Abstract
Significant research has been invested in correlating genetic variations with different disease probabilities. Recently, it has become apparent that other DNA modifications, such as the addition of a methyl or hydroxymethyl group to cytosine, can also play a role. While these modifications do not change the sequence, they can negatively impact the function. Therefore, it is critical to be able to both read the genetic code and identify these modifications. Currently, the detection of hydroxymethylated cytosine (5′hmC) and the two closely related variants, cytosine (C) and 5′methylcytosine (5′mC), relies on a combination of nucleotide modification steps, followed by PCR and gene sequencing. However, this approach is not ideal because transcription errors which are inherent to the PCR process can be misinterpreted as fluctuations in the relative C:5′mC:5′hmC concentrations. As such, an alternative method which does not rely on PCR or nucleotide modification is desirable. One approach is based on label-free optical resonant cavity sensors. In the present work, toroidal resonant cavity sensors are functionalized with antibodies to enable label-free detection and discrimination between C, 5′mC, and 5′hmC in real-time without PCR. Specifically, epoxide chemistry is used to covalently attach the 5′hmC antibody to the surface of the cavity. Subsequently, to thoroughly characterize the sensor platform, detection of C, 5′mC, and 5′hmC is performed over a concentration range from pM to nM. At low (pM) concentrations, the hydroxymethylated cytosine produces a significantly larger signal than the structurally similar epigenetic markers; thus demonstrating the applicability of this platform.
Keywords: optical sensor, methylation, label-free detection, 5′ hydroxymethyl cytosine, epigenetic markers
1. Introduction
By combining advances in computational power and our improved understanding of genetics, researchers are beginning to link specific DNA sequences to a wide range of diseases. However, emerging evidence links many diseases to both the sequence and the methylation state of the DNA (absence or presence of a methyl group to the DNA backbone)(Berger et al. 2009; Bird 2002; Esteller 2007; Pastor et al. 2011; Portela and Esteller 2010; Rando and Verstrepen 2007; Surani et al. 2007; Thu et al. 2010). Most notably, although the fundamental genetic code is not altered by the presence of the methyl group, the function can be significantly altered, disrupting normal cell behavior (Branco et al. 2012; Chen and Riggs 2005; Esteller 2007; Li and O’Neill 2013; Mariani et al. 2013; Riggs 2002). The regulation of cellular function can be traced to guanine-cytosine rich sequences known as CpG islands interspersed throughout the genome but more prominent in the promoter regions (Berger et al. 2009; Bird 2002). Methylation of the cytosine within the CpG islands affects gene expression by silencing the nearby associated gene and even genes many kilobases away by prohibiting the binding of transcription factors(Surani et al. 2007). This point of gene regulation becomes problematic when seen in the methylation of CpG islands for tumor suppression factors because it can result in the decrease or loss of function of tumor suppression.
Recently, a variation on methylation, called hydroxylmethylation, was discovered (Figure 1). In hydroxylmethylation, the methyl cytosine is oxidized forming a methyl hydroxyl group on the number 5 carbon cytosine (5′hmC) (Branco et al. 2012). 5′hmC has gained significant attention in the last few years because it has strong associations with embryonic stem cells. Namely, it plays an important role in maintaining pluripotency which supports earlier findings of 5′hmC nucleotides in the mammalian brain development and neuronal plasticity (Ficz et al. 2011; Szulwach et al. 2011). 5′hmC is also thought of as the intermediate cytosine analog in the demethylation process and basically all three conformations (unmethylated, methylated, and hydroxymethylated) are not only considered important for gene signaling and gene silencing, but they also play an important role in the development, differentiation, and disease states of cells.
Figure 1.
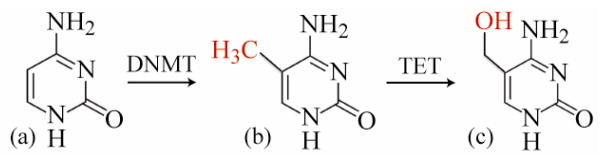
Schematic representation of the nucleotides cytosine (C), 5′ methyl cytosine (5′mC), and 5′ hydroxymethyl cytosine (5′hmC). Cytosine methylation is catalyzed by methyltransferases, DNMT. 5′hydroxymethyl cytosine results from the oxidation of 5′methylcyotsine by the TET family of enzymes(Pastor et al. 2011).
