Abstract
Although expression of the human liver fatty acid binding protein (FABP1) T94A variant alters serum lipoprotein cholesterol levels in human subjects, nothing is known whereby the variant elicits these effects. This issue was addressed by in vitro cholesterol binding assays using purified recombinant wild-type (WT) FABP1 T94T and T94A variant proteins and in cultured primary human hepatocytes expressing the FABP1 T94T (genotyped as TT) or T94A (genotyped as CC) proteins. The human FABP1 T94A variant protein had 3-fold higher cholesterol-binding affinity than the WT FABP1 T94T as shown by NBD-cholesterol fluorescence binding assays and by cholesterol isothermal titration microcalorimetry (ITC) binding assays. CC variant hepatocytes also exhibited 30% higher total FABP1 protein. HDL- and LDL- mediated NBD-cholesterol uptake was faster in CC variant than TT WT human hepatocytes. VLDL- mediated uptake of NBD-cholesterol did not differ between CC and TT human hepatocytes. The increased HDL- and LDL- mediated NBD-cholesterol uptake was not associated with any significant change in mRNA levels of SCARB1, LDLR, CETP, and LCAT encoding the key proteins in lipoprotein cholesterol uptake. Thus, the increased HDL- and LDL- mediated NBD-cholesterol uptake by CC hepatocytes may be associated with higher affinity of T94A protein for cholesterol and/or increased total T94A protein level.
Keywords: Human FABP1, hepatocytes, cholesterol uptake, lipoproteins
1. INTRODUCTION
Cholesterol is a very hydrophobic, poorly aqueous soluble molecule as indicated by critical micellar concentrations near 30 nM [1]. High density lipoprotein (HDL) transports cholesterol from peripheral sites through the vasculature to the liver for biliary elimination [2–5]. A major gap in our knowledge of this reverse-cholesterol-transport (RCT) is the role of intracellular factors mediating the rapid intracellular transport of HDL- cholesterol for biliary excretion [3–5].
Increasing evidence, primarily with murine liver fatty acid binding protein (FABP1), suggests that FABP1 may contribute to this function. FABP1 is present at high levels in murine (2–6% of cytosol protein; 200–400 μM) and human (7–10% of cytosolic protein; 700–1000 μM) [6,7]. Murine FABP1 binds cholesterol and oxidized lipids as well as fatty acids in vitro [6,8–13]. Furthermore, murine FABP1 enhances in vitro cholesterol transfer between the plasma membrane and microsomes to stimulate cholesterol esterification by acyl-CoA cholesterol acyltransferase in vitro [8,14–16] and in cultured fibroblasts overexpressing FABP1 [17]. Finally, murine FABP1 significantly colocalizes with and is in close proximity (10 angstroms) of the plasma membrane HDL receptor (scavenger receptor B1, SRB1) as shown by double immunogold electron microscopy colocalization [18]. Although human FABP1 differs significantly from murine FABP1 with respect to amino acid sequence and structure as well as ligand binding cavity size and affinity for fatty acid and fibrates, almost nothing is known about human FABP1’s ability to interact with cholesterol [13,19–22].
Human genetic variations in FABP1 and sexual dimorphism in FABP1 expression are associated with significant changes in serum lipoprotein cholesterol and hepatic lipid metabolism. In humans, FABP1 expression is sexually dimorphic with females expressing higher levels of FABP1 [23–26]. Furthermore, the human FABP1 T94A variant is one of the most frequently occurring polymorphism in the FABP protein family, with a 26–38% minor allele frequency in all populations examined to date: Northern Europeans, Non-Hispanic White Americans, Hispanic Americans, African Americans, Africans, Japanese, Han Chinese, and others (MAF for 1000 genomes in NCBI dbSNP database; ALFRED database). In these groups, the homozygous variant occurs with a mean frequency of 8.3±1.9% (range 1–17%). Plasma lipoproteins are significantly altered in FABP1 T94A variant expressing subjects: i) Plasma low-density lipoprotein (LDL) cholesterol is elevated in T94A variants [27,28]; ii) Although plasma total HDL cholesterol is unaltered, FABP1 T94A variants have increased plasma triglycerides [28,29], which are known to elicit HDL remodeling and HDL subfraction redistribution (HDL2 vs 3) to impact HDL function in RCT [30–32]. The importance of altered HDL composition is underscored by the fact that singularly raising total HDL level genetically or therapeutically does not necessarily alter RCT or CVD [2–5]. The human FABP1 T94A variant is associated with increased CVD [28,29], atherothrombotic cerebral infarction [33], and non-alcohol fatty liver disease (NAFLD) [27]. Expression of the human FABP1 T94A variant, but not the WT human FABP1 T94T, increased cholesterol accumulation in cultured primary human hepatocytes and in cultured ‘Chang’ liver cells [34,35].
It is not known whether either the WT or T94A variant human FABP1 binds cholesterol and whether the T94A variant impacts cholesterol uptake from HDL and/or other serum lipoproteins. These issues were addressed using human FABP1 WT and T94T variant recombinant proteins and cultured primary human hepatocytes expressing these proteins. Data showed for the first time that human FABP1 bound cholesterol, the T94A substitution increased FABP1’s affinity for cholesterol, T94A expression increased total FABP1 protein, and selectively increased uptake of cholesterol from HDL and LDL, but not very low-density lipoprotein (VLDL).
2. MATERIALS AND METHODS
2.1. Materials
NBD-cholesterol [22-(N-(7-nitrobenz-2-oxa-1, 3-diazol-4-yl)-amino)-23, 24-bisnor-5-cholen-3β-ol] was purchased from Life Technologies (Grand Island, NY). Purified human HDL, LDL, and VLDL were obtained from EMD Biosciences (San Diego, CA). Williams’ Medium E (1x, no phenol red), Cryopreserved Hepatocytes Recovery Medium (CHRM®), Hepatocyte Plating Supplement Pack (Serum-containing), and Hepatocyte Maintenance Supplement Pack (Serum-free) were purchased from Life Technologies (Grand Island, NY). Nunc LabTek chambered coverglass used in confocal imaging was purchased from VWR (Radnor, PA). BD Gentest™ High Viability CryoHepatocyte Recovery Medium and BD Gentest™ CryoHepatocyte Plating Medium were purchased from BD Biosciences (San Jose, CA). RNEasy® kit and RNase-Free DNase® set were from Qiagen Sciences (Maryland, USA) and Qiagen GmbH (Hilden, Germany), respectively. TaqMan®, One-Step RT-PCR Master Mix reagents, TaqMan® Gene Expression Assays for mRNAs encoding the following human proteins were purchased from Life Technologies (Grand Island, NY): peroxisome proliferator activated receptor-α (PPARα), (PPARA, Hs00947539), CCAAT/enhancer-binding protein α (C/EBPα), (CEBPA, Hs00269972_s1), Scavenger receptor class B member 1 (SRB1), (SCARB1, Hs00969821_m1), LDL Receptor, (LDLR, Hs00181192_m1), Cholesteryl ester transfer protein (CETP), (CEPT, Hs00163942_m1), Lecithin-Cholesterol Acyltransferase (LCAT), (LCAT, Hs01068069_m1). Rabbit polyclonal antibody against carnitine palmitoyl transferase IA (CPT1A), carnitine palmitoyl-CoA transferase 2 (CPT2), fatty acid transport protein-4 (FATP4), hepatic lipase, and lipoprotein lipase (LPL) were from Santa Cruz Biotechnology, Inc, (Dallas, TX). Antibodies against fatty acid transport protein-2 (FATP2) and fatty acid transport protein-5 (FATP5) were from Abcam plc (Cambridge, MA). Rabbit polyclonal antibodies against mitochondrial glutamate oxaloacetate transaminase (GOT), SCP-2, and rat WT FABP1 were made in this lab [36]. Antibodies against housekeeping proteins glyceraldehyde-3-phosphate dehydrogenase (GAPDH, from skeletal rabbit muscle, host species – mouse, Millipore, Billerica, MA), and β-actin (mouse monoclonal antibody raised against gizzard actin of avian origin. Santa Cruz Biotechnology, Dallas, TX).
2.2. Recombinant human FABP1 T94T and human T94A and rat FABP1 proteins
The cDNA encoding the human FABP1 was purchased from OriGene (Rockville, MD). Sequencing of this cDNA (Research Technology Support Facility, Michigan State University, East Lansing, MI) revealed it to code for the human FABP1 T94A. Recombinant human FABP1 T94A variant protein was prepared from this cDNA using a standard procedure in our labs [9,37]. Standard mutagenesis procedures were used to create cDNA coding for the human WT FABP1 T94T and the recombinant protein was prepared [9,37]. Recombinant rat FABP1 was obtained as described earlier[19]. Identity, purity and concentration of the recombinant proteins were determined by amino acid analysis and mass spectrometry (Protein Chemistry Laboratory, Texas A&M University) [19,20].
2.3. FABP1 NBD-cholesterol fluorescence binding assay
Since murine FABP1 [9] as well as human intracellular cholesterol binding proteins such as SCP-2 and NPC1 bind NBD-cholesterol similar to cholesterol [9,38–41], the fluorescent NBD-cholesterol binding assay was applied to determine the affinity of the human WT FABP1 T94T and T94A variant proteins for cholesterol. The binding of rat FABP1 was carried out for comparison with the human proteins. Briefly, NBD-cholesterol binding was performed by holding the NBD-cholesterol at low concentration (20nM) below the critical micellar concentration similarly as described previously [1,9,42] modified as follows: NBD-cholesterol fluorescence intensity was recorded in the presence of increasing amount of human FABP1 (0–3.6 μM) using a Varian Cary Eclipse Fluorescence Spectrophotometer (Varian, Inc., Palo Alto, CA). NBD-cholesterol was excited at 490 nm and fluorescence emission spectra were recorded between 515 and 600 nm at 24°C with a circulating water bath. Bmax and Kd were determined by curve fitting the binding curve to hyperbola equation as described [9].
