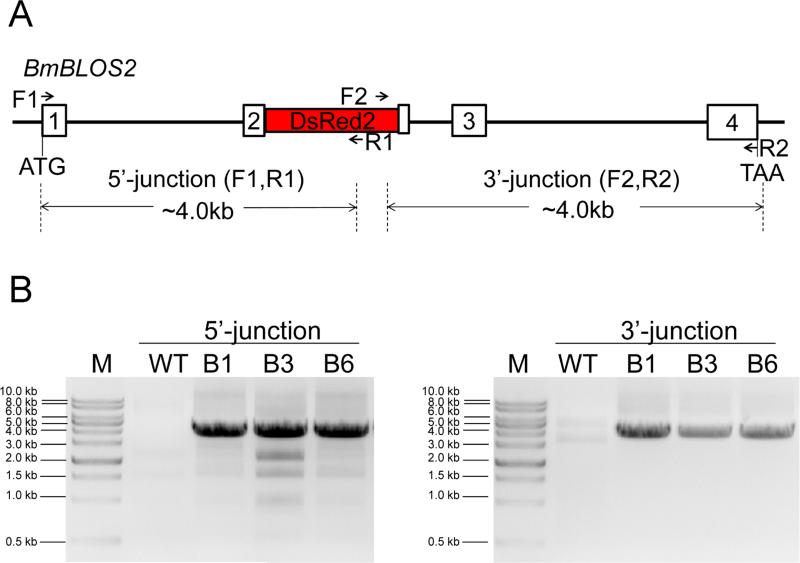
Fig.3.
Gene amplification analyses of transgene insertions in transformed silkworms. (A) Primer positions for amplification. The primer pairs F1/R1 and F2/R2 were used to amplify the 5’- and 3’-end insertion junctions, respectively. (B) The diagnostic DNA fragments for 5’-and 3’-end junctions were detected in brood-1 (B1), brood-3 (B3) and brood-6 (B6).
3.4. Integration of exogenous marker gene is genetic heritable
To confirm heritable germline transformation in G2, heterozygous transgenic male G1 moths of brood 1, which showed wild type skin color, were mated with wild type moths. In G2 animals, females showing OSP and DsRed2-positive and males showing no OSP but DsRed2-positive were recovered (Fig. 4A and 4B), suggesting the site-specific germline integration was stable. For broods 2, 4 and 5, we could not detect the diagnostic fragments, suggesting the potential off-target effect. We performed inverse PCR to investigate the exact genomic integration sites. Although the transgene integration could not be validated in brood 4 and 5, we localized the transgene of brood 2 at Chromosome 17, indicating that off-target effects should be considered in TALEN-mediated genetic transformation.
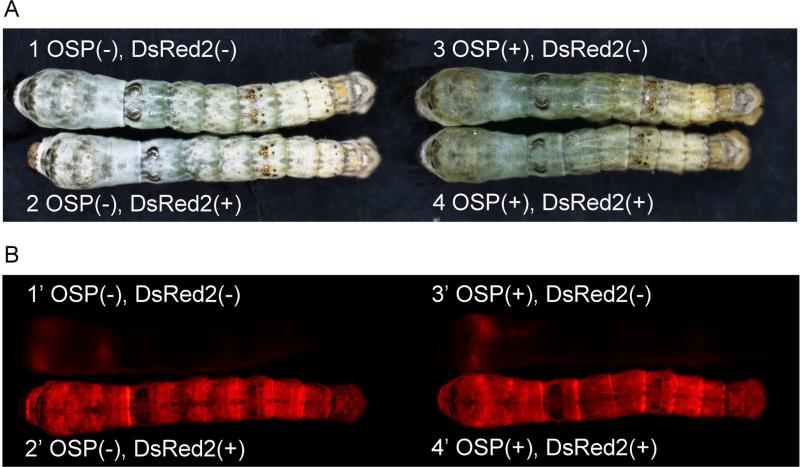
Fig.4.
