Summary
The discovery of the metazoan-specific Integrator Complex represented a breakthrough in our understanding of noncoding U-rich small nuclear RNA (UsnRNA) maturation and has triggered a reevaluation of their biosynthesis mechanism. In the decade since, significant progress has been made to understand the details of its recruitment, specificity, and assembly. While some discrepancies remain on how it interacts with the carboxy-terminal domain of the RNA polymerase II (RNAPII) and the details of its recruitment to UsnRNA genes, preliminary models have emerged. Recent provocative studies now implicate Integrator in the regulation of protein-coding gene transcription initiation and RNA Polymerase II pause-release thereby broadening the scope of Integrator functions in gene expression regulation. Here, we discuss the implications of these findings while putting them into the context of what is understood about Integrator function at UsnRNA genes.
Keywords: Integrator, UsnRNA processing, transcriptional activation, pause-release, RNAPII CTD
Initial discovery of integrator
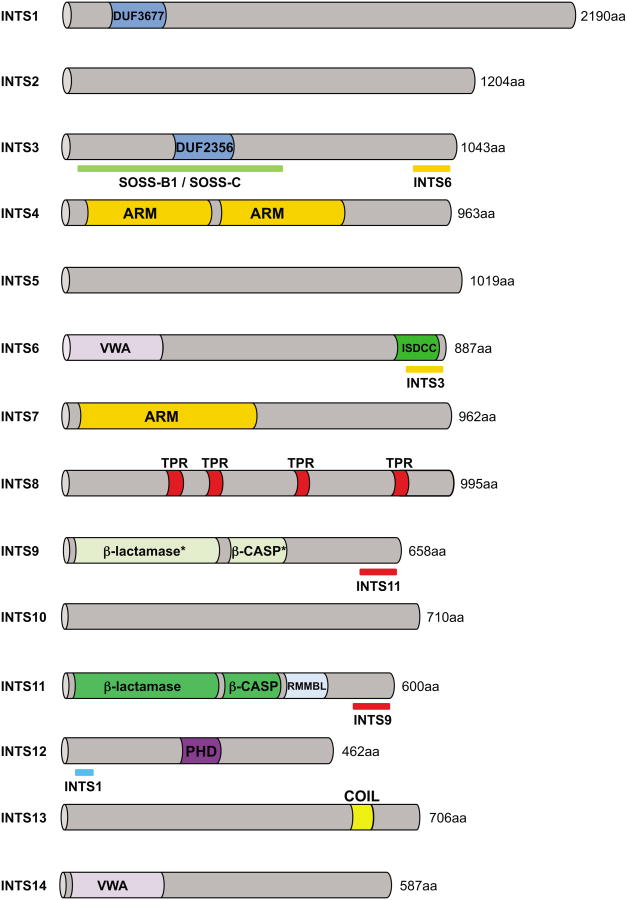
The Integrator complex (INT) was discovered serendipitously while searching for protein partners of Deleted in Split hand/Split foot protein 1 (DSS1). The initial affinity purification of the complex [1] identified twelve Integrator subunits (INTS1 to INTS12, see Figure 1) and demonstrated its association with the C-terminal domain (CTD see glossary, Box 1) of RPB1, the largest subunit of RNA polymerase II (RNAPII). Subsequent proteomic analyses [2-4], while confirming its composition and association with RNAPII, identified possible additional subunits as well as new potential cofactors. Among these proteins, only C12orf11 (also known as Asunder) and C15orf44 (also known as VWA9 or CG4785) have since been proven to be functionally associated with Integrator and renamed INTS13 and INTS14, respectively [5].
Figure 1. Integrator subunit domain schematic.
Predicted protein domains of all 14 Integrator subunits are illustrated and the length of the human orthologues is indicated (in amino acids, aa). DUF=domain of unknown function, ARM=armadillo like repeats, VWA=von Willebrand type A like domain, ISDCC=INTS6/SAGE1/DDX26B/CT45 C-terminus, TPR=tetratricopeptide repeats, β-lactamase/β-CASP (see glossary) “*” indicates the presence of an inactive β-lactamase/β-CASP domain, PHD=plant homeodomain finger, COIL=Coiled coil domain. Identified interacting domains with other proteins are underlined.
Box 1. CTD phosphorylation cycle.
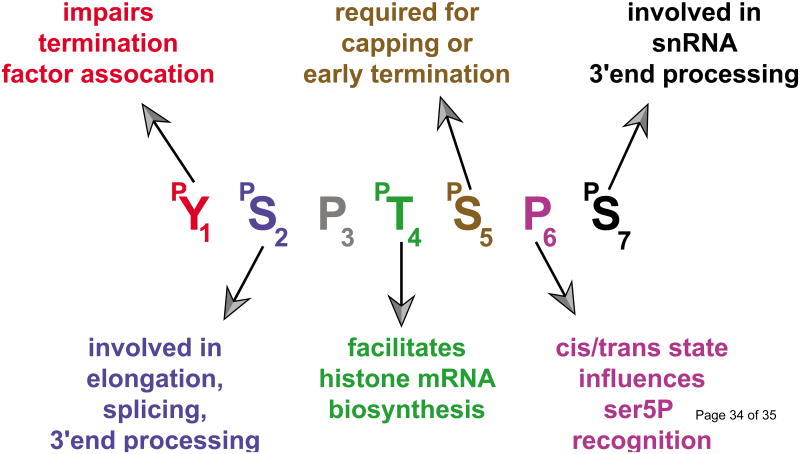
The RNAPII CTD is composed of heptad repeats of consensus amino acid sequence Y1S2P3T4S5P6S7. The exact numbers of repeats (e.g. 26 in yeast, 44 in drosophila and 52 in human) as well as the extent of deviation from the heptad consensus sequence depend on the species [53]. Until recently it was thought that CTD phosphorylation was restricted to Ser2, Ser5 and Ser7, but it has recently become clear that Tyr1 and Thr4 phosphorylation also have a role to play in the regulation of transcription by RNAPII [57-60]. Given that Pro3 and Pro6 can also adopt a cis or trans conformation that is actively regulated by prolyl-isomerases [61,62], all residues of the heptad repeat can be modified, dramatically increasing the complexity of what has been sometimes called the “CTD code” [53]. This combinatorial regulation orchestrates the recruitment of the many factors involved in all the successive steps of transcription: initiation, mRNA 5′ capping, chromatin remodeling, elongation, splicing, 3′ end processing, termination and mRNA export.