2.4. Isothermal titration calorimetry (ITC) analysis of cholesterol binding to FABP1
Equilibrium dissociation constant (Kd), binding stoichiometry (n), and thermodynamic parameters (ΔH, TΔS, ΔG) of cholesterol binding to either rat FABP1 or human WT or T94A variant FABP1 were determined by ITC utilizing an iTC 200 microcalorimeter (MicroCal, Northampton, MA) essentially as described [9]. Briefly, cholesterol (Avanti Polar Lipids, Alabaster, AL) at a final concentration of 300 μM was prepared in a buffer containing 20 mM Tris-HCl (pH 8.0), 20% glycerol, and 4.5% (2-hydroxypropyl)-β-cyclodextrin (Sigma, St. Louis, MO). The cholesterol ligand solution was titrated (2.0 μL per injection) into FABP1 [20 μM protein in 20 mM Tris-HCl (pH 8.0)/20% glycerol] at 25 °C and the amount of heat released or absorbed upon ligand interaction with protein was measured. Control injections of cholesterol into buffer or buffer into protein were performed and these values were subtracted from the experimental value. Each protein/ligand experiment was repeated five times (n = 5). The binding data were analyzed utilizing Origin 7 SR4 as supplied with the iTC 200 microcalorimeter. Statistical analysis was performed by the unpaired t-test utilizing SigmaPlot (Systat Software, San Jose, CA). Data are expressed as the mean ± SEM (n = 5).
2.5. Human hepatocyte culture and genotyping
Cryopreserved plateable primary human hepatocytes from 30 different lots were purchased from Life Technologies (Grand Island, NY) and BD Biosciences (San Jose, CA). The hepatocytes were shipped with cryo shippers containing adsorbed liquid nitrogen. The hepatocytes were immediately stored in a −135°C freezer upon arrival until ready to use. The purchased lots of cryopreserved hepatocytes were isolated from livers of female Caucasian subjects (49.5 ± 2.7 yr) according to the information provided by companies. All personnel working with the human hepatocytes received BL1 and BL2 training and certification for work with human cells in our approved laboratory (Institutional Biosafety Committee, Office of Research Compliance and Biosafety, Division of Research, Texas A&M University). Hepatocytes were cultured and genotyped as described earlier [34] to determine which lots were from carriers of FABP1 T > C single nucleotide polymorphism (SNP) alleles. Out of the total 30 lots from individual donors, 13 lots (representing 13 different individuals) were shown to be carriers of the FABP1 TT alleles (n=13), and 3 lots (representing 3 different individuals) to be carriers of CC SNP alleles (n=3).
2.6. Lipoprotein mediated NBD-cholesterol uptake by human hepatocytes
2.6.1. Preparation of NBD-cholesterol containing lipoproteins
Both in vitro and in vivo findings have shown NBD-cholesterol to be useful as a fluorescent probe for real-time imaging of lipoprotein-mediated cholesterol uptake because NBD-cholesterol is: i) readily incorporated similarly as cholesterol into lipoproteins (HDL, LDL, VLDL) without significantly altering their lipid class composition and functional properties [18,43–46]; ii) an established fluorescent probe for examining lipoprotein-mediated cholesterol uptake and efflux by living murine cultured cells and primary hepatocytes [18,44–47]; iii) taken up and effluxed from HDL-NBD-cholesterol via the murine scavenger receptor B1 (SRB1) similar to HDL-cholesterol [18,46,47]; iv) absorbed and esterified in vivo and in vitro similar to cholesterol [44,45,48]. Therefore, human lipoprotein-mediated NBD-cholesterol uptake by cultured primary human hepatocytes was measured by first incorporating NBD-cholesterol into human HDL, LDL, and VLDL similarly as previously described [18,43–46]. Lipoprotein protein level was quantitated with Bio-Rad protein assays, and the amount of NBD-cholesterol incorporated into the respective lipoproteins was determined by measuring the fluorescence intensity in ethanol and comparison to NBD-cholesterol standard in ethanol.
2.6.2. Lipoprotein mediated NBD-cholesterol uptake using 96-well microplate reader
Cryopreserved human hepatocytes were genotyped as described in Section 2.5., TT (n=13), CC (n=3). Hepatocytes were cultured at 6000 cells/well in collagen-coated 96-well microplates (replicates = 6) and cultured over night as described above. For dose response experiments, cultured primary human hepatocytes were incubated with NBD-cholesterol-labeled human HDL (10, 20, 50, 100mg), LDL (2, 5, 10, 25mg), or VLDL (2, 5, 10, 25mg) in PBS for 2 hours in the 37°C incubator. Then NBD-cholesterol fluorescence intensities were read with a microplate reader. To obtain the NBD-cholesterol uptake curves, fluorescence intensity measurements were started immediately after addition of NBD-cholesterol-labeled HDL (10μg), LDL (2μg), or VLDL (2 μg) to human hepatocytes. Fluorescence intensity was recorded at 37°C every 2min for 2 hours. Because LDL and VLDL are much more lipid-rich, less LDL and VLDL protein were added as compared to HDL protein to avoid overloading the hepatocytes with NBD-cholesterol or saturating the photomultipliers detecting NBD-cholesterol fluorescence. On each microplate, hepatocytes with PBS only were used as controls for light scatter and hepatocyte autofluorescence, while empty wells with NBD-cholesterol-labeled HDL, LDL, or VLDL were used as controls for NBD-cholesterol background fluorescence and photo bleaching. After all the corrections, the uptake curves were fitted to Y= a(1−e−bt), therefore the initial rate IR=ab, and half time t1/2 = ln2/b. A Synergy 2 microplate reader (BioTek Instruments, Inc. Winooski, Vermont) was used to record NBD-cholesterol fluorescence intensity using 460/40 excitation filter, 540/35 emission filter, bottom optics position, and tungsten as the light source.
2.6.3. Confocal imaging study of Lipoprotein-mediated NBD-cholesterol uptake in cultured primary human hepatocytes
Lipoprotein-mediated uptake of NBD-cholesterol by cultured primary human hepatocytes was similar as described for murine fibroblasts, pre-adipocytes, and primary hepatocytes [18,44–47]. Briefly, genotyped human hepatocytes (described in section 2.5.) were plated at a density of 1.5 × 105 cells per well on collagen-coated two-chamber cover glasses (Nalge Nunc Lab-Tek). After overnight incubation with maintenance media, cells were washed twice with warm (37°C) PBS, incubated for 15 min in PBS at 37°C under 5% CO2, and transferred to a 37°C heated microscope stage for confocal time course image acquisition as described earlier for primary mouse hepatocytes [18,46]. Metamorph 4.0 (Molecular Devices, Sunnyvale, CA) was used to quantify the fluorescence intensities of individual cells throughout each time course. Excel (Microsoft, Redmond, WA) and SigmaPlot were used to calculate the average relative intensity of NBD fluorescence at each time point and to graph the resulting values, respectively. Each cell type and treatments were repeated 3 times and the average ± SE was plotted.
2.7. RNA isolation and gene expression analysis by quantitative real time PCR
Genotyped primary human hepatocytes (described in section 2.5.) were cultured in triplicates, total mRNA collected, and mRNA levels of PPARA, CEBPA, SCARB1, LDLR, CEPT, and LCAT determined by quantitative real time PCR (qRT-PCR) as described earlier [34].
2.8. Western blotting
Genotyped primary human hepatocytes (described in Section 2.5.) were cultured overnight in triplicates, homogenized, and protein was determined using a Bio-Rad Protein Assay (Bio-Rad laboratories, Inc., Hercules, CA) as described previously [49,50]. Each was then subjected to SDS-PAGE gel electrophoresis and Western blotting with rabbit anti-human carnitine palmitoyl transferase IA (CPT1A, dilution factor 1/250), carnitine palmitoyl-CoA transferase 2 (CPT2, 1/250), fatty acid transport protein-5 (FATP5, 1/500), fatty acid transport protein-4 (FATP4, 1/250), fatty acid transport protein-2 (FATP2, 1/250), mitochondrial glutamate oxaloacetate transaminase (GOT, 1/2000), and sterol carrier protein-2 (SCP-2, 1/250), FABP1 (1/500), hepatic lipase (1/250), lipoprotein lipase (LPL, 1/250), and against either of the housekeeping proteins GAPDH (1/4000) or β-actin (1/1000) as described previously [49,50].
2.9. Statistics
Statistical analysis was performed by one-way ANOVA combined with the Newman-Keuls multiple-comparisons test (GraphPad Prism, San Diego, CA) unless otherwise specified. P < 0.05 was chosen as threshold for statistical significance.
3. RESULTS
3.1. NBD-cholesterol binding to FABP1
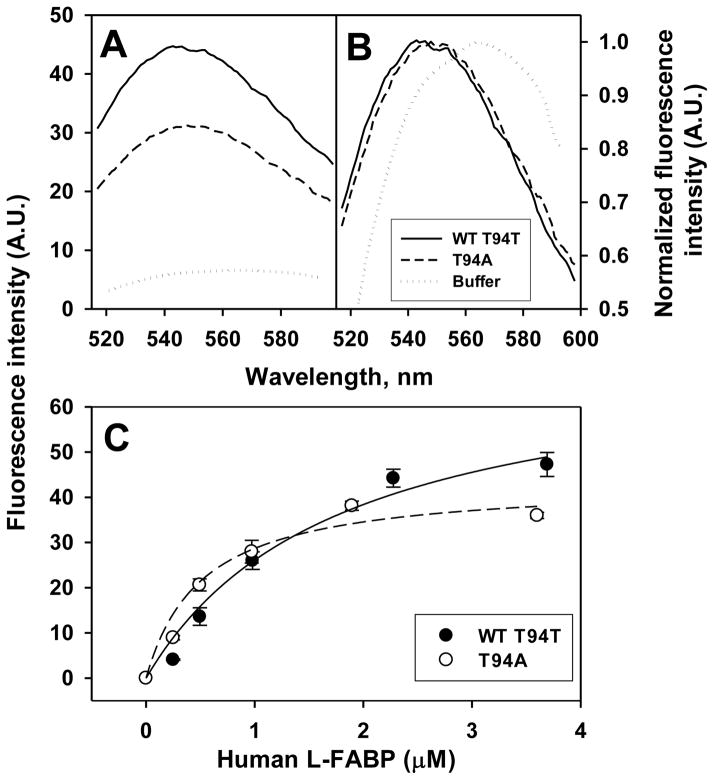
Since murine FABP1 binds NBD-cholesterol similarly as cholesterol [9], this probe was used to examine cholesterol binding to human WT FABP1 T94T, T94A variant and rat FABP1. While NBD-cholesterol fluoresces only weakly in aqueous buffer (Fig 1A dotted line), fluorescence intensity is significantly increased when bound to the recombinant human WT FABP1 T94T and T94A variant proteins (Fig 1A, solid and dashed lines respectively). When normalized, the emission maximum of WT FABP1 bound NBD-cholesterol is blue-shifted nearly 21 nm versus NBD-cholesterol in aqueous buffer (Fig. 1B, solid versus dotted line) while that of NBD-cholesterol bound to the T94A variant appeared red-shifted versus NBD-cholesterol bound to WT FABP1 T94T (Fig 1B dashed versus solid line). Quantitative analysis of multiple spectra shows that the maximum emission of NBD-cholesterol bound to the T94A variant is shifted 5 nm as compared to the WT FABP1 T94T protein i.e. from 544 in the WT FABP1 T94T protein to 549 nm in T94A variant (Table 1). While both the recombinant human WT FABP1 T94T and T94A variant proteins bound NBD-cholesterol, they also differed significantly in maximal fluorescence intensity upon NBD-cholesterol binding. Both proteins displayed saturation binding curves for NBD-cholesterol, but that for the T94A variant protein rose more rapidly and leveled off at lower intensity (Fig 1C, open circles). Quantitative analysis of multiple binding curves showed that the maximal fluorescence intensity of NBD-cholesterol bound to the T94A variant protein was nearly 40% lower than when bound to the WT FABP1 T94T protein (Table 1). This was consistent with NBD-cholesterol exhibiting a lower quantum yield when fully bound to T94A variant than when fully bound to WT T94T. Finally, human WT FABP1 T94T and T94A variant proteins bound NBD-cholesterol with different affinities. Both fit a single exponential binding curve, consistent with each having a single NBD-cholesterol binding site. Analysis of multiple binding curves showed that WT FABP1 T94T protein bound NBD-cholesterol with moderate affinity as evidenced by a Kd of 1.77 μM (Table 1). However, the T94A variant bound NBD-cholesterol with much higher affinity as shown by 3.5-fold lower Kd (Table 1). Finally, human FABP1 T94A variant had very similar NBD-cholesterol binding affinity as the rat FABP1, while human WT FABP1 had significantly lower NBD-cholesterol binding affinity (higher Kd) (Table 1) than rat FABP1.