Additionally, during cell differentiation and embryonic development, as the cell transforms and proliferates, the levels of 5′hmC steadily decrease as the levels of 5′mC increase (Ficz et al. 2011; Pastor et al. 2011). Therefore, the current hypothesis is that it is not only the presence, but the relative concentration of methylation and hydroxymethylation that are important to biological processes such as imprinting, cellular reprogramming, plasticity, tissue and cellular repair. As a result, accurate methods for detecting 5′mC and 5′hmC are critically needed.
Bisulfite sequencing, a prominent standard assay for detecting and quantifying DNA methylation, has provided a tool for comprehensive genome wide analysis of methylated cytosine for over 20 years (Fraga and Esteller 2002). Recently, the reliability and accuracy for detection of 5′mC is now challenged by the occurrence of 5′ hydroxymethyl cytosine. Specifically, when 5′hmC is exposed to the standard bisulfite protocol, it is converted to a stable 5′ methyl sulfonate derivative that is often misread as methylated cytosine when sequenced. Additionally, the bulkiness of the 5′methyl sulfonate intermediate hinders PCR amplification. Therefore, the use of bisulfite sequence analysis is not suitable to accurately identify or distinguish 5′hmC from 5′mC molecules (Cadet and Wagner 2014; Olkhov-Mitsel and Bapat 2012).
While overlapping attempts to find an assay that will distinguish 5′mC from 5′hmC have been successful, many require cumbersome nucleotide chemical modifications steps such as the glycosylation of the hydroxyl group on the number 5 carbon of 5′hmC. This approach has proven successful for isolating 5′hmC from 5′mC and other methylated intermediate molecules. However, due to the size of the glycosyl molecule, it is not possible to glycosylate all of the available hydroxyl methylated molecules, especially when they occur within CpG islands (Mariani et al. 2013).
The introduction of methylation specific antibodies have broadened the range of research of 5′mC and 5′hmC by allowing for direct interrogation and detection of methylated or hydroxymethylated DNA sequences without the need for chemical modifications. Although this advancement has allowed researchers to detect 5′hmC methylation patterns on a genome wide scale, there are still limitations in the detection of these molecules below nanomolar levels. As such, relatively large quantities of DNA are still required (Borgel et al. 2012; Thu et al. 2010). In complementary work, immunoprecipitation experiments such as methylated DNA immunoprecipitation (MeDip) and hydroxymethylated DNA precipitation (hMeDhip) followed by PCR or DNA sequencing have also been explored. However, these assays usually have a starting sample range of 0.1μg of material (Huang et al. 2010; Nestor et al. 2010).
Label free biosensors have gained attention as useful devices for a wide variety of biodetection applications (Hunt and Armani 2010). For example, microcantilever biosensors have demonstrated detection of single viruses and cells, optical resonator sensors have demonstrated detection of cancer biomarkers in serum, and nanowire sensors have detected individual proteins. Because the sensitivity of these integrated devices arises from the fundamental transducer element, only extremely small amounts of analyte are required to produce a detectable signal. However, sensitivity is not sufficient alone for biosensing; specificity is also critical in any biodetection application (Hunt and Armani 2014). Therefore, by combining a high sensitivity transducer with the recently developed antibodies, a sensor platform which is ideal for this challenge can be designed.
One such transducer is based on a whispering gallery mode optical microcavity (Matsko and Ilchenko 2006). This device confines light of specific optical frequencies, or resonant frequencies. When a molecule binds to the surface of the microtoroid device, the refractive index of the cavity changes, and the resonant wavelength shifts. The detection resolution or sensitivity is related to the photon lifetime within the cavity or the cavity quality factor (Q). Namely, higher Q factor devices have higher sensitivity. Previously, optical microcavities have demonstrated specific and sensitive detection of a wide range of biological molecules, including DNA and proteins (Hawk et al. 2013; Kindt and Bailey 2013; Kindt et al. 2013; Shao et al. 2013; Soteropulos et al. 2011; Vollmer and Yang 2012; Wu et al. 2014).
In the present work, a silica optical microtoroid resonant cavity (Figure 2) is used to detect hydroxymethylated cytosine without the need for PCR amplification a priori. We used epoxy-silane coupling to attach previously developed 5′hmC specific antibodies to the microtoroid devices with minimal impact on the device quality factor. Using this approach, the detection and discrimination of 5′mC from 5′hmC is possible at picomolar levels and thus an on-chip assay that is both sensitive and specific without the need for any additional modifications, application steps, or labeling.
