Abstract
Objective
Automatic monitoring of Adverse Drug Reactions (ADRs), defined as adverse patient outcomes caused by medications, is a challenging research problem that is currently receiving significant attention from the medical informatics community. In recent years, user-posted data on social media, primarily due to its sheer volume, has become a useful resource for ADR monitoring. Research using social media data has progressed using various data sources and techniques, making it difficult to compare distinct systems and their performances. In this paper, we perform a methodical review to characterize the different approaches to ADR detection/extraction from social media, and their applicability to pharmacovigilance. In addition, we present a potential systematic pathway to ADR monitoring from social media.
Methods
We identified studies, describing approaches for ADR detection from social media from the Medline, Embase, Scopus and Web of Science databases, and the Google Scholar search engine. Studies that met our inclusion criteria were those that attempted to utilize ADR information posted by users on any publicly available social media platform. We categorized the studies into various dimensions such as primary ADR detection approach, size of data, source(s), availability, evaluation criteria, and so on.
Results
Twenty-two studies met our inclusion criteria, with fifteen (68.2%) published within the last two years. The survey revealed a clear trend towards the usage of annotated data with eleven of the fifteen (73.3%) studies published in the last two years relying on expert annotations. However, publicly available annotated data is still scarce, and we found only six (27.3%) studies that made the annotations used publicly available, making system performance comparisons difficult. In terms of algorithms, supervised classification techniques to detect posts containing ADR mentions, and lexicon-based approaches for extraction of ADR mentions from texts have been the most popular.
Conclusion
Our review suggests that interest in the utilization of the vast amounts of available social media data for ADR monitoring is increasing with time. In terms of sources, both health-related and general social media data have been used for ADR detection— while health-related sources tend to contain higher proportions of relevant data, the volume of data from general social media websites is significantly higher. There is still very limited publicly available annotated data available, and, as indicated by the promising results obtained by recent supervised learning approaches, there is a strong need to make such data available to the research community.
Keywords: Adverse drug reaction, pharmacovigilance, social media
Background
Harmful reactions that are caused by the intake of medication are known as Adverse Drug Reactions (ADRs). Early detection of ADRs associated with drugs in their post-approval periods is a crucial challenge for pharmacovigilance research. Pharmacovigilance is defined as “the science and activities relating to the detection, assessment, understanding and prevention of adverse effects or any other drug problem” [1]. The process of pharmacovigilance begins during the pre-approval clinical trials conducted for a drug and continues after the drug is released into the market. Due to the various limitations of pre-approval clinical trials, it is not possible to fully assess the consequences of the use of a particular drug before it is released [2]. Recent studies have shown that adverse reactions caused by drugs following their release into the market is a major public health problem: with deaths and hospitalizations numbering in millions (up to 5% hospital admissions, 28% emergency visits, and 5% hospital deaths), and associated costs of about seventy-five billion dollars annually [3, 4, 5]. Thus, post-marketing surveillance of drugs is of paramount importance for drug manufacturers, national bodies such as the U.S. Food and Drug Administration (FDA), and international organizations such as the World Health Organization (WHO) [6].
Government agencies, like the FDA or the European Medicines Agency (EMA), have expanded their pharmacovigilance efforts in various ways to reduce the costs associated with ADRs. In the U.S., post marketing surveillance of drugs occurs actively and passively once they have been approved by the FDA. Methods to accomplish this include Phase IV studies, in addition to voluntary and mandatory reporting through the FDA’s Adverse Event Reporting System (FAERS), MedWatch, and the Institute of Safe Medication Practices’ Medication Error Reporting System (MERP) [7]. The MedWatch program, for example, allows the public (patients and providers) to report ADRs which they suspect or observe [8]. While it is mandatory for manufacturers to report adverse events, reporting by healthcare professionals and the public is voluntary. Due to the voluntary nature of these systems, reporting and detection of adverse events may not be timely and is incomplete. Recent research has exposed the various inadequacies of spontaneous reporting systems, prompting researchers to explore additional sources for ADR monitoring [9, 2, 10]. These systems, for example, suffer from under-reporting, over-reporting of known ADRs, incomplete data, duplicated reporting, and unspecified causal links. Various additional techniques have been utilized for post-marketing monitoring of ADRs, including retrospective chart analysis, prospective surveillance, and information extraction from electronic health records, clinical narratives and case reports. These approaches have their own associated challenges. For example, electronic health records generally face challenges associated with the pervasiveness of confounding variables, and the definition and ascertainment of exposures and outcomes [2]. Clinical narratives present the problem of limited access, as typically, only researchers a liated with medical centers can access the data. The rapid growth of electronically available health related information, and the ability to process large volumes of them automatically, using natural language processing (NLP) and machine learning algorithms, have opened new opportunities for pharmacovigilance that could address some of the above-mentioned limitations.
Relevance of social media to public health
One domain where data has grown by massive proportions in recent years, and continues to grow, is social media. Social networks have seen an unprecedented growth in terms of users worldwide (e.g., as of 11th July 2014, Twitter has over 645,750,000 users and grows by an estimated 135,000 users every day, generating 9,100 tweets per second2). The social networks form a platform for people to share, discuss, and update their views and opinions. The Pew Research Center’s survey [11] has elucidated the relevance of social media in modern day public health, explaining that 34% of caregivers and 20% of patients read or watch someone else’s commentary or experience online. Additionally, 11% of caregivers and 6% of patients share experiences and post questions online. A large population of patients are actively involved in sharing and posting health related information in social media and particularly health social networks [12]. Health related social networks, those focusing specifically on issues related to health, have been attracting many users. In such platforms (e.g., DailyStrength [13], MedHelp [14]), users discuss their health-related experiences, including use of prescription drugs, side effects and treatments. Users tend to share their views with others facing similar problems/results, which makes such social networks unique and robust sources of information about health, drugs and treatments. Due to the emergence of such platforms, and the abundance of data available through them, research on public health monitoring, including ADR monitoring, has focused on exploiting data from these sources in recent times [15, 16, 17].
In terms of monitoring public health, this has included studying smoking cessation patterns on Facebook [18], identifying user social circles with common medical experiences (like drug abuse) [19], and monitoring malpractice [20]. When different patients that suffer from a common disease, or use a specific medication, share information about their symptoms, treatments or drug outcomes, this volume of health information can provide valuable clinical insights for both patients and health-related industries that are beyond what could be achieved by traditional communication methods [21]. Infectious/viral disease monitoring, specifically, can benefit strongly from utilizing social media. For example, traditional systems may miss new or rare events (like a new viral strain), and lack the real time capabilities and demographics that social media would provide, including data from people that do not access healthcare through formal channels [22]. Although specific information (e.g., age and gender) about a single user may not be usable for privacy reasons, various resources are currently available to perform demographic analysis using social media data3. Furthermore, since 1994, a number of social media based surveillance systems have been developed, reviewed, and implemented. These have been implemented locally, nationally, and globally [23]. Recent advances in ADR monitoring have seen significant strides towards the use of automatic NLP techniques for mining drugs and associated reactions from social media. User posts in social media contain information about treatment outcomes or provide early access to reported ADRs that can be beneficial for health and pharmaceutical industries. The type and volume of ADR information that social media makes available to the health industry may not be easily accessible by other means. This includes the ADRs experienced by those with special conditions, such as patients with rare diseases, pregnant/nursing women, elderly people or patients with comorbidities who are usually excluded from clinical trials [24].
Social media from the perspective of regulatory authorities and the industry
From the perspective of regulatory authorities, the intent of mining social media is to obtain additional data from the general public that may be used to supplement existing voluntary systems. For example, the Association of the British Pharmaceutical Industry (ABPI) published a set of guidance notes in 2013, which help researchers and stakeholders manage ADR complaints on digital media [25]. Though the document was created for information purposes only, rather than regulatory/legal advice, it offers instructions on how to handle such ADR reports. It clearly defines a minimal information set needed to report the ADR, which includes an identifiable patient, suspect drug, adverse event, and identifiable reporter. The contact details required for the identifiable reporter are compatible with the social media domain, and include emails or screen names. It also states that this information should be collected “if possible,” which leaves room for incomplete data [25]. The U.S. FDA has not published explicit guidelines for social media based pharmacovigilance, but it has issued regulations for publishing promotional material [26] and risk/benefit information [27] on social media. Despite the absence of a formal guidance, ADRs from social media can still be reported to the FDA. The minimum data set for an ADR report is the same as that for the ABPI. Moreover, a recent FDA presentation stated that social media ADR reports are reviewed like any other spontaneous reporting systems, while acknowledging variability in the quality of the reports submitted [28].