Figure 1. Interaction of human FABP1 with NBD-cholesterol: fluorescence emission spectra and binding curves.
Panel A, representative fluorescence emission spectra of NBD-cholesterol (20nM) in buffer and when bound to human WT FABP1 T94T or T94A variant protein (3.6 μM). Panel B, normalized NBD-cholesterol fluorescence emission spectra. Panel C, maximal fluorescence emission intensity of NBD-cholesterol, presented as Mean ± SE (n=3–5), titrated with increasing amount of human FABP1 (T94T and T94A). Panels A-C, WT FABP1 T94T: solid line and dark circles; FABP1 T94A Variant: dashed line and open circles; Buffer: dotted lines.
Table 1.
Binding constants of FABP1 with NBD-cholesterol a.
| Rat | WT | T94A | |
|---|---|---|---|
| Emission max b (nm) | 540.7 ± 0.7 | 543.9 ± 0.5 d | 548.9 ± 1.0 d, e |
| Bmax (A.U.) c | 68.2 ± 7.1 | 72.4 ± 7.7 | 43.4 ± 3.3 d, e |
| Kd (μM) c | 0.48 ± 0.12 | 1.77 ± 0.43 d | 0.51 ± 0.15 e |
Data are presented as Mean ± SE, (n=3–5).
Emission maxima were determined from spectral analysis using the instrument software.
Dissociation constant (Kd) and fluorescence intensity at saturation (Bmax) were calculated by fitting the binding curves to hyperbola equation as described in section 2.3.
P < 0.05, significantly different from rat FABP1.
P < 0.05, significantly different from WT FABP1.
In summary, while both the human WT FABP1 T94T and T94A variant proteins appeared to bind NBD-cholesterol at a single binding site, the T94A variant bound NBD-cholesterol with higher affinity with the NBD-cholesterol localized therein in a slightly different orientation, or the variant protein was folded differently upon NBD-cholesterol binding.
3.2. Thermodynamic Binding Parameters of Cholesterol to human WT FABP1 and T94A variant
In order to confirm that binding of NBD-cholesterol was not simply due to presence of the NBD fluorophore, ITC measured cholesterol binding and in addition resolved the enthalpic and entropic contributions to cholesterol. Human WT FABP1 and T94A variant bound approximately one mole of cholesterol per mole of protein with high affinity, 1.3±0.4 and 0.5±0.1 μM (Table 2). Cholesterol binding to both WT FABP1 and T94A variant was energetically favorable as shown by the negative Gibbs free energy changes (ΔG, Table 2). Cholesterol binding to T94A variant was significantly more favorable enthalpically than the binding of cholesterol to WT FABP1 (ΔH, Table 2). However, the binding of cholesterol to WT FABP1 was not significantly different entropically than the binding of cholesterol to T94A variant (ΔS, Table 2). The net result of these contributions to ligand binding was Gibbs free energy change for the cholesterol/T94A variant interaction trended more negative, even though it was not statistically significant, than was observed for the cholesterol/WT FABP1 interaction (ΔG, Table 2).
Table 2.
Thermodynamic parameters of cholesterol binding to rat or human FABP1 a.
| Parameter | Rat | WT | T94A |
|---|---|---|---|
| Kd, μmol/L | 0.6 ± 0.2 | 1.3 ± 0.4 | 0.5 ± 0.1 |
| n, mol/mol | 0.9 ± 0.1 | 0.8 ± 0.2 | 0.7 ± 0.2 |
| ΔH, kJ/mol | −15 ± 1 | −17 ± 2 | −23 ± 4 b |
| TΔS, kJ/mol | 25 ± 3 | 21 ± 2 | 22 ± 1 |
| ΔG, kJ/mol | −41 ± 2 | −38 ± 3 | −45 ± 3 |
Thermodynamic parameters of cholesterol binding to either rat FABP1 or human WT or T94A variant FABP1 were determined by ITC as described in Materials and Methods, section 2.4. Kd, binding dissociation constant; n, binding stoichiometry; ΔH, enthalpy change; ΔS, entropy change; ΔG, Gibbs free energy change. Values represent the means ± SEM (n = 5).
P < 0.05 for T94A vs Rat.
In summary, ITC binding assay confirmed the higher cholesterol binding affinity of human T94A variant protein as compared to human WT FABP1 T94T.
3.3. Expression of FABP1 and Stability of Primary Human Hepatocytes in Culture
FABP1 protein levels of human hepatocyte (genotyped TT and CC) were quantitated by western blot analysis on day 0 and day 2 in primary culture. CC hepatocytes expressed 30% more FABP1 on both day 0 and day 2 (Table 3). 2-way ANOVA statistical analysis showed significant differences between CC and TT (P = 0.03), but no significant differences between day 0 and day 2 (P= 0.08). FABP1 expression levels of CC hepatocytes on day 0 and day 2 are shown in Fig 2J. Human hepatocytes maintained stable expression of multiple markers for up to 2 days in culture as determined by mRNA levels of CEBPA and PPARA not being significantly different between day 0 and day 2 (Fig 2A,B). Western analysis also showed that protein levels of CPT1A, CPT2, FATP2, FATP5, FATP4, GOT, SCP-2, and FABP1 were not significantly different between day 0 and day 2 (Fig 2C–J). Thus, all studies were performed with primary human hepatocytes maintained in culture ≤2 days.
Table 3.
FABP1 protein expression levels of human hepatocytes (TT and CC).
| FABP1 protein expression level | |||
|---|---|---|---|
| TT | CC | CC/TT ratio | |
| Day 0 | 1.00 ± 0.06 | 1.33 ± 0.30 | 1.33 |
| Day 2 | 0.81 ± 0.08 | 1.06 ± 0.15 | 1.31 |
Total 30 lots of cryopreserved human hepatocytes from Life Technologies and BD Biosciences were genotyped, TT (n=13) and CC (n=3), as described in section 2.5. The hepatocytes were cultured in 6-well plates in triplicates and FABP1 protein levels were determined by western blots (see Section 2.8.). Data are normalized to TT day 0 as 1, and are presented as mean ± SE. The statistical analysis with two-way ANOVA showed significant differences between TT and CC (P = 0.03), but no significant differences between day 0 and day 2 (P = 0.08).
Figure 2. Markers of primary human hepatocyte stability in culture.
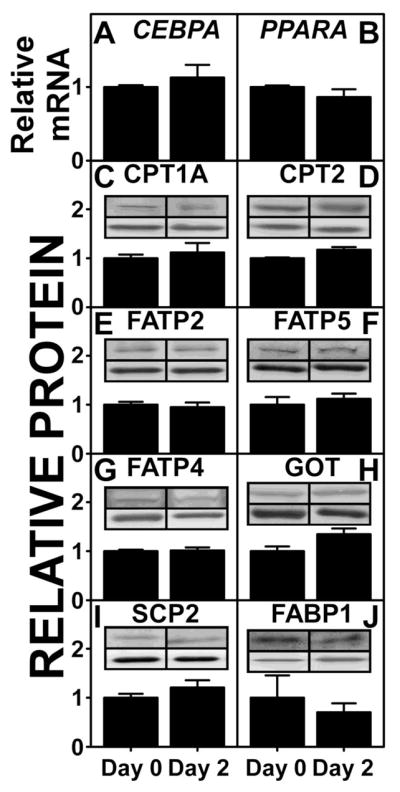
Primary human hepatocytes, genotyped as CC (n=3) were cultured in triplicates in 6-well plates. mRNA and protein levels of marker proteins at day 0 and day 2 were determined as described in MATERIALS AND METHODS. mRNA levels of CEBPA (A) and PPARA (B) were determined by qRT-PCR. Protein levels of CPT1A (C), CPT2 (D), FATP2 (E), FATP5 (F), FATP4 (G), GOT (H), SCP-2 (I), and FABP1 (J) were determined by western analysis relative to housekeeping proteins GAPDH or β-actin. Each inset shows a representative blot of the respective protein (upper band) and housekeeping protein (lower band). Data were presented as Mean ± SE.