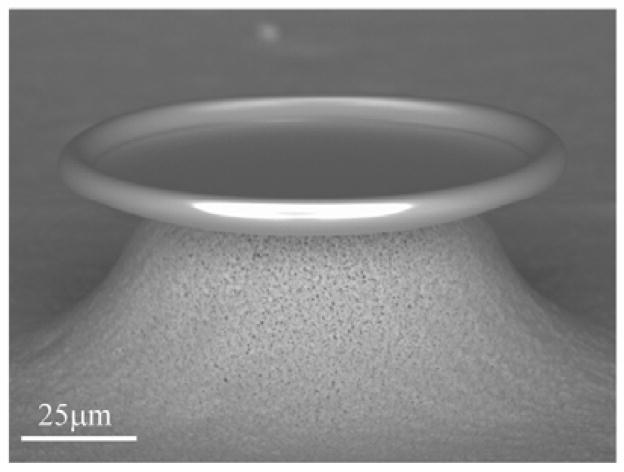
Figure 2.
Scanning electron micrograph of a toroidal optical microcavity.
2. Experimental procedure
2.1. Materials and reagents
The following materials were used for the microtoroid fabrication and device characterization. Silicon wafers with 2μm thermal oxide, WRS Materials (San Jose, CA), Shipley 1813 photoresist, Microchem (Newton MA), Microcrome developer MF-321, Microchem (Newton MA), buffered oxide etchant, Transgene (Danvers, MA) and Xenon Difloride XeF2, Alfa Aesar, (Ward Hill, MA). Optical fiber FSE 780 nm for taper fabrication was purchased from Newport (Irvine, CA.).
To functionalize the surface of the sensor, 5′hydroxymethylcytosine rabbit polyclonal antibody, Active motif (Carlsbad CA.), is used. The methylated, unmethylated, and hydroxymethylated double stranded oligonucleotides are also purchased from Active Motif (Carlsbad CA) to maximize compatibility. In all three oligonucleotides, 36% of the chain is either unmethylated, methylated, or hydroxymethylated cytosine (C, 5′mC or 5′hmC). The precise sequence is located in the supplement. For the bioconjugation verification fluorescent experiments, a 20 mer oligonucleotide with methylated cytosine and Cy5 fluorophore attachment, IDT (Coraville, Iowa) is used. To bioconjugate the antibody to the sensor surface, 3-glycidoxypropyltrimethoxysilane, Sigma Aldrich (St. Louis, MO), is used. The buffers used in the present work are: 10X Phosphate buffer, Invitrogen (Irvine CA.), Tween-20, Abcam (Cambridge, MA) and Tris buffered Saline, Abcam (Cambridge MA).
The antibody’s activity is verified using a dot blot. For this measurement, Bovine serum albumin, Thermo Scientific (Rockland, IL), Nitrocellulose paper, Bio-Rad (Hercules, CA.), Immuno-detection chemoluminescent kit Bio-Rad (Hercules, CA.).
2.2 Optical resonator fabrication
The toroidal cavities were fabricated using the standard three step method which is detailed elsewhere (Armani et al. 2003). Briefly, 160μm diameter 2μm thick circular oxide pads on a silicon wafer are patterned using a combination of photolithography and buffered oxide etching. They are subsequently undercut using XeF2, an isotropic silicon etchant. Finally, the oxide is reflowed using a CO2 laser, forming the toroidal resonant cavity (Figure 2).
A tapered optical fiber waveguide is fabricated using the flame pulling method. In this approach, the cladding layer of single mode optical fiber is removed using wire strippers, and the fiber is mounted on a pair of motorized stages, which pull in opposite directions. While heating the fiber with an oxyhydride flame, the fiber is pulled until the waist region of the taper is approximately 600nm in diameter, or smaller than the wavelength of light.
2.3 Epoxy surface functionalization and verification
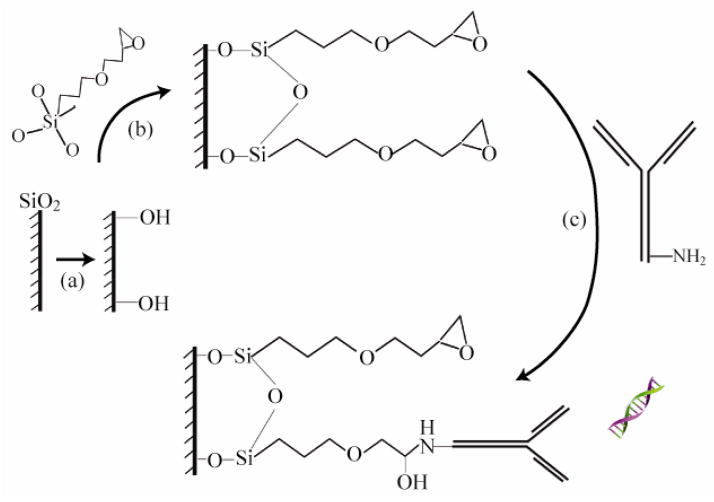
To immobilize the polyclonal antibodies specific for hydroxymethylated cytosine to the surface of our devices, we used a bifunctional linker with an epoxide ring (Figure 3). Specifically, the epoxide ring on the 3-glycidoxypropyltrimethoxysilane linker (GPTMS) forms a covalent bond with the amines on lysine side chains of antibodies while the silane covalently attaches to the hydroxyl groups on the silica toroid surface.