Figure I. The “CTD code”.
For each amino acid of the heptad repeat of the C-terminal domain (CTD) of the largest subunit of the RNA polymerase II (RNAPII) is indicated the known functions of the corresponding modification.
Unlike the Mediator Complex, a multi-subunit complex required for regulated transcription of most RNAPII dependent genes [6,7], Integrator is restricted to metazoans [8]. Its molecular weight is estimated by size exclusion chromatography to be greater than 1 MDa [1,9]. The size of its subunits, in humans, ranges from 49 kDa for INTS12 to 244 kDa for INTS1, with the majority (eight out of fourteen) possessing molecular weights greater than 100 kDa (Figure 1). Few subunits have identifiable paralogs within the human genome and, despite their number and relatively large size, their sequence is strikingly devoid of readily recognizable domains. The most common predicted motifs within Integrator subunits are alpha-helical repeats such as HEAT, ARM and TPR, or VWA domains [10]. These structures are suggestive of protein-protein interaction surfaces, but fail to provide insight into the function of their respective subunit in the complex or into a potential interaction partner. Contrastingly, two subunits, INTS11 and INTS9 [11], are clearly homologous with CPSF73 and CPSF100, proteins that are involved in the cleavage of pre-messenger RNAs [12] and belong to a large group of zinc-dependent nucleases called the β-CASP family [13] (CPSF, Artemis, SMN1/PSO2, see glossary). This relationship was instrumental in implicating Integrator in the 3′end formation of cellular RNA.
A long sought UsnRNA 3′end processing factor
Apart from U6, a typical RNAPIII-dependent transcript whose 3′end is generated by transcription termination driven by a thymidine stretch [14], all UsnRNAs (see glossary) are synthesized by RNAPII. Prior to the discovery of Integrator, extensive work had defined the three requirements governing RNAPII-dependent UsnRNA 3′end formation: i) an UsnRNA-type promoter containing two characteristic elements: a distal sequence element (DSE) that recruits the transcription factors Oct1 and Sp1, and a proximal sequence element (PSE) that is bound by the snRNA activating protein complex (SNAPc, see glossary) [15,16], ii) the CTD of RNAPII [17,18] and iii) a consensus sequence GTTTN0-3AAARNNAGA called the 3′box, which is located 9-19 nucleotides downstream of the 3′end of the UsnRNA [19]. These requirements led to the hypothesis of a co-transcriptional mechanism: a unique factor is recruited to UsnRNA promoters where it associates with the RNAPII CTD and cleaves the nascent pre-UsnRNA once the 3′box is transcribed and recognized.
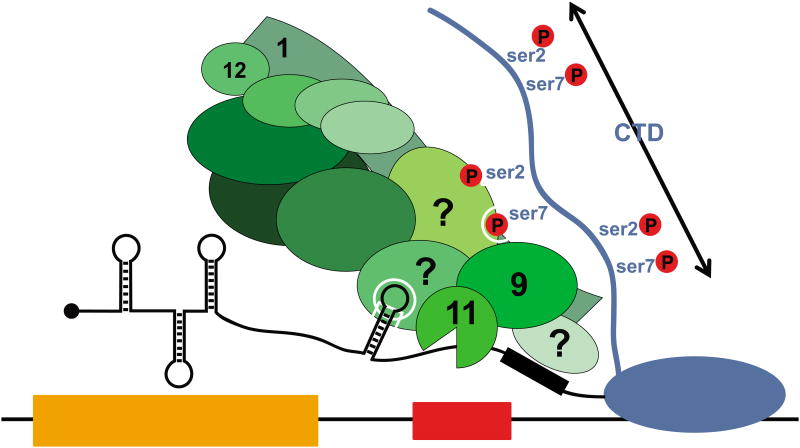
Integrator proved to be this long sought factor (Figure 2). Multiple biochemical purifications indicated that it associates with the RNAPII CTD [1,20]. Chromatin immunoprecipitation (ChIP) experiments showed that Integrator is present at the promoter, body and 3′end of the UsnRNA genes in a pattern suggesting that it travels along with the RNAPII as it transcribes the UsnRNA [1,9]. Finally, RNAi-mediated knock-down of various Integrator subunits leads to the accumulation of elongated misprocessed pre-UsnRNA [1,21]. Given that INTS11 is paralogous to CPSF73 [11], and that the overexpression of an INTS11 mutant predicted to be catalytically dead interferes with UsnRNA 3′end processing [1], it is presumed that INTS11 is the enzyme responsible for the pre-UsnRNA cleavage. Integrator knockdown also results in increased RNAPII density downstream of the 3′end cleavage site, indicating that on UsnRNA genes 3′end processing is linked to transcription termination [22]. Whether the termination event is coupled with 3′end cleavage through a mechanism similar to the torpedo model for mRNA genes [23] or if Integrator directly regulates termination remains to be determined.
Figure 2. Model of Integrator function in UsnRNA processing.
Integrator (INT, green) is recruited early in the UsnRNA transcription cycle and is loaded onto the RNAPII C-terminal domain (CTD) through recognition of the ser7P/ser2P dyad. The identity of the INT subunit(s) that recognize these specific CTD phosphorylations is not known. Once the UsnRNA terminal stem loop and 3′box element emerge from the elongating RNAPII, the RNA is recognized through an unknown mechanism. This event precedes UsnRNA cleavage, which is carried out by the heterodimeric cleavage factor composed of INTS9 and INTS11.