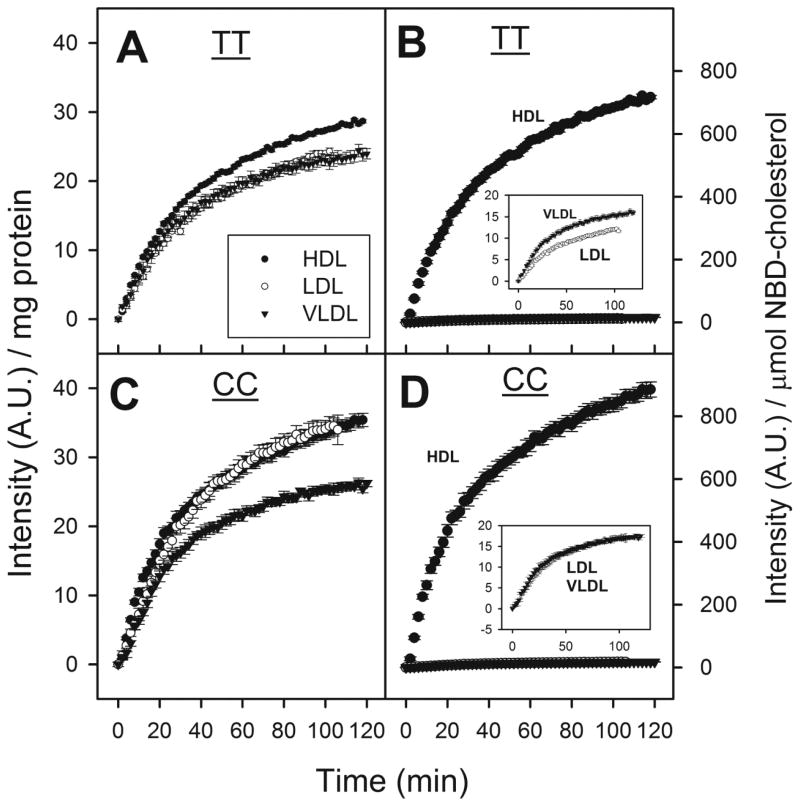
3.4. Lipoprotein mediated NBD-cholesterol uptake by human hepatocytes expressing WT FABP1 (TT) and T94A variant (CC)
3.4.1. HDL mediated NBD-cholesterol uptake
The finding that human WT FABP1 T94T protein bound NBD-cholesterol and this binding affinity was significantly increased in the T94A variant suggested a potential role in lipoprotein-mediated cholesterol uptake in human hepatocytes. Therefore, the impact of the FABP1 T94A variant on HDL-mediated cholesterol uptake was examined by both a fluorescence microplate assay of a large number of hepatocytes and confirmed by confocal imaging of a smaller number of hepatocytes.
NBD-cholesterol uptake experiments with 96-well plates made it possible to perform experiments with different cell types, blanks, controls, and different treatments on the same plate at the same time, therefore greatly reduced the time variations. Furthermore, the uptake curves obtained from preliminary HDL mediated NBD-cholesterol uptake experiments with TT hepatocytes at the beginning and the end of the day showed no significant differences (data not shown).
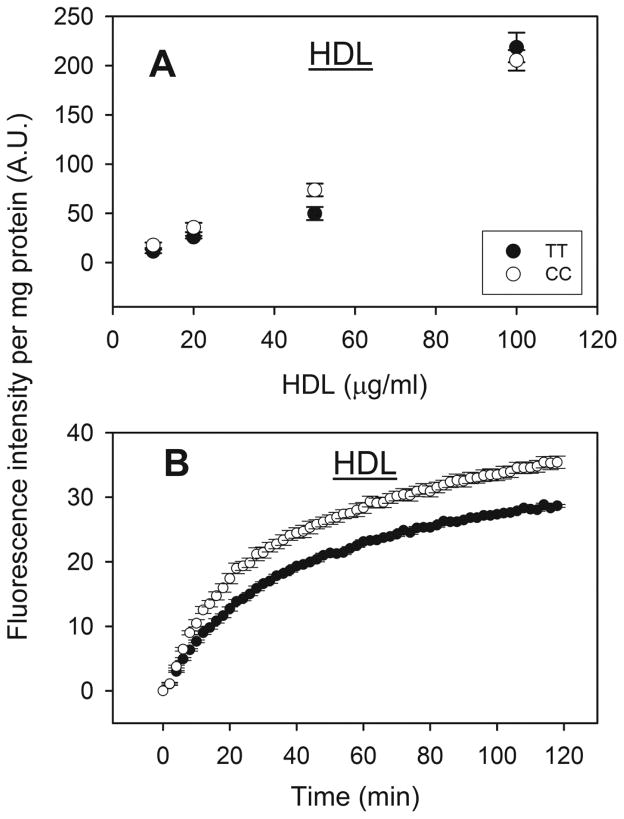
First, in both TT and CC hepatocytes the HDL-mediated uptake of NBD-cholesterol examined using a fluorescence microplate assay (6,000 hepatocytes/well) increased nearly linearly with increasing HDL concentration up to 100 μg HDL protein/ml culture medium (Fig 3A). Although HDL mediated uptake of NBD-cholesterol was not saturated even at the highest HDL protein concentration tested in either genotype, all subsequent studies were performed using a lower dose (10μg/ml) of NBD-cholesterol labeled HDL.
Figure 3. Effect of human FABP1 T94A variant expression on HDL-mediated cholesterol uptake: fluorescence microplate assay.
Human hepatocytes (genotyped as TT and CC) from Table 3 were cultured in 96 well plates, NBD-cholesterol labeled HDL was added to cells and fluorescence intensity determined in a microplate reader at 37°C. Panel A: Dose response of HDL-mediated NBD-Cholesterol uptake by human hepatocytes over the range of 10–100μg HDL protein/ml. The fluorescence intensities were measured at the end of 2 hour incubation. Panel B: Time curve of HDL-mediated NBD-cholesterol uptake (10μg HDL protein/ml) by cultured primary human hepatocytes. Data shown were from a representative donor of each genotype (TT and CC), presented as Mean ± SE (replicate = 6), and were corrected for hepatocyte autofluorescence, photo-bleaching, scatter, and NBD-cholesterol fluorescence in buffer (section 2.6.2.).
Second, the T94A variant significantly increased HDL-mediated NBD-cholesterol uptake. HDL-mediated uptake of NBD-cholesterol by WT TT primary human hepatocytes was rapid, increasing linearly for the first 5 min and slowing thereafter without reaching a maximum even after 2 h (Fig 3B, solid circles). Quantitative analysis of NBD-cholesterol fluorescence in multiple wells in the microplate assay showed that the initial rate of HDL-mediated uptake by CC hepatocytes was significantly faster than by TT hepatocytes, 1.1 versus 0.8 arbitrary fluorescence units (AU)/min (Table 4). Consequently, the half-time of HDL-mediated uptake by CC hepatocytes was significantly shorter than by TT hepatocytes, 22 versus 26 min (Table 4).
Table 4.
HDL, LDL, and VLDL mediated uptake of NBD-Cholesterol by human hepatocytes (genotyped as TT and CC) a.
| Initial rate (A.U./min) | Half time (t1/2) (min) | |||
|---|---|---|---|---|
| TT | CC | TT | CC | |
| HDL | 0.79 ± 0.05 | 1.08 ± 0.09 b | 25.7 ± 0.4 | 22.3 ± 0.6 b |
| LDL | 0.63 ± 0.02 | 0.94 ± 0.02 b | 28.5 ± 0.7 | 27.3 ± 0.4 |
| VLDL | 0.71 ± 0.01 | 0.81 ± 0.01 b | 23.5 ± 0.4 | 22.5 ± 0.4 |
Genotyped human hepatocytes from Table 3 were cultured in 96-well plates (replicates = 6).
Data are presented as Mean ± SE.
P < 0.05, CC significantly different from TT.
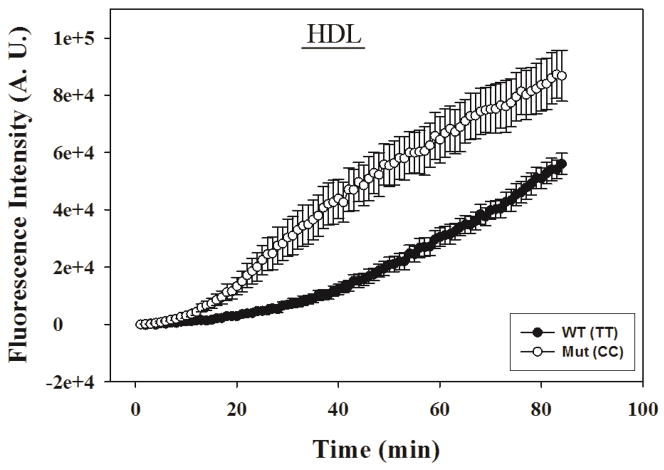
Confocal imaging of a much smaller number of individual hepatocytes’ HDL-mediated uptake of NBD-cholesterol suggested a slower, more delayed uptake by both genotypes (Fig 4), as compared to that in the fluorescence microplate assay (Fig 3B). Imaging method also allowed the monitoring of cell morphology, which showed the cells did not swell or change volume during the 2 hr period that uptake experiments were taking place (data not shown). Slower uptake in the confocal imaging assay was likely due to slower mixing after addition of the NBD-cholesterol-labeled HDL than in the microplate assay. Regardless, however, the confocal imaging assay also confirmed that the CC hepatocytes took up NBD-cholesterol from HDL more rapidly than did the WT TT hepatocytes (Fig 4).
Figure 4. Effect of human FABP1 T94A variant expression on HDL-mediated cholesterol uptake: confocal imaging.
Genotyped human hepatocytes (TT and CC) from Table 3 were cultured in triplicate in 2 chamber coverglasses, treated with NBD-cholesterol labeled HDL (10 μg HDL protein/ml), and fluorescence increase of 5–8 cells per field was monitored by confocal fluorescence imaging as described in Section 2.6.3. Imaging analysis was done with Metamorph software. Data shown were from a representative donor of each genotype (TT and CC), presented as Mean ± SE (replicate = 3).
Taken together these findings showed that the CC human hepatocytes exhibited faster and higher HDL-mediated uptake of NBD-cholesterol than TT hepatocytes.