In addition to regulatory authorities, signals identified through social media could be used by pharmaceutical manufacturers, the healthcare system, or healthcare researchers to fulfill requirements of mandatory reporting. While the intent of social media mining is to provide early signals, it could potentially be used by the interested parties to validate or reject signals that have arisen in other reporting systems. Pharmaceutical manufacturers, such as AstraZeneca, have considered the use of social media from an industry perspective: focusing on manufacturers’ responsibilities to provide accurate and quality information regarding drugs [29]. Because regulatory authorities and pharmaceutical manufacturers play a role in public safety, both may utilize social media to fulfill the safety mission.
Pros and cons of social media monitoring
Various pros and cons of using social media for automatic ADR monitoring [30, 31], and more generally, for public health monitoring, have been mentioned in recent literature— a full discussion of which is outside the scope of this paper. In this subsection, we briefly outline the opportunities that social media presents, and the obstacles associated with its use for health-related research.
As already mentioned earlier in this section, the size and growth of data on social media is unparalleled. Recent advances in the data processing capabilities of machines, and machine learning and NLP research present the possibility of utilizing this massive data source for a variety of purposes, including public health. The fact that it is a direct source of users’ personal experiences makes it a lucrative resource. According to Harpaz et al. [32], social media offers new opportunities for public health monitoring due to the availability of large amounts of data that is internet-based, patient-generated, unsolicited, and up-to-date. The use of social media for health-related and other tasks is, however, not without drawbacks and difficulties. The drawbacks found when utilizing the user generated content of social media may include issues with the credibility, recency, uniqueness, frequency, and salience of the data [33]. Abbasi and Adjeroh [33] demonstrate the potential downside of each of these five points and the importance of selecting the right media channel for social media analytics. For example, the potential salience of Twitter with its short text limits. In addition to these general problems related to the data generated within social media, there are difficulties and challenges posed by the processing and extraction of relevant information using NLP techniques. A frequently encountered challenge is due to the fact that the data is generated by consumers, and as such they tend to use non-medical, descriptive terms to discuss health issues. This reduces a system’s ability to extract the terms automatically from text [17, 34, 35]. Traditional methods that are used on longer texts have proven to be inadequate when applied to short texts, such as those found in Twitter [36].
From the perspective of pharmacovigilance and NLP specifically, existing literature presents the various obstacles associated with the use of social media. First of all, user posts on social media contain colloquial language and also misspellings. Especially when using lexicon-based approaches, these present problems as the accuracy of direct matches decreases. Furthermore, colloquial and informal language is more difficult to parse and tokenize, and thus recent research tasks have focused on developing NLP tools specifically for data from social media [37]. Secondly, some recent articles (e.g., [38, 39, 40, 41]) have reported the imbalance that exists in data coming from social media. Only a small proportion of drug-associated data collected from social media tend to contain information associated with ADRs. This results in problems associated with annotations, since large volumes of data need to be annotated for the inclusion of su cient numbers of posts containing ADRs. This data imbalance issue is a major problem for supervised machine learning approaches, particularly because it is the smaller class that is of primary interest for the research. Thirdly, while access to users’ personal experiences with drugs is one of the key advantages of social media, automatic determination of personal and non-personal experiences is extremely challenging. In addition to these, there are also technical, policy, and privacy challenges associated with the use of social media for pharmacovigilance, as pointed out by Edwards and Lindquist [30].
In this paper, we present a methodological review, which we conducted on studies that attempt to overcome some of the obstacles, and detect/extract ADRs from social media data using NLP-based techniques. The primary intent of this paper is to categorize the studies across various dimensions such as primary aim, technique/algorithm, size of data, availability of data, source and so on. Despite the recent flurry of work, there is no established evaluation framework for ADR detection. Neither is there any resource that contains the common information from various research efforts and attempts to unify them across various aspects. Thus, we believe that a review, such as this, will provide the necessary information to drive the development and evaluation of future approaches. The rest of the paper is organized as follows: the Methods section discusses the data search, selection and abstraction approaches for the survey; the Results section elaborates the various findings of the survey, including summaries of all the studies that met the inclusion criteria; finally, the Discussion section summarizes the main findings from the Results section, and concludes the paper by proposing a possible pipeline for the development and evaluation of social media based ADR monitoring systems using publicly available resources.
Methods
Data search and selection
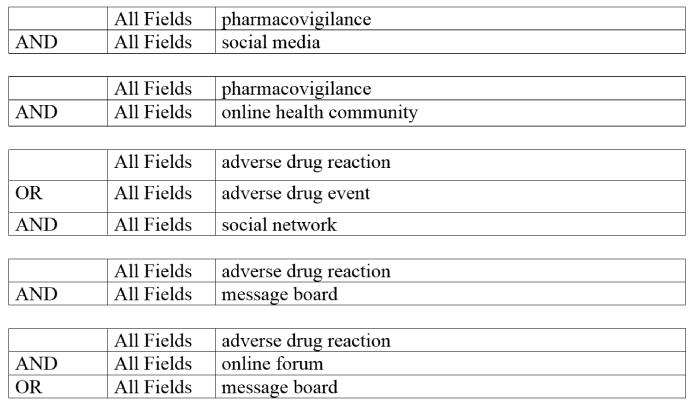
Pharmacovigilance using electronic data is a relatively recent research topic, and the use of social media data has only started receiving significant research attention in the last few years. As such, when collecting data, we searched for articles published in the last ten years only. We searched the databases Medline and Embase, and also the citation databases Scopus and Web of Science. We obtained relevant citations from the Medline and Embase databases by using the advanced search options. When searching, besides enforcing the constraint associated with the year of publication, we added several keyword-based constraints. To summarize, we attempted to obtain publications that contain indications of ADR detection or Pharmacovigilance AND social networks, social media, online forums, online health communities or message boards. Figure 1 presents some example search queries that we used for searching Medline (using the PubMed4 interface). Since ADR detection from social media generally involves the use of natural language processing (NLP), computational linguistics or text mining techniques, we suspected that there might be publications that are more computer science oriented rather than medical informatics. Thus, the publication venues for such articles may not be indexed in Medline. We, therefore, searched Google Scholar using the same keywords to identify publications that may not have been indexed in the more medical focused databases such as Medline.
Figure 1.
Sample search queries used for article retrieval from Medline.
For all the search engines, we sorted our search results by relevance. We filtered a total of thirty-nine publications for manual review and obtained their full texts. We added articles to this list if their titles or abstracts suggested that the investigators utilized data from social media for detecting ADRs or for monitoring drug safety in general. Studies that met our inclusion criteria were those that presented original data, utilized any internet-based resource (e.g., forums, message boards, social networks), and indicated the use of automatic algorithms for ADR detection (e.g., NLP techniques, and/or rule-based or machine learning approaches). In our initial shortlist, we included studies for which we could not determine if the data in the internet-based resource consisted of user posts, or if we could not immediately determine whether the detection algorithms and analyses were automatic or manual. Our exclusion criteria included studies that utilized data sources that emerged from the following information systems: laboratory, pharmacy, radiology, or administrative. Studies were also excluded if they focused exclusively on drug-drug interactions, detected ADRs in randomized controlled trials, drug labels, or were not published in English.
Data abstraction
For all included studies, we abstracted data on study characteristics including study size, research aim(s), primary ADR identification/extraction approach, data source, availability of data, and the type of evaluation performed. For the study size factor, we focused on two aspects— size of data and number of drugs. We also attempted to categorize if the study focused on a specific sub-domain of drugs (e.g., diabetes) or included a more general set of drugs. Classifying the primary identification/extraction technique was slightly more challenging because some articles describe the whole pipeline— from data collection to ADR detection. For these studies, we focused on the general approach that was employed at the final stage of detection. For example, we found that a number of techniques relied on ADR lexicons, while another set of techniques relied on detecting linguistic patterns for the ADR detection task.