Figure 3.
Schematic representation of the immobilization of 5′hmC antibody. (a) Hydroxyl groups are added to the microtoroid devices through O2 plasma etching. (b) The epoxy linker (GPTMS) is added by vapor deposition. (c) The amino groups on the lysine side chain (ε-NH3+) present on the antibodies are able to covalently linked to the epoxy end of GPTMS at 37°C in a humid environment (Weetall 1993).
While the conventional approach for increasing the density of hydroxyl groups on a silica surface is a wet chemical process (piranha etch), previous work has shown that this method degrades the device performance (Hunt et al. 2010). Therefore, in the present work, an alternative method based on an oxygen plasma surface treatment is used. Specifically, the devices are exposed to the O2 plasma at 120 watts, 200 mTorr for 5 minutes at 30 sccm. Subsequently, the epoxide coupling reagent, 3′glycidoxypropyltrimethoxysilane (GPTMS), is deposited onto the surface by vapor deposition for 30 minutes. The 5′hmC rabbit polyclonal antibodies were diluted in 1X Phosphate buffer (pH 7.4) to a final concentration of 50μg/ml. The antibody solution was applied as a 5μl microdroplet to the surface of the microtoroid and incubated for a minimum of 3 hrs. at 37° C with 50%–75% humidity (Cha et al. 2005; Hawk et al. 2013; Weetall 1993). The devices were then washed twice in 1x PBS pH 7.4 for 10 minutes, followed by one brief rinse in distilled water.
To verify the uniformity of the surface chemistry on the sensor surface and the bioactivity of the immobilized antibodies, fluorescent microscopy is performed using a Nikon Eclipse LV100D with a 50X objective lens. First, the 5′mC antibody is attached to the microtoroid devices, as explained above. This antibody is used instead of 5′hmC due to the availability of a fluorescently labeled analyte specific for this antibody. A 20mer oligonucleotide (IDT) with 20% methylated cytosine and a Cy5 fluorophore conjugated on the 5′ end is diluted to 1 μM in 1X PBS and used as the analyte.
The microtoroid with the immobilized antibody is immersed in the oligonucleotide solution for 30 min before washing twice with 1X PBS for 10 minutes followed with a quick rinse in double distilled water. As a control, a microtoroid device array underwent the same functionalization process without the silanization step.
In the imaging experiments, the Cy5 fluorophore is excited using a liquid light guide from a broadband lamp with the Chroma Cy5 Filter set (passband filters). Image parameters and fluorescence excitation wavelength are held constant to enable direct comparison of the fluorescence intensity between the control and the functionalized samples.
2.4 Device Characterization
To determine the quality factor (Q) of the toroidal cavity, the resonant linewidth (Δλ) is measured using a tunable laser centered at λ=765nm. Light from the laser is coupled into the cavity using the tapered optical fiber waveguide (Armani et al. 2005; Spillane et al. 2003). Tapered fibers have demonstrated very high efficiency (low loss) coupling from the visible through the near-IR in both air and water environments (Armani et al. 2005). The taper is aligned with the cavity using nanopositioning stages and a machine vision system. The signal is recorded on a computer using a high-speed digitizer/oscilloscope for subsequent data analysis. To ensure that non-linear effects did not distort the measurement, the resonant linewidths were measured at very low input powers in the under-coupled regime. Additionally, the scan rate and scan frequency of the laser is optimized to minimize thermal effects. The measured linewidths are fit to a Lorentzian, and, using the expression for Q (Q=λ/Δλ), the loaded Q of the cavity is determined. All reported Q values are for functionalized devices immersed in 1x PBS. A diagram of the sensor characterization set-up is included in the supplemental material.