Relation between Integrator and the RNAPII CTD
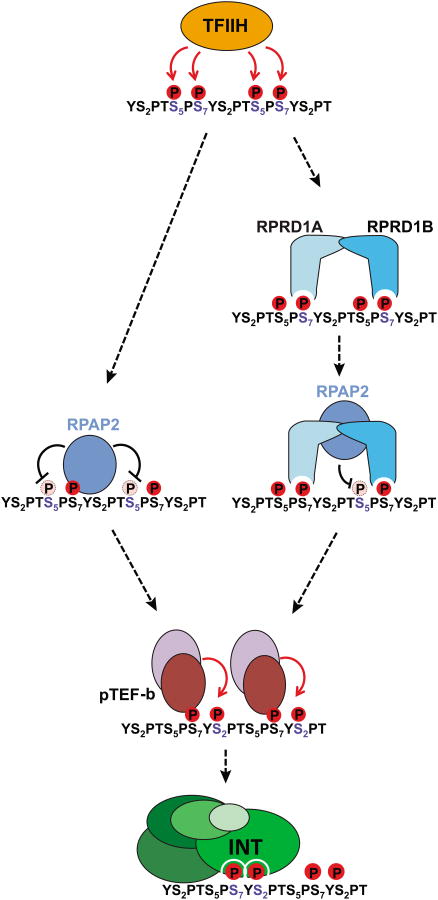
Of the three functions postulated for Integrator at the UsnRNA genes [recognition of the UsnRNA promoter, specific binding to the RNAPII CTD (Box 1), and cleavage of the nascent UsnRNA, (Figure 2)], the interaction with the RNAPII CTD has been the most thoroughly investigated. It has been clearly established that Integrator shows a strong preference for a ser7P/ser2P dyad (YS2PTS5PS7YS2PTS5PS7) while ser5 phosphorylation appears to be detrimental to Integrator recruitment [20]. Experimental evidence indicated that the RNAPII Associated Protein 2 (RPAP2, homolog of the yeast atypical phosphatase rtr1 [24]) removes the ser5P mark on UsnRNA genes. RPAP2 affinity purified from mammalian cells co-elutes with both Integrator and RNAPII and its interaction with the RNAPII CTD is ser7P-dependent. Moreover, purified human RPAP2 protein exhibits ser5P phosphatase activity in vitro and its knockdown in mammalian cells results in elevated levels of ser5P, decreased Integrator occupancy on the UsnRNA genes and accumulation of misprocessed UsnRNAs [9]. Altogether, these results were coalesced into a model (Figure 3, left pathway) where RNAPII is initially phosphorylated on ser5 and ser7 by TFIIH (Transcription Factor IIH, see glossary) at the UsnRNA promoter [25]. This phosphorylation pattern, in turn, recruits RPAP2 through ser7P and results in the removal of the ser5P mark. Finally, ser2 phosphorylation by p-TEFb (Positive Transcription Elongation Factor b, see glossary) creates the substrate required for optimal Integrator recruitment and subsequent 3′end processing.
Figure 3. CTD phosphorylation cycle at UsnRNA genes.
The CTD of RNAPII is first phosphorylated by TFIIH on ser5/7 positions coinciding with transcription initiation and UsnRNA capping. Two possible paths are then taken. This first (left) involves RPAP2 binding to ser7P and dephosphorylation of ser5P. The second scenario (right) involves the binding of a RPRD1A/B dimer to two ser7P, which in turn recruits RPAP2 and positions it to dephosphorylate ser5P. Either of these events is followed by ser2 phosphorylation by p-TEFb leading to the proper dyad modification pattern (ser7P/ser2P) required for Integrator interaction.
Nevertheless, the role of RPAP2 as well as the exact mechanism by which the ser5P mark is removed has been debated. Recent studies established that RPAP2 phosphatase activity is low [26] (with a turnover rate several orders lower than other known CTD serine phosphatases) and that the enzyme lacks a proper grove or pocket that could fulfill the role of an active site [27]. However, these shortcomings have been mitigated by the recent characterization of two RNAPII CTD binding proteins, Regulation Of Nuclear Pre-MRNA Domain Containing 1A and 1B (RPRD1A and RPRD1B). These proteins form homo- or heterodimers through a coiled-coil domain, bind to the ser2P or ser7P marks on the RNAPII CTD and stimulate RPAP2 phosphatase activity toward ser5P through protein-protein interactions [28]. These findings suggests a possible alternative model (Figure 3, right panel) where an RPRD1A/RPRD1B dimer binds two ser7P marks bracketing a ser5P mark in order to recruit and position RPAP2 optimally toward its substrate.
Although this model presents a parsimonious solution to explain RPAP2 function in regulating RNAPII CTD phosphorylation, it probably overlooks other roles for RPAP2 in the RNAPII transcription cycle. Recent cellular biology experiments demonstrated that, similar to its yeast homolog rtr1, RPAP2 cellular localization is predominantly cytoplasmic [29]. Considering that two known RPAP2 interacting proteins, GPN-loop GTPase 1 (GPN1) and GPN-loop GTPase 3 (GPN3), play an important role in RNAPII biogenesis and nuclear import [30,31], RPAP2 cytoplasmic localization raises the possibility of a similar role. Moreover it was shown that RPAP2 binds not only the CTD of RPB1 but also to its N-terminal domain [29]. This interaction could correspond to a different function for RPAP2 or could participate in the ser5P mark removal by stabilizing the interaction between RPB1 and RPAP2 to compensate for its slow phosphatase activity.
Beyond their involvement in Integrator recruitment, there is a general question about ser7P and RPAP2 role in UsnRNA transcription and 3′end processing. Two independent studies using an inducible knockout system in chicken cells investigated the role of the Ssu72 phosphatase and of ser7P in transcription [32,33]. It was found that, similar to RPAP2, knocking out Ssu72 results both in increased ser5P marks on UsnRNA genes and defective UsnRNA processing, indicating a possible redundancy between Ssu72 and RPAP2 [32]. In addition, the substitution of ser7 to alanine in the RNAPII CTD, hence preventing its phosphorylation, showed little effect on UsnRNA processing [33], complicating our interpretation of ser7P and RPAP2 role in UsnRNA processing. The initial work identifying ser7P role in UsnRNA processing [9] relied on α-amanitin resistant RNAPII mutant complementation. Although this method has proven to be a powerful tool to study transcription, prolonged α-amanitin exposure is not without consequences and results obtained through this approach should be interpreted with caution. For example, the elongation factor DSIF (DRB Sensitivity Inducing Factor, see glossary), whose knockdown negatively affects RNAPII recruitment on snRNA genes [34] and that directly interacts with Integrator (see below), is targeted for rapid degradation by α-amanitin even in the presence of the α-amanitin resistant RNAPII mutant [35]. Finally, there is a broader question about ser7P and RPAP2 function in general transcription. Indeed, in eukaryotes ser7P is present on all RNAPII transcribed genes and despite its presumed importance, ser7 to alanine substitution is not lethal in yeast (or chicken cells) and does not result in increased global ser5 phosphorylation as would be predicted [33,36]. Similarly, recruitment of RPAP2 to protein coding genes appears to be independent of ser7 phosphorylation [9].