For the data source factor, we categorized approaches based on the social network or type of social media from which the data was extracted. In terms of availability of data, we categorized studies based on whether the data used for the study were publicly available for research purposes or not. Furthermore, we also abstracted studies based on whether they utilized annotated data, which may be utilized for supervised machine learning, and is invariably more useful than unannotated data. Finally, for the evaluation criterion, we categorized articles based on the type of evaluation performed to assess performance. At a high-level, this included determining if the studies presented qualitative or quantitative evaluations. For quantitative evaluations, we further categorized on the specific evaluation approaches used.
Results
In this section, we provide details of our methodical survey of the literature. We first present a summary of the data collection process. Following that, we summarize our review of the selected literature using the criteria mentioned in the previous section. We elaborate on the studies in the Discussion section.
Data collection results
Our data search using the various search engines resulted in more than 1,500 citations, of which thirty-nine articles were retrieved and reviewed in full. The false positives consisted of a variety of topics including research on social media (e.g., trust and security), NLP approaches for social media mining, and pharmacovigilance studies focusing on non-social media data. We excluded articles for the various reasons mentioned in the Data search and selection subsection. Our final set consisted of twenty-two publications, which describe automatic methods for ADR detection from user posted data on social media. This set consists of journal articles, and conference and workshop proceedings. The earliest, pioneering work we identified was from 2010 [17], which employs a lexicon-based approach and manually annotated data for evaluation. Following this work, this research topic has received more attention with three publications in the years 2011 and 2012, four in 2013, and eleven in 2014.
Dimensions of characterization
We now present two tables summarizing some of the key information associated with the studies that we reviewed. In addition, we present some statistics and explanations regarding the contents shown in the table.
Table 1 summarizes crucial characteristics of the studies. In addition to the publication years, it shows the data sources, sub-domains of focus (if any), number of drugs involved in the studies, the sizes of the data used, and annotations and availability of the data. The table illustrates some key information regarding what pharmacovigilance research has covered over the years, and how research has evolved. The study by Leaman et al. [17] utilized data from the health related social network DailyStrength [13] and exploited expert annotated data. The number of drugs studied, however, is only six. Table 1 suggests that DailyStrength is a relatively popular source of health-related user posted data, and it is used by six studies in total. The table also suggests that early, exploratory research generally focused on a small number of drugs for ADR investigation. Prior to 2014, there is only one study that involved more than ten drugs for investigation. Very recent studies, tend to go beyond investigating ADRs associated with a small set of drugs, as depicted by the last few studies in the table. Furthermore, while some studies focused on specific domains of drugs (e.g., breast cancer, diabetes, etc.), most studies, particularly very recent ones, tend to concentrate on a range of drugs not specific to a domain.
Table 1.
Articles published on social media mining for ADR detection, their years of publication, data sources, domains of focus, numbers of drugs, size of data, and annotations and public availability of the data.
| Study | Year | Source | Sub-domain | # drugs |
# instances | Annotations/ Available |
|---|---|---|---|---|---|---|
| Leaman et al. [17] | 2010 | DailyStrength [13] |
- | 6 | 3,600 | Yes/No |
| Nikfarjam and Gonzalez [34] |
2011 | DailyStrength [13] |
- | 4 | 1,200 | Yes/No |
| Chee et al. [40] | 2011 | Yahoo! groups | - | 435 | 1,200,000 | No |
| Benton et al. [42] | 2011 | various forums5 | Breast cancer | 4 | 1,100,000 | No |
| Hadzi-puric and Grmusa [43] |
2012 | eight parenting forums |
Pediatrics | 9 | 1,290 | Yes/No |
| Yang et al. [44] | 2012 | MedHelp [14] | 5 ADRs | 10 | 6,2446 | No |
| Bian et al. [45] | 2012 | Cancer | 5 | 2,000,000,000 | No | |
| Liu and Chen [46] | 2013 | American Diabetes Association [47] |
Diabetes | - | 1,348,364 | No7 |
| Yang et al. [48] | 2013 | Yahoo! groups | - | 2 | 6,400 | Yes/No |
| Jiang and Zheng [49] |
2013 | - | 5 | 885 | Yes/No | |
| Yates and Goharian [50] |
2013 | AskAPatient [51], Drugs.com [52], DrugRatingZ [53] |
Breast cancer | 5 | 2,5008 | Yes/Yes |
| Yeleswarapu et al.9 [54] |
2014 | PatientsLikeMe [55], Daily Strength [13], MediGuard [56] |
- | 1210 | 13,50011 | No |
| Freifeld et al. [57] | 2014 | - | 23 | 60,000 | No | |
| Segura et al. [58] | 2014 | ForumClinic [59] |
- | 18712 | 400 | Yes/Yes |
| Ginn et al. [38] | 2014 | - | 74 | 10,822 | Yes/Yes | |
| Liu et al. [60] | 2014 | MedHelp [14] | Heart disease | - | 600 | Yes/No |
| Patki et al. [39] | 2014 | DailyStrength | - | 38 | 10,617 | Yes/No |
| O'Connor et al. [35] |
2014 | - | 54 | 1,873 | Yes/Yes | |
| Yang et al. [61] | 2014 | MedHelp [14] | - | 10, 5 ADRs |
- | No/No |
| Sampathkumar et al. [62] | 2014 |
Medications.com [63], Steady- Health [64] |
- | - | 2,000 | Yes/No |
| Sarker and Gonzalez 13 [41] |
2014 | Twitter, Daily Strength [13] |
- | 74 for Twitter, 56 for Daily Strength |
10,822 for Twitter, 10,617 for Daily Strength |
Yes/Yes |
| Nikfarjam et al. [65] |
2014 | Twitter, Daily Strength [13] |
- | 81 | 6,279 for Daily Strength, 1,784 for |
Yes/Yes |
In terms of data sizes, the studies presented in Table 1 can be divided into two important categories— large data sets without any expert annotations, and relatively small data sets which contain expert annotations. Among the twenty-two publications included in this review, fourteen (63.6%) utilized expert annotated data and eight did not. Among the fifteen papers published since 2013, eleven (73.3%) exploit annotated data. The table suggests that there is an increasing trend towards the use of annotated data for ADR detection. Some of the studies [40, 42, 45, 46] utilize very large volumes of data and derive statistics via unsupervised techniques. In contrast, studies that rely on annotated data, are capable of applying supervised approaches and also evaluation against goldstandards prepared by human experts. Perhaps as a consequence of the benefits of using annotated data, the recent efforts at creating annotated corpora have been executed. However, the public availability of annotated data is still a concern. We only found four data sets that have been made publicly available [50, 58, 38, 35, 41, 65], all of them published since 2013. The data set14 released by Yates and Goharian [50] contains only 247 posts containing ADRs. The data set15 released by Segura-Bedmar et al. [58] contains only 400 posts in Spanish. The latter data set, therefore, is unlikely to be suitable for future research tasks in English, but is the first of its kind in languages other than English. Both these data sets contain binary annotations only, and are also quite small, meaning that their use in supervised learning technique is likely to be minimal. The data set16 discussed in [38, 35] contains only binary annotations indicating whether a Twitter post contains an ADR or not, and includes over 7,000 instances (70% of the full set used in the study). While this data set, as published, is not suitable for supervised extraction of ADRs from text, it is suitable for training algorithms to detect ADR assertive text— a task that has already received attention within and outside of social media [66], and will be crucial to explore within this domain as well. In a more recent publication, span and concept normalization annotations for the same data set, containing over 1,500 instances, have been released to the public [65], and this data set can be utilized for ADR extraction tasks.