3.4.2. LDL-mediated NBD-cholesterol uptake
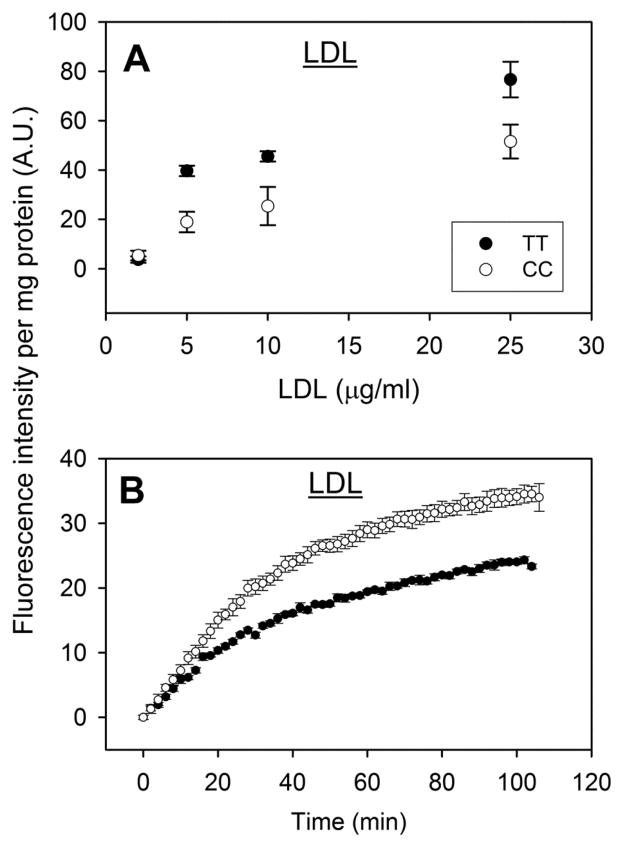
The dose response curve for LDL-mediated NBD-cholesterol uptake by both WT TT and CC variant expressing hepatocytes was linear only at low concentrations (i.e. less than 5 μg LDL protein/ml culture medium), slowly approaching saturation thereafter (Fig 5A). At ≥5 μg LDL protein/ml culture medium, the CC variant hepatocytes were more readily saturated by LDL-mediated NBD-cholesterol uptake (Fig 5A). Therefore, all subsequent LDL-mediated NBD-cholesterol uptake experiments were performed with 2 μg LDL protein/ml culture medium. T94A variant expression significantly increased LDL-mediated NBD-cholesterol uptake (Fig 5B). Quantitative analysis of NBD-cholesterol fluorescence in multiple wells in the microplate assay showed that the initial rate of LDL-mediated uptake by CC hepatocytes was again significantly faster than by TT hepatocytes, 0.9 versus 0.6 arbitrary fluorescence units (AU)/min (Table 4). However, due to higher maximal uptake the half-time of LDL-mediated uptake of NBD-cholesterol by CC and TT hepatocytes did not significantly differ (Table 4).
Figure 5. Effect of human FABP1 T94A variant expression on LDL-mediated cholesterol uptake: fluorescence microplate assay.
LDL-mediated uptake of NBD-cholesterol was determined in genotyped human hepatocytes (TT and CC) cultured in 96 well plates. Panel A: Dose response of LDL-mediated NBD-Cholesterol uptake by human hepatocytes over the range of 2–25 μg LDL protein/ml. Panel B: Time curve of LDL-mediated NBD-cholesterol uptake (2 μg LDL protein/ml) by cultured primary human hepatocytes. Data shown were from a representative donor of each genotype (TT and CC), presented as Mean ± SE (replicate = 6).
In summary, LDL-mediated uptake of NBD-cholesterol by WT TT primary human hepatocytes was only slightly slower and exhibited longer half-time than that of NBD-cholesterol uptake from HDL. Expression of the T94A variant significantly increased the LDL-mediated uptake of NBD-cholesterol, primarily the initial rate.
3.4.3. VLDL mediated NBD-cholesterol uptake
VLDL-mediated NBD-cholesterol uptake by both WT TT and CC variant expressing hepatocytes was linear from 0 – 25 μg/ml (Fig 6A). However, all subsequent VLDL-mediated NBD-cholesterol uptake experiments were performed with 2 μg LDL protein/ml culture medium. Expression of the T94A variant modestly but significantly increased VLDL-mediated NBD-cholesterol uptake (Fig 6B). Quantitative analysis of multiple uptake curves showed that the initial rate of VLDL-mediated NBD-cholesterol uptake by CC variant hepatocytes was significantly faster than by WT TT hepatocytes, 0.8 versus 0.7 arbitrary fluorescence units (AU)/min (Table 4). However, due to higher saturation the half-times of VLDL-mediated uptake of NBD-cholesterol by CC hepatocytes were not significantly altered as compared to that by TT hepatocytes (Table 4).
Figure 6. Effect of human FABP1 T94A variant expression on VLDL-mediated cholesterol uptake: fluorescence microplate assay.
VLDL-mediated uptake of NBD-cholesterol was determined in genotyped human hepatocytes (TT and CC) that were cultured in 96 well plates. Panel A: Dose response of VLDL-mediated NBD-Cholesterol uptake by human hepatocytes over the range of 2–25 μg VLDL protein/ml. Panel B: Time curve of VLDL-mediated NBD-cholesterol uptake (2 μg VLDL protein/ml) by cultured primary human hepatocytes. Data shown were from a representative donor of each genotype (TT and CC), presented as Mean ± SE (replicate = 6).
Thus, the initial rate of VLDL-mediated NBD-cholesterol uptake in WT TT hepatocytes was intermediate between that of HDL- and LDL-mediated uptake. While CC hepatocytes had increased initial rate of VLDL-mediated NBD-cholesterol uptake, the magnitude of increase was smaller than that mediated by HDL and LDL. Consequently, the initial rate of NBD-cholesterol mediated by VLDL was slower than that by LDL and even slower than that mediated by HDL.
3.4.4. Impact of FABP1 T94A expression on specificity of lipoprotein mediated NBD-cholesterol uptake by cultured primary human hepatocytes
The measured NBD-cholesterol content in the respective lipoproteins, i.e. 0.04, 2.0, and 1.5 μmol/mg protein for NBD-cholesterol-labeled HDL, LDL, and VLDL, respectively, generally reflected that of the free cholesterol content/mg protein of these lipoproteins [18]. Therefore, for direct comparisons of the specificity of lipoprotein-mediated uptake of NBD-cholesterol, uptake curves were normalized based on protein and cholesterol content. When expressed on the basis of protein content, maximal lipoprotein-mediated NBD-cholesterol uptake by WT TT genotype human hepatocytes appeared slightly faster and to a greater extent for HDL than for LDL and VLDL which were similar to each other (Fig 7A). In contrast, when normalized for NBD-cholesterol content the WT TT genotype hepatocytes preferentially took up NBD-cholesterol in the order HDL ≫ VLDL > LDL (Fig 7B). T94A variant significantly altered the specificity of lipoprotein-mediated NBD-cholesterol uptake, when expressed on the basis of both protein and NBD-cholesterol contents. Based on protein content, maximal lipoprotein-mediated NBD-cholesterol uptake by CC variant genotype human hepatocytes appeared in the order: HDL, LDL > VLDL (Fig 7C). When uptake was normalized for NBD-cholesterol content such that CC variant genotype hepatocytes preferentially took up NBD-cholesterol in the order HDL ≫ LDL, VLDL. (Fig 7D).
Figure 7. Impact of FABP1 T94A variant expression on specificity of lipoprotein mediated NBD-Cholesterol uptake.
Panels A and C: NBD-cholesterol uptake curves from Figures 3B, 5B, and 6B were normalized for lipoprotein protein content. Panels B and D: NBD-cholesterol uptake curves from Figures 3B, 5B, and 6B were normalized for lipoprotein NBD-cholesterol content. Data represent the Mean ± SE (replicates = 6).
Thus, based on NBD-cholesterol content the uptake was fastest and most extensively facilitated by HDL, consistent with the known role of HDL in reverse cholesterol transport (RCT). Regardless of whether expressed on the basis of protein or NBD-cholesterol content, the T94A variant increased HDL-mediated NBD-cholesterol uptake. However, when normalized to NBD-cholesterol content, expression of the FABP1 T94A variant subtly abolished the difference in specificity of LDL vs VLDL mediated NBD-cholesterol uptake.
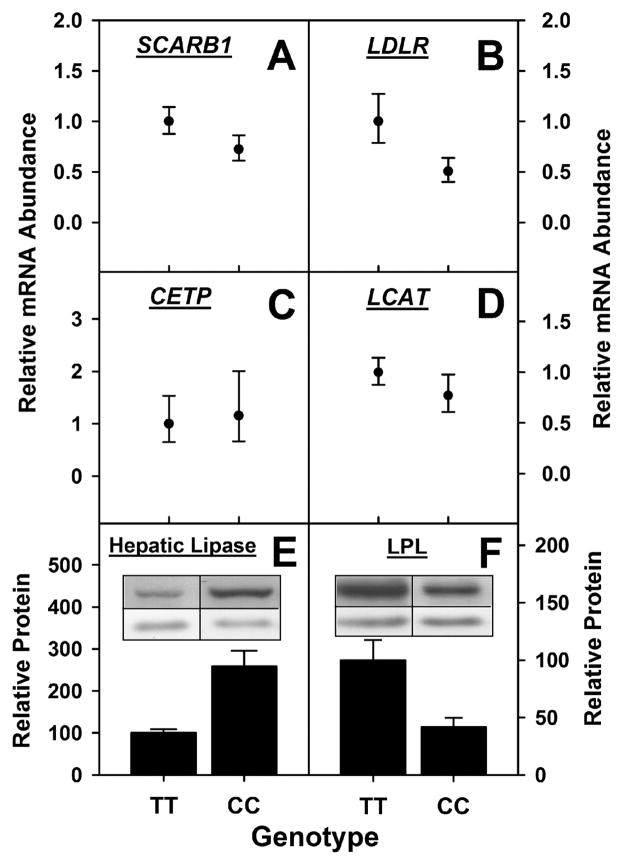
3.5. Hepatic expression of proteins involved in lipoprotein-mediated cholesterol uptake (SCARB1, LDLR), lipoprotein cholesterol esterification/transfer (CETP, LCAT), and lipoprotein TG hydrolysis (hepatic lipase, LPL)
Since expression of the T94A variant increased lipoprotein-mediated uptake of NBD-cholesterol by cultured primary human hepatocytes, it was important to determine if this was associated with concomitant upregulation of key proteins involved in lipoprotein cholesterol uptake and esterification/transfer. The mRNA level of key receptors for HDL and LDL mediated cholesterol uptake was not significantly altered in CC hepatocytes. The mRNA level of the scavenger receptor B1 (SCARB1) instead tended to decrease, albeit not significantly in CC hepatocytes (Fig 8A). Likewise, mRNA level of the LDL receptor (LDLR) tended to decrease, again not significantly, in CC hepatocytes (Fig 8B). Expression of the VLDL receptor was not examined because, unlike SCARB1 and LDLR, the VLDL receptor is a peripheral tissue receptor barely detectable in liver [51]. The mRNA level of proteins involved in lipoprotein cholesterol esterification and transfer was not altered in CC hepatocytes. The mRNA levels of cholesterol ester transfer protein, CETP (Fig 8C), and lecithin cholesterol acyltransferase, LCAT (Fig 8D) were not significantly altered in CC hepatocytes as compared to their WT TT counterparts. Hepatocyte protein level of hepatic lipase (hydrolyzes TG in HDL but not VLDL or LDL) was significantly higher (Fig 8E) in CC hepatocytes as compared to their WT TT counterparts. In contrast, hepatocyte level of LPL (hydrolyzes TG in VLDL) was significantly lower in CC hepatocytes (Fig 8F).