Altogether, these data indicate that RPAP2 is most likely involved in removing the RNAPII CTD ser5P mark to facilitate Integrator recruitment, in particular on UsnRNA genes. However, there does not appear to be a direct relation between ser7 phosphorylation, RPAP2 recruitment, and ser5P removal genome-wide; possibly because other protein partners and redundant mechanisms are affecting this relationship. Identifying these factors represents an upcoming challenge to understand how Integrator is temporally and spatially recruited to a specific gene. Conversely, identifying the Integrator subunit(s) involved in the RNAPII CTD recognition will also be critical to answer this question.
Integrator interacts with SPT5 and NELF
A fascinating development in the study of the Integrator biology is the recent discovery of its relationship with the transcription elongation machinery. ChIP experiments revealed that the negative elongation factor NELF (Negative Elongation Factor, see glossary) accumulates at the 3′end of UsnRNA genes. Its knockdown results in increased RNAPII occupancy downstream of the 3′box and in the accumulation of long readthrough transcripts, reflective of a termination defect and of a potential functional interaction with Integrator [34,37,38]. This observation is consistent with a recent affinity purification of SPT5 (see DSIF in glossary) and of the NELF subunit NELF-E that revealed a physical interaction between Integrator and these factors [34]. The interplay between Integrator, SPT5, and NELF on UsnRNA genes is particularly interesting for several reasons. SPT5 appears to function early in the transcription cycle as its knockdown results in a reduction of RNAPII, NELF, and Integrator density at the UsnRNA genes [34]. Conversely, knockdown of NELF-E or Integrator both results in accumulation of RNAPII and SPT5 on the 3′end of the genes and in the accumulation of long misprocessed transcripts [34,37,38]. Therefore, even if both SPT5 and NELF interact with Integrator, their role in relation to the complex seems functionally distinct with SPT5 playing a possible role in transcription initiation and Integrator recruitment while NELF most likely functions in UsnRNA 3′end processing and transcription termination.
Role of Integrator in RNAPII transcriptional pause-release
The connection between Integrator, SPT5/NELF and elongation revealed its full significance with the recent evidence for Integrator function at mRNA coding genes [39–41]. The initial study of Integrator using conventional ChIP analysis was limited by the small repertoire of high quality antibodies available at the time and failed to demonstrate its presence on protein coding genes [1]. As more antibodies became available, Gardini et al. used ChIP-seq to revisit Integrator occupancy genome-wide and uncovered its association with active mRNA transcription and enrichment at Immediate Early Genes (IEG, see glossary) after epidermal growth factor (EGF) stimulation [39]. Transcriptional regulation of these genes functionally resembles that of the Drosophila Heat Shock gene (HSP70), which is the archetype for RNAPII pause-release (Box 2). Interestingly, the authors observed that under starvation conditions, low levels of Integrator were specifically detected at IEG transcription start sites (TSSs); however after EGF stimulation Integrator occupancy markedly increased at the TSS and within the body of the gene. This localization proved functionally relevant as the knockdown of Integrator subunits (INTS1 and INTS11) abrogates responsiveness of IEGs to EGF stimulation. Importantly, upon Integrator depletion there is a failure of the RNAPII to escape pausing and progress into productive elongation. Mechanistically, the role for Integrator in transcriptional pause-release appears to stem from its capacity to recruit the positive elongation factors p-TEFb and SEC to promoter proximal paused genes upon activation.
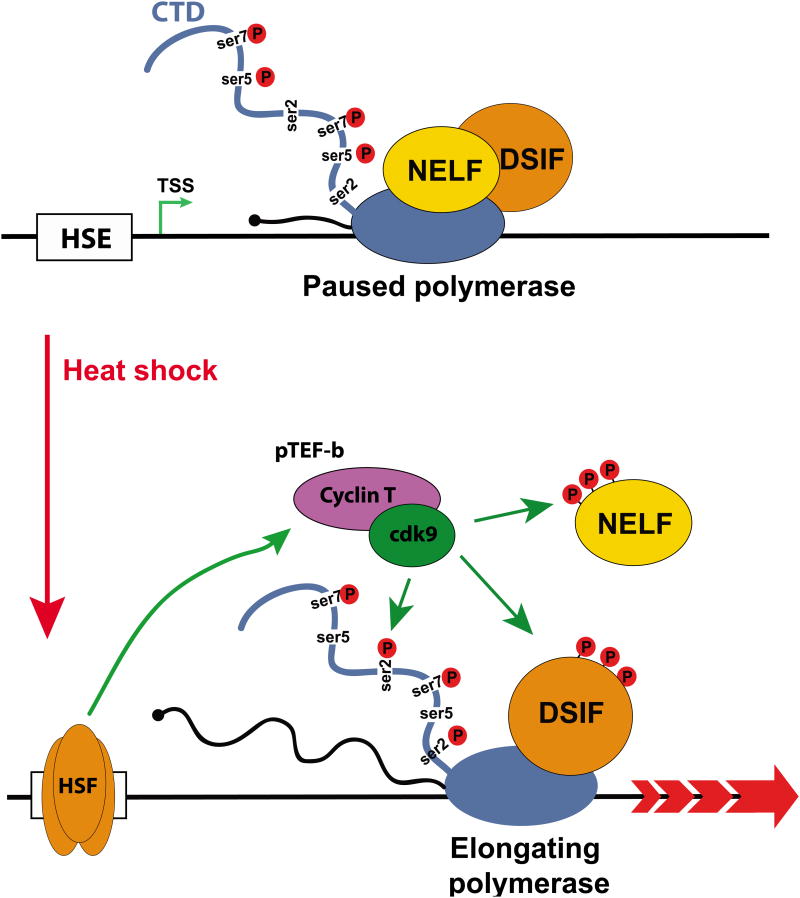
Box 2. RNA polymerase II pause-release at Drosophila heat shock genes.