2.5 Biodetection Experiments
To perform the biosensing experiments, the functionalized optical resonator is placed in a sample chamber containing ~100μL of 1x PBS buffer and a tapered fiber waveguide. By changing the relative position between the taper and the resonator, the amount of the power coupled into the device is increased, approaching 100% power coupled into the device or critical coupling. Using a syringe pump, the oligonucleotide solution is controllably injected into the chamber at 50 μL/min. To thoroughly characterize the device response, a range of solution concentrations spanning 1x10−15M to 1x10−9M are used in these experiments. The wavelength position is continuously recorded and analyzed by a LabView software program.
Additionally, a pair of control experiments is performed: 1) the signal in the absence of fluid flow and 2) the signal when only buffer is injected. The second control experiment is used as the baseline background noise level and is plotted as “buffer” in the subsequent graphs. The error reported is the 3-sigma (3σ) error value and is determined by analyzing >500 data points at the final shift location in the presence of fluid flow. Therefore, this error takes into account all possible sources of noise in the system.
2.6 Preparation of antibodies and oligonucleotides
Goat anti-rabbit polyclonal antibodies specific for 5′hydroxymethycytosine are diluted to a final concentration of 50 μg/ml in 1x PBS before they are immobilized onto the microtoroid sensors. For analyte detection, a series of solutions containing the double stranded oligonucleotides in 1x PBS buffer are made. The concentration of the oligonucleotide ranges from 1fM to 1nM.
2.7 Antibody sensitivity analysis by dot blot
To prepare the nitrocellulose paper for the dot blot analysis, it is pre-soaked in 6x SSC. Then, 10ng of each of the three oligonucleotides are blotted onto the paper using a vacuum pump followed by UV crosslinking for 5 minutes. Before the oligonucleotides are blotted, they are thermally denatured by heating for 5 minutes at 95° and then cooling on ice. After a 10 minute wash in TBST (20 mM Tris-HCL, 150 mM NaCL, 0.5% Tween20), the membrane is soaked in a prehybridization solution (TBST, 0.1% BSA) for 30 minutes, followed by a 4 hr. incubation with anti-rabbit HRP linked 5′hmC antibodies at a 1:500 dilution at room temperature. After three 15 min washes in TBST, the membrane is incubated with a secondary antibody specific for HRP at 1:5000 dilution and allowed to incubate at room temperature for 45 minutes followed by three washes with 1X TBS for 15 minutes each. The blots are treated with Bio-Rad ECL reagents for 1 minute and then analyzed by Chemiluminescence Imager (Bio-Rad) to detect the bands.
3. Results and discussion
3.1 Verification of the Epoxy Chemistry
The bright field image (Fig. 4a) shows that our biosensing devices are not damaged during the functionalization process. In Figure 4(b), the fluorescent intensity around the toroidal cavity is uniform, indicating that the oligonucleotide-antibody complex is bound uniformly on the surface of the microtoroid and that there are minimal vacancies in the surface chemistry.
Figure 4.
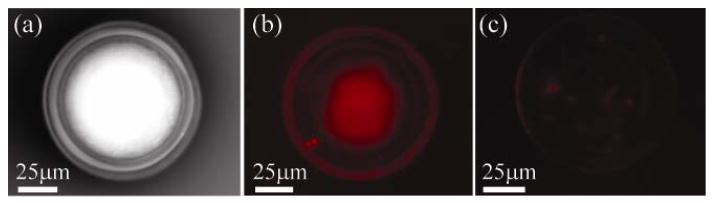
Brightfield and fluorescent microscopy is used to verify the attachment and uniformity of antibodies to the microtoroid sensors. (a) Bright field image of a microtoroid with 5′methylcytosine polyclonal antibody immobilized on the surface. (b) Fluorescent image of the same microtoroid shown in part (a), after incubation with the Cy5-labeled 20mer oligonucleotide. (c) Fluorescent image of a control biosensor in which the GPTMS reaction step was left out of the procedure. There is minimal attachment of the oligonucleotide without the epoxy-silane linker.
As a control experiment, the GPTMS step is left out of the bioconjugation process. As can be observed in Fig. 4(c), the fluorescent image changes considerably when the epoxide group is not present. Most notably, the overall fluorescent signal decreases significantly, as the only fluorescence which is present arises from non-specific binding to the silica surface.
These results confirm an epoxide-based antibody attachment strategy, which leaves the bioactivity of the antibody intact. It is also important to note that the oligonucleotide analyte can efficiently bind to the antibodies at room temperature.