DsRed2 and oily-skin phenotypes following germline transformated silkworms. Insects in (A) and (B) photographed with normal light and UV fluorescence, respectively. The wild-type larva has the normal skin color (OSP-) and no red fluorescence (DsRed2-) (1 and 1’). The heterozygous transformed male was OSP- but DsRed2+ (2 and 2’). The TALEN-induced mutant without transgene integration was OSP+ but DsRed2- (3 and 3’). The transformed female or male with both chromosomes modified was OSP+ and DsRed2+ (4 and 4’).
4. Discussion
A transposon-free, site-specific germline transformation system with a technically- useful transformation efficiency (7.9%) is established successfully in the silkworm, B. mori. Although there was a report on successful transgene integration event in B. mori mediated by ZFNs, the integration efficiency was extremely low (Daimon et al., 2013). Although the detailed mechanisms of the site-specific integration processes are unclear, we provide evidence that the insertions recovered in our work result from NHEJ, HR and SSA. A circular donor plasmid was impossible to be processed to generate a single-strand DNA (ssDNA) tail during DSB repair; thus, integration of the circular donor most likely occurred only through the HR repair mechanism (Symington and Gautier, 2011). Circular donor molecules function as templates and this type of exogenous donor usually cannot be integrated directly at the target site. DNA strand invasion is indispensable in HR-mediated integration and we anticipate that circular donors with longer homologous arms may have higher probabilities of facilitating this process (Beumer et al., 2013; San Filippo et al., 2008). The linearizable donors (donor B) can be resected to generate ssDNA tails during the DSB repair process. These donors contain long homologous ssDNA tails with the broken DNA ends and therefore may anneal with the ssDNA tail of broken DNA ends, similar to the SSA repair process, and facilitate efficient integration(Sugawara et al., 2000) (Supplementary Fig. 3). In addition, the unexpected off-target events suggesting that the linearizable donors could also be directly integrated into organism genome through NHEJ mechanism. In this case, one should be aware of off-target effects when using linearizable donors although it increases integration efficiency.
The major merits of a transposon-free targeted IT system minimize concerns about variability of transgene activity in transgenic animals; also none of the hypothetical risks articulated in relation to the use of transgenic technology in agriculture apply in targeted IT (Dafa'alla et al., 2006). The current targeted IT system relies on NHEJ, SSA and HR repair in the presence of a linearizable homologous donor after TALENs-induced DSB. We propose that this system is extendable to other custom-engineered nuclease systems, such as ZFNs and CRISPR/Cas, to enhance efficiency of site-specific integration. Introduction of exogenous marker genes such as a fluorescent protein in targeted loci will facilitate screening mutagenesis schemes in most loci for which conveniently-assessable phenotypes do not occur. A reliable transposon-free targeted IT system will be an invaluable contribution to functional genetics of important insect species.
Supplementary Material
Highlights.
Site-specific integration was achieved by co-injection of TALENs mRNA and a donor DNA construct;
Developing a solution to the problem of screening mutations induced by engineered nuclease injection;
Linearizeable donor DNAs with TALENs cleavage sites are more effective than circular donors at targeted gene knock-in.
