Abstract
Objective
Structured data on mammographic findings are difficult to obtain without manual review. We developed and evaluated a rule-based natural language processing (NLP) system to extract mammographic findings from free-text mammography reports.
Materials and Methods
The NLP system extracted four mammographic findings: mass, calcification, asymmetry, and architectural distortion, using a dictionary look-up method on 93,705 mammography reports from Group Health. Status annotations and anatomical location annotation were associated to each NLP detected finding through association rules. After excluding negated, uncertain, and historical findings, affirmative mentions of detected findings were summarized. Confidence flags were developed to denote reports with highly confident NLP results and reports with possible NLP errors. A random sample of 100 reports was manually abstracted to evaluate the accuracy of the system.
Results
The NLP system correctly coded 96 to 99 out of our sample of 100 reports depending on findings. Measures of sensitivity, specificity and negative predictive values exceeded 0.92 for all findings. Positive predictive values were relatively low for some findings due to their low prevalence.
Discussion
Our NLP system was implemented entirely in SAS Base, which makes it portable and easy to implement. It performed reasonably well with multiple applications, such as using confidence flags as a filter to improve the efficiency of manual review. Refinements of library and association rules, and testing on more diverse samples may further improve its performance.
Conclusion
Our NLP system successfully extracts clinically useful information from mammography reports. Moreover, SAS is a feasible platform for implementing NLP algorithms.
Keywords: natural language processing, SAS-based, evaluation, mammographic findings
1. BACKGROUND
Mammographic findings, such as a mass, calcifications, asymmetry, or architectural distortion may increase breast cancer risk.[1,2] Definitions for and examples of these findings are clearly outlined in the American College of Radiology’s Breast Imaging Reporting And Data System (ACR BI-RADS®) manual.[3] However, they are often not collected in structured fashion in mammography reporting systems, such as Centricity Radiology Information System (RIS).[4] The BI-RADS manual provides a standardized data form for mammography facilities to use when recording mammography data,[5] but mammographic findings are not included in the data form. In many settings, mammographic findings are only reported in free-text mammography reports.
Traditionally manual abstraction has to be performed in order to convert information contained in free text to structured data. While this may be feasible for small-scale studies, manual abstraction is expensive and even infeasible to use in studies with large sample sizes. Natural language processing (NLP) is a field of study focused on understanding the meaning of spoken or written text, using various computational techniques.[6] NLP has been widely tested in research and clinical settings to overcome the limitations of manual data abstraction. For example, Jain and Friedman extracted suspicious findings from mammography reports through their NLP system MedLEE. [7] However, the corpus they used was known to have suspicious findings and normal mammograms were not examined. More importantly, the NLP system they used has not been made publicly available. Using various open source or commercial software, other informatics researchers identified breast cancer recurrence through clinical documents,[8] classified breast density,[9] identified results of mammograms and Pap smears,[10] extracted recommendations for radiology reports, [11] and annotated mammography reports.[12] While sophisticated clinical NLP systems can be very powerful, a disadvantage of working with them is that they require specialized informatics knowledge, which is uncommon in many research settings. Moreover, most commercial NLP systems are not readily portable and can be very expensive. Therefore, we developed and evaluated a relatively simple rule-based NLP system implemented entirely in SAS Base (V.9.2, SAS Institute) to extract mammographic findings (mass, calcification, asymmetry and architectural distortion) from free-text mammography reports.
2. METHODS
2.1 Data
We undertook this study within Group Health, a large integrated health care delivery system based in Seattle, Washington and one of 6 sites in the Breast Cancer Surveillance Consortium (BCSC).[13] The BCSC is the largest breast cancer screening research database in the US with over 10.7 million mammograms on 2.4 million women. Our data included 93,705 deidentified screening and diagnostic mammography exams interpreted by 31 radiologists from Group Health in 2008 and 2009. We also used BI-RADS assessments from Centricity Radiology Information System (RIS) for checking consistencies of the NLP detected mammographic findings. We received approval from the Group Health Institutional Review Board for this study.