3.2 Device Characterization
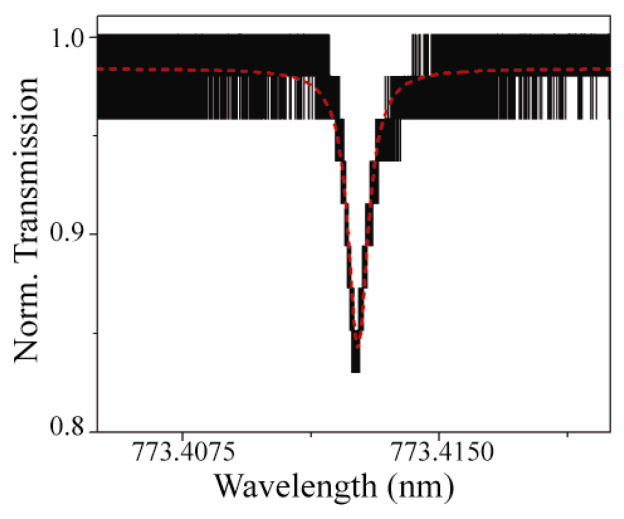
Figure 5 shows a normalized linewidth measurement of the functionalized device at 778nm in buffer. The loaded quality factor is 2.5x105. In this measurement, approximately 18% of the optical power is coupled into the device. The Q is decreased from above 108 to 105 because of the moderate optical loss of water at 780nm and the device degradation that occurs during the surface functionalization process.
Figure 5.
The graph displays a resonance spectrum that is fitted to a Lorentzian curve based on the full width half maximum obtained in 1X PBS. The Quality factor is 2.5 x 105.
3.3 Characterization of functionalized microtoroids
A summary of all results is shown in Figure 6a, and three key transition concentrations are highlighted in the bar graph in Figure 6b. From Figure 6a, it is immediately apparent that these devices are able to differentiate 5′hmC from both C and 5′mC, across a wide range of solution concentrations. This ability is derived from both the high specificity of the antibodies and the high sensitivity of the optical sensors. However, from these results, a more in depth analysis of the sensor performance is also possible. Namely, the limit of detection (related to signal to noise and specificity), and linear working range can be determined.
Figure 6.
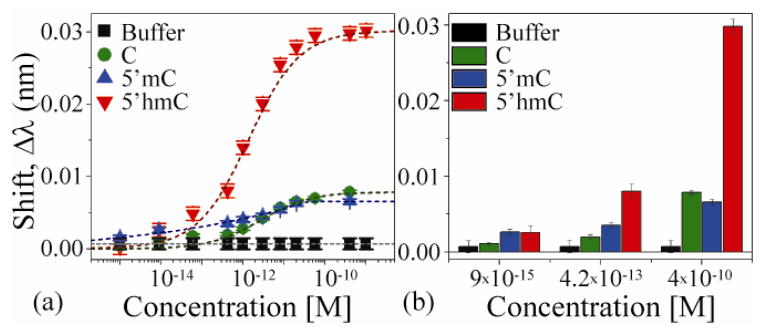
Biosensing analysis of unmethylated methylated, and hydroxymethylated cytosine within a DNA sequence. (a) The working range of the optical sensor, and (b) the detection results at three distinct concentrations: below detection threshold (9x10−15M), at the limit of detection (4.2x10−13), and at device saturation (4x10−10M).
The limit of detection (LOD) is the smallest quantity of an analyte which can be reliably detected. These values are typically determined by comparing sensing data either to a blank (buffer) solution or to a competing molecule. However, clearly, the latter approach is a better representation of the sensor’s behavior in a real-world scenario. Comparing the signals from the 5′mC and the 5′hmC, the experimentally demonstrated limit of detection is approximately 4.2x10−13M. At this concentration, the signal, including the error in the signal, from the 5′hmC is twice that of the 5′mC.
As expected, as the concentration of the 5′hmC increases, the detection signal increases. However, at high concentrations, the signal from the non-specific binders (C and 5′mC) also increases. This increase could be the result of non-specific interactions with the binding site or with the device surface. However, the signals from these false-positive events are always significantly lower than the true-positive signals.
The linear working range of a sensor is an equally important metric, particularly for a device with applications in diagnostics. From Figure 6a, the linear working range of the device is nearly two orders of magnitude (3x10−13M to 1x10−11M).
As already discussed, the most similar biodetection technique is the enzyme linked immunosorbant assay. Currently, the limit of detection for ELISA-based methods is between 10ng–100ng (Fraga et al., 2002; Tellez et al., 2014). Based on the sample volumes used in the present work, the amount of DNA needed to generate a reliable signal using resonant cavities is 0.46ng. This quantity is over an order of magnitude smaller than ELISA. It is also important to note that ELISA requires a secondary fluorophore or signal amplification (e.g. horse radish peroxidase) which increases the time and cost of the method. The present approach is label-free which both accelerates the time to decision and reduces the number of reagents needed.