Table 2 provides a brief summary of the ADR detection/extraction approaches proposed by the studies, their primary research aims, and how the evaluations were designed. Note that in this context, ADR refers to adverse reactions only, as well as drug and adverse reaction pairs. The table follows on from the information provided in Table 1, and enables us to achieve an understanding of the success of different classes of approaches for ADR detection/extraction problems. The table illustrates that two of the most frequently addressed problems have been the detection of comments/sentences discussing ADRs, and the extraction of specific ADRs from sentences. This suggests that these two problems are perhaps the most important for systems attempting to propose end-to-end pharmacovigilance solutions. The evaluation approaches, however, vary more between systems. When annotated data is available, generally standard measures such as Recall, Precision and F-score are used. In the absence of annotated data, evaluation approaches and metrics tend to be varied. Later in this section, we discuss some of the evaluation methodologies mentioned in 2. For the readers, this will serve as a resource of compiled information for this task.
Table 2.
A summary table showing primary ADR detection approaches and evaluation methodologies.
| Study | Research Aim | Primary Approach(es) | Evaluation Methodology |
|---|---|---|---|
| Leaman et al. (2010) [17] |
Concept/relation extraction |
Lexicon-based (450 comments for system development). |
Quantitative. Against manually annotated data (3,150 instances). |
| Nikfarjam and Gonzalez (2011) [34] |
Concept/relation extraction |
Lexical pattern-matching (2,400 comments for pattern building). Association rule mining to identify patterns. |
Quantitative. Against manually annotated data (1,200 instances). |
| Chee et al. (2011) [40] |
Drug classification | Ensemble classification using drug categories as classes. |
Mixed. Classification results are combined to generate drug scores for 3 drugs, which are compared against scores for drugs (12) with known adverse effects. |
| Benton et al. (2011) [42] |
Concept/relation extraction |
Lexicon-based. Association rule mining to identify drug reaction pairs. |
Quantitative. Adverse reactions associated with drugs obtained from product labels and compared against system reported adverse events. |
| Hadzi-puric and Grmusa (2012) [43] |
Concept/relation extraction |
Lexicon-based approach for ADR detection. Statistical scoring for identifying drug relation associations. |
Mixed. Qualitative analysis of identified ADRs against known ADRs. Recall, precision and F-score computed for evaluation against annotated data. |
| Yang et al. (2012) [44] |
Concept/relation extraction |
Lexicon-based. Association rule mining to identify drug reaction pairs. |
Quantitative. FDA AERS used as the gold standard. Lift, Leverage, and Proportional Reporting Ratio used as metrics. |
| Bian et al. (2012) [45] |
ADR classification | Classification of tweets using Support Vector Machine (SVM) classifiers. Two class ifiers built: one to predict if a user has used a drug (based on the tweets), and the second to classify if a post contains an adverse effect. |
Mixed. Evaluation and training is performed on the same data. Only classification accuracies reported. Analysis describes the limitations introduced by noise in Twitter. |
| Liu and Chen (2013) [46] |
Concept/relation extraction |
Lexicon-based approach for ADR and drug detection. Shortest dependency path based machine learning algorithm for relation extraction. |
Quantitative. Separate evaluations for entity extraction, ADR detection and classification of patient experiences using 200 manually annotated comments. |
| Yang et al. (2013) [48] |
ADR Classification | A combination of supervised and unsupervised approaches for training binary classifiers. A mixture of syntactic, semantic, and sentiment features are used to train SVM and Naïve Bayes classifiers. |
Quantitative. Evaluation performed on 1,600 annotated instances. Evaluation demonstrates that the combination of supervised and unsupervised training performs significantly better than using supervised training only. |
| Jiang and Zheng (2013) [49] |
Concept/relation extraction and classification |
Supervised classification of tweets using a Maximum Entropy classifier trained on a data set of 600 tweets only. MetaMap [67] to identify drug and ADR categories. |
Mixed. 285 tweets for testing the classification accuracy. ADR extraction accuracy is evaluated against known adverse reactions. |
| Yates and Goharian (2013) [50] |
Concept/relation extraction |
Pattern-based. 7 patterns used for extracting ADRs from approximately 125 manually annotated comments. |
Quantitative. Against manually annotated data (125 instances). |
| Yeleswarapu et al. (2014) [54] |
Concept/relation extraction |
Lexicon-based. Prepared lexicon used for drug and ADR detection. Association rule mining and BCPNN used for identifying drug-symptom and drug-disease pairs. |
Qualitative. Evaluation is performed via comparative analysis with findings from previous studies. Primary conclusion of evaluation is that combining social media data with other sources such as medical literature and ADR databases can improve ADR detection performance. |
| Freifeld et al. (2014) [57] |
Concept/relation extraction |
Lexicon-based. A prepared lexicon is used to detect ADRs. Aggregated frequencies are used to compare drug-reaction pairs. |
Quantitative. Aggregated frequency of identified product-event pairs compared with data from AERS. Correlation between the two sources computed to assess the effectiveness of social media as a resource for ADR monitoring. |
| Segura et al. (2014) [58] |
Concept/relation extraction |
Lexicon-based. A prepared lexicon was used in a multilingual text analysis engine to detect drugs and ADRs in text. |
Quantitative. Against manually annotated data (400 instances). Drug and ADR detection evaluated separately. |
| Ginn et al. (2014) [38] |
Corpus presentation/ description. Supervised learning experiments to illustrate utility of corpus. |
Supervised classification of ADR assertive tweets using 10-fold cross validation over a large annotated data set of 10,822 tweets. Data set artificially balanced to lower ADR- noADR class imbalance. |
Quantitative. Evaluated against annotated data on the artificially balanced data set. |
| Liu et al. (2014) [60] |
Medical entity extraction, adverse event extraction, report source classification. |
Lexicon-based approach for entity extraction and ADR extraction. Rule-based approach for relation classification. |
Quantitative. Against manually annotated data (600). Same set of instances used for the tasks of events and treatments recognition, ADR identification, and patient report extraction. |
| Patki et al. (2014) [39] |
ADR/drug classification | Supervised classification of ADR assertive comments using SVMs and a rich set of features extracted via NLP techniques. Probabilities of all comments associated with each drug combined to predict if drug should be categorized as normal or blackbox. |
Mixed. Annotated data used for evaluating the classification task. Accuracy values used for evaluating drug categorization strategy. |
| O’Connor et al. (2014) [35] |
Concept/relation extraction |
Lexicon-based approach for detecting ADR mentions in Twitter data. Lexicon created by combining several existing ADR lexicons. |
Quantitative. Against manually annotated data (1,873 instances). |
| Yang et al. (2014) [61] |
Drug-ADR relation extraction |
Lexicon-based approach for detecting ADR mentions. Association rule mining to identify relationships between drugs and ADRs. |
Quantitative. Lift and Proportional Reporting Ratio for scoring association of ADRs with drugs. Recall, precision and F-measure used to compare the performance against three publicly available systems.17 |
| Sampathkumar et al. (2014) [62] |
Concept/relation extraction and relationship (causal) identification. |
Lexicon-based approach for detecting mentions of ADRs. Hidden Markov Model applied to detect relationship between drug-ADR pairs. |
Mixed. 10-fold cross validation against manually annotated data (2,000 instances). Extracted ADRs compared against drug package labels to verify performance and to identify unknown ADRs. |
| Sarker and Gonzalez (2014) [41] |
ADR classification. | Supervised classification to detect ADR assertive texts. Features incorporated from distinct research areas such as sentiment analysis, polarity classification and topic modeling. Multiple corpora combined to boost classification performance. |
Quantitative. F-score for the ADR class is computed against gold standard annotations. |
| Nikfarjam et al. (2014) [65] |
Concept/relation extraction |
Concept extraction is performed using supervised learning via conditional random fields (CRF). Word clusters, learnt from large unlabeled data, are used as features. |
Quantitative. Against manually annotated data (1,559 and 444 instances for two data sources). |
A Summary of methodologies and resources
Lexicons and knowledge bases
Our survey revealed that ADR lexicons and knowledge bases have been the most widely used resource for pharmacovigilance techniques from social media. These resources contain lists of ADR mentions, collected from various sources ranging from drug labels, clinical trials, caregivers, and even user posts on social media. Significant efforts have been made for the creation of new knowledge sources and the combination of existing ones. From the studies that utilized lexicons, we have compiled a list of resources containing ADR mentions, which is as follows:
FDA AERS.18. This is the FDA adverse event reporting system and database that is designed to support FDA’s post-marketing surveillance of drugs. Healthcare professionals and consumers are voluntarily capable of reporting adverse reactions. This database has been widely used for pharmacovigilance research, including those involving social media [42, 46, 54, 57, 60].