2.2 Preprocess
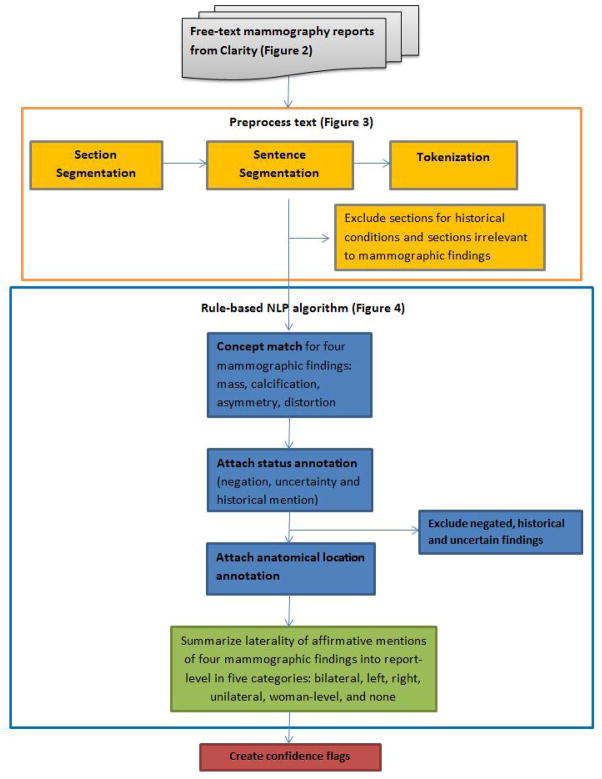
An overview of the NLP system is shown in Figure 1. We extracted electronic mammography reports from Clarity, the reporting database of EPIC (EPIC2010, Epic Systems Corporation), Group Health’s electronic medical record system. Each mammography report spans multiple lines and may split at locations other than sentence boundaries. For example, Figure 2 shows the original text from part of a mammography report from Clarity. The line breaks occur at unpredictable places, such as line 2, which breaks in the middle of a sentence.
Figure 1.
Process diagram for a SAS-based NLP system for detecting mammographic findings
Figure 2.
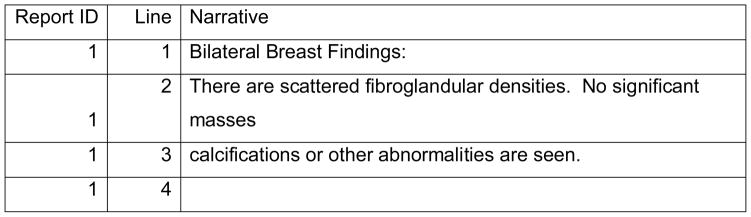
Original text from Clarity
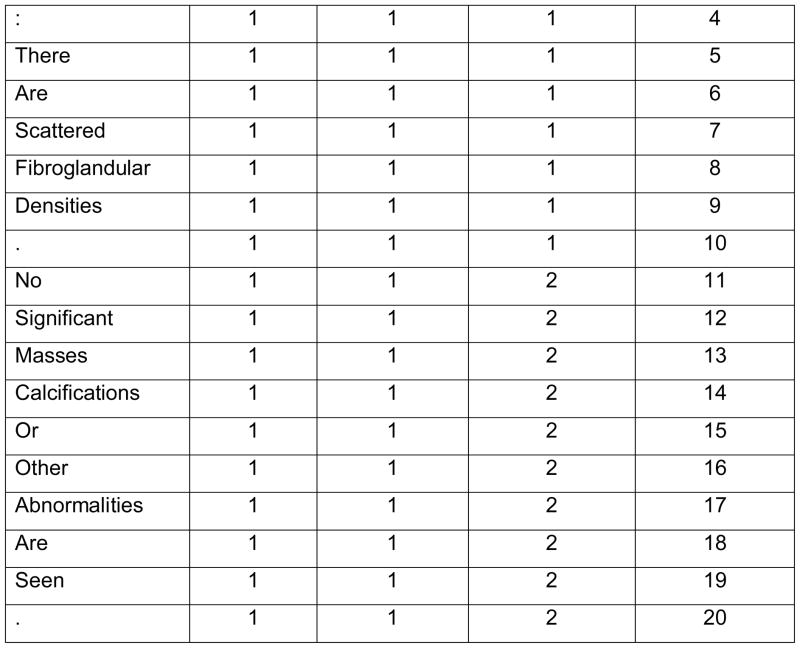
To facilitate NLP analysis, text reports were preprocessed. First, section boundaries were noted. For our text reports, each section began with a section heading and ended with one or more empty lines. The section headings were phrases from the beginning of a new line; they were either in square brackets or ended with a colon, such as “[HST]”, “Bilateral Breast Findings:”. Second, sentence boundaries were noted. Sentence segmentation is a complex task because periods can also be used for decimal places and abbreviations. We first replaced periods that stood for decimal places or abbreviations with another symbol. Then, we used periods, question marks, exclamation marks and section boundaries as boundaries between sentences. Third, tokenization was done by using spaces and certain symbols (such as slashes, comma, semicolons, etc.) as separators. With the three steps above, each report was converted into a list of words and each word was flagged with its section number, sentence number, and order of its position in the report. For example, Figure 3 shows how the original text of the report in Figure 2 was converted and ordered into individual words within each section and sentence of the report.