4. Conclusion
In conclusion, we have demonstrated PCR-free detection and differentiation of 5′hmC at sub-pM concentrations using optical resonant cavities in combination with 5′hmC specific antibodies. The specificity for 5′hmC over chemically similar variants, 5′mC and C, was extremely good at both low and high concentrations. The sensor system also demonstrated a nearly two order of magnitude linear working range. This research involved advances in surface chemistry and in sensor design to accomplish this combination of sensitivity and specificity. This type of assay is not only promising in expanding the scope of research due to the ability to monitor biomarkers at minute levels in real-time, it also displays progress towards developing a lab on a chip device capable of early diagnosis and prognosis of cancer and other debilitating diseases.
Supplementary Material
Highlights.
Detection and differentiation of two structural methylated cytosine analogs, 5′mC and 5′hmC, at sub-pM concentration levels
PCR-free detection based on a label-free, real-time integrated optical sensor.
Epoxy-silane based antibody bioconjugation method developed to enable selective detection
Specificity greater than 3:1 at nM concentrations.
Acknowledgments
The authors would like to thank Kelvin Kuo, Ashley Maker, and Soheil Soltani (University of Southern California) for the toroidal cavity fabrication. We would also like to thank Simin Mehrabani (University of Southern Caifornia) for the SEM image of the microtoroid. This work was supported by the National Institutes of Health through NIH Director’s New Innovator Award Program (1DP2OD007391-01), a NIH Diversity Supplement, and the Congressionally Directed Medical Research Program (W81XWH-10-1-0406).
Appendix A. Supporting Information
Supplementary data associated with the verification of the antibody activity and the oligonucleotide sequence can be found in the online version at http//XXX.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armani AM, Armani DK, Min B, Vahala KJ, Spillane SM. Ultra-high-Q microcavity operation in H2O and D2O. Applied Physics Letters. 2005;87(15):151118. [Google Scholar]
- Armani DK, Kippenberg TJ, Spillane SM, Vahala KJ. Ultra-high-Q toroid microcavity on a chip. Nature. 2003;421(6926):925–928. doi: 10.1038/nature01371. [DOI] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes & Development. 2009;23(7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Development. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Borgel J, Guibert S, Weber M. Methylated DNA Immunoprecipitation (MeDIP) from Low Amounts of Cells. Springer; New York: 2012. [DOI] [PubMed] [Google Scholar]
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nature Reviews Genetics. 2012;13(1):7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- Cadet J, Wagner JR. TET enzymatic oxidation of 5-methylcytosine, 5-hydroxymethylcytosine and 5-formylcytosine. Mutation Research-Genetic Toxicology and Environmental Mutagenesis. 2014;764:18–35. doi: 10.1016/j.mrgentox.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Cha T, Guo A, Zhu XY. Enzymatic activity on a chip: The critical role of protein orientation. Proteomics. 2005;5(2):416–419. doi: 10.1002/pmic.200400948. [DOI] [PubMed] [Google Scholar]
- Chen ZX, Riggs AD. Maintenance and regulation of DNA methylation patterns in mammals. Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire. 2005;83(4):438–448. doi: 10.1139/o05-138. [DOI] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nature Reviews Genetics. 2007;8(4):286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–U589. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Fraga M, Esteller M. DNA Methylation: A Profile of Methods and Applications. Biotechniques. 2002;33(3):632–649. doi: 10.2144/02333rv01. [DOI] [PubMed] [Google Scholar]
- Hawk RM, Chistiakova MV, Armani AM. Monitoring DNA hybridization using optical microcavities. Optics Letters. 2013;38(22):4690–4693. doi: 10.1364/OL.38.004690. [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 6-hydroxymethylcytosine in bisulfie sequencing. PLoS One. 2010;5(1):e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt HK, Armani AM. Label-Free Biological and Chemical Sensors. Nanoscale. 2010;2(9):1544–1559. doi: 10.1039/c0nr00201a. [DOI] [PubMed] [Google Scholar]
- Hunt HK, Armani AM. Bioconjugation strategies for label-free optical microcavity sensors. IEEE Journal of Selected Topics in Quantum Electronics. 2014;20(2):6900213. [Google Scholar]