COSTART19 (Coding Symbols for a Thesaurus of Adverse Reaction Terms). This resource is used for coding, filing, and retrieving post-marketing ADRs. It is organized in a hierarchical structure. This resource contained a total of 3,787 ADRs, and has been superseded by MedDRA. Used by: [17, 34].
CHV20 (Consumer Health Vocabulary). This database was created as an initiative to map words and phrases representing ADRs from lay persons to technical terms used by health professionals. Since this resource contains terms and phrases used by non-experts, it has become a very useful for pharmacovigilance research using social media data [42, 46, 60, 61].
MedEffect.21 MedEffect Canada provides consumers, health professionals and patients with access to adverse drug reaction reporting, obtain safety information, and learn and better understand the importance of reporting side effects. The adverse reaction database contains reports of suspected adverse drug reactions. Used by: [17].
UMLS22 (Unified Medical Language System). UMLS is a broad metathesaurus containing a large collection of biomedical vocabulary. It categorizes medical terms into broad and fine-grained categories, and these categorizations have been used to detect mentions of ADRs [46].
MedDRA23 (Medical Dictionary for Regulatory Activities). This is a rich, highly specific, multilingual, standardized medical terminology to facilitate sharing of regulatory information internationally for medical products. It has also been a popular resource for building lexicons for ADR detection [40, 54, 57].
SIDER24 (Side Effect Resource). This is a knowledge base that uses MedDRA and contains ADR information on marketed medicines from public documents and package inserts. The resource contains a total of 4,192 ADRs associated with 996 drugs. Used by: [17, 34, 62].
Automatic classification of ADR containing user posts
A number of the studies we reviewed focus on the automatic classification of user posts to determine if ADRs are mentioned in the posts [40, 45, 48, 49, 38, 41]. The motivation for such classification approaches arises from the fact that most drug related posts on social media are not associated with ADRs, and thus, filtering out irrelevant posts is a crucial step in identifying ADRs. Such supervised classification approaches require manually annotated data, and large numbers of annotated posts are required to make reliable evaluations. The recent preparation of large, annotated data sets (e.g., the data set described in [38, 41]), will invariably be crucial future resources.
Research tasks have designed supervised classification efforts using broad categories of drugs as classes, and the comments associated with them for training (e.g., [40]). Very small training data have also been applied (e.g., [45]) with common machine learning algorithms such as Näive Bayes, Support Vector Machines and Maximum Entropy. One important challenge that has been constantly discussed in supervised learning tasks is the data imbalance in social media text [40, 38, 39]. The research by Ginn et al. [38], which is, to the best of our knowledge, the largest annotated data set from a generic social media website (in this case Twitter), suggests that only a very small amount of drug related posts contain ADRs (approximately 10%). Recent annotation work on health-related social networks [39] suggests that the proportion of ADR associated information in such networks is higher (approximately 20-25%). However, this imbalance is still a challenge from the perspective of machine learning, and this problem has been addressed in detail in very recent research [41]. In the mentioned study, the authors employed a number of strategies including the use of weighted classifiers, incorporation of features from other text classification problems, and the combination of multiple corpora for training.
ADR mention extraction
A majority of the papers that met our inclusion criteria focused on identifying specific ADR mentions from user posts and extracting them. Most of the approaches (54.5%) mentioned in Table 2 are lexicon-based, meaning that their primary technique is to identify ADRs using a list of precompiled ADR mentions [17, 42, 44, 46, 54, 57, 58, 35, 61, 62]. Considering the availability of several extensive ADR resources, applying lexicon-based NLP techniques can successfully identify a subset of the ADR mentions in user posts. However, pure lexiconbased approaches do not address some important challenges. Consumers do not always use technical terms found in the existing lexicons. Instead, they use creative phrases, descriptive symptom explanations, and idiomatic expressions. For example, the phrase ‘messed up my sleeping patterns’ was used to report the well-known ADR ‘sleep disturbance’ in the data set made available by [38, 35]. Even when a mention in a user sentence is matched with a lexicon term, it is not necessarily an adverse effect. The terms used to describe ADRs can also be used for indications, beneficial effects, or other mention types. Finally, the various properties of user generated text mentioned earlier (e.g., misspellings, abbreviations, and phrase construction irregularities) limit the performance of lexicon-based approaches.
In addition to extracting ADR mentions, some studies have focused on identifying the relations between ADRs and drugs. Following the study by Nikfarjam and Gonzalez [34], a popular approach for the discovery of drug-ADR pairs, in lexicon-based and other techniques, has been the use of association rule mining [68]— a class of techniques by which associations between entities are discovered. In general, following the identification of ADRs and drugs, association rule mining is used to identify if a drug and ADR pair is associated or not. Frequent occurrence of drug-ADR pair mentions in close proximity within user posts are considered to be indications of ADRs associated with the drugs, and these associations are detected by association rule mining in unannotated data.
While most approaches use lexicons for detecting drug and ADR mentions in text, some attempt to discover patterns in texts which are likely to be indicative of ADRs [34, 50]. An advantage of pattern-based approaches over lexicon-based approaches is that they are capable of detecting inexact matches. This is particularly useful for mining social media where users frequently use colloquial terms and the texts contain misspellings. The hypothesis behind using pattern-based approaches for social media mining is that, although users tend to use highly informal language, they tend to use some converging patterns, which can be used to detect ADR mentions. One of the main drawbacks of such approaches, however, is the need for very large amounts of data for the generation of patterns. With the generation of annotated data in recent times, supervised learning approaches are becoming increasingly popular, and they have also shown promising performances in quantitative evaluations [62, 65].
A summary of evaluation techniques and metrics
Our review of the approaches for ADR detection/extraction techniques from social media suggests that despite the increasing interest in this research area, a common evaluation approach that can be applied across systems is still absent. This is primarily due to the absence of common data sets which can be utilized for performing comparative evaluations of systems. As such, research tasks generally design their own evaluation approaches, and either propose new evaluation techniques or use existing evaluation techniques compatible with their proposed approaches. We now briefly discuss some of the evaluation approaches that have been applied. We group them into two broad categories: Qualitative and Quantitative.25
Qualitative evaluation
The end goal of ADR detection from social media sources is to be able to identify drugs that are either frequently related to ADRs or those that are associated with serious ADRs. Therefore, some past research has focused on devising strategies for computing scores for drugs, with known ADRs, based on various criteria, and perform the final evaluation in a qualitative manner. For example, Chee et al. [40] use an ensemble classification approach to classify drugs into two predetermined categories: watchlist and normal. Following this, a score is computed based on the number of times a drug is classified as watch-list, and the scores are compared to drugs already withdrawn from the market for associated ADRs. The final evaluation is qualitative, with a comparison of the scores obtained by withdrawn drugs and some watchlist drugs, which, according to the authors should be scrutinized. Similar qualitative discussions accompany quantitative analyses in [45, 49, 39]. Patki et al. [39] utilize supervised machine learning to classify comments associated with drugs belonging to two categories blackbox26 and normal. For evaluation, the authors combine the classification probabilities of comments associated with each drug and suggest that the combined probabilities may act as indicators for the detection of drugs containing important ADRs. The evaluation, however, is not fully quantitative, and primarily compares and discusses the reasons for misclassified drugs.
Quantitative evaluation
As already mentioned, comparing all the different systems that have been reviewed in this paper is not possible, since most research tasks utilized inhouse data that have not been made available to the research community. Most research tasks have been designed such that evaluations could be performed using existing metrics such as Recall, Precision, F-score and Accuracy [17, 34, 42, 43, 45, 46, 50, 57, 58, 38, 39, 41, 62, 65]. In the absence of manually annotated data, these metrics have been computed using various gold standards, such as known adverse reactions from FDA product labels [42] or databases [44, 57]. We found ten studies that used manually annotated data for the evaluation of the drug-ADR extraction task. Table 3 presents the results, and illustrates the difficulty of comparing the various systems because of the use of different data sets of varying sizes. Considering the small amount of annotated data based on which most of these systems were built, it is likely that the overall performances will improve as more annotated data become available. Other metrics for quantitative evaluations have also been used, though less frequently. They include: lift, leverage, proportional reporting ratio [44]; and matching rate [49].