Figure 3.
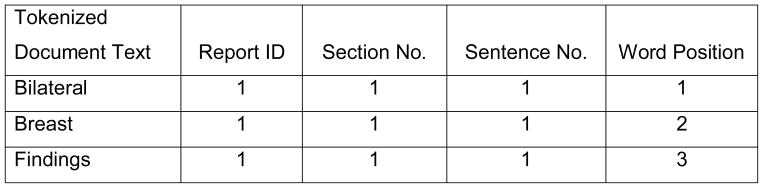
The text in Figure 1 after section segmentation, sentence segmentation, and tokenization
Identifying the section, sentence, and position of each word is useful in many ways. For example, it can allow for sections to be deleted before analyses to avoid unnecessary confusion or potential NLP errors. These sections either contain historical mentions of findings that should be excluded (such as sections of history, indication, symptom, comparison, note, etc.) or would not contain mammographic finding information (such as sections for exam type, exam date and time, physician signature, etc.). Also, annotations should be within a certain distance to the finding word in order to be considered valid. Therefore, associating valid annotations with findings requires knowing the sentence and position of each word.
2.3 Concept match
Using Perl Regular Expressions in SAS, we built a library covering text strings that match various phrases radiologists used to describe four mammographic findings (Table 1). Text strings were provided by two clinical researchers with over 10 years combined experience reading and abstracting breast pathology reports. Stem-words were used to cover all possible word form; for example, “microcalcification”, “macrocalcification”, “recalcification”, and misspellings like “mecrocalcification”, were all matched to the “calcification” concept including plural forms. For the concept of “asymmetry”, we created two additional rules to improve the accuracy of our NLP system based on empirical investigations. First, we allowed 0 to 6 words between “focal” and “density”. We created this rule after reviewing an additional hold-out set of 200 randomly selected reports that contained the words “focal” and “density” with 1 to 10 words between them (20 reports for each distance). Then we manually reviewed the relevant sentences from these reports. We found that when the distance between “focal” and “density” was 6 words or fewer, it was quite likely that they were related; when the distance was greater than 6 words, it was much less likely that they were related. Our second rule disallowed specific nouns and verbs between the words “focal” and “density”. If nouns like “compression” or “view”, or verbs like “is” or “reveal” occurred between “focal” and “density”, it was unlikely that “focal” and “density” were related to each other.
Table 1.
Library of text strings for concepts
| Concept | Text String |
|---|---|
| Mass | mass(es)?; lump(s)? |
| Calcification | *calcification(s)? |
| Asymmetry | asymmetr*; [(fibro)?nodular|focal] (0 to 6 words in between, excluding some nouns and verbs) densit* |
| Distortion | distortion(s)?; deformity; architectural |
Note: The symbol * means any string before or after a word stem.
The symbols “( )?” and “[ ]?” mean the strings within the parentheses are optional.
The symbol “[ | ]” means one of the strings separated by the pipe is used.
2.4 Status annotations
The NLP system attached status annotations for each detected finding. First, it detected text strings that denote negation, uncertainty or historical mention. The SAS code (appendix A) contained a complete list of text strings we used as cues for negation, uncertainty and historical mention. We used Chapman’s NegEx list as the foundation for negation cues, and revised the list based on empirical examination of the list of all unique words in the corpus, ordered by frequency. [14] Cues for uncertainty and historical mention were also gathered through empirical examination of the same word list. Then, we applied distance restrictions to associate negation words and uncertainty words with the corresponding finding words. Each negation (or uncertainty word) was assigned values limiting the maximum distance before and after the corresponding finding word within the same sentence. These values were decided based on empirical investigations using the same approach used to determine number of words allowed between “nodular” and “density” (described above). For example, we found the negation word “absence” could occur up to eight words before a finding word within the same sentence with minimal false positive and false negative rates, while the uncertainty word “may” could only occur up to five words before a finding word within the same sentence with similar accuracy. For most of the negation or uncertainty words, the direction was more sensitive than the distance. For example, in most cases, the negation word “no” only negated the finding words when it showed before the finding words in the same sentence. How far away the negation word “no” was before the finding words within a sentence usually did not matter. We imposed a rule of 20 words as the default distance for “no” and limited the direction to only before a finding word.