After formation at the promoter of the pre-initiation complex (PIC) under the influence of sequence-specific DNA-binding transcription factors and of the basal transcription machinery, RNAPII is released from the promoter and initiates transcription. Shortly after, about 40 to 60 nucleotides downstream of the transcription start site, the RNAPII encounters a second rate-limiting transcription barrier halting transcription. This RNAPII transcriptional paused state is further stabilized by the association with two negative elongation factors, DSIF and NELF, which are able to inhibit early elongation. RNAPII transcriptional pausing is relieved by the recruitment of the positive elongation factor p-TEFb. Upon recruitment, p-TEFb phosphorylates DSIF, NELF and the serine 2 residue of the RNAPII CTD. This leads to the dissociation of NELF and to the transition of DSIF from a negative to a positive elongation factor. Productive elongation ensues.
Figure I. Heat Shock Response, a model of promoter proximal pause-release.
Top. On promoter proximal paused gene such as HSP70, RNAPII initiates transcription and, in absence of further transcription activation, is stalled 40-60 nucleotides downstream of the transcription start site by the association with the negative elongation factors DSIF and NELF. Bottom. Following heat shock, the Heat Shock Factor (HSF) is recruited to the HSP70 promoter, leading to pTEFb recruitment. The CDK9 component of pTEFb phosphorylates DSIF, NELF and the serine 2 residue of the RNAPII CTD. This cascade of event results in NELF release and progression into productive elongation.
The second study linking Integrator to RNAPII pause-release on mRNA coding genes originates from the investigation of NELF function in Tat-activated transcription of the HIV-1 long terminal repeat (LTR, see glossary) [41]. While purifying the NELF complex from HeLa cells, Stadelmeyer et al. detected low but significant amounts of Integrator, prompting them to explore its function in HIV transcription. They found that Integrator is recruited along with NELF to the HIV-1 TAR element (see glossary) and that knocking down either INTS11 or INTS9 (but not INTS3) resulted in loss of promoter proximal pausing. They then observed that Integrator function is not restricted to the HIV LTR as a common set of genes (>2000) were differentially expressed in response to NELF, INTS3 or INTS11 knockdown in asynchronously growing cells. Consistent with this finding, the analysis of RNAPII, Integrator and NELF ChIP-seq read densities at the TSS of these genes revealed that Integrator and NELF binding closely correlates with the amount of RNAPII pausing at the TSS. Furthermore, in genes bound by NELF and Integrator, knockdown of INTS11 resulted in increased RNAPII occupancy and RNA-seq read density on the gene body, reflective of a promoter proximal pausing defect. Interestingly, INTS3 knockdown had the opposite effect on both LTR and mRNA-coding gene transcription resulting in decreased RNAPII occupancy and RNA-seq read density on the gene body. Whether this effect reflects of antagonistic roles for INTS3 and INTS11 within the complex or of the existence of functionally distinct Integrator subcomplexes remains to be determined.
Although both studies clearly implicate Integrator in the regulation of pause-release and elongation (Figure 4), an apparent discrepancy exists between the observed phenotypes. In Gardini et al., INTS11 knockdown decreases RNAPII density as well as RNA-seq reads on the body of IEGs while the work by Stadelmeyer et al. describes the opposite behavior. This possibly reflects the dual role of NELF dependent pausing that attenuates transcription under non-induced conditions while at the same time maintaining an active open chromatin state at the promoter. Indeed, the study conducted by Gardini et al. focused on the transcriptional response of IEGs in serum starved cells after EGF induction which is affected mostly by RNAPII pausing and release. In contrast, the work conducted by Stadelmeyer et al. uses asynchronously growing cells and considered a wider range of transcriptional responses, in particular genes whose transcription is stimulated after NELF and Integrator depletion. Regardless of the differences, the data presented in both of these studies indicate that there is a role for Integrator in the transcriptional regulation of protein encoding genes. The details of this function are likely going to depend on the cellular context and the nature of the signal produced to alter gene expression.
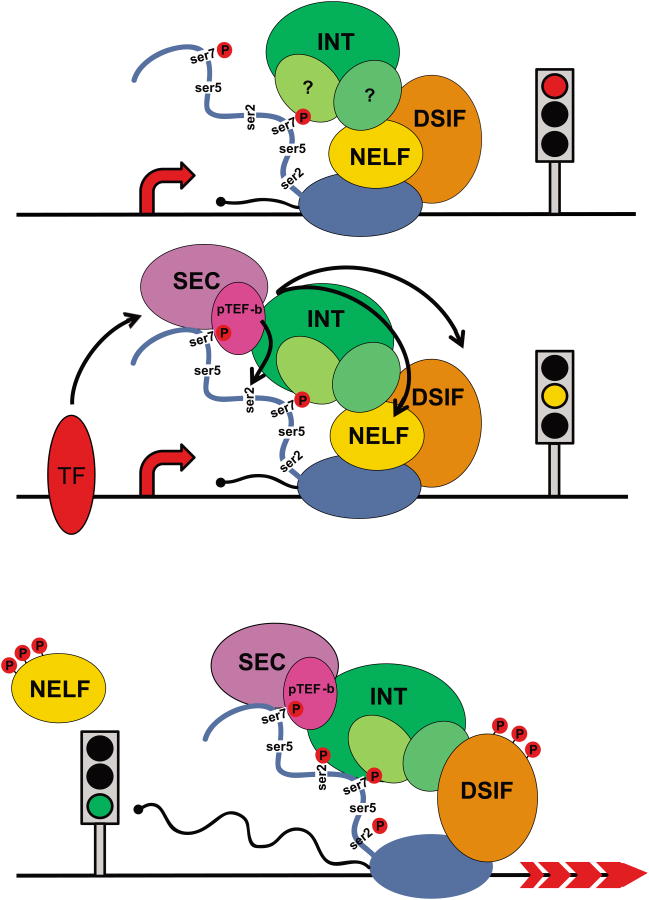
Figure 4. Integrator role in RNAPII promoter proximal pause-release.