Table 3.
Comparison of system recalls, precisions and F-scores when manually annotated data is used for evaluation.
| Study | Size | Recall | Precision | F-score |
|---|---|---|---|---|
|
| ||||
| Leaman et al. [17]* | 3,150 | 0.70 | 0.78 | 0.74 |
| Nikfarjam and Gonzalez* [34] | 1,200 | 0.66 | 0.70 | 0.68 |
| Hadzi-puric and Grmusa [43] | 990 | 0.65 | 0.75 | 0.70 |
| Liu and Chen [46] | 200 | 0.80 | 0.87 | 0.84 |
| Yates and Goharian [50] | 125 | 0.89 | 0.69 | 0.78 |
| Freifeld et al. [57] | 437 | 0.86 | 0.72 | 0.78 |
| Segura-Bedmar et al. [58] | 400 | 0.56 | 0.85 | 0.68 |
| O'Connor et al. [35] | 1,873 | 0.62 | 0.54 | 0.58 |
| Sampathkumar et al. [62] | 2,000 | 0.74 | 0.79 | 0.76 |
| Nikfarjam et al. (DailyStrength) [65] | 1,559 | 0.78 | 0.86 | 0.82 |
| Nikfarjam et al. (Twitter) [65] | 444 | 0368 | 0.77 | 0.72 |
indicates systems using the same (or subsets of the same) data set.
Discussion and conclusions
Our survey covers research efforts for automatic pharmacovigilance techniques from social media data. The review includes carefully selected articles, published over the last four years, starting with the pioneering work of Leaman et al. [17]. The studies included in the study show the growing attention that the utilization of social media data is receiving. Moreover, while early research tasks have been mostly exploratory, recent approaches have illustrated the need and interest for structured standardized approaches and annotated data. All but six studies in our sample used data that is publicly unavailable for system development and evaluation. As such, at this point, performing a direct comparison of existing detection/extraction approaches is impossible. At the same time, evaluations of systems have also progressed in various directions, without the development of any standard evaluation criteria. A transition in research methodologies is however clearly visible, as large annotated data sets are gradually becoming available.
Most extraction approaches relied on using lexicons for identifying/extracting ADR mentions in text, while pattern-matching-based approaches have also been applied. Lexicon-based approaches face specific obstacles when applied to social media data, whereas pattern-based methods require large amounts of data for system development. Only recently, there has been a trend in supervised learning approaches that attempt to utilize annotated data, and it is likely that comprehensive supervised classification approaches will be used more frequently in the near future.
A pathway for ADR detection and extraction from social media data
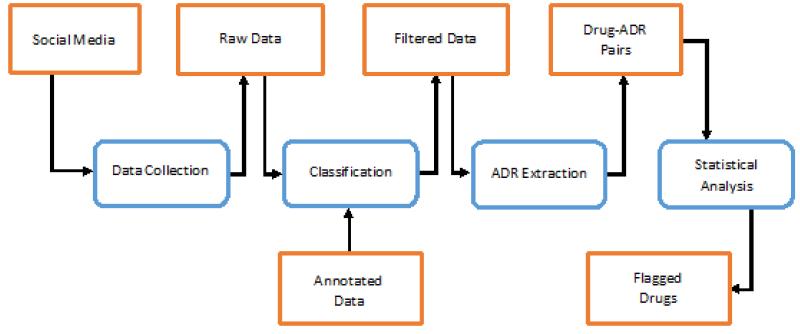
Building on this review, we propose a possible framework for future ADR detection efforts from social media. Considering the recent developments of annotated data and large-scale annotation efforts, much of future research will invariably attempt to utilize supervised learning approaches. In the proposed framework, we only referred to data that is publicly available for performing ADR detection from social media. Figure 2 presents a high level illustration of the framework.
Figure 2.
A pathway for ADR detection and extraction from social media data.
The first step in working with social media data is the collection of the data. All the papers discussed in this review perform data collection from various sources. For health related social networks, such as Dailystrength, the collection of relevant data is generally easy since the data is categorized according to various criteria (e.g., drug name). For generic social networks, such as Twitter, the collection problem is harder. It is possible to collect posts by using drug names as search keywords, but drug names are often misspelt by users. To address this problem recent research [38, 35] has utilized phonetic spelling filters to generate common misspellings for drug names [69]. These recent advances in NLP will aid future data collection processes.
Following data collection, the challenge is to filter data. As explained earlier, data imbalance is an important problem in ADR mining from social media text, which has resulted in various research tasks on classification of ADR assertive text [49, 48, 38, 39]. With the creation of recent publicly available corpora (e.g. [50, 38, 35, 58], learning algorithms can be trained and optimized to detect ADR assertive instances with high accuracies. Most classification research, however, have only used very basic linguistic features for classification (e.g., bag of words), and only very recent research has focused on exploring deep linguistic and semantic features and advanced machine learning techniques [41].
Effective filtering/classification techniques are likely to aid the process of ADR mention extraction by removing the majority of irrelevant information. We have discussed various ADR extraction approaches in the paper, the most popular being lexicon-based ones. Lexicon-based approaches have benefited from recent expansions and merging of existing lexicons, and the incorporation of colloquial terms. Recent release of publicly available annotated data [65] will inevitably popularize supervised learning approaches for this task.
The last step in the pipeline is to perform statistical analysis on extracted drug-ADR pairs to identify potentially harmful drugs. This step has hardly received any research attention till date, and we only identified some exploratory research attempting to perform this task on social media data [40, 39]. Progress in ADR extraction and classification research is likely to raise the research focus on the analysis of drug-ADR signals generated from social media data. Considering the rapid growth of social media data, this source of information is likely to have a massive impact on pharmacovigilance research.
Highlights.
We present a review of pharmacovigilance techniques from social media (SM) data
Our review discusses twenty-two studies, comparing them across various axes
We present a possible pathway for automated pharmacovigilance research from SM
Acknowledgements
We thank the NIH/NLM support for this project (Award: 1R01LM011176-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
http://www.statisticbrain.com/twitter-statistics/. Accessed on: 26th August, 2014.
Some resources for demographic information analysis: http://textalytics.com/core/userdemographics-info, http://www.demographicspro.com/, http://smallbiztrends.com/2013/04/research-twitter-followers-demographics.html.
http://www.ncbi.nlm.nih.gov/pubmed. Accessed: 6/10/2014.
The following social media sites were involved: breastcancer.org, komen.org, csn.cancer.org, bcsupport.org, healthboards.com, cancercompass.com, webmd.com, dailystrength.org, revolutionhealth.com, ehealthforum.com, oprah.com
This is the number of threads included, not the number of comments.
Only 200 comments are annotated for evaluation.
Only 10% of this data is annotated.
Study also includes data from AERS and Medline.
Includes 6 drugs from Leaman et al.
This is the data that is obtained from the three sources mentioned. The study utilized additional non-social media data.
Not unique drugs. The number of unique drugs is not mentioned.
Study also includes a corpus from outside social media.
ir.cs.georgetown.edu/data/adr/. Accessed on 06/12/2014.
http://labda.inf.uc3m.es/doku.php?id=en:labda_spanishadrcorpus. Accessed on 04/12/2014.
diego.asu.edu/downloads/. Accessed on 06/12/2014.
The three systems are: Treato.com, sideeffective.com, and AdverseEvents.com.
http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/default.htm Accessed on 06/16/2014.
http://bioportal.bioontology.org/ontologies/COSTART. Accessed: 06/13/2014.
http://www.consumerhealthvocab.org/. Accessed 06/16/2014.
http://www.hc-sc.gc.ca/dhp-mps/medeff/index-eng.php. Accessed on 06/13/2014.
http://www.nlm.nih.gov/research/umls/. Accessed 06/16/2014.
http://www.meddra.org/. Accessed 06/16/2014.
http://sideeffects.embl.de/. Accessed: 06/13/2014.