For historical mentions, we set a rule that all findings in the same sentence as the words denoting historical mentions were historical findings.
2.5 Anatomical location annotation
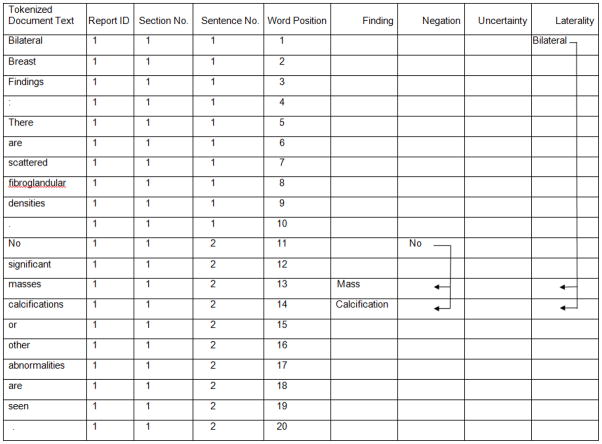
The NLP system attached laterality to each finding. First, it detected text strings that denoted bilateral, right, left and unilateral. If right and left were both mentioned in the same sentence with a finding word, that finding was coded as “bilateral”. Otherwise, finding words were coded as “left” or “right” as long as the laterality word “left” or “right” was found in the same sentence. If no laterality word was found in the same sentence as finding words, the laterality word before and closest to the finding word within the same section was attributed to that finding word.
Figure 4 shows an example of how the words were matched with each concept and how status annotations and laterality were attached to each finding. Words “masses” and “calcifications” from the original text report (Figure 1) were matched to the corresponding finding categories in the “Finding” column; the word “No” was associated with both “masses” and “calcifications” because it was within 20 words before finding words in the same sentence. The word “Bilateral” was also associated with “masses” and “calcifications” because it was the closest laterality word before the finding words in the same section. There were no words matched to the concepts of history or uncertainty in Figure 4 because they did not exist in this example.
Figure 4.
preprocessed text with matched concepts
2.6 NLP results summarization
We summarized NLP outcomes into report-level results. We excluded findings with status annotations indicating negation, uncertainty or historical reference. The remaining affirmative mentions of the same finding in the same report were summarized into one of six categories: bilateral, left, right, unilateral, woman-level (meaning laterality could not be determined) or none. If a finding was not mentioned at all in a mammography report, it was coded as “No”. Table 2 shows 4 examples of original text, their corresponding NLP outputs, and whether the assigned codes were correct. In the original text, we underlined the phrases that our NLP system used to make the coding decisions.
Table 2.
Examples of original text, its corresponding NLP output and accuracy
| Original Text | Mass | Calcification | Asymmetry | Distortion | |
|---|---|---|---|---|---|
| FINDINGS: There are scattered fibroglandular densities. There are vascular calcifications in both breasts and there is stable parenchymal asymmetry in the upper outer left breast. No dominant mass, clustered calcifications, or areas of distortion are noted. | NLP output | No | bilateral | bilateral | No |
| Accuracy | Correct | Correct | Incorrect1 | Correct | |
| IMPRESSION: Right breast mass for which additional imaging spot compression views and ultrasound is recommended. | NLP output | Right | No | No | No |
| Accuracy | Correct | Correct | Correct | Correct | |
| LEFT BREAST: The breast is heterogeneously dense, which can lower the sensitivity of mammography. In the subareolar tissues, the previously noted mass has been removed. Tissue asymmetry with mild architectural change and associated surgical clips are present, consistent with postoperative sequela. Stable, benign macrocalcifications and tissue asymmetry are unchanged. There are no new masses, microcalcifications or architectural distortion to suggest malignancy. | NLP output | No | Left | Left | Left |
| Accuracy | Correct | Correct | Correct | Correct | |
| RIGHT BREAST: The breast is extremely dense, which can lower the sensitivity of mammography. In the right breast at six o’clock location, 4 cm from the nipple, there is a cluster of microcalcifications. Recommend spot magnification views. Additionally, the patient complains of palpable lump in the right breast. Recommend ultrasound evaluation of the palpable lump. | NLP output | Right | Right | No | No |
| Accuracy | Incorrect2 | Correct | Correct | Correct |
Note:
Asymmetry should be on the right breast only. However, the NLP system incorrectly associated the words “both breasts” (for mass) with asymmetry based on the association rule that all laterality words were taken for mammographic findings in the same sentence.