Top, under non-stimulated conditions, RNAPII initiates transcription and pauses 40-60 nucleotides downstream of the TSS. This paused complex includes the negative elongation factors NELF, DSIF, and likely INT through its association with the RNAPII CTD through ser7P recognition. Middle, upon activation, INT is further enriched at the pause site and recruits p-TEFb and SEC (see glossary), which phosphorylates DSIF, NELF, and ser2. Bottom, once phosphorylated, NELF is displaced, DSIF transitions into a positive regulator of elongation, and the polymerase is converted into an elongation competent state.
Is Integrator a modular complex?
While it can be biochemically purified as a single entity, Integrator appears to act as a modular complex on the genome. ChIP experiments conducted on UsnRNA genes in human cells or on the HSP70 gene in fly show that different subunits give distinct occupancy patterns. Human INTS5 shows a predominant occupancy from the promoter region through the 3′end of the U2 snRNA while INTS11 is mostly present at the 3′end of the gene [9]. Similarly, Drosophila INTS12 is present at the HSP70 promoter and peaks at the transcriptional pausing site while INTS9 occupancy is shifted toward the 3′end of the gene with a marked peak in the body of the gene [39]. These observations could indicate a sequential recruitment and different functions during the transcription cycle. Early recruitment of INTS5 or INTS12 could reflect the existence of a module with a primary role in transcription initiation and pausing while later recruitment of INTS9 and INTS11 could identify a module with a role in elongation and 3′end processing. Alternatively, Integrator could exist as a single complex but would be subject to significant conformational remodeling during the transcription cycle resulting in a change in accessibility by ChIP. On mRNA coding genes, the opposing effects of INTS3 and INTS11 knockdown on transcription also suggests the existence of functionally distinct modules [41]. Currently, our understanding of Integrator occupancy throughout the genome is fragmentary because only a limited number of subunits have been fully mapped. Determining the localization of more subunits and the impact of their knockdown on transcription will be essential to clearly identify functional and structural submodules within Integrator. Moreover, such studies might reveal additional unsuspected functions for the Integrator complex.
Another interesting aspect of Integrator is the association of some of its subunits into functionally unrelated complexes as exemplified by the association of INTS3 and INTS6 with the SOSS (Sensor of single stranded DNA, see glossary) complex [42-46]. We can therefore speculate that the presence of Integrator and potentially SOSS, through its interaction with INTS3 and INTS6, at transcriptional pause sites might also serve a role in maintaining genome integrity. Indeed, regions of the genome with an open chromatin state such as UsnRNA genes or proximal promoter pause sites are more fragile and prone to genome instability [47-49]. UsnRNA genes have been shown to be particularly sensitive to Ad12-induced chromosome instability and display a weak constitutive fragility in cells defective in transcription-coupled nucleotide excision repair [47,49]. This genome instability is transcription-dependent, as inactive UsnRNA pseudo genes do not display such fragility. Similarly, promoter proximal pausing maintains a constitutively open chromatin state that is favorable to the formation of R-loop structures where the nascent RNA transcript falls back on the template DNA strand, leaving the single stranded non-template strand exposed [50,51]. Similar structures also form at the transcriptional pausing site downstream of the polyadenylation signal and help in the transcription termination process [52]. While R-loops can have a positive effect on transcription initiation and termination, they also present a risk for genome stability if not properly resolved as the exposed non-template strand becomes more susceptible to DNA damage. Therefore, the presence of Integrator, through the ribonuclease INTS11 or its SOSS-interaction subunits INTS3 and INTS6, might have a part to play in the prevention of DNA damage induced by R-loops or constitutively open chromatin states.
Concluding remarks
Altogether, recent biochemical, genomic and functional data elevates the Integrator complex to the status of a primary RNAPII cofactor involved in many steps of the transcription cycle: initiation, pause-release, elongation, 3′end processing and termination. Nevertheless, many aspects of the recruitment of Integrator to the RNAPII remain to be elucidated (see Box 3, Outstanding Questions). Indeed, the model of recruitment of Integrator to UsnRNA versus mRNA genes appears to be in open conflict. The most obvious discrepancy lies in the role played by the RNAPII CTD phosphorylation and the responsible and/or associated kinases, which includes the particularly concerning example of ser2 phosphorylation. It is established that ser2 phosphorylation is necessary in vivo and in vitro for efficient binding of Integrator to the RNAPII CTD. Furthermore, on UsnRNA genes ser2 phosphorylation appears to coincide with INTS11 recruitment, leading to the current model where ser2 phosphorylation by p-TEFb actually triggers the recruitment of Integrator (at least INTS11) leading to efficient UsnRNA 3′end processing. On the contrary, the work conducted on mRNA coding genes tends to demonstrate that INTS11 recruitment precedes and is necessary for the recruitment of p-TEFb and subsequent ser2 phosphorylation. The convenient interpretation, but also the least intellectually satisfactory, is that completely distinct mechanisms govern the recruitment of Integrator, p-TEFb and ser2 phosphorylation on UsnRNA and mRNA coding genes. Due to the short size of the UsnRNA transcription units, ChIP based techniques are probably unable to precisely analyze the interplay between these factors on these genes. Only the precise characterization of how Integrator interacts physically and temporally with the different actors of the transcription cycle (RNAPII, DSIF, NELF, p-TEFb) will bring a clear answer to these essential questions.
Box 3. Outstanding Questions.
-Does Integrator directly bind to the CTD (i.e. is there a CTD binding protein within the Integrator Complex)?
-How is Integrator differentially recruited to UsnRNA promoters versus mRNA promoters? -How does Integrator recognize promoters with paused polymerase?
-How does Integrator cleave RNA specifically (i.e. is there a RNA binding protein within the Integrator Complex?)
-Is the endonuclease activity of Integrator involved in RNAPII pause-release?
-Is Integrator modular and, if so, what are the constituents and the function of those modules?
Highlights.
-The current model of UsnRNA biogenesis is discussed
-Perspective is provided on how the RNAPII CTD is recognized by Integrator.
-CTD kinases and binding proteins influence RNAPII CTD-Integrator association
-Integrator has a new role in the pause-release of RNAPII at mRNA-encoding genes
-Integrator has modular properties opening the possibility of new roles
Acknowledgments
We thank Ambro van Hoof for his critical reading of this manuscript. Work in the Wagner laboratory is supported through funding from the National Institute of Health (CA167752 and CA166274 to EJW).