Figure 8. Effect of human FABP1 T94A variant expression on levels of lipoprotein receptors and cholesterol esterification/transfer proteins, and lipases.
Primary human hepatocytes genotyped as TT (n=13) and CC (n=3) from Table 3 were cultured in triplicate and mRNA levels of SCARB1 (panel A), LDLR (panel B), CETP (panel C), and LCAT (panel D) were determined by qRT-PCR as described in Section 2.7. The data are presented as mean value of CC relative to TT (set as 1) with error bars showing the upper and lower range. Protein levels of hepatic lipase (panel E) and LPL (panel F) were determined by western blots as described in Section 2.8. The data are presented as Mean ± SE, relative to TT (set as 100).
Thus, the increased lipoprotein-mediated uptake of NBD-cholesterol by CC hepatocytes was not due to: i) upregulation of SRB1 or LDL receptor; ii) upregulation of the two key proteins involved in cholesterol conversion to cholesteryl ester and cholesteryl ester transfer between lipoproteins (CETP, LCAT); iii) increased levels of lipase that hydrolyzes VLDL since LPL was decreased.
4. DISCUSSION
While low level and reduced function of HDL correlate with increased risk of cardiovascular disease (CVD), some genetic and therapeutic mechanisms that raise HDL cholesterol do not appear to stimulate reverse cholesterol transport (RCT) nor lower risk of CVD [2–5]. Not only altered HDL composition, but also yet poorly understood intracellular factors, impact HDL cholesterol clearance [3–5]. Recent intriguing clinical studies of the human FABP1 T94A variant suggest a potential role for the human FABP1 T94A variant [27–29,33]. Despite the limited scope of this investigation, the results presented herein provide the following new insights regarding potential mechanism(s) whereby the human FABP1 T94A variant elicits these clinical abnormalities:
First, the T94A substitution significantly alters the polarity of the NBD-cholesterol binding site in the human FABP1. The NBD fluorophore is highly sensitive to the polarity of the surrounding microenvironment [52]. The maximal emission wavelength of NBD-cholesterol bound to the human FABP1 T94A variant was red shifted 5 nm and its intensity was lower as compared to that in the WT FABP1 T94T protein, suggesting increased polarity of the binding site. The T94A substitution occurs at position 94 of the β-barrel comprising the ligand binding cavity and results in the replacement of a medium-sized, polar, uncharged threonine (T) residue with a smaller, nonpolar, aliphatic alanine (A) residue [20]. Since alanine is less polar and could potentially make the binding site less polar locally, the increased polarity is likely the result of the significantly altered secondary structure of the T94A protein [19,20]. Furthermore, the altered secondary structure of the T94A protein significantly alters this variant’s conformational stability as well as impairs its ability to facilitate ligand activation of PPARα in cultured primary human hepatocytes [19,20].
Second, the human FABP1 T94A variant has more than 3-fold higher affinity for NBD-cholesterol than WT. The increased ability to bind cholesterol (confirmed by ITC) appears highly specific for this ligand since the human WT FABP1 and T94A variant proteins do not or only very little differ in affinity for the fluorescent NBD-stearic acid and other lipidic ligands [19,20].
Third, the human FABP1 T94A variant increased HDL-mediated uptake of NBD-cholesterol in cultured primary human hepatocytes. The increased uptake by CC hepatocytes was associated with a three-fold higher binding affinity of the T94A protein for cholesterol, 30% increased total FABP1 protein, and increased protein level of hepatic lipase, a lipase specific for TG in HDL (but not VLDL or LDL) [53,54]. In vivo alterations in hydrolysis of VLDL triglycerides cannot be ruled out since the hydrolysis of VLDL triglycerides also occurs in peripheral tissue. However, these findings are consistent with clinical studies showing that, while the T94A variant does not alter total HDL cholesterol, nevertheless expression of the T94A variant increases plasma triglycerides [28,29]. Increased plasma triglyceride level is known to induce HDL remodeling and thereby impact HDL function in RCT [30–32].
Fourth, the human FABP1 T94A variant increased LDL-mediated uptake of NBD-cholesterol in cultured primary human hepatocytes. This increased LDL-mediated NBD-cholesterol uptake was not due to concomitant upregulation of human LDL receptor LDLR, LCAT, CETP, or LPL (actually decreased) in CC variant human hepatocytes. It is important to note, however, that additional factors not measured herein may also contribute to the increased LDL-mediated NBD-cholesterol uptake including altered expression of proteins involved in cholesterol efflux to serum (e.g. ABCA1 and ABCG1) and excretion into bile (e.g. ABCG5 and ABCG8). Importantly, functional and clinical studies support a role for the FABP1 T94A variant in altered LDL cholesterol metabolism. For example, Chang liver cells overexpressing the human FABP1 T94A variant accumulated more cholesterol than did WT human FABP1 overexpressors [35]. Clinical studies show that the human FABP1 T94A mutation is associated with elevated plasma TG and LDL cholesterol [27,28].
Fifth, expression of the FABP1 T94A variant significantly altered the specificity of lipoprotein-mediated NBD-cholesterol uptake. On the basis of lipoprotein NBD-cholesterol content, WT TT hepatocytes took up NBD-cholesterol from HDL ≫ VLDL > LDL, reflecting HDL’s primary role in RCT [18]. The order of specificity was also consistent with the specificity of the SRB1 receptor which binds not only HDL but also LDL and VLDL with Kds of 2–35, 5–20, and 0.7 μg/ml, respectively [55–60]. Furthermore, SRB1 mediates selective uptake of cholesterol not only from HDL but also LDL and VLDL, albeit less efficiently [55,56,61–63]. SRB1 binding affinities for these lipoproteins were in the similar range with Kds of 10–19 nM (LDL) [56]. Finally, T94A expression increased human hepatocyte secretion of apoB and cholesteryl ester, indicating that T94A also increased secretion of nascent VLDL [34].
In summary, the increased cholesterol binding affinity of the human FABP1 T94A variant protein provides possible new insights whereby expression of this variant may impact lipoprotein mediated cholesterol uptake and accumulation in hepatic cells. Analogous studies of genetic polymorphisms in other FABPs [FABP2 and FABP4 (adipocyte fatty acid binding protein)]—also linked to dyslipidemias, type 2 diabetes, and CVD—have spurred creation of new types of anti-diabetic and lipid-lowering drugs, such as small molecule inhibitors of FABP4 [64] and SCP-2 [65,66]. While small molecule activators/inhibitors of FABP1 have not yet been reported, the proceeding studies suggest the potential for such an approach. Finally, the studies presented herein contribute new concepts whereby intracellular cholesterol binding proteins may facilitate the uptake and/or intracellular disposition of cholesterol derived from plasma lipoproteins.
Highlights.
The human FABP1 T94A variant protein bound cholesterol 3-fold higher than the WT.
HDL and LDL-mediated cholesterol uptake was faster in the CC genotyped hepatocytes.
VLDL-mediated cholesterol uptake was unchanged in the CC genotyped hepatocytes.
Key proteins in lipoprotein cholesterol uptake were unchanged among the genotypes.
Acknowledgments
This work was supported in part by the USPHS National Institutes of Health Grants DK41402 (FS, ABK) and DK70965 (BPA).