Even though the NLP system correctly identified a lump, coding this as a mass was incorrect in this case. From the original text provided, the lump was identified by the patient, not on the mammogram. We were interested in identifying mammographic findings, thus the lump would not count as a mass in this case.
2.7 Confidence flags
We developed five confidence flags to denote reports with highly confident NLP results (that would not need manual review) and reports with possible NLP errors (that would need manual review). (1) When our NLP system did not detect any mention of a finding in a report and coded the finding as “none”, the confidence flag was set to “no review required”. (2) When NLP detected findings were discordant with BI-RADS mammography assessments, (e.g., when NLP identified an affirmative mention of a mass, but the BI-RADS assessment was normal) the NLP results were presumed incorrect and the confidence flag was set to “needs review”. (3) When both negative and affirmative mentions of the same finding with the same laterality appeared in one report, the confidence flag was set to “needs review”. (4) A finding in a sentence with two negation words could be positive (double negative) or could still be negative, such as “There is no mass and there is no calcification.” Therefore, wherever a sentence contained more than one negation word, we flagged the finding as “needs review”. (5) When NLP detected mentions of “recent”, “previous” or “prior” in the same sentence as an affirmative mention of a finding, the finding was likely to be historical condition and the confidence flag was set to “needs review”.
2.8 Evaluation
In order to evaluate the accuracy of our NLP system to identify mammographic findings, an experienced abstractor manually reviewed a stratified random sample of 100 reports (25 screening exams and 75 diagnostic exams). We compared the results of each finding from the abstraction with the corresponding NLP results. To calculate overall performance metrics, results were combined into a 2*2 table. A true positive meant the NLP system and manual review both detected the same finding and laterality was also correct. If both NLP and manual review detected the same finding but the laterality disagreed, this was considered a false negative. We also compared mammographic findings to BI-RADS assessments and coded their consistencies with one of the confidence flags.
3. RESULTS
Table 3 shows the distribution of mammographic findings from 76,049 screening and 17,656 diagnostic mammography reports. In general, screening exams had fewer affirmative mentions of findings than diagnostic exams. Calcifications (24.7%) and asymmetry (22.2%) were the most common findings in diagnostic exams, while architectural distortion (3.7%) was the least common finding. For screening exams, calcifications (13.2%) were the most common finding, while mass (1.5%) and architectural distortion (1.6%) were the least common findings. There was no difference in the distribution of mammographic findings by year of the report (data not shown).
Table 3.
NLP results: mammography findings from Group Health in 2008 and 2009
| Diagnostic Exams (N=17656) | Screening Exams (N=76049) | |||
|---|---|---|---|---|
| Mass | N | % | N | % |
| Bilateral | 236 | 1.3 | 117 | 0.2 |
| Left | 977 | 5.5 | 519 | 0.7 |
| Right | 899 | 5.1 | 509 | 0.7 |
| Unilateral | 7 | 0.0 | 0 | 0.0 |
| Woman-level | 86 | 0.5 | 33 | 0.0 |
| No | 15451 | 87.5 | 74871 | 98.5 |
| Calcification | ||||
| Bilateral | 862 | 4.9 | 4342 | 5.7 |
| Left | 1672 | 9.5 | 1912 | 2.5 |
| Right | 1636 | 9.3 | 1984 | 2.6 |
| Unilateral | 9 | 0.1 | 0 | 0.0 |
| Woman-level | 191 | 1.1 | 1831 | 2.4 |
| No | 13286 | 75.3 | 65980 | 86.8 |
| Asymmetry | ||||
| Bilateral | 526 | 3.0 | 1167 | 1.5 |
| Left | 1654 | 9.4 | 2309 | 3.0 |
| Right | 1615 | 9.2 | 2156 | 2.8 |
| Unilateral | 3 | 0.0 | 0 | 0.0 |
| Woman-level | 117 | 0.7 | 426 | 0.6 |
| No | 13741 | 77.8 | 69991 | 92.0 |
| Distortion | ||||
| Bilateral | 41 | 0.2 | 128 | 0.2 |
| Left | 296 | 1.7 | 494 | 0.7 |
| Right | 284 | 1.6 | 501 | 0.7 |
| Unilateral | 1 | 0.0 | 0 | 0.0 |
| Woman-level | 36 | 0.2 | 69 | 0.1 |
| No | 16998 | 96.3 | 74857 | 98.4 |
The NLP system incorrectly coded 1 report out of 100 reports for mass, 1 report for calcification, 4 reports for asymmetry, and 2 reports for architectural distortion. The NLP system reached at least 0.92 for sensitivity, specificity and negative predictive value for each finding. Positive predictive value for architectural distortion was relatively low (0.5) due to the low prevalence of architectural distortion in mammographic findings (only 2 out of 100 reports had true positive architectural distortion and the NLP system correctly identified both of them) (Table 4).