Glossary
- β-CASP family
a large group of zinc dependent nucleases within the β-lactamase fold acting on DNA and RNA substrates. The family is named after its four founding members: CPSF73 (involved in the cleavage of pre-messenger RNAs [12]), Artemis (endonuclease involved in V(D)J recombination), SMN1 (involved in DNA crosslink repair), and its yeast homolog PSO2 [13]
- DRB Sensitivity Inducing Factor (DSIF)/SPT5
a protein complex composed of the SPT4 and SPT5 subunits. Interacts with the RNAPII and acts first in the transcription cycle as a negative elongation factor in association with NELF and then as a positive elongation factor once NELF has been released after its phosphorylation by p-TEFb
- Immediate early genes (IEGs)
a class of genes that are rapidly and transiently activated in response to various extracellular stimuli. Their transcription is independent of de novo protein synthesis and in most cases is regulated by RNAPII promoter proximal pausing, ensuring a fast and coordinated response upon activation. Prototypical IEGs are c-fos, c-jun and c-myc.
- Long terminal repeat (LTR)
repeated sequences flanking retrotransposons and proviral DNAs. They contain all the regulatory elements required for viral gene expression (enhancer, promoter, terminator and polyA signal) and also mediate the integration of the provirus into the host genome
- Negative Elongation Factor (NELF)
a four-subunit (NELF-A,-B,-C/D and -E) protein complex that negatively regulates RNAPII transcription at the promoter proximal pause site located 40-60 nucleotides downstream of the transcription start site
- Positive Transcription Elongation Factor-b (p-TEFb)
a cyclin dependent kinase containing the catalytic subunit CDK9 and one of T1, T2 or K cyclin subunit. It phosphorylates the C-terminal domain of the largest subunit of the RNA polymerase II as well as the DRB sensitivity inducing factor (DSIF) and the negative elongation factor (NELF), which leads to the transition from promoter proximal pausing to productive elongation
- RNAPII C-terminal Domain (CTD)
an elongated domain located at the C-terminus of the largest subunit of RNA polymerase II that is composed of a repetition of the amino acid heptad Y1S2P3T4S5P6S7. (reviewed in[53], see Box 1)
- Super Elongation Complex (SEC)
a large protein complex composed of the eleven-nineteen Lys-rich leukemia (ELL) protein and elongation factors ELL1, ELL2 and ELL3, the scaffold proteins AFF1 and AFF4, the cyclin dependent kinase p-TEFb, ENL/MLLT1 and AF9/MLLT3. SEC is required for rapid transcription in response to external stimuli such as heat shock, retinoic acid or serum treatment as well as for LTR-driven HIV-1 provirus transcription
- snRNA Activating Protein Complex (SNAPc)
a UsnRNA specific basal transcription factor (also called PTF) that binds to the PSE element in the promoter of RNAPII- and RNAPIII-dependent UsnRNAs. It consists of 5 subunits in mammals and 3 subunits in fly (reviewed in [15,16])
- Sensor of Single Stranded DNA (SOSS) complex
a multiprotein complex composed of one oligonucleotide/oligosaccharide-binding (OB) fold domain-containing protein hSSB1 (SOSS-B1) or hSSB2 (SOSS-B2), the Integrator complex subunits INTS3 (SOSS-A) and INTS6, and the uncharacterized protein C9orf80 (SOSS-C) [43-45]. The SOSS complex is recruited to double stranded DNA breaks and was shown to be important for DNA damage response, homologous recombination and genome stability [42-45,54]
- HIV Trans-Activation Response (TAR) element and Trans-Activator of Transcription (TAT) protein (TAR/TAT)
transcription initiating from the HIV-1 LTR is regulated by RNAPII promoter proximal pausing. The TAR sequence, located near the 5′ of the nascent transcript, acts as a cis-acting RNA regulatory element by binding the protein TAT. This activator protein is required for transcriptional pause-release and efficient transcription through the recruitment of p-TEFb and the SEC complex
- Transcription Factor II Human (TFIIH)
a general transcription factor involved in basal transcription and transcription-coupled nucleotide excision repair. It contains 9 subunits including the kinase CDK7 and the regulatory cyclin H
- U-rich small nuclear RNAs (UsnRNAs)
a family of small non-coding RNAs, ranging from 60 to 200 nucleotides. With the exception of U7, which is involved in cell cycle-dependent histone pre-mRNA 3′end processing [55], UsnRNAs are the RNA component of the major (U1, U2, U4, U5 and U6) and minor (U11, U12, U4atac, U5 and U6atac) spliceosome [56]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Baillat D, et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–76. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 2.Jeronimo C, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–74. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malovannaya A, et al. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci U S A. 2010;107:2431–6. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malovannaya A, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–99. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, et al. An RNAi screen identifies additional members of the Drosophila Integrator complex and a requirement for cyclin C/Cdk8 in snRNA 3′-end formation. RNA. 2012;18:2148–56. doi: 10.1261/rna.035725.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poss ZC, et al. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. 2013;48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlsten JOP, et al. The multitalented Mediator complex. Trends Biochem Sci. 2013;38:531–7. doi: 10.1016/j.tibs.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Peart N, et al. Non-mRNA 3′ end formation: how the other half lives. Wiley Interdiscip Rev RNA. 2013;4:491–506. doi: 10.1002/wrna.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egloff S, et al. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell. 2012;45:111–22. doi: 10.1016/j.molcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Wagner EJ. snRNA 3′ end formation: the dawn of the Integrator complex. Biochem Soc Trans. 2010;38:1082–7. doi: 10.1042/BST0381082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albrecht TR, Wagner EJ. snRNA 3′ end formation requires heterodimeric association of integrator subunits. Mol Cell Biol. 2012;32:1112–23. doi: 10.1128/MCB.06511-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang K, et al. Delineating the structural blueprint of the pre-mRNA 3′-end processing machinery. Mol Cell Biol. 2014;34:1894–910. doi: 10.1128/MCB.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callebaut I, et al. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–69. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung KH, Stumph WE. Regulation of snRNA gene expression by the Drosophila melanogaster small nuclear RNA activating protein complex (DmSNAPc) Crit Rev Biochem Mol Biol. 2011;46:11–26. doi: 10.3109/10409238.2010.518136. [DOI] [PubMed] [Google Scholar]