ABBREVIATIONS
- C/EBPα
CCAAT/enhancer binding protein alpha
- CETP
Cholesteryl ester transfer protein
- CPT1A
carnitine palmitoyl transferase IA, liver
- CPT2
carnitine palmitoyl-CoA transferase II
- CVD
cardiovascular disease
- FABP1
human liver fatty acid binding protein
- FABP1 T94T
wild type (WT) human FABP1
- FABP1 T94A
human FABP1 T94A variant
- FABP2
intestinal fatty acid binding protein
- FABP4
adipocyte fatty acid binding protein
- FATP2
fatty acid transport protein-2
- FATP4
fatty acid transport protein-4
- FATP5
fatty acid transport protein-5
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GOT
mitochondrial glutamate oxaloacetate transaminase (aspartate aminotransferase)
- HDL
high-density lipoprotein
- ITC
isothermal titration calorimetry
- LCAT
Lecithin-Cholesterol Acyltransferase
- LDL
low-density lipoprotein
- LDLR
Low-Density Lipoprotein (LDL) Receptor
- LPL
lipoprotein lipase
- LSCM
laser scanning confocal microscopy
- NAFLD
non-alcohol fatty liver disease
- NBD-cholesterol
22-(N-(7-nitrobenz-2-oxa-1, 3-diazol-4-yl)-amino)-23, 24-bisnor-5-cholen-3β-ol
- PPARα-β/δ, or -γ
peroxisome proliferator-activated receptor alpha, beta/delta, or gamma
- RCT
reverse-cholesterol-transport
- SCP-2
sterol carrier protein-2
- SNP
single nucleotide polymorphism
- SRB1
Scavenger receptor class B member 1
- VLDL
very low-density lipoprotein
- WT
wild type
Footnotes
CONFLICT OF INTEREST. The authors declare that there is no conflict of interest, financial or otherwise.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haberland ME, Reynolds JA. Self-association of cholesterol in aqueous solution. Proc Natl Acad Sci. 1973;70:2313–2318. doi: 10.1073/pnas.70.8.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, et al. Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. The Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navab M, Anantharamaiah GM, Reddy ST, Van Lenten BJ, Fogelman AM. HDL as a biomarker, potential therapeutic target, and therapy. Diabetes. 2009;58:2711–2717. doi: 10.2337/db09-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaefer EJ, Santos RD, Asztalos BF. Marked HDL deficiency and premature coronary heart disease. Curr Opin Lipidol. 2010;21:289–297. doi: 10.1097/MOL.0b013e32833c1ef6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frikke-Schmidt R. Genetic variation in the ABCA1 gene, HDL cholesterol, and risk of ischemic heart disease in the general population. Atherosclerosis. 2010;208:305–316. doi: 10.1016/j.atherosclerosis.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 6.McArthur MJ, Atshaves BP, Frolov A, Foxworth WD, Kier AB, Schroeder F. Cellular uptake and intracellular trafficking of long chain fatty acids. J Lipid Res. 1999;40:1371–1383. [PubMed] [Google Scholar]
- 7.Favretto F, Assfalg M, Gallo M, Cicero DO, D’Onofrio M, Molinari H. Ligand binding promiscuity and human liver fatty acid binding protein: structural and dynamic insights from an interaction study with glycocholate and oleate. ChemBioChem. 2013;14:1807–1819. doi: 10.1002/cbic.201300156. [DOI] [PubMed] [Google Scholar]
- 8.Nemecz G, Schroeder F. Selective binding of cholesterol by recombinant fatty acid-binding proteins. J Biol Chem. 1991;266:17180–17186. [PubMed] [Google Scholar]
- 9.Martin GG, Atshaves BP, Huang H, McIntosh AL, Williams BW, Pai P-J, Russell DH, Kier AB, Schroeder F. Hepatic phenotype of liver fatty acid binding protein (L-FABP) gene ablated mice. Am J Physiol. 2009;297:G1053–G1065. doi: 10.1152/ajpgi.00116.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer RT, Cowlen MS, Dempsey ME, Schroeder F. Fluorescence of delta 5,7,9(11),22-ergostatetraen-3 beta-ol in micelles, sterol carrier protein complexes, and plasma membranes. Biochemistry. 1985;24:3322–3331. doi: 10.1021/bi00334a037. [DOI] [PubMed] [Google Scholar]
- 11.Yan J, Gong Y, She Y-M, Wang G, Roberts MS, Burczynski FJ. Molecular mechanism of recombinant liver fatty acid binding protein’s antioxidant activity. J Lipid Res. 2009;50:2445–2454. doi: 10.1194/jlr.M900177-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan J, Gong Y, She YM, Wang G, Robertes MS, Burczynski FJ. Molecular mechanism of recombinant L-FABP’s antioxidant activity. J Lipid Res. 2010;50:2445–2454. doi: 10.1194/jlr.M900177-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richieri GV, Ogata RT, Kleinfeld AM. Equilibrium constants for the binding of fatty acids with fatty acid binding proteins from adipocyte, intestine, heart, and liver measured with the flourescent probe ADIFAB. J Biol Chem. 1994;269:23918–23930. [PubMed] [Google Scholar]
- 14.Frolov A, Woodford JK, Murphy EJ, Billheimer JT, Schroeder F. Spontaneous and protein-mediated sterol transfer between intracellular membranes. J Biol Chem. 1996;271:16075–16083. doi: 10.1074/jbc.271.27.16075. [DOI] [PubMed] [Google Scholar]
- 15.Frolov AA, Woodford JK, Murphy EJ, Billheimer JT, Schroeder F. Fibroblast membrane sterol kinetic domains: modulation by sterol carrier protein 2 and liver fatty acid binding protein. J Lipid Res. 1996;37:1862–1874. [PubMed] [Google Scholar]
- 16.Woodford JK, Behnke WD, Schroeder F. Liver fatty acid binding protein enhances sterol transfer by membrane interaction. Mol Cell Biochem. 1995;152:51–62. doi: 10.1007/BF01076463. [DOI] [PubMed] [Google Scholar]
- 17.Jefferson JR, Slotte JP, Nemecz G, Pastuszyn A, Scallen TJ, Schroeder F. Intracellular sterol distribution in transfected mouse L-cell fibroblasts expressing rat liver fatty acid binding protein. J Biol Chem. 1991;266:5486–5496. [PubMed] [Google Scholar]
- 18.Storey SM, McIntosh AL, Huang H, Martin GG, Landrock KK, Landrock D, Payne HR, Kier AB, Schroeder F. Intracellular cholesterol binding proteins enhance HDL-mediated cholesterol uptake in cultured primary mouse hepatocytes. Am J Physiol Gastrointest and Liver Phys. 2012;302:G824–G839. doi: 10.1152/ajpgi.00195.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H, McIntosh AL, Martin GG, Landrock K, Landrock D, Gupta S, Atshaves BP, Kier AB, Schroeder F. Structural and functional interaction of fatty acids with human liver fatty acid binding protein (L-FABP) T94A variant. FEBS J. 2014;281:2266–2283. doi: 10.1111/febs.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin GG, McIntosh AL, Huang H, Gupta S, Atshaves BP, Kier AB, Schroeder F. Human liver fatty acid binding protein (L-FABP) T94A variant alters structure, stability, and interaction with fibrates. Biochemistry. 2013;52:9347–9357. doi: 10.1021/bi401014k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maatman RG, van de Westerlo EM, Van Kuppevelt TH, Veerkamp JH. Molecular identification of the liver- and the heart-type fatty acid-binding proteins in human and rat kidney. Use of the reverse transcriptase polymerase chain reaction. Biochem J. 1992;288:285–290. doi: 10.1042/bj2880285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfrum C, Borchers T, Sacchettini JC, Spener F. Binding of fatty acids and perooxisome proliferators to orthologous fatty acid binding proteins from human, murine, and bovine liver. Biochemistry. 2000;39:1469–1474. doi: 10.1021/bi991638u. [DOI] [PubMed] [Google Scholar]
- 23.Atshaves BP, Payne HR, McIntosh AL, Tichy SE, Russell D, Kier AB, Schroeder F. Sexually dimorphic metabolism of branched chain lipids in C57BL/6J mice. J Lipid Res. 2004;45:812–830. doi: 10.1194/jlr.M300408-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Atshaves BP, McIntosh AL, Payne HR, Mackie J, Kier AB, Schroeder F. Effect of branched-chain fatty acid on lipid dynamics in mice lacking liver fatty acid binding protein gene. Am J Physiol. 2005;288:C543–C558. doi: 10.1152/ajpcell.00359.2004. [DOI] [PubMed] [Google Scholar]
- 25.Atshaves BP, McIntosh AL, Landrock D, Payne HR, Mackie J, Maeda N, Ball JM, Schroeder F, Kier AB. Effect of SCP-x gene ablation on branched-chain fatty acid metabolism. Am J Physiol. 2007;292:939–951. doi: 10.1152/ajpgi.00308.2006. [DOI] [PubMed] [Google Scholar]
- 26.Atshaves BP, McIntosh AL, Martin GG, Landrock D, Payne HR, Bhuvanendran S, Landrock K, Lyuksyutova OI, Johnson JD, Macfarlane RD, Kier AB, Schroeder F. Overexpression of sterol carrier protein-2 differentially alters hepatic cholesterol accumulation in cholesterol-fed mice. J Lipid Res. 2009;50:1429–1447. doi: 10.1194/jlr.M900020-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng X-E, Wu YL, Lu Q-Q, Ju Z-J, Lin X. Two genetic variants in FABP1 and susceptibility to non-alcoholic fatty liver disease in a Chinese population. Gene. 2012;500:54–58. doi: 10.1016/j.gene.2012.03.050. [DOI] [PubMed] [Google Scholar]
- 28.Fisher E, Weikert C, Klapper M, Lindner I, Mohlig M, Spranger J, Boeing H, Schrezenmeir J, Doring F. L-FABP T94A is associated with fasting triglycerides and LDL-cholesterol in women. Mol Gen and Metab. 2007;91:278–284. doi: 10.1016/j.ymgme.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Brouillette C, Bose Y, Perusse L, Gaudet D, Vohl M-C. Effect of liver fatty acid binding protein (FABP) T94A missense mutation on plasma lipoprotein responsiveness to treatment with fenofibrate. J Hum Gen. 2004;49:424–432. doi: 10.1007/s10038-004-0171-2. [DOI] [PubMed] [Google Scholar]
- 30.Verges B. Lipid modificaiton in type 2 diabetes: the role of LDL and HDL. Fundamental and Clin Pharmacol. 2009;23:685. doi: 10.1111/j.1472-8206.2009.00739.x. [DOI] [PubMed] [Google Scholar]
- 31.Quintao ECR, Medina WL, Passarelli M. Reverse cholesterol transport in diabetes mellitus. Diabetes Metabolism Research Rev. 2000;16:237–250. doi: 10.1002/1520-7560(200007/08)16:4<237::aid-dmrr127>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 32.Passarelli M, Shimabukuro AFM, Catanozi S, Nakandakare ER, Rocha JC, Carrilho AJF, Quintao ECR. Diminished rate of mouse peritoneal macrophage cholesterol efflux is not related to the degree of HDL glycation in diabetes mellitus. Clin Chim Acta. 2000;301:119–134. doi: 10.1016/s0009-8981(00)00336-3. [DOI] [PubMed] [Google Scholar]