Table 4.
Accuracy of NLP results by mammographic finding
| Gold standard from manual review | Accuracy | Sensitivity | Specificity | PPV | NPV | |||
|---|---|---|---|---|---|---|---|---|
| Results from NLP algorithm | Positive | Negative | ||||||
| Mass | positive | 5 | 1 | 0.99 | 1.00 | 0.99 | 0.83 | 1.00 |
| negative | 0 | 94 | ||||||
| Calcification | positive | 17 | 0 | 0.99 | 0.94 | 1.00 | 1.00 | 0.99 |
| negative | 1 | 82 | ||||||
| Asymmetry | positive | 11 | 3 | 0.96 | 0.92 | 0.97 | 0.79 | 0.99 |
| negative | 1 | 85 | ||||||
| Architectural distortion | positive | 2 | 2 | 0.98 | 1.00 | 0.98 | 0.50 | 1.00 |
| negative | 0 | 96 | ||||||
Note: PPV, positive predictive value; NPV, negative predictive value
Error Analysis
Several typical NLP errors occurred. First, the NLP system could not distinguish between current and historical findings unless words such as “history” or “hx” were found in the sentence. For example, the words “recalled” and “recent” were used in the following text to express historical findings: “The patient was recalled because of parenchymal asymmetry and possible microcalcifications seen in the right breast on the exaggerated CCL view only on the most recent mammogram.” The NLP system could not tell that “asymmetry” referred to the previous exam, not a finding from the current exam. Second, the NLP system did not attempt to detect hypothetical statements. For example, for the following text: “study was done to evaluate developing architectural distortion in the retroareolar area of the left breast. Today’s 90 degree lateral film shows no abnormally increased density or persistent abnormality.” The first sentence was a hypothetical statement, because it only explained that the reason for the mammogram, which was to determine whether architectural distortion existed. The second sentence confirmed there was no abnormality. However, the NLP system could not tell that “architectural distortion” mentioned in the first sentence was a hypothetical statement, not a real finding. Some researchers have worked out algorithms that can annotate hypothetical status, in addition to negation and historical mentions. [15] We may be able to add such features in SAS in future studies. Third, because our concept library was not comprehensive, infrequently used words were overlooked; for example, in one of our reports, a radiologist used the word “efface” to denote negation; but this word was not in our library of negation. Fourth, the association rules were imperfect. For example, in the following text: “The small round architectural distortion … in the mid outer breast is no longer seen on the mammogram.”, the negation word “no” occurred after the finding word, but the current association rule only counted the negation word “no” if it occurred before finding words.
Table 5 summarizes each confidence flag for our sample. Most reports in our sample (82,051; 87.5%) did not contain any mention of asymmetry and would require no subsequent manual review. Over two-thirds of reports (64,083; 68.4%) did not contain any text string for architectural distortion; 19,836 (21.2%) reports did not contain any text string for mass; and 17,978 (19.2%) reports did not contain any text string for calcification.
Table 5.
Results of confidence flags
| Confidence Flags | Mass | Calcification | Asymmetry | Architectural distortion | |
|---|---|---|---|---|---|
| No review required: NLP did not detect any mention of a finding in a report | N | 19836 | 17978 | 82051 | 64083 |
| % | 21.2 | 19.2 | 87.5 | 68.4 | |
| Needs review: NLP detected findings discordant with BI-RADS assessments | N | 531 | NA | NA | NA |
| % | 0.6 | NA | NA | NA | |
| Needs review: both negative and affirmative mentions of a finding in one report | N | 1022 | 5136 | 198 | 359 |
| % | 1.1 | 5.5 | 0.2 | 0.4 | |
| Needs review: more than one negation word with a finding in a sentence | N | 102 | 122 | 11 | 119 |
| % | 0.1 | 0.1 | 0.0 | 0.1 | |
| Needs review: sentences with phrases of “recent”, “previous” or “prior”. | N | 244 | 905 | 1699 | 661 |
| % | 0.3 | 1.0 | 1.8 | 0.7 |
The reports with the following flags indicate the NLP results were suspicious and merited manual review. First, 286 (0.3%) reports in our sample found masses, but the assessments were normal; 245 (0.3%) reports did not find a mass, but the assessments were abnormal. Second, 1022 (1.1%) reports in our sample found both negative and confirmative phrases for mass, 5136 (5.5%) reports for calcification, 198 (0.2%) reports for asymmetry, and 359 (0.4%) reports for architectural distortion. Third, 102 (0.1%) reports in our sample contained sentences with two negation words and a finding for mass; 122 (0.1%) reports for calcification; 11 (0.0%) reports for asymmetry; and 119 (0.1%) reports for architectural distortion. Fourth, 244 (0.3%) reports in our sample contained sentences with the phrases of “recent”, “previous” or “prior” and a positive finding of mass; 905 (1.0%) reports for calcification; 1699 (1.8%) reports for asymmetry; and 661 (0.7%) for architectural distortion.