For evaluation approaches applying a mixture of quantitative and qualitative evaluations, we categorize them into one of the categories based on the primary evaluation ideology.
Drugs containing boxed warnings regarding ADRs.
Contributor Information
Abeed Sarker, Department of Biomedical Informatics, Arizona State University, Scottsdale, Arizona.
Rachel Ginn, Department of Biomedical Informatics, Arizona State University, Scottsdale, Arizona.
Azadeh Nikfarjam, Department of Biomedical Informatics, Arizona State University, Scottsdale, Arizona.
Karen O’Connor, Department of Biomedical Informatics, Arizona State University, Scottsdale, Arizona.
Karen Smith, Rueckert-Hartman College for Health Professions, Regis University, Denver, Colorado.
Swetha Jayaraman, School of Computing, Informatics, and Decision Systems Engineering, Arizona State University, Tempe, Arizona.
Tejaswi Upadhaya, School of Computing, Informatics, and Decision Systems Engineering, Arizona State University, Tempe, Arizona.
Graciela Gonzalez, Department of Biomedical Informatics, Arizona State University, Scottsdale, Arizona.
References
- [1].World Health Organization The Importance of Pharmacovigilance - Safety Monitoring of Medicinal Products. 2002 URL http://apps.who.int/medicinedocs/en/d/Js4893e/1.html.
- [2].Harpaz R, DuMouchel W, Shah NH, Madigan D, Ryan P, Friedman C. Novel data-mining methodologies for adverse drug event discovery and analysis. Clin Pharmacol Ther. 2012;91(3):1010–1021. doi: 10.1038/clpt.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lazarou J, Pomeranz BH, Corey PN. Incidence of Adverse Drug Reactions in Hospitalized Patients: A Meta-analysis of Prospective Studies. JAMA. 1998;279(15):1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- [4].Ahmad SR. Adverse Drug Event Monitoring at the Food and Drug Administration - Your Report Can Make a Difference. Journal of Internal Medicine. 2003;18(1):57–60. doi: 10.1046/j.1525-1497.2003.20130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Xu R, Wang Q. Large-scale combining signals from both biomedical literature and FDA Adverse Event Reporting System (FAERS) to improve post-marketing drug safety signal detection. BMC Bioinformatics. 15(17) doi: 10.1186/1471-2105-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gurulingappa H, Fluck J, Hofmann-apitius M, Toldo L. Identification of Adverse Drug Event Assertive Sentences in Medical Case Reports; Proceedings of ECML PKDD 2-11 Workshop on Knowledge Discovery in Health Care and Medicine; 2011. [Google Scholar]
- [7].FDA Adverse Event Reporting System (FAERS) http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/
- [8].MedWatch: The FDA Safety Information and Adverse Event Reporting Program. http://www.fda.gov/Safety/MedWatch/default.htm
- [9].Forster AJ, Jennings A, Chow C, Leeder C, van Walraven C. A syst-system review to evaluate the accuracy of electronic adverse drug event detection. Journal of the American Medical Informatics Association (JAMIA) 2011;19:31–38. doi: 10.1136/amiajnl-2011-000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kumar V. Challenges and Future Considerations for Pharmacovigilance. Journal of Pharmacovigilance. 2013;1(1):1–3. [Google Scholar]
- [11].The Pew Research Center The Social Life of Health Information. http://www.pewinternet.org/2011/05/12/the-social-life-of-health-information-2011/
- [12].ying Sylvia Chou W, Hunt YM, Beckjord EB, Moser RP, Hesse BW. Social Media Use in the United States: Implications for Health Communication. Journal of Medical Internet Research. 2009;11(4):e48. doi: 10.2196/jmir.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Online Support Groups and Forums at DailyStrength. http://www.dailystrength.org
- [14].MedHelp Medical Support Communities http://www.medhelp.org/forums/list
- [15].Corley CD, Cook DJ, Mikler AR, Singh KP. Advances in Computational Biology. Springer; New York: 2010. pp. 559–564. Ch. using Web and Social Media for Influenza Surveillance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kass-Hout T, Alhinnawi H. Social media in public health. British Medical Bulletin. 2013;108(1):5–24. doi: 10.1093/bmb/ldt028. [DOI] [PubMed] [Google Scholar]
- [17].Leaman R, Wojtulewicz L, Sullivan R, Skariah A, Yang J, Gonzalez G. Towards Internet-Age Pharmacovigilance: Extracting Adverse Drug Reactions from User Posts to Health-Related Social Networks; Proceedings of the 2010 Workshop on Biomedical Natural Language Processing; 2010.pp. 117–125. [Google Scholar]
- [18].Struik LL, Baskerville NB. The Role of Facebook in Crush the Crave, a Mobile- and Social Media-Based Smoking Cessation Intervention: Qualitative Framework Analysis of Posts. Journal of Medical Internet Research. 16(7) doi: 10.2196/jmir.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hanson CL, Cannon B, Burton S, Giraud-Carrier C. An Exploration of Social Circles and Prescription Drug Abuse Through Twitter. Journal of Medical Internet Research. 15(9) doi: 10.2196/jmir.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nakhasi A, Passarella RJ, Bell SG, Paul MJ, Dredze M, Provost PJ. Information Retrieval and Knowledge Discovery in Biomedical Text. Johns Hopkins University; 2012. Malpractice and Malcontent: Analysing Medical Complaints in Twitter, AAAI Technical Report. [Google Scholar]
- [21].Sarasohn-Kahn J. The Wisdom of Patients: Health Care Meets Online Social Media. 2008 http://www.chcf.org/~/media/MEDIA%20LIBRARY%20Files/%20PDF/H/PDF%20HealthCareSocialMedia.pdf
- [22].Velasco E, Agheneza T, Denecke K, Kirchner G, Eckmanns T. Social Media and Internet-Based Data in Global Systems for Public Health Surveillance: A Systematic Review. The Milbank Quarterly. 2014;92:7–33. doi: 10.1111/1468-0009.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hartley DM. Using Social Media and Internet Data for Public Health Surveillance: The Importance of Talking. The Milbank Quarterly. 2014;92:34–39. doi: 10.1111/1468-0009.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stricker BH, Psaty BM. Detection, verification, and quantification of adverse drug reactions. BMJ. 329(44) doi: 10.1136/bmj.329.7456.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Guidance notes on the management of adverse events and product complaints from digital media. 2013 http://www.abpi.org.uk/our-work/library/guidelines/Pages/safety-data-websites.aspx
- [26].Guidance for Industry Fulfilling Regulatory Requirements for Post-marketing Submissions of Interactive Promotional Media for Prescription Human and Animal Drugs and Biologics. 2014 http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM381352.pdf
- [27].Guidance for Industry Internet/Social Media Platforms with Character Space Limitations — Presenting Risk and Benefit Information for Prescription Drugs and Medical Devices. 2014 http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm401087.pdf
- [28].Pan GJD. Adverse Event Reporting and Medication Safety Considerations: A View from CDER’s Office of Surveillance and Epidemiology. 2013 May; http://www.fda.gov/downloads/AboutFDA/CentersOffices/OfficeofMedicalProductsandTobacco/CDER/UCM352766.pdf
- [29].White Paper: Social Media in the Pharmaceutical Industry. http://www.brandchannel.com/images/papers/522_2011-02_AZ_Social_Media.pdf
- [30].Edwards IR, Lindquist M. Social Media and Networks in Pharmacovigilance: Boon or Bane. Drug Safety. 2011;34(4):267–271. doi: 10.2165/11590720-000000000-00000. [DOI] [PubMed] [Google Scholar]
- [31].Franzen W. Can Social Media Benefit Drug Safety. Drug Safety. 2012;34(9):793. doi: 10.2165/11595510-000000000-00000. [DOI] [PubMed] [Google Scholar]