ACKNOWLEDGMENTS
We thank Dr. David Stanley (USDA-ARS, Columbia MO, USA) for comments on earlier drafts of this paper. This work was supported by grants from National Science Foundation of China (31030060), National Basic Research Program of China (2012CB114101), the External Cooperation Program of BIC, Chinese Academy of Sciences (Grant No. GJHZ201305) and the Shanghai Committee of Science and Technology, China (11JC1413900). AAJ was supported in part by a grant from the NIH NIAID (AI29746).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Baena-Lopez LA, Alexandre C, Mitchell A, Pasakarnis L, Vincent JP. Accelerated homologous recombination and subsequent genome modification in Drosophila. Development. 2013;140:4818–4825. doi: 10.1242/dev.100933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Trautman JK, Mukherjee K, Carroll D. Donor DNA Utilization during Gene Targeting with Zinc-finger Nucleases. G3 (Bethesda) 2013 doi: 10.1534/g3.112.005439. doi: 10.1534/g3.112.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat. Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- Dafa'alla TH, Condon GC, Condon KC, Phillips CE, Morrison NI, Jin L, Epton MJ, Fu G, Alphey L. Transposon-free insertions for insect genetic engineering. Nat. Biotechnol. 2006;24:820–821. doi: 10.1038/nbt1221. [DOI] [PubMed] [Google Scholar]
- Daimon T, Kiuchi T, Takasu Y. Recent progress in genome engineering techniques in the silkworm, Bombyx mori. Dev. Growth. Differ. 2014;56:14–25. doi: 10.1111/dgd.12096. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Daimon T, Uchino K, Banno Y, Katsuma S, Sezutsu H, Tamura T, Shimada T. Transgenic analysis of the BmBLOS2 gene that governs the translucency of the larval integument of the silkworm, Bombyx mori. Insect. Mol. Biol. 2010;19:659–667. doi: 10.1111/j.1365-2583.2010.01020.x. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends. Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler AM, Zimowska GJ, Horn C. Post-integration stabilization of a transposon vector by terminal sequence deletion in Drosophila melanogaster. Nat. Biotechnol. 2004;22:1150–1154. doi: 10.1038/nbt1002. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- Katsuyama T, Akmammedov A, Seimiya M, Hess SC, Sievers C, Paro R. An efficient strategy for TALEN-mediated genome engineering in Drosophila. Nucleic. Acids. Res. 2013;41:e163. doi: 10.1093/nar/gkt638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador M, Corces VG. Transposable element-host interactions: regulation of insertion and excision. Annu. Rev. Genet. 1997;31:381–404. doi: 10.1146/annurev.genet.31.1.381. [DOI] [PubMed] [Google Scholar]
- Ma S, Zhang S, Wang F, Liu Y, Xu H, Liu C, Lin Y, Zhao P, Xia Q. Highly efficient and specific genome editing in silkworm using custom TALENs. PloS One. 2012;7:e45035. doi: 10.1371/journal.pone.0045035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brochta DA, Handler AM. Perspectives on the state of insect transgenics. Adv Exp Med Biol. 2008;627:1–18. doi: 10.1007/978-0-387-78225-6_1. [DOI] [PubMed] [Google Scholar]
- Robinson AS, Franz G, Atkinson PW. Insect transgenesis and its potential role in agriculture and human health. Insect. Biochem. Mol. Biol. 2004;34:113–120. doi: 10.1016/j.ibmb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Schetelig MF, Gotschel F, Viktorinova I, Handler AM, Wimmer EA. Recombination technologies for enhanced transgene stability in bioengineered insects. Genetica. 2011;139:71–78. doi: 10.1007/s10709-010-9494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetelig MF, Scolari F, Handler AM, Kittelmann S, Gasperi G, Wimmer EA. Site-specific recombination for the modification of transgenic strains of the Mediterranean fruit fly Ceratitis capitata. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18171–18176. doi: 10.1073/pnas.0907264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Takasu Y, Kobayashi I, Beumer K, Uchino K, Sezutsu H, Sajwan S, Carroll D, Tamura T, Zurovec M. Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect. Biochem. Mol. Biol. 2010;40:759–765. doi: 10.1016/j.ibmb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Tan A, Fu G, Jin L, Guo Q, Li Z, Niu B, Meng Z, Morrison NI, Alphey L, Huang Y. Transgene-based, female-specific lethality system for genetic sexing of the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 2013;110:6766–6770. doi: 10.1073/pnas.1221700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Bellen HJ, Gehring WJ. Position effects on eukaryotic gene expression. Annu. Rev. Cell. Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Z, Xu J, Zeng B, Ling L, You L, Chen Y, Huang Y, Tan A. The CRISPR/Cas System mediates efficient genome engineering in Bombyx mori. Cell Res. 2013;23:1414–1416. doi: 10.1038/cr.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Hu Y, Luo Z, Huang P, Wu Q, Zhu Z, Zhang B, Lin S. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods. 2013;10:329–331. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.