4. DISCUSSION
We developed a high-performing NLP system that accurately extracts mammographic findings with laterality from free-text mammography reports. While several papers have been published on extracting clinically useful information from mammography reports through various NLP systems, [7,9,12] we are unaware of any implemented as publicly available SAS code. Our work is similar to the SAS-based NLP system for identifying cancer diagnoses in pathology reports reported by Dr. Strauss’ team but their SAS code has not been publicly available yet. [16]
We developed our NLP system entirely in SAS Base, a widely used data analytic programming software. This system implements relatively simple versions of language processing tasks, including (a) named entity recognition (i.e., identifying mass, calcification, asymmetry and architectural distortion in a dictionary-look-up step); (b) status annotation (i.e., flags indicating when recognized named entities are negated, qualified by uncertainty, or historical references), (c) anatomical location annotation (i.e., flags indicating laterality of recognized named entities), and (d) confidence flagging. Our evaluation showed that the SAS-based NLP system performed reasonably well in extracting mammographic findings from free-text mammography reports. While additional information on BI-RADS assessments was helpful in developing confidence flags and confirming our results, they are not necessary for the sole task of obtaining findings from mammography reports. Compared with more sophisticated open source clinical NLP systems, which might out-perform our system, our SAS-based NLP system has the advantage of being more portable and easier to implement because SAS can be used by individuals without specialized informatics or machine learning knowledge. [17] For applications like mammographic findings, which present reasonably straightforward information extraction tasks, this trade-off seems worthwhile.
Though manual abstraction can be more accurate, it is often not feasible for large-scale research. Before we developed this NLP system, we were unable to submit structured mammographic findings data to the BCSC because we had limited resources to do manual abstraction. This NLP system automatically extracted mammographic findings from 617,912 free-text mammography reports in several hours and therefore we were able to submit structured mammographic findings to the BCSC. If an abstractor reviewed the same number of reports taking 30 seconds per record, we estimate the abstractor would have required over 5,000 hours of work.
Confidence flags assess the likely accuracy of the information extracted and are useful in applications intended to improve the efficiency of manual review. They are suitable for use as a filter to determine which reports need subsequent manual review and which can solely rely on NLP results. For mammography reports that our NLP system did not detect any mention of a finding, we are highly confident in saying that there is no such finding. In our database, 87.5% reports do not have any NLP detected mention of asymmetry, and we can be highly confident in coding these reports without further review. For reports with “needs review” flags, it is more likely that the NLP system did not correctly extract findings and manual review should be done. In this corpus, the percentages of reports needing review for any reason were low.
Some of limitations of the NLP system are shown from error analysis, such as an incomplete library for clinical concepts and imperfect association rules. Future work includes refinement of library as well as the association rules to increase accuracy. Additional work in this area should explore improving annotation of historical mentions and hypothetical statements to reduce false positives. Though our NLP system performed well on our own reports, evaluating it on corpora from other institutions is needed. To this end we have provided the SAS code for the NLP system in Appendix A and invite others to test it on local reports.
5. CONCLUSIONS
Our SAS-based NLP system performed well in automatically processing free-text mammography reports and accurately identifying four categories of mammographic findings. Our results suggest that mammographic findings can be successfully extracted from free-text mammography reports using NLP implemented in SAS Base.
Supplementary Material
Highlights.