- [32].Harpaz R, Callahan A, Tamang S, Low Y, Odgers D, Finlayson S, Jung K, LePendu P, Shah NH. Text Mining for Adverse Drug Events: the Promise, Challenges, and State of the Art. Drug Safety. doi: 10.1007/s40264-014-0218-z. doi:10.1007/s40264-014-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Abbasi A, Adjeroh D. Social Media Analytics for Smart Health. Intelligent Systems. 2014:60–80. [Google Scholar]
- [34].Nikfarjam A, Gonzalez G. Pattern Mining for Extraction of Mentions of Adverse Drug Reactions from User Comments; Proceedings of the American Medical Informatics Association (AMIA) Annual Symposium; 2011; pp. 1019–1026. [PMC free article] [PubMed] [Google Scholar]
- [35].O’Connor K, Nikfarjam A, Ginn R, Pimpalkhute P, Sarker A, Smith K, Gonzalez G. Pharmacovigilance on Twitter? Mining Tweets for Adverse Drug Reactions; Proceedings for the American Medical Informatics Association (AMIA) Annual Symposium; 2014; [PMC free article] [PubMed] [Google Scholar]
- [36].Tuarob S, Tucker CS, Salathe M, Ram N. An ensemble heterogeneous classification methodology for discovering health-related knowledge in social media messages. Journal of Biomedical Informatics. 2014;49:255–268. doi: 10.1016/j.jbi.2014.03.005. [DOI] [PubMed] [Google Scholar]
- [37].Owoputi O, O’Connor B, Dyer C, Gimpel K, Schneider N, Smith NA. Improved Part-of-Speech Tagging for Online Conversational Text with Word Clusters. Proceedings of the NAACL-HLT. 2-13:380–390. [Google Scholar]
- [38].Ginn R, Pimpalkhute P, Nikfarjam A, Patki A, O’Connor K, Sarker A, Smith K, Gonzalez G. Mining Twitter for Adverse Drug Reaction Mentions: A Corpus and Classification Benchmark; Proceedings of the Fourth Workshop on Building and Evaluating Resources for Health and Biomedical Text Processing; 2014. [Google Scholar]
- [39].Patki A, Sarker A, Pimpalkhute P, Nikfarjam A, Ginn R, O’Connor K, Smith K, Gonzalez G. Proceedings of BioLinkSig 2014. 2014. Mining Adverse Drug Reaction Signals from Social Media: Going Beyond Extraction. [Google Scholar]
- [40].Chee BW, Berlin R, Schatz B. Predicting Adverse Drug Events from Personal Health Messages; Proceedings of the American Medical Informatics Association (AMIA) Annual Symposium; 2011; pp. 217–226. [PMC free article] [PubMed] [Google Scholar]
- [41].Sarker A, Gonzalez G. Portable automatic text classification for adverse drug reaction detection via multi-corpus training. Journal of Biomedical Informatics. doi: 10.1016/j.jbi.2014.11.002. doi:doi:10.1016/j.jbi.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Benton A, Ungar L, Hill S, Hennessy S, Mao J, Chung A, Leonard CE, Holmes JH. Identifying potential adverse effects using the web: A new approach to medical hypothesis generation. Journal of Biomedical Informatics. 2011;44:989–996. doi: 10.1016/j.jbi.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hadzi-Puric J, Grmusa J. Automatic drug adverse reaction discovery from parenting websites using disproportionality methods; IEEE/ACM Conference on Advances in Social Networks Analysis and Mining; 2012.pp. 792–797. [Google Scholar]
- [44].Yang CC, Yang H, Jiang L, Zhang M. Social media mining for drug safety signal detection; Proceedings of the 2012 international workshop on smart health and wellbeing; 2012.pp. 33–44. [Google Scholar]
- [45].Bian J, Topaloglu U, Yu F. Towards large-scale twitter mining for drug-related adverse events; Proceedings of the 2012 international workshop on Smart health and wellbeing; 2012; pp. 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu X, Chen H. AZDrugMiner: an information extraction system for mining patient-reported adverse drug events in online patient forums; Proceedings of the 2013 international conference on smart health; 2013.pp. 134–150. [Google Scholar]
- [47].American Diabetes Association Community community.diabetes.org
- [48].Yang M, Wang X, Kiang M. Identification of Consumer Adverse Drug Reaction Messages on Social Media; Proceedings of the Pacific Asia Conference on Information Systems (PACIS); 2013; URL http://aisel.aisnet.org/pacis2013/193. [Google Scholar]
- [49].Jiang K, Zheng Y. Mining Twitter Data for Potential Drug Effects. Advanced Data Mining and Applications. 2013;8346:434–443. [Google Scholar]
- [50].Yates A, Goharian N. ADRTrace: detecting expected and unexpected adverse drug reactions from user reviews on social media sites; Proceedings of the 35th European conference on Advances in Information Retrieval; 2013.pp. 816–819. [Google Scholar]
- [51].Ask a Patients. http://www.askapatient.com
- [52]. Drugs.com: Know more. Be Sure. Drugs.com http://www.drugs.com
- [53].DrugRatingz: Find, rate and review Drugs and Medications. http://www.drugratingz.com
- [54].Yeleswarapu S, rao A, Joseph T, Saipradeep VG, Srinivasan R. A pipeline to extract drug-adverse event pairs from multiple data sources. BMC Medical Informatics and Decision Making. 14(13) doi: 10.1186/1472-6947-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].PatientsLikeMe: Live better, together! http://www.patientslikeme.com
- [56].Mediguard: Medication Monitoring Made Simple. https://www.mediguard.org
- [57].Freifeld CC, Brownstein JS, Menone CM, Bao W, Felice R, Kass-Hout T, Dasgupta N. Digital Drug Safety Surveillance: Monitoring Pharmaceutical Products in Twitter. Drug Safety. 2014;37(5):343–350. doi: 10.1007/s40264-014-0155-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Segura-Bedmar I, Revert R, Martinez P. Detecting drugs and adverse events from Spanish health social media streams; Proceedings of the 5th International Workshop on Health Text Mining and Information Analysis (LOUHI); 2014.pp. 106–115. [Google Scholar]
- [59].Forumclinic: Programa interactivo para pacientes. https://www.forumclinic.org
- [60].Liu X, Liu J, Chen H. Identifying Adverse Drug Events from Health Social Media: A Case Study on Heart Disease Discussion Forums; Proceedings of the International Conference on Smart Health (ICSH); 2014.pp. 25–36. [Google Scholar]
- [61].Yang CC, Yang H, Jiang L. Postmarketing drug safety surveillance using publicly available health-consumer-contributed content in social media. ACM Trans. Manage. Inf. Syst. 2014;5(1):2:1–2:21. doi:10.1145/2576233. URL http://doi.acm.org/10.1145/2576233. [Google Scholar]
- [62].Sampathkumar H, wen Chen X, Luo B. Mining Adverse Drug Reactions from online healthcare forums using Hidden Markov Model. BMC Medical Informatics and Decision Making. 14(91) doi: 10.1186/1472-6947-14-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].The premier community to talk about health. http://www.medications.com/
- [64].SteadyHealth — Ask, Share, Contribute. http://www.steadyhealth.com/
- [65].Nikfarjam A, Sarker A, O’Connor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. Journal of the American Medical Informatics Association (JAMIA)-(−) 2014 doi: 10.1093/jamia/ocu041. article in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Gurulingappa H, Mateen-Rajput A, Toldo L. Extraction of potential adverse drug eevent from medical case reports. Journal of Biomedical Semantics. 3(15) doi: 10.1186/2041-1480-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Aronson AR. Effective mapping of biomedical text to the UMLS metathesaurus: The metamap program; Proceedings of the American Medical Informatics Association Annual Symposium; 2001; pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- [68].Piatetsky-Shapiro G. Knowledge Discovery in Databases. AAAI/MIT Press; Cambridge, MA: 1991. Ch. Discovery, analysis, and presentation of strong rules. [Google Scholar]
- [69].Pimpalkhute P, Patki A, Gonzalez G. Phonetic Spelling Filter for Keyword Selection in Drug Mention Mining from Social Media; Proceedings of the American Medical Informatics Association (AMIA) Annual Symposium; 2013; [PMC free article] [PubMed] [Google Scholar]