- 33.Yamada Y, Kato K, Oguri M, Yoshida T, Yokoi K, Watanabe S, Metoki N, Yoshida H, Satoh K, Ichihara S, Aoyagi Y, Yasunaga A, Park H, Tanaka M, Nozawa Y. Association of genetic variants with atherothrombotic cerebral infarction in Japanese individuals with metabolic syndrome. Int J Mol Med. 2008;21:801–808. [PubMed] [Google Scholar]
- 34.McIntosh AL, Huang H, Storey SM, Landrock K, Landrock D, Petrescu AD, Gupta S, Atshaves BP, Kier AB, Schroeder F. Human FABP1 T94A variant impacts fatty acid metabolism and PPARa activation in cultured human female hepatocytes. Am J Physiol Gastrointest and Liver Phys. 2014;307:G164–G176. doi: 10.1152/ajpgi.00369.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao N, Qu X, Yan J, Huang Q, Yuan HY, Ouyang D-S. L-FABP T94A decreased fatty acid uptake and altered hepatic triglyceride and cholesterol accumulation in Chang liver cells stably transfected with L-FABP. Mol Cell Biochem. 2010;345:207–214. doi: 10.1007/s11010-010-0574-7. [DOI] [PubMed] [Google Scholar]
- 36.Atshaves BP, Petrescu A, Starodub O, Roths J, Kier AB, Schroeder F. Expression and Intracellular Processing of the 58 kDa Sterol Carrier Protein 2/3-Oxoacyl-CoA Thiolase in Transfected Mouse L-cell Fibroblasts. J Lipid Res. 1999;40:610–622. [PubMed] [Google Scholar]
- 37.Frolov A, Cho TH, Murphy EJ, Schroeder F. Isoforms of rat liver fatty acid binding protein differ in structure and affinity for fatty acids and fatty acyl CoAs. Biochemistry. 1997;36:6545–6555. doi: 10.1021/bi970205t. [DOI] [PubMed] [Google Scholar]
- 38.Avdulov NA, Chochina SV, Igbavboa U, Warden CH, Schroeder F, Wood WG. Lipid binding to sterol carrier protein-2 is inhibited by ethanol. Biochim Biophys Acta. 1999;1437:37–45. doi: 10.1016/s0005-2760(98)00178-7. [DOI] [PubMed] [Google Scholar]
- 39.Schroeder F, Frolov A, Schoer J, Gallegos A, Atshaves BP, Stolowich NJ, Scott AI, Kier AB. Intracellular Sterol Binding Proteins, Cholesterol Transport and Membrane Domains. In: Chang TY, Freeman DA, editors. Intracellular Cholesterol Trafficking. Kluwer Academic Publishers; Boston: 1998. pp. 213–234. [Google Scholar]
- 40.Stolowich NJ, Frolov A, Petrescu AD, Scott AI, Billheimer JT, Schroeder F. Holo-sterol carrier protein-2: 13C-NMR investigation of cholesterol and fatty acid binding sites. J Biol Chem. 1999;274:35425–35433. doi: 10.1074/jbc.274.50.35425. [DOI] [PubMed] [Google Scholar]
- 41.Liu R, Lu P, Chu JWK, Sharom FJ. Characterization of fluorescent sterol binding to purified human NPC1. J Biol Chem. 2009;284:1840–1852. doi: 10.1074/jbc.M803741200. [DOI] [PubMed] [Google Scholar]
- 42.Avdulov NA, Chochina SV, Igbavboa U, Warden CS, Vassiliev A, Wood GW. Lipid Binding to Amyloid B-Peptide Aggregates: Preferential Binding of Cholesterol as Compared with Phosphatidylcholine and Fatty Acids. Journal of Neurochemistry. 1997;69:1746–1752. doi: 10.1046/j.1471-4159.1997.69041746.x. [DOI] [PubMed] [Google Scholar]
- 43.Reaven E, Tsai L, Azhar S. Cholesterol uptake by the selective pathway of ovarian granulosa cells: early intracellular events. J Lipid Res. 1995;36:1602–1617. [PubMed] [Google Scholar]
- 44.Frolov A, Petrescu A, Atshaves BP, So PTC, Gratton E, Serrero G, Schroeder F. High density lipoprotein mediated cholesterol uptake and targeting to lipid droplets in intact L-cell fibroblasts. J Biol Chem. 2000;275:12769–12780. doi: 10.1074/jbc.275.17.12769. [DOI] [PubMed] [Google Scholar]
- 45.Atshaves BP, Starodub O, McIntosh AL, Roths JB, Kier AB, Schroeder F. Sterol carrier protein-2 alters HDL-mediated cholesterol efflux. J Biol Chem. 2000;275:36852–36861. doi: 10.1074/jbc.M003434200. [DOI] [PubMed] [Google Scholar]
- 46.Storey SM, Atshaves BP, McIntosh AL, Landrock KK, Martin GG, Huang H, Johnson JD, Macfarlane RD, Kier AB, Schroeder F. Effect of sterol carrier protein-2 gene ablation on HDL-mediated cholesterol efflux from primary cultured mouse hepatocytes. Am J Physiol. 2010;299:244–254. doi: 10.1152/ajpgi.00446.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dagher G, Donne N, Klein C, Ferre P, Dugail I. HDL-mediated cholesterol uptake and targeting to lipid droplets in adipocytes. J Lipid Res. 2003;44:1811–1820. doi: 10.1194/jlr.M300267-JLR200. [DOI] [PubMed] [Google Scholar]
- 48.Sparrow CP, Patel S, Baffic J, Chao Y-S, Hernandez M, Lam M-H, Montenegro J, Wright SD, Detmers PA. A fluorescent cholesterol analog traces cholesterol absorption in hamsters and is esterified in vivo and in vitro. J Lipid Res. 1999;40:1747–1757. [PubMed] [Google Scholar]
- 49.Atshaves BP, McIntosh AL, Lyuksyutova OI, Zipfel WR, Webb WW, Schroeder F. Liver fatty acid binding protein gene ablation inhibits branched-chain fatty acid metabolism in cultured primary hepatocytes. J Biol Chem. 2004;279:30954–30965. doi: 10.1074/jbc.M313571200. [DOI] [PubMed] [Google Scholar]
- 50.McIntosh AL, Atshaves BP, Hostetler HA, Huang H, Davis J, Lyuksyutova OI, Landrock D, Kier AB, Schroeder F. Liver type fatty acid binding protein (L-FABP) gene ablation reduces nuclear ligand distribution and peroxisome proliferator activated receptor-alpha activity in cultured primary hepatocytes. Arch Biochem Biophys. 2009;485:160–173. doi: 10.1016/j.abb.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi S, Sakai J, Fujino T, Hattori H, Zenimary Y, Suzuki J, Miyamori I, Yamamoto TT. The very low density lipoprotein (VLDL) receptor: characterization and functions as a peripheral lipoprotein receptor. J Atheroscler Thromb. 2004;11:200–208. doi: 10.5551/jat.11.200. [DOI] [PubMed] [Google Scholar]
- 52.Schroeder F, Myers-Payne SC, Billheimer JT, Wood WG. Probing the ligand binding sites of fatty acid and sterol carrier proteins: effects of ethanol. Biochemistry. 1995;34:11919–11927. doi: 10.1021/bi00037a033. [DOI] [PubMed] [Google Scholar]
- 53.Galan X, Peinado-Onsurbe J, Julve J, Ricart-Jane D, Robert MQ, Llobera M, Ramirez I. Inactive hepatic lipase in rat plasma. J Lipid Res. 2003;44:2250–2256. doi: 10.1194/jlr.M300131-JLR200. [DOI] [PubMed] [Google Scholar]
- 54.Perret B, Mabile L, Martinez L, Terce F, Barabas R, Collet X. Hepatic lipase: structure/function relationship, synthesis, and regulation. J Lipid Res. 2002;43:1163–1169. [PubMed] [Google Scholar]
- 55.Connelly MA, Williams DL. Scavenger receptor B1: A scavenger receptor with a mission to transport high density lipoprotein lipids. Cur Opin Lipidology. 2004;15:287–295. doi: 10.1097/00041433-200406000-00008. [DOI] [PubMed] [Google Scholar]
- 56.Rigotti A, Miettinen H, Kreiger M. The role of the high-density lipoprotein receptor SR-B1 in the lipid metabolism of endocrine and other tissues. Endocrine Rev. 2003;23:357–383. doi: 10.1210/er.2001-0037. [DOI] [PubMed] [Google Scholar]
- 57.Calvo M, Gomez-Coronado D, Lasuncion MA, Vega MA. CLA-1 is an 85 kD plasma membrane glycoprotein that acts as a high affinity receptor for both native (HDL, LDL, and VLDL) and modified (OxLDL and AcLDL) lipoproteins. Arterioscler Thromb Vasc Biol. 1997;17:2341–2349. doi: 10.1161/01.atv.17.11.2341. [DOI] [PubMed] [Google Scholar]
- 58.Acton SL, Scherer PE, Lodish HF, Krieger M. Expression cloning of SR-B1, a CD36-related class B scavenger receptor. J Biol Chem. 1994;269:21003–21009. [PubMed] [Google Scholar]
- 59.de la Llera-Moya M, Rothblat GH, Connelly MA, Kellner-Weibel G, Sakr SW, Phillips MC, Williams DL. Scavenger receptor B1 (SRB1) mediates free cholesterol flux independently of HDL tethering to the surface. J Lipid Res. 1999;40:575–580. [PubMed] [Google Scholar]
- 60.Murao K, Terpstra V, Green SR, Kondratenko N, Steinberg D, Quehenberger O. Characterization of CLA-1, a human homologue of rodent scavenger receptor B1, as a receptor for high density lipoprotein and apoptotic thymocytes. J Biol Chem. 1997;272:17551–17557. doi: 10.1074/jbc.272.28.17551. [DOI] [PubMed] [Google Scholar]
- 61.Krieger M. Charting the fate of the “good cholesterol”: identification and characterization of the high density lipoprotein receptor SR-B1. Ann Rev Biochem. 1999;68:523–558. doi: 10.1146/annurev.biochem.68.1.523. [DOI] [PubMed] [Google Scholar]
- 62.Van Eck M, Hoekstra M, Out R, Bos IST, Van Kruijt JK, Hildebrand RB, Van Berkel TJC. SRB1 facilitates the metabolism of VLDL lipoproteins in vivo. J Lipid Res. 2008;49:136–146. doi: 10.1194/jlr.M700355-JLR200. [DOI] [PubMed] [Google Scholar]
- 63.Rohrl C, Fruhwurth S, Schreier SM, Lohninger A, et al. SRB1 provides an alternative means for b-VLDL uptake independent of the LDL receptor in tissue culture. Biochim Biophys Acta. 2010;1801:198–204. doi: 10.1016/j.bbalip.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Furuhashi M, Tuncman G, Gorgun CZ, Makowski L, Atsumi G, Vallaincourt E, Kono K, Babaev VR, Fazio S, Linton MF, Sulsky R, Robl JA, Parker RA, Hotamisligil GS. Treatment of diabetes and atherosclerosis by inhibiting fatty acid binding protein aP2. Nature. 2008;447:959–965. doi: 10.1038/nature05844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kriska T, Pilat A, Schmitt JC, Girotti AW. Sterol carrier protein-2 (SCP-2) involvement in cholesterol hydroperoxide cytotoxicity as revealed by SCP-2 inhibitor effects. J Lipid Res. 2010;51:3184. doi: 10.1194/jlr.M008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim M-S, Wessely V, Lan Q. Identification of mosquito sterol carrier protein-2 inhibitors. Journal of Lipid Research. 2005;46:650–657. doi: 10.1194/jlr.M400389-JLR200. [DOI] [PubMed] [Google Scholar]