We developed and evaluated a rule-based natural language processing system
The NLP system extracts mammographic findings from free-text mammography reports
Manual review showed that the NLP system performs reasonably well
We developed confidence flags to facilitate further manual review
The NLP system was implemented entirely in SAS Base with SAS code available
Acknowledgments
This work was supported by a grant from the National Cancer Institute (CA146917), a National Cancer Institute-funded Program Project (P01CA154292) and the Breast Cancer Surveillance Consortium (HHSN261201100031C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Timmers JM, Verbeek AL, IntHout J, Pijnappel RM, Broeders MJ, den Heeten GJ. Breast cancer risk prediction model: a nomogram based on common mammographic screening findings. Eur Radiol. 2013;23(9):2413–9. doi: 10.1007/s00330-013-2836-8. [DOI] [PubMed] [Google Scholar]
- 2.Venkatesan A, Chu P, Kerlikowske K, Sickles EA, Smith-Bindman R. Positive predictive value of specific mammographic findings according to reader and patient variables. Radiology. 2009;250(3):648–57. doi: 10.1148/radiol.2503080541. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American College of Radiology (ACR) ACR BI-RADS - Mammography. ACR Breast Imaging and Reporting and Data System, Breast Imaging Atlas. 4. Reston, VA: American College of Radiology; 2003. [Google Scholar]
- 4.GE Healthcare. Centricity RIS-IC. [accessed July 3 2014];Secondary Centricity RIS-IC updated: 2014. http://www3.gehealthcare.com/en/Products/Categories/Healthcare_IT/Medical_Imaging_Informatics_-_RIS-PACS-CVIS/Centricity_RIS-IC#tabs/tab0142BFC079304C1391DE7FA0016AF568.
- 5.American College of Radiology. ACR BI-RADS Atlas-Mammography Section II. [accessed: July 3 2014];Reporting System. updated: 2013. http://www.acr.org/~/media/ACR/Documents/PDF/QualitySafety/Resources/BIRADS/01%20Mammography/02%20%20BIRADS%20Mammography%20Reporting.pdf.
- 6.Lacson R, Khorasani R. Natural language processing for radiology (part 2) J Am Coll Radiol. 2011;8(8):583–4. doi: 10.1016/j.jacr.2011.04.019. S1546-1440(11)00218-3 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Jain N, Friedman C. Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proc AMIA Annu Fall Symp. 1997;1997:829–33. [PMC free article] [PubMed] [Google Scholar]
- 8.Carrell DS, Halgrim S, Tran DT, et al. Using natural language processing to improve efficiency of manual chart abstraction in research: the case of breast cancer recurrence. Am J Epidemiol. 2014;179(6):749–58. doi: 10.1093/aje/kwt441[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Percha B, Nassif H, Lipson J, Burnside E, Rubin D. Automatic classification of mammography reports by BI-RADS breast tissue composition class. J Am Med Inform Assoc. 2012;19(5):913–6. doi: 10.1136/amiajnl-2011-000607[pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore CR, Farrag A, Ashkin E. Using Natural Language Processing to Extract Abnormal Results From Cancer Screening Reports. Journal of Patient Safety. Post Author Corrections. 2014 Jul 14; doi: 10.1097/PTS.0000000000000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yetisgen-Yildiz M, Gunn ML, Xia F, Payne TH. A text processing pipeline to extract recommendations from radiology reports. J Biomed Inform. 2013;46:354–362. doi: 10.1016/j.jbi.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Bozkurt S, Gulkesen KH, Burin D. Annotation for information extraction from mammography reports. Stud Health Technol Inform. 2013;190:183–5. [PubMed] [Google Scholar]
- 13.National Cancer Institute. Breast Cancer Surveillance Consortium Homepage. [accessed: July 3 2014];Secondary Breast Cancer Surveillance Consortium Homepage updated. 2014 Feb 7; http://breastscreening.cancer.gov/
- 14.Chapman W, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of Biomedical Informatics. 2001;34:301–10. doi: 10.1006/jbin.2001.1029[pii]. [DOI] [PubMed] [Google Scholar]
- 15.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: An algorithm for determining negation, experiencer, and temporal status from clinical reports. Journal of Biomedical Informatics. 2009;42(5):839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strauss JA, Chao CR, Kwan ML, Ahmed SA, Schottinger JE, Quinn VP. Identifying primary and recurrent cancers using a SAS-based natural language processing algorithm. J Am Med Inform Assoc. 2013;20(2):349–55. doi: 10.1136/amiajnl-2012-000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uzuner O, Zhang X, Sibanda T. Machine learning and rule-based approaches to assertion classification. J Am Med Inform Assoc. 2009;16(1):109–15. doi: 10.1197/jamia.M2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.