- 16.Jawdekar GW, Henry RW. Transcriptional regulation of human small nuclear RNA genes. Biochim Biophys Acta. 2008;1779:295–305. doi: 10.1016/j.bbagrm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medlin JE, et al. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 2003;22:925–34. doi: 10.1093/emboj/cdg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medlin J, et al. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 2005;24:4154–65. doi: 10.1038/sj.emboj.7600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez N. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985;4:1827–37. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egloff S, et al. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J Biol Chem. 2010;285:20564–9. doi: 10.1074/jbc.M110.132530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ezzeddine N, et al. A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3′-end formation. Mol Cell Biol. 2011;31:328–41. doi: 10.1128/MCB.00943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Reilly D, et al. Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucleic Acids Res. 2014;42:264–75. doi: 10.1093/nar/gkt892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 24.Mosley AL, et al. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell. 2009;34:168–78. doi: 10.1016/j.molcel.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akhtar MS, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009;34:387–93. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu PL, et al. Rtr1 is a dual specificity phosphatase that dephosphorylates Tyr1 and Ser5 on the RNA polymerase II CTD. J Mol Biol. 2014;426:2970–81. doi: 10.1016/j.jmb.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang K, et al. The yeast regulator of transcription protein Rtr1 lacks an active site and phosphatase activity. Nat Commun. 2012;3:946. doi: 10.1038/ncomms1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni Z, et al. RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nat Struct Mol Biol. 2014;21:686–95. doi: 10.1038/nsmb.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forget D, et al. Nuclear import of RNA polymerase II is coupled with nucleocytoplasmic shuttling of the RNA polymerase II-associated protein 2. Nucleic Acids Res. 2013;41:6881–91. doi: 10.1093/nar/gkt455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carre C, Shiekhattar R. Human GTPases Associate with RNA Polymerase II To Mediate Its Nuclear Import. Mol Cell Biol. 2011;31:3953–3962. doi: 10.1128/MCB.05442-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staresincic L, et al. GTP-dependent binding and nuclear transport of RNA polymerase II by Npa3 protein. J Biol Chem. 2011;286:35553–35561. doi: 10.1074/jbc.M111.286161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wani S, et al. Vertebrate Ssu72 regulates and coordinates 3′-end formation of rNas transcribed by RNA polymerase II. PLoS One. 2014;9:e106040. doi: 10.1371/journal.pone.0106040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsin JP, et al. Function and Control of RNA Polymerase II C-Terminal Domain Phosphorylation in Vertebrate Transcription and RNA Processing. Mol Cell Biol. 2014;34:2488–98. doi: 10.1128/MCB.00181-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto J, et al. DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nat Commun. 2014;5:4263. doi: 10.1038/ncomms5263. [DOI] [PubMed] [Google Scholar]
- 35.Tsao DC, et al. Prolonged α-amanitin treatment of cells for studying mutated polymerases causes degradation of DSIF160 and other proteins. RNA. 2012;18:222–9. doi: 10.1261/rna.030452.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell. 2011;43:311–8. doi: 10.1016/j.molcel.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Reilly D, et al. Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egloff S, et al. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Mol Cell Biol. 2009;29:4002–13. doi: 10.1128/MCB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardini A, et al. Integrator Regulates Transcriptional Initiation and Pause Release following Activation. Mol Cell. 2014;1:1–12. doi: 10.1016/j.molcel.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skaar JR, et al. The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res. 2015 doi: 10.1038/cr.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stadelmayer B, et al. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun. 2014;5:5531. doi: 10.1038/ncomms6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skaar JR, et al. INTS3 controls the hSSB1-mediated DNA damage response. J Cell Biol. 2009;187:25–32. doi: 10.1083/jcb.200907026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, et al. HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J Biol Chem. 2009;284:23525–31. doi: 10.1074/jbc.C109.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang F, et al. A core hSSB1-INTS complex participates in the DNA damage response. J Cell Sci. 2013;126:4850–5. doi: 10.1242/jcs.132514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang J, et al. SOSS complexes participate in the maintenance of genomic stability. Mol Cell. 2009;35:384–93. doi: 10.1016/j.molcel.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu P, et al. Single strand DNA binding proteins 1 and 2 protect newly replicated telomeres. Cell Res. 2013;23:705–19. doi: 10.1038/cr.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu a, et al. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol Cell. 2000;5:801–10. doi: 10.1016/s1097-2765(00)80320-2. [DOI] [PubMed] [Google Scholar]
- 48.Yu a, et al. Metaphase fragility of the human RNU1 and RNU2 loci is induced by actinomycin D through a p53-dependent pathway. Hum Mol Genet. 1998;7:609–17. doi: 10.1093/hmg/7.4.609. [DOI] [PubMed] [Google Scholar]
- 49.Li Z, et al. Adenovirus type 12-induced fragility of the human RNU2 locus requires p53 function. J Virol. 1998;72:4183–91. doi: 10.1128/jvi.72.5.4183-4191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ginno PA, et al. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–25. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ginno PA, et al. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23:1590–600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skourti-Stathaki K, et al. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev. 2013;113:8456–90. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 54.Yang SH, et al. The SOSS1 single-stranded DNA binding complex promotes DNA end resection in concert with Exo1. EMBO J. 2013;32:126–39. doi: 10.1038/emboj.2012.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marzluff WF, et al. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–54. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matera aG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–21. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hintermair C, et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012;31:2784–97. doi: 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mayer A, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–5. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 59.Hsin JP, et al. RNAP II CTD tyrosine 1 performs diverse functions in vertebrate cells. Elife. 2014;3:e02112. doi: 10.7554/eLife.02112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsin JP, et al. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334:683–6. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kubicek K, et al. Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. 2012 doi: 10.1101/gad.192781.112.displacement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hanes SD. The Ess1 prolyl isomerase: traffic cop of the RNA polymerase II transcription cycle. Biochim Biophys Acta. 2014;1839:316–33. doi: 10.1016/j.bbagrm.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]