- Hunt HK, Soteropulos C, Armani AM. Bioconjugation strategies for microtoroidal optical resonator. Sensors. 2010;10(10):9317–9336. doi: 10.3390/s101009317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt JT, Bailey RC. Biomolecular analysis with microring resonators: applications in multiplexed diagnostics and interaction screening. Current Opinion in Chemical Biology. 2013;17(5):818–826. doi: 10.1016/j.cbpa.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt JT, Luchansky MS, Qavi AJ, Lee SH, Bailey RC. Subpicogram Per Milliliter Detection of Interleukins Using Silicon Photonic Microring Resonators and an Enzymatic Signal Enhancement Strategy. Analytical Chemistry. 2013;85(22):10653–10657. doi: 10.1021/ac402972d. [DOI] [PubMed] [Google Scholar]
- Li Y, O’Neill C. 5 ′-methylcytosine and 5 ′-hydroxymethylcytosine Each Provide Epigenetic Information to the Mouse Zygote. Plos One. 2013;8(5) doi: 10.1371/journal.pone.0063689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani C, Madzo J, Moen E, Yesilkanal A, Godley L. Alterations of 5-Hydroxymethylcytosine in Human Cancers. Cancers. 2013;5(3):786–814. doi: 10.3390/cancers5030786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsko AB, Ilchenko VS. Optical Resonators with Whispering-Gallery Modes-Part I: Basics. IEEE Journal of Selected Topics in Quantum Electronics. 2006;12(1):3–14. [Google Scholar]
- Nestor C, Ruzov A, Meehan R, Dunican D. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 2010;48(4):317–319. doi: 10.2144/000113403. [DOI] [PubMed] [Google Scholar]
- Olkhov-Mitsel E, Bapat B. Strategies for discovery and validation of methylated and hydroxymethylated DNA biomarkers. Cancer Medicine. 2012;1(2):237–260. doi: 10.1002/cam4.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A, Esteller M. Epigenetic modifications and human disease. Nature Biotechnology. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128(4):655–668. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Riggs AD. X chromosome inactivation, differentiation, and DNA methylation revisited, with a tribute to Susumu Ohno. Cytogenetic and Genome Research. 2002;99(1–4):17–24. doi: 10.1159/000071569. [DOI] [PubMed] [Google Scholar]
- Shao LB, Jiang XF, Yu XC, Li BB, Clements WR, Vollmer F, Wang W, Xiao YF, Gong QH. Detection of Single Nanoparticles and Lentiviruses Using Microcavity Resonance Broadening. Advanced Materials. 2013;25(39):5616. doi: 10.1002/adma201302572. [DOI] [PubMed] [Google Scholar]
- Soteropulos CE, Hunt HK, Armani AM. Determination of binding kinetics using whispering gallery mode microcavities. Applied Physics Letters. 2011;99(10) doi: 10.1063/1.3634023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane SM, Kippenberg TJ, Painter OJ, Vahala KJ. Ideality in a fiber-taper-coupled microresonator system for application to cavity quantum electrodynamics. Physical Review Letters. 2003;91(4):043902. doi: 10.1103/PhysRevLett.91.043902. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128(4):747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Szulwach KE, CH, Xuekun Li, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, Vasanthakumar A, Godley LA, Chang Q, Cheng X, He C, Jin P. 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature Neuroscience. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez-Plaza M, Tang WY, Shang Y, Umans JG, Francesconi KA, Goessler W, Ledesma M, Leon M, Laclaustra M, Pollak J, Guallar E, Cole SA, Fallin MD, Navas-Acien A. Association of Global DNA Methylation and Global DNA Hydroxymethylation with Metals and other Exposures in Human Blood DNA Samples. Environ Health Perspect. 2014;122(9):946–954. doi: 10.1289/ehp.1306674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu KL, Pikor LA, Kennett JY, Alvarez CE, Lam WL. Methylation Analysis by DNA Immunoprecipitation. Journal of Cellular Physiology. 2010;222(3):522–531. doi: 10.1002/jcp.22009. [DOI] [PubMed] [Google Scholar]
- Vollmer F, Yang L. Label-free detection with high-Q microcavities: a review of biosensing mechanisms for integrated devices. Nanophotonics. 2012;1(3–4):267–291. doi: 10.1515/nanoph-2012-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetall HH. Preparation of immobilized proteins covalently coupled through silane coupling agents to inorganic supports. Applied Biochemistry and Biotechnology. 1993;41(3):157–188. doi: 10.1007/BF02916421. [DOI] [PubMed] [Google Scholar]
- Wu YQ, Zhang DY, Yin P, Vollmer F. Ultraspecific and Highly Sensitive Nucleic Acid Detection by Integrating a DNA Catalytic Network with a Label-Free Microcavity. Small. 2014;10(10):2067–2076. doi: 10.1002/smll.201303558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.